Super-Resolution Fluorescence Microscopy Reveals Clustering Behaviour of Chlamydia pneumoniae’s Major Outer Membrane Protein
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
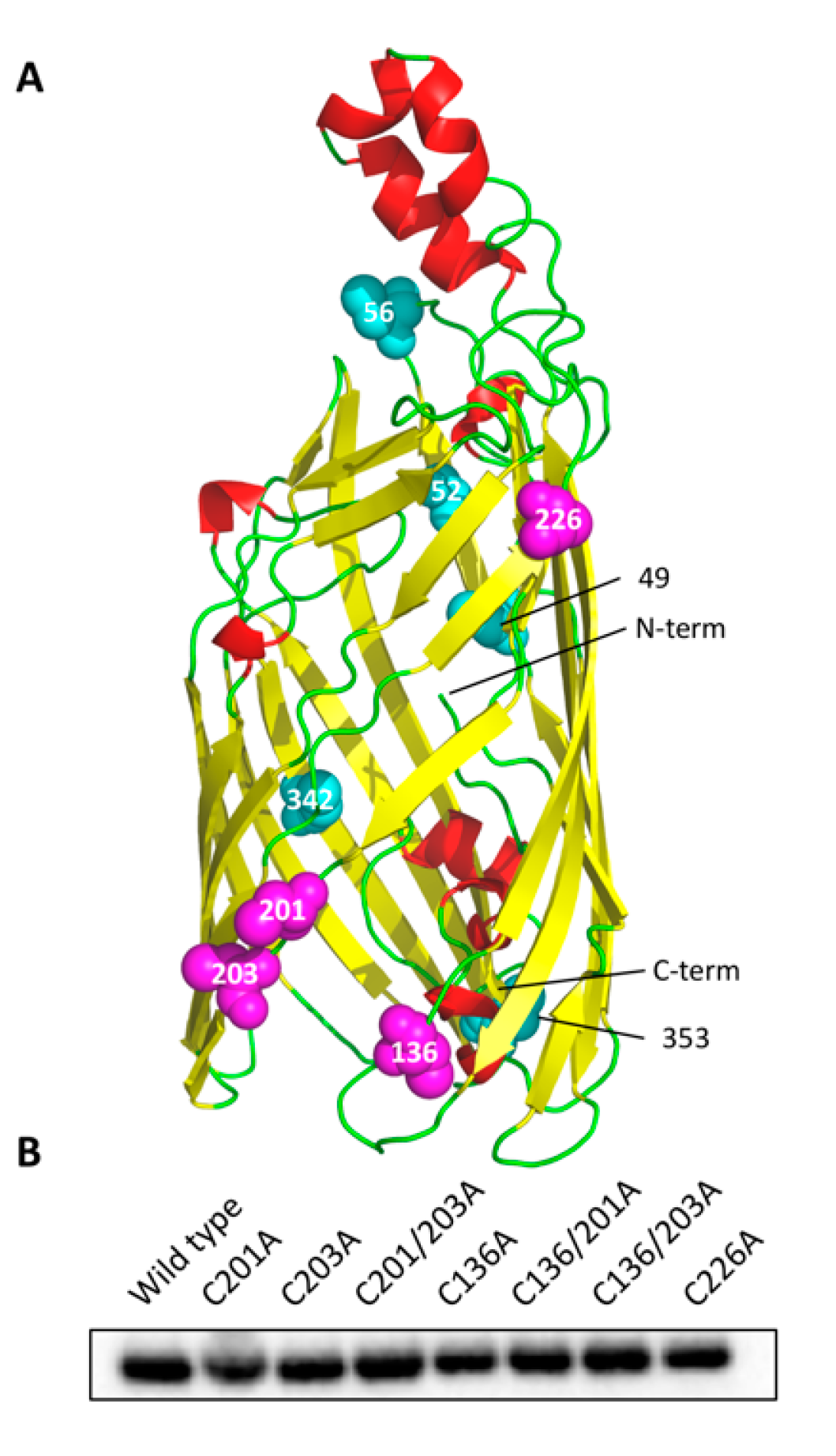
2.1. Construct Design
2.2. Bacterial Growth Conditions and Induction of rMOMP
2.3. Slide Preparation
2.4. Epifluorescence and TIRF Imaging Conditions
2.5. dSTORM Immunostaining Procedure
2.6. dSTORM Imaging Conditions
2.7. Clustering Analysis of dSTORM Images
3. Results
3.1. Cysteine Mutant Design
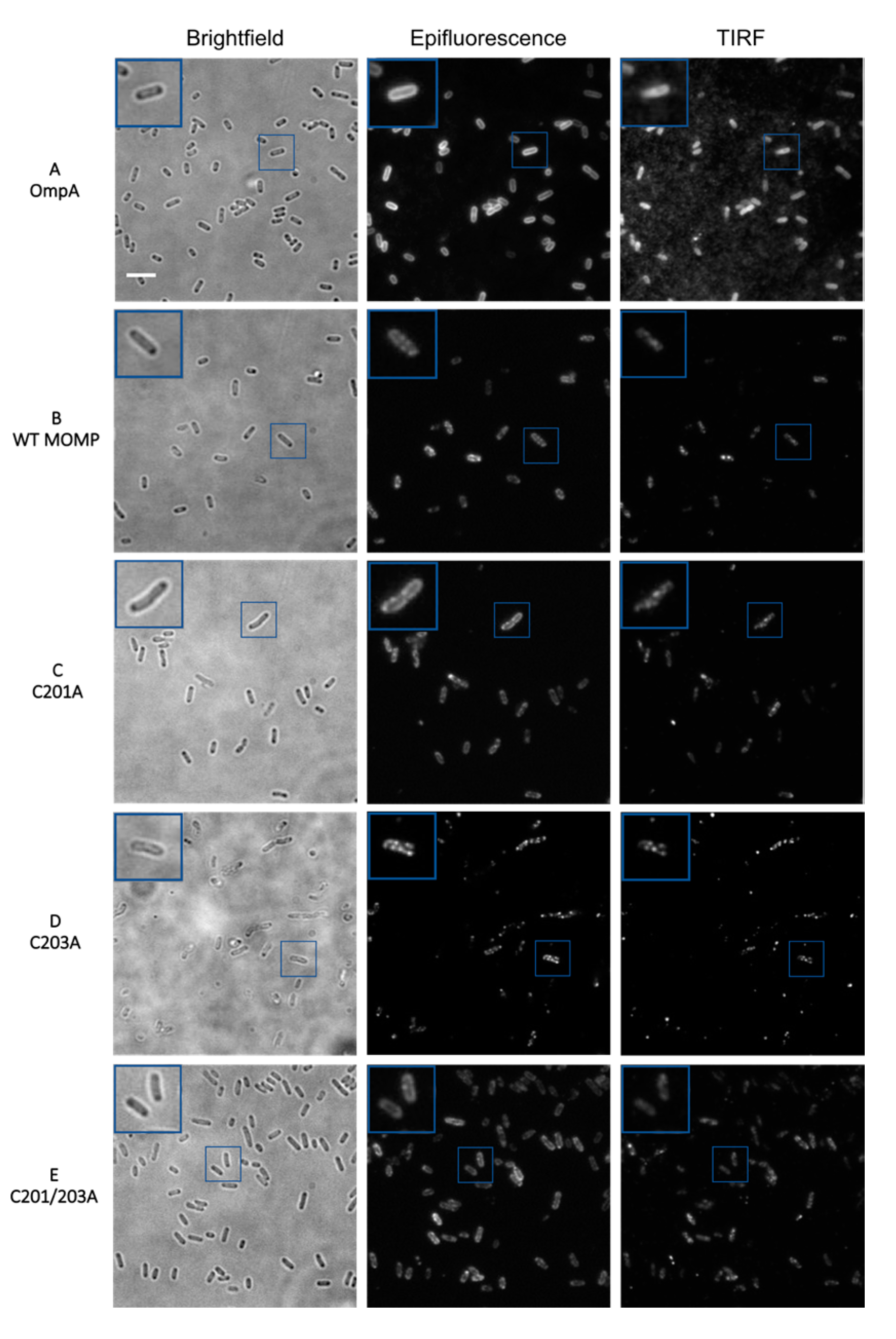
3.2. Epifluorescence and TIRF Imaging Demonstrated Differences in Membrane Distribution of rMOMP Mutants
3.3. Quantitative Analysis of Direct Stochastic Optical Reconstruction Microscopy (dSTORM)
3.4. Significant Decreases in Clustering Were Observed Between Wild-Type rMOMP and Cysteine Mutants C201/203A, C136/201A, and C226A
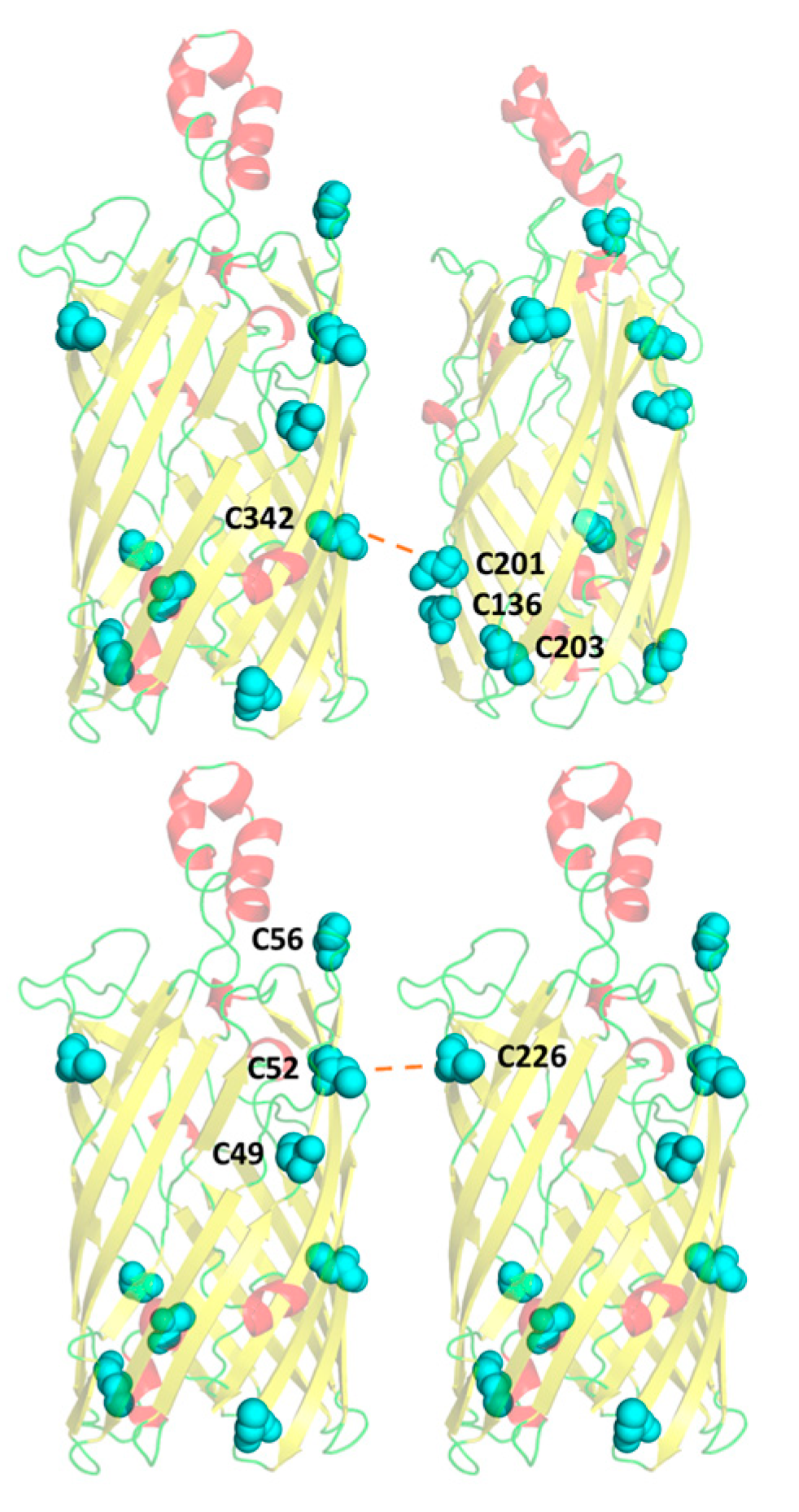
3.5. A Compensatory Mechanism of Disulphide Bond Formation between Clusters
3.6. Residue C226 May Play a Key Role in Intermolecular Disulphide Bonding with Pocket C49–52–56
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Grayston, J.T.; Kuo, C.C.; Campbell, L.A.; Wang, S.P. Chlamydia Pneumoniae Sp. Nov. for Chlamydia Sp. Strain TWAR. Int. J. Syst. Bacteriol. 1989, 39, 88–90. [Google Scholar] [CrossRef]
- Saikku, P.; Mattila, K.; Nieminen, M.S.; Huttunen, J.K.; Leinonen, M.; Ekman, M.R.; Mäkelä, P.H.; Valtonen, V. Serological evidence of an association of a novel chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet 1988, 332, 983–986. [Google Scholar] [CrossRef]
- Belland, R.J.; Ouellette, S.P.; Gieffers, J.; Byrne, G.I. Chlamydia Pneumoniae and Atherosclerosis. Cell. Microbiol. 2004, 117–127. [Google Scholar] [CrossRef]
- Sorrentino, R.; Yilmaz, A.; Schubert, K.; Crother, T.R.; Pinto, A.; Shimada, K.; Arditi, M.; Chen, S. A Single Infection with Chlamydia Pneumoniae Is Sufficient to Exacerbate Atherosclerosis in ApoE Deficient Mice. Cell. Immunol. 2015, 294, 25–32. [Google Scholar] [CrossRef]
- Rahman, M.U.; Hudson, A.P.; Schumacher, H.R. Chlamydia and Reiter’s Syndrome (Reactive Arthritis). Rheum. Dis. Clin. North. Am. 1992, 18, 67–79. [Google Scholar] [PubMed]
- Carter, J.D.; Inman, R.D.; Whittum-Hudson, J.; Hudson, A.P. Chlamydia and Chronic Arthritis. Ann. Med. 2012, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Zeidler, H.; Hudson, A.P. Causality of Chlamydiae in Arthritis and Spondyloarthritis: A Plea for Increased Translational Research. Curr. Rheumatol. Rep. 2016, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Hahn, D.L.; McDonald, R. Can Acute Chlamydia Pneumoniae Respiratory Tract Infection Initiate Chronic Asthma? Ann. Allergy Asthma Immunol. 1998, 81, 339–344. [Google Scholar] [CrossRef]
- Webley, W.C.; Hahn, D.L. Infection-Mediated Asthma: Etiology, Mechanisms and Treatment Options, with Focus on Chlamydia Pneumoniae and Macrolides. Respir. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Liechti, G.W.; Kuru, E.; Hall, E.; Kalinda, A.; Brun, Y.V.; Vannieuwenhze, M.; Maurelli, A.T. A New Metabolic Cell-Wall Labelling Method Reveals Peptidoglycan in Chlamydia Trachomatis. Nature 2014, 506, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Bavoil, P.; Ohlin, A.; Schachter, J. Role of Disulfide Bonding in Outer Membrane Structure and Permeability in Chlamydia Trachomatis. Infect. Immun. 1984, 44, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Hatch, T.P.; Allan, I.; Pearce, J.H. Structural and Polypeptide Difference between Envelopes of Infective and Reproductive Life Cycle Forms of Chlamydia Spp. J. Bacteriol. 1984, 157, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Hatch, T.P. Disulfide Cross-Linked Envelope Proteins: The Functional Equivalent of Peptidoglycan in Chlamydiae? J. Bacteriol. 1996, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Newhall, W.J.; Jones, R.B. Disulfide-Linked Oligomers of the Major Outer Membrane Protein of Chlamydiae. J. Bacteriol. 1983, 154, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Afrane, M.; Clemmer, D.E.; Zhong, G.; Nelson, D.E. Identification of Chlamydia Trachomatis Outer Membrane Complex Proteins by Differential Proteomics. J. Bacteriol. 2010, 192, 2852–2860. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, H.D.; Kromhout, J.; Schachter, J. Purification and Partial Characterization of the Major Outer Membrane Protein of Chlamydia Trachomatis. Infect. Immun. 1981, 31, 1161–1176. [Google Scholar] [CrossRef]
- Christensen, S.; McMahon, R.M.; Martin, J.L.; Huston, W.M. Life inside and out: Making and Breaking Protein Disulfide Bonds in Chlamydia. Crit. Rev. Microbiol. 2019, 33–50. [Google Scholar] [CrossRef]
- Atanu, F.O.; Oviedo-Orta, E.; Watson, K.A. A Novel Transport Mechanism for MOMP in Chlamydophila Pneumoniae and Its Putative Role in Immune-Therapy. PLoS ONE 2013, 8, e61139. [Google Scholar] [CrossRef]
- Feher, V.A.; Randall, A.; Baldi, P.; Bush, R.M.; de la Maza, L.M.; Amaro, R.E. A 3-Dimensional Trimeric β-Barrel Model for Chlamydia MOMP Contains Conserved and Novel Elements of Gram-Negative Bacterial Porins. PLoS ONE 2013, 8, e68934. [Google Scholar] [CrossRef]
- Pourhajibagher, M.; Bahador, A. Designing and in Silico Analysis of PorB Protein from Chlamydia Trachomatis for Developing a Vaccine Candidate. Drug Res. 2016, 66, 479–483. [Google Scholar] [CrossRef]
- Yen, T.Y.; Pal, S.; De La Maza, L.M. Characterization of the Disulfide Bonds and Free Cysteine Residues of the Chlamydia Trachomatis Mouse Pneumonitis Major Outer Membrane Protein. Biochemistry 2005, 44, 6250–6256. [Google Scholar] [CrossRef]
- Wang, Y. Identification of Surface-Exposed Components of MOMP of Chlamydia Trachomatis Serovar, F. Protein Sci. 2006, 15, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Findlay, H.E.; McClafferty, H.; Ashley, R.H. Surface Expression, Single-Channel Analysis and Membrane Topology of Recombinant Chlamydia Trachomatis Major Outer Membrane Protein. BMC Microbiol. 2005, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Schwarzer, C.; Hybiske, K.; Machen, T.E.; Stephens, R.S. Developmental Stage Oxidoreductive States of Chlamydia and Infected Host Cells. MBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.; Halili, M.A.; Strange, N.; Petit, G.A.; Huston, W.M.; Martin, J.L.; McMahon, R.M. Oxidoreductase Disulfide Bond Proteins DsbA and DsbB Form an Active Redox Pair in Chlamydia Trachomatis, a Bacterium with Disulfide Dependent Infection and Development. PLoS ONE 2019, 14, e222595. [Google Scholar] [CrossRef]
- Meehan, B.M.; Landeta, C.; Boyd, D.; Beckwith, J. The Disulfide Bond Formation Pathway Is Essential for Anaerobic Growth of Escherichia Coli. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef]
- Domínguez-Escobar, J.; Chastanet, A.; Crevenna, A.H.; Fromion, V.; Wedlich-Söldner, R.; Carballido-López, R. Processive Movement of MreB-Associated Cell Wall Biosynthetic Complexes in Bacteria. Science 2011, 333, 225–228. [Google Scholar] [CrossRef]
- Jovanovic, G.; Mehta, P.; McDonald, C.; Davidson, A.C.; Uzdavinys, P.; Ying, L.; Buck, M. The N-Terminal Amphipathic Helices Determine Regulatory and Effector Functions of Phage Shock Protein A (PspA) in Escherichia Coli. J. Mol. Biol. 2014, 426, 1498–1511. [Google Scholar] [CrossRef]
- Heilemann, M.; Van De Linde, S.; Schüttpelz, M.; Kasper, R.; Seefeldt, B.; Mukherjee, A.; Tinnefeld, P.; Sauer, M. Subdiffraction-Resolution Fluorescence Imaging with Conventional Fluorescent Probes. Angew. Chem. Int. Ed. 2008, 47, 6172–6176. [Google Scholar] [CrossRef]
- Godin, A.G.; Lounis, B.; Cognet, L. Super-Resolution Microscopy Approaches for Live Cell Imaging. Biophys. J. 2014, 1777–1784. [Google Scholar] [CrossRef]
- Bates, M.; Jones, S.A.; Zhuang, X. Stochastic Optical Reconstruction Microscopy (STORM): A Method for Superresolution Fluorescence Imaging. Cold Spring Harb. Protoc. 2013, 8, 498–520. [Google Scholar] [CrossRef]
- Shroff, H.; Galbraith, C.G.; Galbraith, J.A.; Betzig, E. Live-Cell Photoactivated Localization Microscopy of Nanoscale Adhesion Dynamics. Nat. Methods. 2008, 5, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-Diffraction-Limit Imaging by Stochastic Optical Reconstruction Microscopy (STORM). Nat. Methods. 2006, 3, 793–795. [Google Scholar] [CrossRef]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef] [PubMed]
- Hess, S.T.; Girirajan, T.P.K.; Mason, M.D. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophys. J. 2006, 91, 4258–4272. [Google Scholar] [CrossRef] [PubMed]
- Filip, C.; Fletcher, G.; Wulff, J.L.; Earhart, C.F. Solubilization of the Cytoplasmic Membrane of Escherichia Coli by the Ionic Detergent Sodium-Lauryl Sarcosinate. J. Bacteriol. 1973, 115, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Willson, B.J.; Kovács, K.; Wilding-Steele, T.; Markus, R.; Winzer, K.; Minton, N.P. Production of a Functional Cell Wall-Anchored Minicellulosome by Recombinant Clostridium Acetobutylicum ATCC 824. Biotechnol. Biofuels 2016, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Pageon, S.V.; Nicovich, P.R.; Mollazade, M.; Tabarin, T.; Gaus, K. Clus-DoC: A Combined Cluster Detection and Colocalization Analysis for Single-Molecule Localization Microscopy Data. Mol. Biol. Cell. 2016, 27, 3627–3636. [Google Scholar] [CrossRef]
- Atanu, F.O. Structural Studies of Chlamydophila Pneumoniae Major Outer Membrane Protein (MOMP) and MOMP-Derived Peptides: Candidate Vaccine Targets for Human Inflammatory Diseases. Ph.D. Thesis, University of Reading, Berkshire, UK, 2014. [Google Scholar]
- Dempsey, G.T.; Vaughan, J.C.; Chen, K.H.; Bates, M.; Zhuang, X. Evaluation of Fluorophores for Optimal Performance in Localization-Based Super-Resolution Imaging. Nat. Methods. 2011, 1027–1040. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. KDD-96 Proc. 1996, 96, 226–231. [Google Scholar]

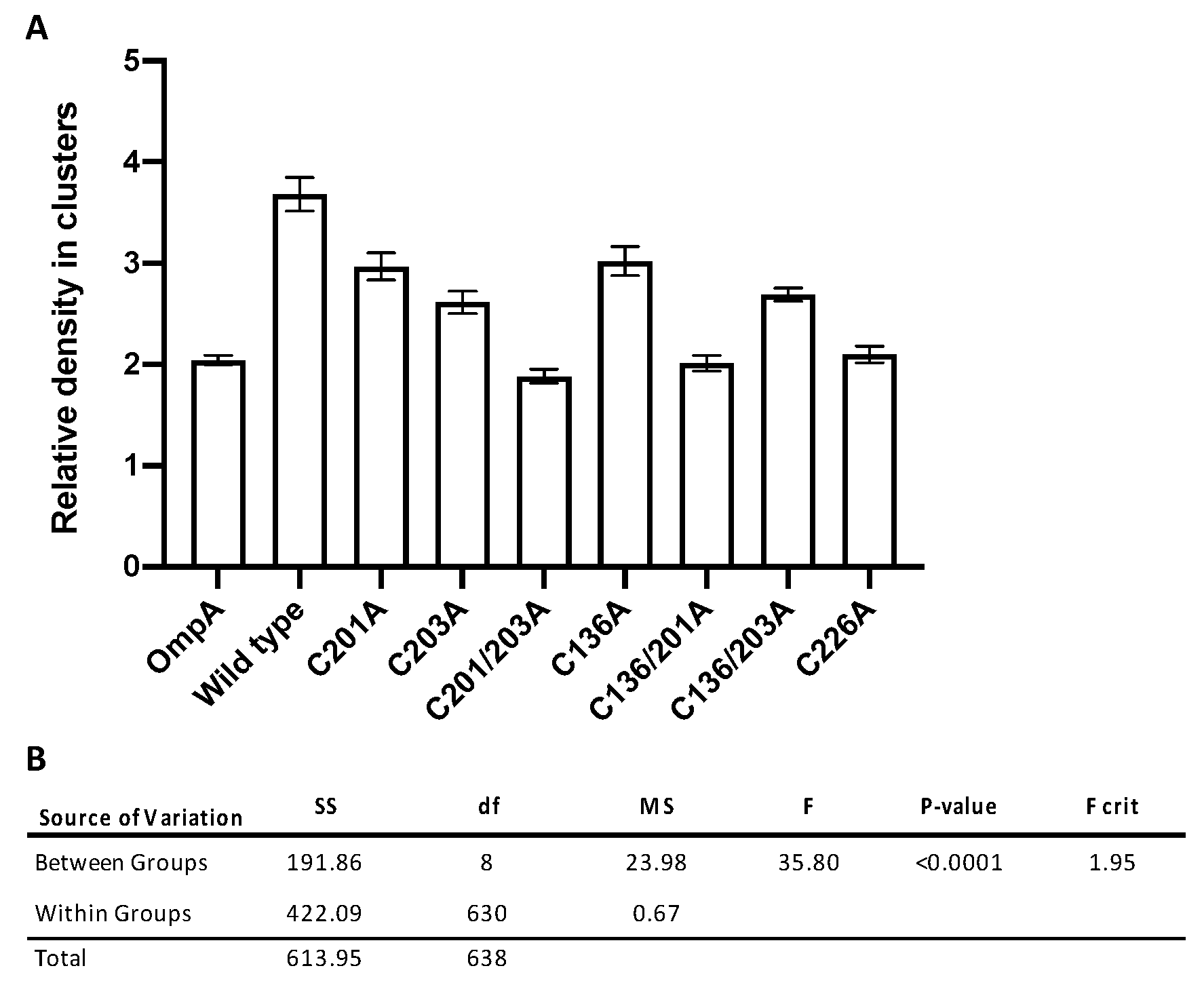
| Construct | Relative Density in Clusters | SE | n |
|---|---|---|---|
| OmpA | 2.04 | 0.05 | 94 |
| Wild type | 3.68 | 0.17 | 59 |
| C201A | 2.97 | 0.13 | 51 |
| C203A | 2.61 | 0.11 | 62 |
| C201/203A | 1.88 | 0.07 | 71 |
| C136A | 3.02 | 0.14 | 69 |
| C136/201A | 2.01 | 0.08 | 78 |
| C136/203A | 2.69 | 0.07 | 90 |
| C226A | 2.10 | 0.08 | 65 |
| Important Pairs | Mean Difference | N (Group 1) | N (Group 2) | SE | q-Crit | Significant | |
|---|---|---|---|---|---|---|---|
| OmpA | Wild type | 1.64 | 94 | 59 | 0.10 | 17.05 | YES |
| Wild type | C201A | 0.71 | 59 | 51 | 0.11 | 6.44 | YES |
| Wild type | C203A | 1.07 | 59 | 62 | 0.11 | 10.15 | YES |
| Wild type | C201/203A | 1.80 | 59 | 71 | 0.10 | 17.63 | YES |
| Wild type | C136A | 0.66 | 59 | 69 | 0.10 | 6.45 | YES |
| Wild type | C136/201A | 1.67 | 59 | 78 | 0.10 | 16.71 | YES |
| Wild type | C136/203A | 0.99 | 59 | 90 | 0.10 | 10.21 | YES |
| Wild type | C226A | 1.58 | 59 | 65 | 0.10 | 15.21 | YES |
| OmpA | C201A | 0.93 | 94 | 51 | 0.10 | 9.20 | YES |
| OmpA | C203A | 0.57 | 94 | 62 | 0.09 | 6.03 | YES |
| OmpA | C201/203A | 0.16 | 94 | 71 | 0.09 | 1.75 | NO |
| OmpA | C136A | 0.98 | 94 | 69 | 0.09 | 10.65 | YES |
| OmpA | C136/201A | 0.03 | 94 | 78 | 0.09 | 0.33 | NO |
| OmpA | C136/203A | 0.65 | 94 | 90 | 0.09 | 7.60 | YES |
| OmpA | C226A | 0.06 | 94 | 65 | 0.09 | 0.60 | NO |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danson, A.E.; McStea, A.; Wang, L.; Pollitt, A.Y.; Martin-Fernandez, M.L.; Moraes, I.; Walsh, M.A.; MacIntyre, S.; Watson, K.A. Super-Resolution Fluorescence Microscopy Reveals Clustering Behaviour of Chlamydia pneumoniae’s Major Outer Membrane Protein. Biology 2020, 9, 344. https://doi.org/10.3390/biology9100344
Danson AE, McStea A, Wang L, Pollitt AY, Martin-Fernandez ML, Moraes I, Walsh MA, MacIntyre S, Watson KA. Super-Resolution Fluorescence Microscopy Reveals Clustering Behaviour of Chlamydia pneumoniae’s Major Outer Membrane Protein. Biology. 2020; 9(10):344. https://doi.org/10.3390/biology9100344
Chicago/Turabian StyleDanson, Amy E., Alex McStea, Lin Wang, Alice Y. Pollitt, Marisa L. Martin-Fernandez, Isabel Moraes, Martin A. Walsh, Sheila MacIntyre, and Kimberly A. Watson. 2020. "Super-Resolution Fluorescence Microscopy Reveals Clustering Behaviour of Chlamydia pneumoniae’s Major Outer Membrane Protein" Biology 9, no. 10: 344. https://doi.org/10.3390/biology9100344
APA StyleDanson, A. E., McStea, A., Wang, L., Pollitt, A. Y., Martin-Fernandez, M. L., Moraes, I., Walsh, M. A., MacIntyre, S., & Watson, K. A. (2020). Super-Resolution Fluorescence Microscopy Reveals Clustering Behaviour of Chlamydia pneumoniae’s Major Outer Membrane Protein. Biology, 9(10), 344. https://doi.org/10.3390/biology9100344






