Membrane Protein Structure Determination and Characterisation by Solution and Solid-State NMR
Simple Summary
Abstract
1. Introduction
2. Nuclear Magnetic Resonance
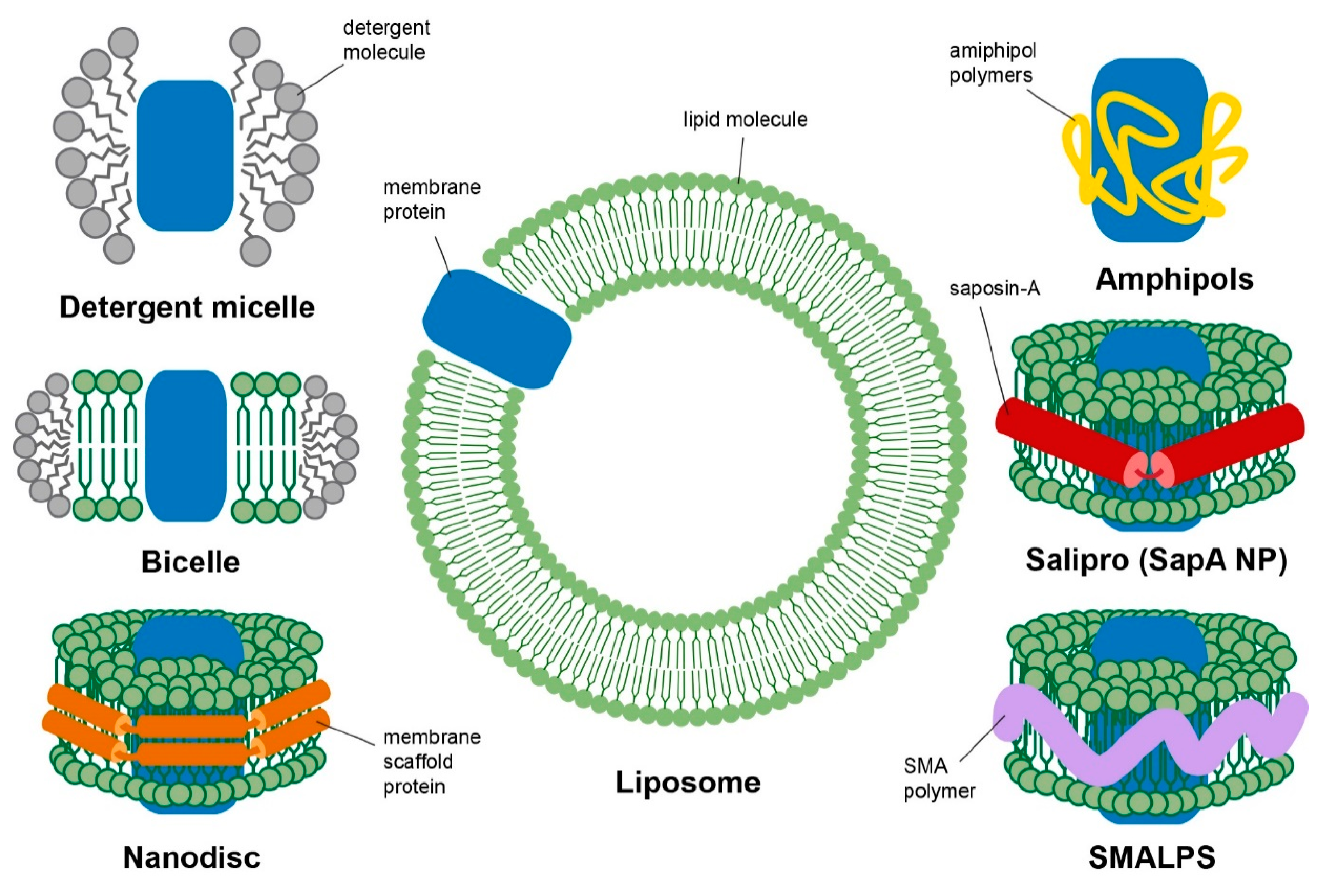
3. Membrane Mimetics
3.1. Detergent Micelles
3.2. Bicelles
3.3. Liposomes
3.4. Nanodiscs
3.5. Alternative Membrane Mimetic
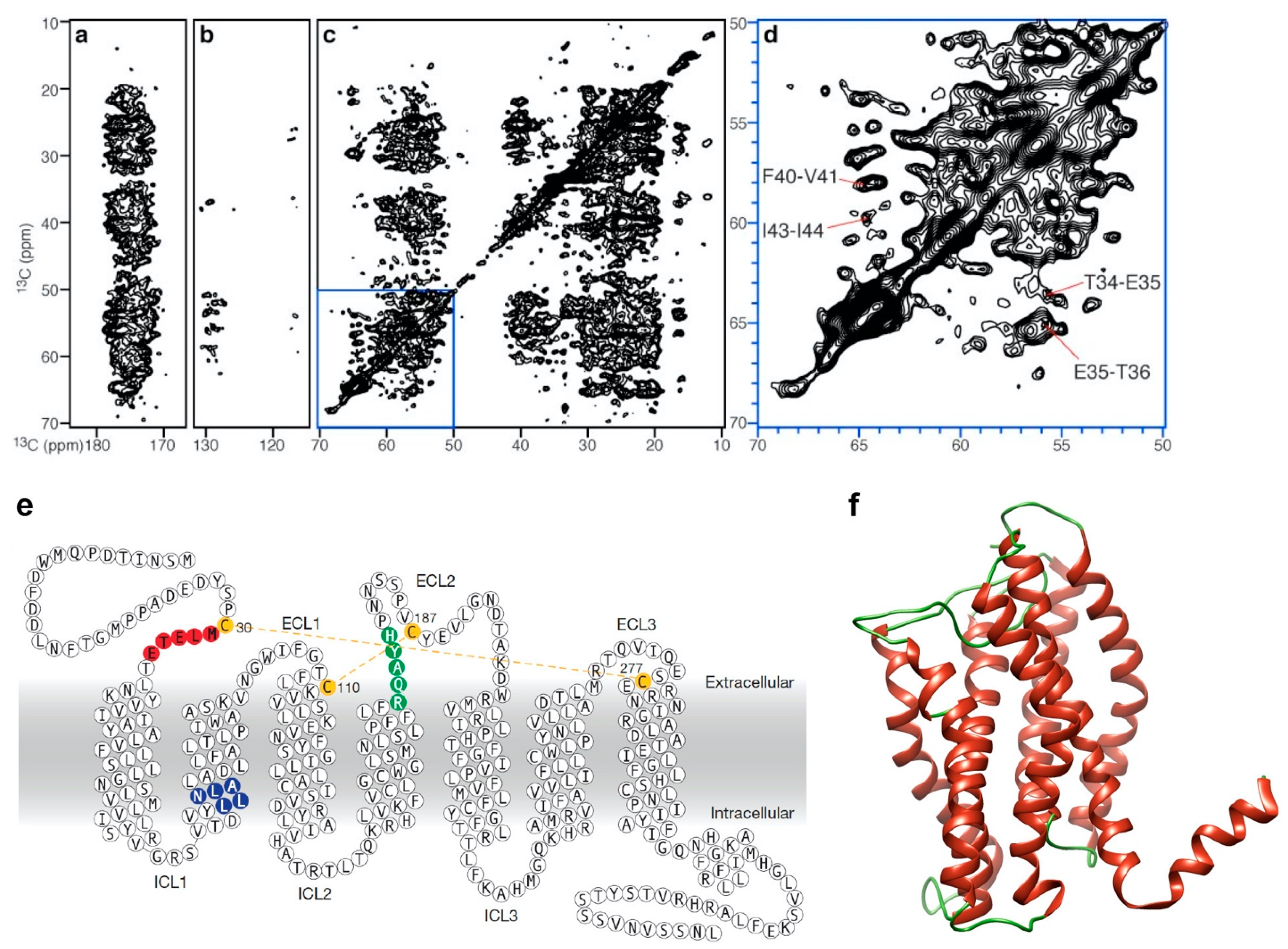
4. Solution NMR
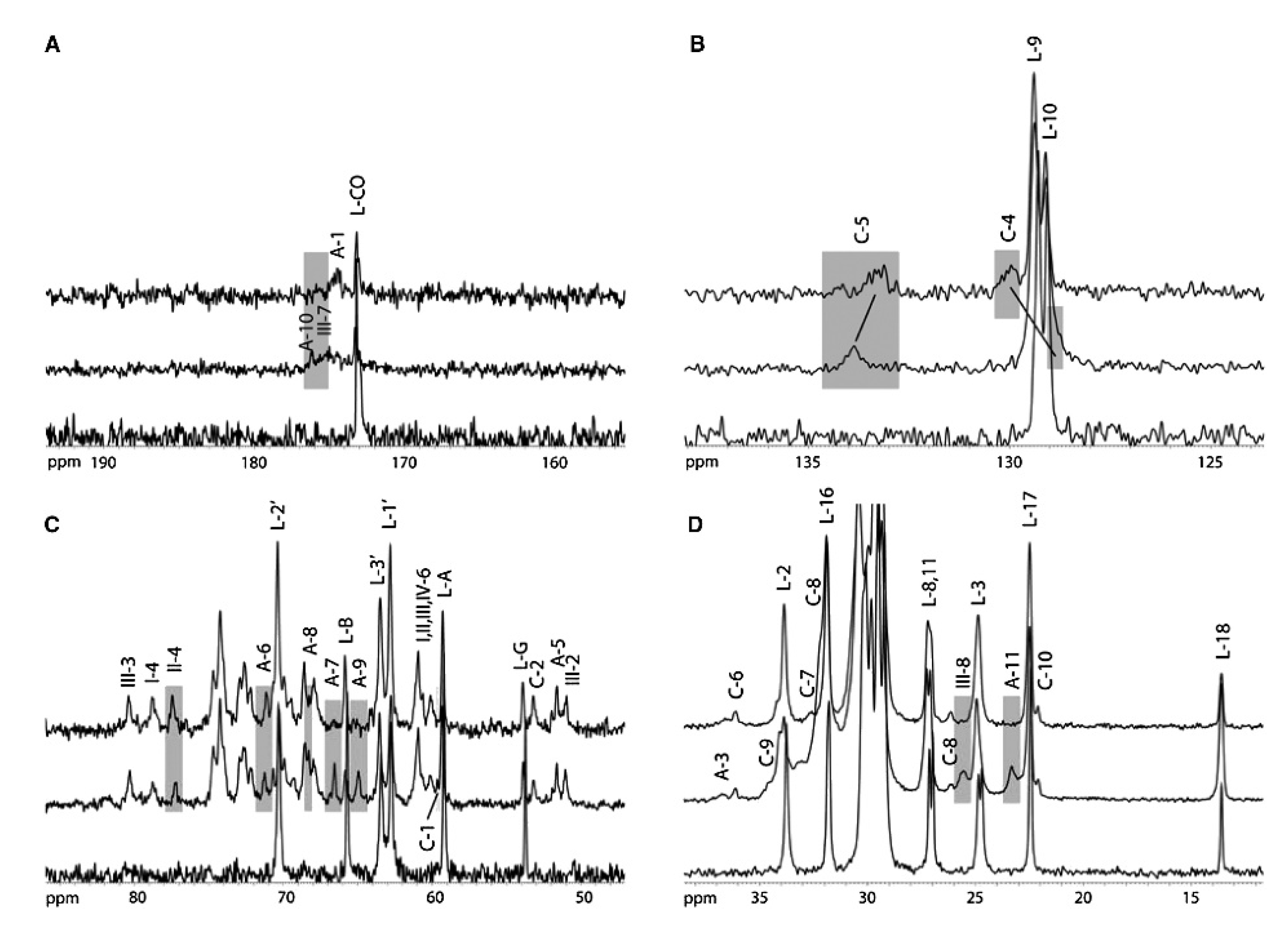
5. Solid-State NMR
6. Dynamic Nuclear Polarisation NMR
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Von Heijne, G. The membrane protein universe: What’s out there and why bother? J. Intern. Med. 2007, 261, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Bakheet, T.M.; Doig, A.J. Properties and identification of human protein drug targets. Bioinformatics 2009, 25, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Whited, A.M.; Johs, A. The interactions of peripheral membrane proteins with biological membranes. Chem. Phys. Lipids 2015, 192, 51–59. [Google Scholar] [CrossRef]
- Kobilka, B.K. G protein coupled receptor structure and activation. Biochim. Biophys. Acta Biomembr. 2007, 1768, 794–807. [Google Scholar] [CrossRef]
- Shimada, I.; Ueda, T.; Kofuku, Y.; Eddy, M.T.; Wüthrich, K. GPCR drug discovery: Integrating solution NMR data with crystal and cryo-EM structures. Nat. Rev. Drug Discov. 2019, 18, 59–82. [Google Scholar] [CrossRef]
- Tamm, L.K.; Hong, H.; Liang, B. Folding and assembly of β-barrel membrane proteins. Biochim. Biophys. Acta Biomembr. 2004, 1666, 250–263. [Google Scholar] [CrossRef]
- Fairman, J.W.; Noinaj, N.; Buchanan, S.K. The structural biology of β-barrel membrane proteins: A summary of recent reports. Curr. Opin. Struct. Biol. 2011, 21, 523–531. [Google Scholar] [CrossRef]
- Tapaneeyakorn, S.; Goddard, A.D.; Oates, J.; Willis, C.L.; Watts, A. Solution- and solid-state NMR studies of GPCRs and their ligands. Biochim. Biophys. Acta Biomembr. 2011, 1808, 1462–1475. [Google Scholar] [CrossRef]
- Yanamala, N.; Dutta, A.; Beck, B.; Fleet, B.V.; Hay, K.; Yazbak, A.; Ishima, R.; Doemling, A.; Klein-Seetharaman, J. NMR-Based Screening of Membrane Protein Ligands. Chem. Biol. Drug Des. 2010, 75, 237–256. [Google Scholar] [CrossRef]
- Purslow, J.A.; Khatiwada, B.; Bayro, M.J.; Venditti, V. NMR Methods for Structural Characterization of Protein-Protein Complexes. Front. Mol. Biosci. 2020, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Puthenveetil, R.; Vinogradova, O. Solution NMR: A powerful tool for structural and functional studies of membrane proteins in reconstituted environments. J. Biol. Chem. 2019, 294, 15914–15931. [Google Scholar] [CrossRef] [PubMed]
- Sattler, M.; Fesik, S.W. Use of deuterium labeling in NMR: Overcoming a sizeable problem. Structure 1996, 4, 1245–1249. [Google Scholar] [CrossRef]
- Bonev, B.; Grieve, S.; Herberstein, M.E.; Kishore, A.I.; Watts, A.; Separovic, F. Orientational order of Australian spider silks as determined by solid-state NMR. Biopolymers 2006, 82, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Lacabanne, D.; Meier, B.H.; Böckmann, A. Selective labeling and unlabeling strategies in protein solid-state NMR spectroscopy. J. Biomol. NMR 2018, 71, 141–150. [Google Scholar] [CrossRef]
- Sugiki, T.; Furuita, K.; Fujiwara, T.; Kojima, C. Amino Acid Selective 13C Labeling and 13C Scrambling Profile Analysis of Protein α and Side-Chain Carbons in Escherichia coli Utilized for Protein Nuclear Magnetic Resonance. Biochemistry 2018, 57, 3576–3589. [Google Scholar] [CrossRef]
- Solt, A.S.; Bostock, M.J.; Shrestha, B.; Kumar, P.; Warne, T.; Tate, C.G.; Nietlispach, D. Insight into partial agonism by observing multiple equilibria for ligand-bound and G s -mimetic nanobody-bound β1-adrenergic receptor. Nat. Commun. 2017, 8, 1795. [Google Scholar] [CrossRef]
- Kofuku, Y.; Ueda, T.; Okude, J.; Shiraishi, Y.; Kondo, K.; Mizumura, T.; Suzuki, S.; Shimada, I. Functional Dynamics of Deuterated β2-Adrenergic Receptor in Lipid Bilayers Revealed by NMR Spectroscopy. Angew. Chem. Int. Ed. 2014, 53, 13376–13379. [Google Scholar] [CrossRef]
- Warschawski, D.E.; Arnold, A.A.; Beaugrand, M.; Gravel, A.; Chartrand, É.; Marcotte, I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim. Biophys. Acta Biomembr. 2011, 1808, 1957–1974. [Google Scholar] [CrossRef]
- Garavito, R.M.; Ferguson-Miller, S. Detergents as Tools in Membrane Biochemistry. J. Biol. Chem. 2001, 276, 32403–32406. [Google Scholar] [CrossRef]
- Newby, Z.E.R.; O’Connell, J.D.; Gruswitz, F.; Hays, F.A.; Harries, W.E.C.; Harwood, I.M.; Ho, J.D.; Lee, J.K.; Savage, D.F.; Miercke, L.J.W.; et al. A general protocol for the crystallization of membrane proteins for X-ray structural investigation. Nat. Protoc. 2009, 4, 619–637. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, D.A.P.; Mizohata, E.; Newstead, S.; Ferrandon, S.; Henderson, P.J.F.; van Veen, H.W.; Byrne, B. A high-throughput method for membrane protein solubility screening: The ultracentrifugation dispersity sedimentation assay. Protein Sci. 2007, 16, 1422–1428. [Google Scholar] [CrossRef] [PubMed]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane proteins, lipids and detergents: Not just a soap opera. Biochim. Biophys. Acta Biomembr. 2004, 1666, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Franzin, C.M.; Gong, X.-M.; Thai, K.; Yu, J.; Marassi, F.M. NMR of membrane proteins in micelles and bilayers: The FXYD family proteins. Methods 2007, 41, 398–408. [Google Scholar] [CrossRef][Green Version]
- Prosser, R.S.; Evanics, F.; Kitevski, J.L.; Al-Abdul-Wahid, M.S. Current Applications of Bicelles in NMR Studies of Membrane-Associated Amphiphiles and Proteins. Biochemistry 2006, 45, 8453–8465. [Google Scholar] [CrossRef]
- Mueller, K. Structural dimorphism of bile salt/lecithin mixed micelles. A possible regulatory mechanism for cholesterol solubility in bile? X-ray structural analysis. Biochemistry 1981, 20, 404–414. [Google Scholar] [CrossRef]
- Sanders, C.R.; Landis, G.C. Reconstitution of Membrane Proteins into Lipid-Rich Bilayered Mixed Micelles for NMR Studies. Biochemistry 1995, 34, 4030–4040. [Google Scholar] [CrossRef]
- Van Dam, L.; Karlsson, G.; Edwards, K. Direct observation and characterization of DMPC/DHPC aggregates under conditions relevant for biological solution NMR. Biochim. Biophys. Acta Biomembr. 2004, 1664, 241–256. [Google Scholar] [CrossRef]
- Angelis, A.A.D.; Opella, S.J. Bicelle samples for solid-state NMR of membrane proteins. Nat. Protoc. 2007, 2, 2332–2338. [Google Scholar] [CrossRef]
- Nieh, M.-P.; Glinka, C.J.; Krueger, S.; Prosser, R.S.; Katsaras, J. SANS Study of the Structural Phases of Magnetically Alignable Lanthanide-Doped Phospholipid Mixtures. Langmuir 2001, 17, 2629–2638. [Google Scholar] [CrossRef]
- Wu, C.H.; Ramamoorthy, A.; Opella, S.J. High-Resolution Heteronuclear Dipolar Solid-State NMR Spectroscopy. J. Magn. Reson. A 1994, 109, 270–272. [Google Scholar] [CrossRef]
- Marassi, F.M.; Opella, S.J. A Solid-State NMR Index of Helical Membrane Protein Structure and Topology. J. Magn. Reson. 2000, 144, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Opella, S.J.; Marassi, F.M. Structure Determination of Membrane Proteins by NMR Spectroscopy. Chem. Rev. 2004, 104, 3587–3606. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tonggu, L. Membrane protein reconstitution for functional and structural studies. Sci. China Life Sci. 2015, 58, 66–74. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Grinkova, Y.V.; Sligar, S.G. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002, 2, 853–856. [Google Scholar] [CrossRef]
- Denisov, I.G.; Grinkova, Y.V.; Lazarides, A.A.; Sligar, S.G. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J. Am. Chem. Soc. 2004, 126, 3477–3487. [Google Scholar] [CrossRef]
- Hagn, F.; Etzkorn, M.; Raschle, T.; Wagner, G. Optimized Phospholipid Bilayer Nanodiscs Facilitate High-Resolution Structure Determination of Membrane Proteins. J. Am. Chem. Soc. 2013, 135, 1919–1925. [Google Scholar] [CrossRef]
- Hagn, F.; Nasr, M.L.; Wagner, G. Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR. Nat. Protoc. 2018, 13, 79. [Google Scholar] [CrossRef]
- Yusuf, Y.; Massiot, J.; Chang, Y.-T.; Wu, P.-H.; Yeh, V.; Kuo, P.-C.; Shiue, J.; Yu, T.-Y. Optimization of the Production of Covalently Circularized Nanodiscs and Their Characterization in Physiological Conditions. Langmuir 2018, 34, 3525–3532. [Google Scholar] [CrossRef]
- Nasr, M.L.; Baptista, D.; Strauss, M.; Sun, Z.-Y.J.; Grigoriu, S.; Huser, S.; Plückthun, A.; Hagn, F.; Walz, T.; Hogle, J.M.; et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods 2017, 14, 49–52. [Google Scholar] [CrossRef]
- Denisov, I.G.; Sligar, S.G. Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 2016, 23, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Denisov, I.G.; Sligar, S.G. Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.-Y.; Raschle, T.; Hiller, S.; Wagner, G. Solution NMR spectroscopic characterization of human VDAC-2 in detergent micelles and lipid bilayer nanodiscs. Biochim. Biophys. Acta Biomembr. 2012, 1818, 1562–1569. [Google Scholar] [CrossRef] [PubMed]
- Raschle, T.; Hiller, S.; Yu, T.-Y.; Rice, A.J.; Walz, T.; Wagner, G. Structural and Functional Characterization of the Integral Membrane Protein VDAC-1 in Lipid Bilayer Nanodiscs. J. Am. Chem. Soc. 2009, 131, 17777–17779. [Google Scholar] [CrossRef] [PubMed]
- Morgado, L.; Zeth, K.; Burmann, B.M.; Maier, T.; Hiller, S. Characterization of the insertase BamA in three different membrane mimetics by solution NMR spectroscopy. J. Biomol. NMR 2015, 61, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Kucharska, I.; Edrington, T.C.; Liang, B.; Tamm, L.K. Optimizing nanodiscs and bicelles for solution NMR studies of two β-barrel membrane proteins. J. Biomol. NMR 2015, 61, 261–274. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Vishnivetskiy, S.A.; McLean, M.A.; Morizumi, T.; Huang, C.; Tesmer, J.J.G.; Ernst, O.P.; Sligar, S.G.; Gurevich, V.V. Monomeric Rhodopsin Is Sufficient for Normal Rhodopsin Kinase (GRK1) Phosphorylation and Arrestin-1 Binding. J. Biol. Chem. 2011, 286, 1420–1428. [Google Scholar] [CrossRef]
- Lee, T.-Y.; Yeh, V.; Chuang, J.; Chan, J.C.C.; Chu, L.-K.; Yu, T.-Y. Tuning the Photocycle Kinetics of Bacteriorhodopsin in Lipid Nanodiscs. Biophys. J. 2015, 109, 1899–1906. [Google Scholar] [CrossRef][Green Version]
- Yeh, V.; Hsin, Y.; Lee, T.-Y.; Chan, J.C.C.; Yu, T.-Y.; Chu, L.-K. Lipids influence the proton pump activity of photosynthetic protein embedded in nanodiscs. RSC Adv. 2016, 6, 88300–88305. [Google Scholar] [CrossRef]
- Yeh, V.; Lee, T.-Y.; Chen, C.-W.; Kuo, P.-C.; Shiue, J.; Chu, L.-K.; Yu, T.-Y. Highly Efficient Transfer of 7TM Membrane Protein from Native Membrane to Covalently Circularized Nanodisc. Sci. Rep. 2018, 8, 13501. [Google Scholar] [CrossRef]
- Tribet, C.; Audebert, R.; Popot, J.-L. Amphipols: Polymers that keep membrane proteins soluble in aqueous solutions. Proc. Natl. Acad. Sci. USA 1996, 93, 15047–15050. [Google Scholar] [CrossRef] [PubMed]
- Gorzelle, B.M.; Hoffman, A.K.; Keyes, M.H.; Gray, D.N.; Ray, D.G.; Sanders, C.R. Amphipols Can Support the Activity of a Membrane Enzyme. J. Am. Chem. Soc. 2002, 124, 11594–11595. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.-T.H.; Helfinger, L.R.; Bostock, M.J.; Solt, A.; Tan, Y.L.; Nietlispach, D. An Adaptable Phospholipid Membrane Mimetic System for Solution NMR Studies of Membrane Proteins. J. Am. Chem. Soc. 2017. [Google Scholar] [CrossRef] [PubMed]
- Frauenfeld, J.; Löving, R.; Armache, J.-P.; Sonnen, A.F.-P.; Guettou, F.; Moberg, P.; Zhu, L.; Jegerschöld, C.; Flayhan, A.; Briggs, J.A.G.; et al. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat. Methods 2016, 13, 345–351. [Google Scholar] [CrossRef]
- Kolter, T.; Sandhoff, K. Lysosomal degradation of membrane lipids. FEBS Lett. 2010, 584, 1700–1712. [Google Scholar] [CrossRef]
- Knowles, T.J.; Finka, R.; Smith, C.; Lin, Y.-P.; Dafforn, T.; Overduin, M. Membrane Proteins Solubilized Intact in Lipid Containing Nanoparticles Bounded by Styrene Maleic Acid Copolymer. J. Am. Chem. Soc. 2009, 131, 7484–7485. [Google Scholar] [CrossRef]
- Orwick-Rydmark, M.; Lovett, J.E.; Graziadei, A.; Lindholm, L.; Hicks, M.R.; Watts, A. Detergent-Free Incorporation of a Seven-Transmembrane Receptor Protein into Nanosized Bilayer Lipodisq Particles for Functional and Biophysical Studies. Nano Lett. 2012, 12, 4687–4692. [Google Scholar] [CrossRef]
- Scheidelaar, S.; Koorengevel, M.; Pardo, J.; Meeldijk, J.; Breukink, E.; Killian, J. Antoinette Molecular Model for the Solubilization of Membranes into Nanodisks by Styrene Maleic Acid Copolymers. Biophys. J. 2015, 108, 279–290. [Google Scholar] [CrossRef]
- Dörr, J.M.; Koorengevel, M.C.; Schäfer, M.; Prokofyev, A.V.; Scheidelaar, S.; van der Cruijsen, E.A.W.; Dafforn, T.R.; Baldus, M.; Killian, J.A. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. USA 2014, 111, 18607–18612. [Google Scholar] [CrossRef]
- Long, A.R.; O’Brien, C.C.; Malhotra, K.; Schwall, C.T.; Albert, A.D.; Watts, A.; Alder, N.N. A detergent-free strategy for the reconstitution of active enzyme complexes from native biological membranes into nanoscale discs. BMC Biotechnol. 2013, 13, 41. [Google Scholar] [CrossRef]
- Smirnova, I.A.; Sjöstrand, D.; Li, F.; Björck, M.; Schäfer, J.; Östbye, H.; Högbom, M.; von Ballmoos, C.; Lander, G.C.; Ädelroth, P.; et al. Isolation of yeast complex IV in native lipid nanodiscs. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2984–2992. [Google Scholar] [CrossRef] [PubMed]
- Danmaliki, G.I.; Hwang, P.M. Solution NMR spectroscopy of membrane proteins. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183356. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Wu, J.; Yao, Y.; Singh, C.; Tian, Y.; Marassi, F.M.; Opella, S.J. Membrane proteins in magnetically aligned phospholipid polymer discs for solid-state NMR spectroscopy. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183333. [Google Scholar] [CrossRef] [PubMed]
- Ravula, T.; Kim, J.; Lee, D.-K.; Ramamoorthy, A. Magnetic Alignment of Polymer Nanodiscs Probed by Solid-State NMR Spectroscopy. Langmuir 2020, 36, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Cierpicki, T.; Liang, B.; Tamm, L.K.; Bushweller, J.H. Increasing the Accuracy of Solution NMR Structures of Membrane Proteins by Application of Residual Dipolar Couplings. High-Resolution Structure of Outer Membrane Protein A. J. Am. Chem. Soc. 2006, 128, 6947–6951. [Google Scholar] [CrossRef]
- Sprangers, R.; Velyvis, A.; Kay, L.E. Solution NMR of supramolecular complexes: Providing new insights into function. Nat. Methods 2007, 4, 697–703. [Google Scholar] [CrossRef]
- Horn, W.D.V.; Kim, H.-J.; Ellis, C.D.; Hadziselimovic, A.; Sulistijo, E.S.; Karra, M.D.; Tian, C.; Sönnichsen, F.D.; Sanders, C.R. Solution Nuclear Magnetic Resonance Structure of Membrane-Integral Diacylglycerol Kinase. Science 2009, 324, 1726–1729. [Google Scholar] [CrossRef]
- Zhou, Y.; Cierpicki, T.; Jimenez, R.H.F.; Lukasik, S.M.; Ellena, J.F.; Cafiso, D.S.; Kadokura, H.; Beckwith, J.; Bushweller, J.H. NMR Solution Structure of the Integral Membrane Enzyme DsbB: Functional Insights into DsbB-Catalyzed Disulfide Bond Formation. Mol. Cell 2008, 31, 896–908. [Google Scholar] [CrossRef]
- Liang, B.; Bushweller, J.H.; Tamm, L.K. Site-Directed Parallel Spin-Labeling and Paramagnetic Relaxation Enhancement in Structure Determination of Membrane Proteins by Solution NMR Spectroscopy. J. Am. Chem. Soc. 2006, 128, 4389–4397. [Google Scholar] [CrossRef]
- Hiller, S.; Garces, R.G.; Malia, T.J.; Orekhov, V.Y.; Colombini, M.; Wagner, G. Solution Structure of the Integral Human Membrane Protein VDAC-1 in Detergent Micelles. Science 2008, 321, 1206–1210. [Google Scholar] [CrossRef]
- Fernández, C.; Hilty, C.; Bonjour, S.; Adeishvili, K.; Pervushin, K.; Wüthrich, K. Solution NMR studies of the integral membrane proteins OmpX and OmpA from Escherichia coli. FEBS Lett. 2001, 504, 173–178. [Google Scholar] [CrossRef]
- Fernández, C.; Hilty, C.; Wider, G.; Güntert, P.; Wüthrich, K. NMR Structure of the Integral Membrane Protein OmpX. J. Mol. Biol. 2004, 336, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Bibow, S.; Carneiro, M.G.; Sabo, T.M.; Schwiegk, C.; Becker, S.; Riek, R.; Lee, D. Measuring membrane protein bond orientations in nanodiscs via residual dipolar couplings. Protein Sci. 2014, 23, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Abildgaard, F.; Bushweller, J.H.; Tamm, L.K. Structure of outer membrane protein a transmembrane domain by NMR spectroscopy. Nat. Struct. Mol. Biol. 2001, 8, 334–338. [Google Scholar] [CrossRef]
- Liang, B.; Tamm, L.K. Structure of outer membrane protein G by solution NMR spectroscopy. Proc. Natl. Acad. Sci. USA 2007, 104, 16140–16145. [Google Scholar] [CrossRef]
- Gautier, A.; Mott, H.R.; Bostock, M.J.; Kirkpatrick, J.P.; Nietlispach, D. Structure determination of the seven-helix transmembrane receptor sensory rhodopsin II by solution NMR spectroscopy. Nat. Struct. Mol. Biol. 2010, 17, 768–774. [Google Scholar] [CrossRef]
- Jaremko, Ł.; Jaremko, M.; Giller, K.; Becker, S.; Zweckstetter, M. Structure of the Mitochondrial Translocator Protein in Complex with a Diagnostic Ligand. Science 2014, 343, 1363–1366. [Google Scholar] [CrossRef]
- OuYang, B.; Xie, S.; Berardi, M.J.; Zhao, X.; Dev, J.; Yu, W.; Sun, B.; Chou, J.J. Unusual architecture of the p7 channel from hepatitis C virus. Nature 2013, 498, 521–525. [Google Scholar] [CrossRef]
- Oestringer, B.P.; Bolivar, J.H.; Claridge, J.K.; Almanea, L.; Chipot, C.; Dehez, F.; Holzmann, N.; Schnell, J.R.; Zitzmann, N. Hepatitis C virus sequence divergence preserves p7 viroporin structural and dynamic features. Sci. Rep. 2019, 9, 8383. [Google Scholar] [CrossRef]
- Oestringer, B.P.; Bolivar, J.H.; Hensen, M.; Claridge, J.K.; Chipot, C.; Dehez, F.; Holzmann, N.; Zitzmann, N.; Schnell, J.R. Re-evaluating the p7 viroporin structure. Nature 2018, 562, E8–E18. [Google Scholar] [CrossRef]
- Chen, W.; Dev, J.; Mezhyrova, J.; Pan, L.; Piai, A.; Chou, J.J. The Unusual Transmembrane Partition of the Hexameric Channel of the Hepatitis C Virus. Structure 2018, 26, 627–634.e4. [Google Scholar] [CrossRef] [PubMed]
- Fisher, L.E.; Engelman, D.M.; Sturgis, J.N. Effect of Detergents on the Association of the Glycophorin A Transmembrane Helix. Biophys. J. 2003, 85, 3097–3105. [Google Scholar] [CrossRef][Green Version]
- Lakomek, N.-A.; Frey, L.; Bibow, S.; Böckmann, A.; Riek, R.; Meier, B.H. Proton-Detected NMR Spectroscopy of Nanodisc-Embedded Membrane Proteins: MAS Solid-State vs Solution-State Methods. J. Phys. Chem. B 2017, 121, 7671–7680. [Google Scholar] [CrossRef] [PubMed]
- Hallock, K.J.; Henzler Wildman, K.; Lee, D.-K.; Ramamoorthy, A. An Innovative Procedure Using a Sublimable Solid to Align Lipid Bilayers for Solid-State NMR Studies. Biophys. J. 2002, 82, 2499–2503. [Google Scholar] [CrossRef][Green Version]
- Opella, S.J.; Marassi, F.M.; Gesell, J.J.; Valente, A.P.; Kim, Y.; Oblatt-Montal, M.; Montal, M. Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy. Nat. Struct. Biol. 1999, 6, 374–379. [Google Scholar] [CrossRef]
- Park, S.H.; Prytulla, S.; De Angelis, A.A.; Brown, J.M.; Kiefer, H.; Opella, S.J. High-Resolution NMR Spectroscopy of a GPCR in Aligned Bicelles. J. Am. Chem. Soc. 2006, 128, 7402–7403. [Google Scholar] [CrossRef]
- Bowie, J.U. Helix packing in membrane proteins. J. Mol. Biol. 1997, 272, 780–789. [Google Scholar] [CrossRef]
- De Planque, M.R.R.; Bonev, B.B.; Demmers, J.A.A.; Greathouse, D.V.; Koeppe, R.E.; Separovic, F.; Watts, A.; Killian, J.A. Interfacial Anchor Properties of Tryptophan Residues in Transmembrane Peptides Can Dominate over Hydrophobic Matching Effects in Peptide−Lipid Interactions †. Biochemistry 2003, 42, 5341–5348. [Google Scholar] [CrossRef]
- Zhou, D.H.; Nieuwkoop, A.J.; Berthold, D.A.; Comellas, G.; Sperling, L.J.; Tang, M.; Shah, G.J.; Brea, E.J.; Lemkau, L.R.; Rienstra, C.M. Solid-state NMR analysis of membrane proteins and protein aggregates by proton detected spectroscopy. J. Biomol. NMR 2012, 54, 291–305. [Google Scholar] [CrossRef]
- Linser, R.; Dasari, M.; Hiller, M.; Higman, V.; Fink, U.; Lopez del Amo, J.-M.; Markovic, S.; Handel, L.; Kessler, B.; Schmieder, P.; et al. Proton-Detected Solid-State NMR Spectroscopy of Fibrillar and Membrane Proteins. Angew. Chem. Int. Ed. 2011, 50, 4508–4512. [Google Scholar] [CrossRef]
- Barbet-Massin, E.; Pell, A.J.; Retel, J.S.; Andreas, L.B.; Jaudzems, K.; Franks, W.T.; Nieuwkoop, A.J.; Hiller, M.; Higman, V.; Guerry, P.; et al. Rapid Proton-Detected NMR Assignment for Proteins with Fast Magic Angle Spinning. J. Am. Chem. Soc. 2014, 136, 12489–12497. [Google Scholar] [CrossRef] [PubMed]
- Andreas, L.B.; Jaudzems, K.; Stanek, J.; Lalli, D.; Bertarello, A.; Marchand, T.L.; Paepe, D.C.-D.; Kotelovica, S.; Akopjana, I.; Knott, B.; et al. Structure of fully protonated proteins by proton-detected magic-angle spinning NMR. Proc. Natl. Acad. Sci. USA 2016, 113, 9187–9192. [Google Scholar] [CrossRef] [PubMed]
- Szeverenyi, N.M.; Sullivan, M.J.; Maciel, G.E. Observation of spin exchange by two-dimensional fourier transform 13C cross polarization-magic-angle spinning. J. Magn. Reson. 1969 1982, 47, 462–475. [Google Scholar] [CrossRef]
- Takegoshi, K.; Nakamura, S.; Terao, T. 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001, 344, 631–637. [Google Scholar] [CrossRef]
- Bennett, A.E.; Rienstra, C.M.; Griffiths, J.M.; Zhen, W.; Lansbury, P.T.; Griffin, R.G. Homonuclear radio frequency-driven recoupling in rotating solids. J. Chem. Phys. 1998, 108, 9463–9479. [Google Scholar] [CrossRef]
- Das, B.B.; Nothnagel, H.J.; Lu, G.J.; Son, W.S.; Tian, Y.; Marassi, F.M.; Opella, S.J. Structure Determination of a Membrane Protein in Proteoliposomes. J. Am. Chem. Soc. 2012, 134, 2047–2056. [Google Scholar] [CrossRef]
- Park, S.H.; Das, B.B.; Casagrande, F.; Tian, Y.; Nothnagel, H.J.; Chu, M.; Kiefer, H.; Maier, K.; De Angelis, A.A.; Marassi, F.M.; et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature 2012, 491, 779–783. [Google Scholar] [CrossRef]
- Wylie, B.J.; Bhate, M.P.; McDermott, A.E. Transmembrane allosteric coupling of the gates in a potassium channel. Proc. Natl. Acad. Sci. USA 2014, 111, 185–190. [Google Scholar] [CrossRef]
- Wang, S.; Munro, R.A.; Shi, L.; Kawamura, I.; Okitsu, T.; Wada, A.; Kim, S.-Y.; Jung, K.-H.; Brown, L.S.; Ladizhansky, V. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nat. Methods 2013, 10, 1007–1012. [Google Scholar] [CrossRef]
- Van der Cruijsen, E.A.W.; Prokofyev, A.V.; Pongs, O.; Baldus, M. Probing Conformational Changes during the Gating Cycle of a Potassium Channel in Lipid Bilayers. Biophys. J. 2017, 112, 99–108. [Google Scholar] [CrossRef]
- Pinto, C.; Mance, D.; Julien, M.; Daniels, M.; Weingarth, M.; Baldus, M. Studying assembly of the BAM complex in native membranes by cellular solid-state NMR spectroscopy. J. Struct. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Hohwy, M.; Jakobsen, H.J.; Eden, M.; Levitt, M.H.; Nielsen, N.C. Broadband dipolar recoupling in the nuclear magnetic resonance of rotating solids: A compensated C7 pulse sequence. J. Chem. Phys. 1998, 108, 2686–2694. [Google Scholar] [CrossRef]
- Carravetta, M.; Edén, M.; Zhao, X.; Brinkmann, A.; Levitt, M.H. Symmetry principles for the design of radiofrequency pulse sequences in the nuclear magnetic resonance of rotating solids. Chem. Phys. Lett. 2000, 321, 205–215. [Google Scholar] [CrossRef]
- Hohwy, M.; Rienstra, C.M.; Jaroniec, C.P.; Griffin, R.G. Fivefold symmetric homonuclear dipolar recoupling in rotating solids: Application to double quantum spectroscopy. J. Chem. Phys. 1999, 110, 7983–7992. [Google Scholar] [CrossRef]
- Lopez, J.J.; Kaiser, C.; Shastri, S.; Glaubitz, C. Double quantum filtering homonuclear MAS NMR correlation spectra: A tool for membrane protein studies. J. Biomol. NMR 2008, 41, 97–104. [Google Scholar] [CrossRef]
- Elkins, M.R.; Sergeyev, I.V.; Hong, M. Determining Cholesterol Binding to Membrane Proteins by Cholesterol 13C Labeling in Yeast and Dynamic Nuclear Polarization NMR. J. Am. Chem. Soc. 2018, 140, 15437–15449. [Google Scholar] [CrossRef]
- Lopez, J.J.; Shukla, A.K.; Reinhart, C.; Schwalbe, H.; Michel, H.; Glaubitz, C. The Structure of the Neuropeptide Bradykinin Bound to the Human G-Protein Coupled Receptor Bradykinin B2 as Determined by Solid-State NMR Spectroscopy. Angew. Chem. Int. Ed. 2008, 47, 1668–1671. [Google Scholar] [CrossRef]
- Gullion, T.; Schaefer, J. Rotational-echo double-resonance NMR. J. Magn. Reson. 1969 1989, 81, 196–200. [Google Scholar] [CrossRef]
- Jia, L.; Liang, S.; Sackett, K.; Xie, L.; Ghosh, U.; Weliky, D.P. REDOR solid-state NMR as a probe of the membrane locations of membrane-associated peptides and proteins. J. Magn. Reson. 2015, 253, 154–165. [Google Scholar] [CrossRef][Green Version]
- Murphy, O.J.; Kovacs, F.A.; Sicard, E.L.; Thompson, L.K. Site-Directed Solid-State NMR Measurement of a Ligand-Induced Conformational Change in the Serine Bacterial Chemoreceptor. Biochemistry 2001, 40, 1358–1366. [Google Scholar] [CrossRef]
- Pines, A.; Gibby, M.G.; Waugh, J.S. Proton-enhanced nuclear induction spectroscopy 13C chemical shielding anisotropy in some organic solids. Chem. Phys. Lett. 1972, 15, 373–376. [Google Scholar] [CrossRef]
- Hartmann, S.R.; Hahn, E.L. Nuclear Double Resonance in the Rotating Frame. Phys. Rev. 1962, 128, 2042–2053. [Google Scholar] [CrossRef]
- Sanghera, N.; Correia, B.E.F.S.; Correia, J.R.S.; Ludwig, C.; Agarwal, S.; Nakamura, H.K.; Kuwata, K.; Samain, E.; Gill, A.C.; Bonev, B.B.; et al. Deciphering the Molecular Details for the Binding of the Prion Protein to Main Ganglioside GM1 of Neuronal Membranes. Chem. Biol. 2011, 18, 1422–1431. [Google Scholar] [CrossRef] [PubMed]
- Sinnige, T.; Houben, K.; Pritisanac, I.; Renault, M.; Boelens, R.; Baldus, M. Insight into the conformational stability of membrane-embedded BamA using a combined solution and solid-state NMR approach. J. Biomol. NMR 2015, 61, 321–332. [Google Scholar] [CrossRef]
- Bhate, M.P.; McDermott, A.E. Protonation state of E71 in KcsA and its role for channel collapse and inactivation. Proc. Natl. Acad. Sci. USA 2012, 109, 15265–15270. [Google Scholar] [CrossRef]
- Varga, K.; Tian, L.; McDermott, A.E. Solid-state NMR study and assignments of the KcsA potassium ion channel of S. lividans. Biochim. Biophys. Acta Proteins Proteom. 2007, 1774, 1604–1613. [Google Scholar] [CrossRef]
- Ader, C.; Pongs, O.; Becker, S.; Baldus, M. Protein dynamics detected in a membrane-embedded potassium channel using two-dimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta Biomembr. 2010, 1798, 286–290. [Google Scholar] [CrossRef]
- Schneider, R.; Ader, C.; Lange, A.; Giller, K.; Hornig, S.; Pongs, O.; Becker, S.; Baldus, M. Solid-State NMR Spectroscopy Applied to a Chimeric Potassium Channel in Lipid Bilayers. J. Am. Chem. Soc. 2008, 130, 7427–7435. [Google Scholar] [CrossRef]
- Park, S.H.; Casagrande, F.; Cho, L.; Albrecht, L.; Opella, S.J. Interactions of Interleukin-8 with the Human Chemokine Receptor CXCR1 in Phospholipid Bilayers by NMR Spectroscopy. J. Mol. Biol. 2011, 414, 194–203. [Google Scholar] [CrossRef]
- Song, C.; Hu, K.-N.; Joo, C.-G.; Swager, T.M.; Griffin, R.G. TOTAPOL: A Biradical Polarizing Agent for Dynamic Nuclear Polarization Experiments in Aqueous Media. J. Am. Chem. Soc. 2006, 128, 11385–11390. [Google Scholar] [CrossRef]
- Sauvée, C.; Rosay, M.; Casano, G.; Aussenac, F.; Weber, R.T.; Ouari, O.; Tordo, P. Highly Efficient, Water-Soluble Polarizing Agents for Dynamic Nuclear Polarization at High Frequency. Angew. Chem. Int. Ed. 2013, 52, 10858–10861. [Google Scholar] [CrossRef] [PubMed]
- Zagdoun, A.; Casano, G.; Ouari, O.; Schwarzwälder, M.; Rossini, A.J.; Aussenac, F.; Yulikov, M.; Jeschke, G.; Copéret, C.; Lesage, A.; et al. Large Molecular Weight Nitroxide Biradicals Providing Efficient Dynamic Nuclear Polarization at Temperatures up to 200 K. J. Am. Chem. Soc. 2013, 135, 12790–12797. [Google Scholar] [CrossRef] [PubMed]
- Van der Cruijsen, E.A.W.; Koers, E.J.; Sauvée, C.; Hulse, R.E.; Weingarth, M.; Ouari, O.; Perozo, E.; Tordo, P.; Baldus, M. Biomolecular DNP-Supported NMR Spectroscopy using Site-Directed Spin Labeling. Chem. Eur. J. 2015, 21, 12971–12977. [Google Scholar] [CrossRef] [PubMed]
- Fernández-de-Alba, C.; Takahashi, H.; Richard, A.; Chenavier, Y.; Dubois, L.; Maurel, V.; Lee, D.; Hediger, S.; De Paëpe, G. Matrix-Free DNP-Enhanced NMR Spectroscopy of Liposomes Using a Lipid-Anchored Biradical. Chem. Eur. J. 2015, 21, 4512–4517. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, V.S.; Mak-Jurkauskas, M.L.; Belenky, M.; Herzfeld, J.; Griffin, R.G. Functional and shunt states of bacteriorhodopsin resolved by 250 GHz dynamic nuclear polarization–enhanced solid-state NMR. Proc. Natl. Acad. Sci. USA 2009, 106, 9244–9249. [Google Scholar] [CrossRef] [PubMed]
- Ni, Q.Z.; Can, T.V.; Daviso, E.; Belenky, M.; Griffin, R.G.; Herzfeld, J. Primary Transfer Step in the Light-Driven Ion Pump Bacteriorhodopsin: An Irreversible U-Turn Revealed by Dynamic Nuclear Polarization-Enhanced Magic Angle Spinning NMR. J. Am. Chem. Soc. 2018, 140, 4085–4091. [Google Scholar] [CrossRef]
- Becker-Baldus, J.; Bamann, C.; Saxena, K.; Gustmann, H.; Brown, L.J.; Brown, R.C.D.; Reiter, C.; Bamberg, E.; Wachtveitl, J.; Schwalbe, H.; et al. Enlightening the photoactive site of channelrhodopsin-2 by DNP-enhanced solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 9896–9901. [Google Scholar] [CrossRef]
- Stöppler, D.; Song, C.; van Rossum, B.-J.; Geiger, M.-A.; Lang, C.; Mroginski, M.-A.; Jagtap, A.P.; Sigurdsson, S.T.; Matysik, J.; Hughes, J.; et al. Dynamic Nuclear Polarization Provides New Insights into Chromophore Structure in Phytochrome Photoreceptors. Angew. Chem. Int. Ed. 2016, 55, 16017–16020. [Google Scholar] [CrossRef]
- Koers, E.J.; van der Cruijsen, E.A.W.; Rosay, M.; Weingarth, M.; Prokofyev, A.; Sauvée, C.; Ouari, O.; van der Zwan, J.; Pongs, O.; Tordo, P.; et al. NMR-based structural biology enhanced by dynamic nuclear polarization at high magnetic field. J. Biomol. NMR 2014, 60, 157–168. [Google Scholar] [CrossRef]
- Ong, Y.S.; Lakatos, A.; Becker-Baldus, J.; Pos, K.M.; Glaubitz, C. Detecting Substrates Bound to the Secondary Multidrug Efflux Pump EmrE by DNP-Enhanced Solid-State NMR. J. Am. Chem. Soc. 2013, 135, 15754–15762. [Google Scholar] [CrossRef]
- Lehnert, E.; Mao, J.; Mehdipour, A.R.; Hummer, G.; Abele, R.; Glaubitz, C.; Tampé, R. Antigenic Peptide Recognition on the Human ABC Transporter TAP Resolved by DNP-Enhanced Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2016, 138, 13967–13974. [Google Scholar] [CrossRef] [PubMed]
- Joedicke, L.; Mao, J.; Kuenze, G.; Reinhart, C.; Kalavacherla, T.; Jonker, H.R.A.; Richter, C.; Schwalbe, H.; Meiler, J.; Preu, J.; et al. The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors. Nat. Chem. Biol. 2018, 14, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Jacso, T.; Franks, W.T.; Rose, H.; Fink, U.; Broecker, J.; Keller, S.; Oschkinat, H.; Reif, B. Characterization of Membrane Proteins in Isolated Native Cellular Membranes by Dynamic Nuclear Polarization Solid-State NMR Spectroscopy without Purification and Reconstitution. Angew. Chem. Int. Ed. 2012, 51, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Viennet, T.; Viegas, A.; Kuepper, A.; Arens, S.; Gelev, V.; Petrov, O.; Grossmann, T.N.; Heise, H.; Etzkorn, M. Selective Protein Hyperpolarization in Cell Lysates Using Targeted Dynamic Nuclear Polarization. Angew. Chem. Int. Ed. 2016, 55, 10746–10750. [Google Scholar] [CrossRef]
- Linden, A.H.; Lange, S.; Franks, W.T.; Akbey, Ü.; Specker, E.; van Rossum, B.-J.; Oschkinat, H. Neurotoxin II Bound to Acetylcholine Receptors in Native Membranes Studied by Dynamic Nuclear Polarization NMR. J. Am. Chem. Soc. 2011, 133, 19266–19269. [Google Scholar] [CrossRef]
- Takahashi, H.; Ayala, I.; Bardet, M.; De Paëpe, G.; Simorre, J.-P.; Hediger, S. Solid-State NMR on Bacterial Cells: Selective Cell Wall Signal Enhancement and Resolution Improvement using Dynamic Nuclear Polarization. J. Am. Chem. Soc. 2013, 135, 5105–5110. [Google Scholar] [CrossRef]
- Yamamoto, K.; Caporini, M.A.; Im, S.-C.; Waskell, L.; Ramamoorthy, A. Cellular solid-state NMR investigation of a membrane protein using dynamic nuclear polarization. Biochim. Biophys. Acta Biomembr. 2015, 1848, 342–349. [Google Scholar] [CrossRef]
- Albert, B.J.; Gao, C.; Sesti, E.L.; Saliba, E.P.; Alaniva, N.; Scott, F.J.; Sigurdsson, S.T.; Barnes, A.B. Dynamic Nuclear Polarization Nuclear Magnetic Resonance in Human Cells Using Fluorescent Polarizing Agents. Biochemistry 2018, 57, 4741–4746. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeh, V.; Goode, A.; Bonev, B.B. Membrane Protein Structure Determination and Characterisation by Solution and Solid-State NMR. Biology 2020, 9, 396. https://doi.org/10.3390/biology9110396
Yeh V, Goode A, Bonev BB. Membrane Protein Structure Determination and Characterisation by Solution and Solid-State NMR. Biology. 2020; 9(11):396. https://doi.org/10.3390/biology9110396
Chicago/Turabian StyleYeh, Vivien, Alice Goode, and Boyan B. Bonev. 2020. "Membrane Protein Structure Determination and Characterisation by Solution and Solid-State NMR" Biology 9, no. 11: 396. https://doi.org/10.3390/biology9110396
APA StyleYeh, V., Goode, A., & Bonev, B. B. (2020). Membrane Protein Structure Determination and Characterisation by Solution and Solid-State NMR. Biology, 9(11), 396. https://doi.org/10.3390/biology9110396




