Abstract
In vivo functionalization of diatom biosilica frustules by genetic manipulation requires careful consideration of the overall structure and function of complex fusion proteins. Although we previously had transformed Thalassiosira pseudonana with constructs containing a single domain antibody (sdAb) raised against the Bacillus anthracis Sterne strain, which detected an epitope of the surface layer protein EA1 accessible in lysed spores, we initially were unsuccessful with constructs encoding a similar sdAb that detected an epitope of EA1 accessible in intact spores and vegetative cells. This discrepancy limited the usefulness of the system as an environmental biosensor for B. anthracis. We surmised that to create functional biosilica-localized biosensors with certain constructs, the biosilica targeting and protein trafficking functions of the biosilica-targeting peptide Sil3T8 had to be uncoupled. We found that retaining the ER trafficking sequence at the N-terminus and relocating the Sil3T8 targeting peptide to the C-terminus of the fusion protein resulted in successful detection of EA1 with both sdAbs. Homology modeling of antigen binding by the two sdAbs supported the hypothesis that the rescue of antigen binding in the previously dysfunctional sdAb was due to removal of steric hindrances between the antigen binding loops and the diatom biosilica for that particular sdAb.
1. Introduction
Diatoms are a group of unicellular microalgae, often with a highly silicified, mesoporous cell wall (frustule) exhibiting nano- to meso-scale hierarchical architecture [1,2,3]. Extraction of diatom frustules can be accomplished through acid washing (e.g., [4,5]) to remove all organic components, or by detergent extraction [6] to retain the protein complement. Acid washed diatom frustules resemble de novo assembled mesoporous silica nanoparticles [7,8]. Whether bioinspired or diatom-derived, these silicate structures can be chemically modified for a wide variety of uses, such as biomedical applications [9] (e.g., cell scaffolding [10,11] and drug delivery [12,13]) or biomass conversion [14]. One attractive method for functionalizing diatom biosilica is through genetic engineering of diatoms, whereby a biosilica-targeting protein or a derivative peptide thereof is fused to a protein of interest that is to be embedded into the biosilica through in vivo self-assembly. Such an approach does not require additional chemicals for attachment, and is scalable to large growth volumes.
The diatom Thalassiosira pseudonana is a model organism for in vivo self-assembly of genetically-modified frustules. As such, a number of proteins successfully have been used to functionalize the biosilica frustule of this diatom species [15,16,17,18]. Recent efforts in our lab focused on functionalization of the biosilica of T. pseudonana with chimeric fusion proteins consisting of the diatom-derived silica targeting peptide Sil3T8 [15,16] and a small synthetic antibody derivative (e.g., a single-chain variable fragment, scFv, or a single domain antibody, sdAb) [18,19]. Functionalization of T. pseudonana biosilica with an sdAb against the surface layer (S-layer) protein extractable antigen 1 (EA1) of Bacillus anthracis [18] by in vivo self-assembly utilized sdAbEA1, clone A1, which recognizes an epitope of EA1 accessible in lysed spores [20]. Preliminary work with sdAbEA1, clone G10, which recognizes an epitope of EA1 that is accessible in intact vegetative cells and spores [20], produced no functional diatom lines when targeted to the biosilica frustule. Noting that the N-terminus of the llama VHH domain sits adjacent to the binding loops (see Figure 2 of [21], for example), we hypothesized that our existing biosilica targeting constructs with the Sil3T8 peptide fused to the N-terminus of the sdAbEA1 might produce a fusion protein whereby the antigen’s access to the sdAb binding loops was occluded. For this particular protein to be functional when tethered to diatom biosilica, fusion protein structure needed to be optimized. Our solution was to uncouple the silica targeting peptide Sil3T8 from its ER targeting sequence and locate Sil3T8 at the C-terminus of the fusion protein. After doing so, the biosilica functionalized with sdAbEA1 clone G10—containing fusion proteins was able to bind its target antigen EGFP-tagged EA1 protein.
2. Materials and Methods
2.1. Diatoms
Native and transformed cultures of T. pseudonana (CCMP1335; Provasoli-Guillard National Center for Marine Algae and Microbiota, East Boothbay, ME, USA) were maintained in artificial seawater (ESAW; https://ncma.bigelow.org/media/pdf/NCMAalgalmedium.ESAW.pdf) supplemented with 100 μg/mL penicillin (VWR, Visalia, CA, USA) and 100 μg/mL streptomycin (Sigma-Aldrich, St. Louis, MO, USA) under continuous illumination on an orbital shaker (~10–40 μmol/m2/s, 20–22 °C) with gentle agitation or in a Caron (Marietta, OH, USA) plant incubator (150 μmol/m2/s, 20 °C) without agitation. Diatoms were transformed by microparticle bombardment with a PDS-1000/He particle delivery system as was previously described [18]. To verify integration of Gateway expression clones into the T. pseudonana genome, PCR was performed directly on 5 μL diatom culture (1/10 volume of PCR). Sequences of PCR primers are described in Table S1. GAPDH was used as the control gene to verify presence of T. pseudonana DNA. Diatom biosilica frustules were isolated as previously described for detergent extraction at 50 °C with acetone rinse [6], but substituting 1% Igepal CA-630 (Sigma-Aldrich, St. Louis, MO, USA) for 1% SDS [18].
2.2. Expression Clone Construction
A diatom-specific destination vector (termed pDDV2 for Diatom Destination Vector #2) was created by restriction cloning to contain the T. pseudonana fcp promoter [22] followed by the endoplasmic reticulum trafficking sequence coding for the peptide MKTSAIVLLAVLATTAATEPR, the Gateway attR1/2 integration cassette with V5 and His6 tags, and the T. pseudonana fcp terminator [22]. The Multi-Site Gateway Pro cloning protocol was used to construct diatom-specific expression clones for biosilica-targeted fusion proteins using either the pDDV2 (this work) or the previously created pDDV1 [18]. Plasmids containing the unmodified sdAbEA1 (either clone A1 or G10) [20] and Sil3T8 [15,16] targeting sequences were used as templates for PCR to create entry clones for insertion into the pDDV vectors. Detailed descriptions of all clonings are available in the Supplementary Information.
2.3. Fluorescent Antigen Synthesis and Binding to Single Domain Antibodies
EA1-EGFP fusion protein was expressed and purified as previously described [18]. Isolated biosilica frustules from untransformed T. pseudonana and T. pseudonana cell lines transformed with various biosilica-targeted sdAbEA1 (either clone A1 or G10) fusion proteins were incubated with a saturating amount of EA1-EGFP (125 nM) in PBS containing 0.05% Tween-20 (Fisher, Hampton, NJ, USA) and 1% BSA Fraction V (Fisher, Hampton, NJ, USA) for 1 h at 4 °C, followed by 1 h at room temperature (20–25 °C). Antigen-bound frustules were washed three times with PBS containing 0.05% Tween-20 prior to imaging in the same buffer on PEI-coated coverslips. Frustule fluorescence was examined with a Leica DM IRB inverted epifluorescence microscope equipped with a mercury metal halide light source and liquid light guide (Leica, Wetzlar, Germany). A 460–500/505/512–542 nm filter cube was used to collect GFP fluorescence and a 635–675/716/696–736 nm filter cube was used to verify the absence of chlorophyll in frustule samples. Images were captured with a CoolSNAP Myo camera (Photometrics, Tucson, AZ, USA) and Metamorph software (v.7.7.11.0; Molecular Devices, San Jose, CA, USA). Frustules lacking detectable chlorophyll fluorescence and not overlapping any other frustules were manually selected and their GFP-channel fluorescence intensity measured using the Metamorph software package (v.7.7.11.0; Molecular Devices, San Jose, CA, USA).
2.4. Protein Modeling
The T. pseudonana silaffin precursor amino acid sequence (Sil3, GenBank: AAU44819.1) in FASTA format was input into SignalP-5.0 [23,24]. Eukarya was selected as the organism group, and long output format with figures was selected. SignalP-5.0 signal peptide prediction results are shown in Figure S1.
Homology modeling of the structures of the sdAbs was conducted using the SWISS-MODEL server [25]. The sdAbEA1 sequences (both clones A1 and G10) were uploaded to the ExPASy web server, and the server searched evolutionary related protein structures against the SWISSMODEL template library (SMTL) using two search methods: BLAST and HHblits. The templates were ranked according to expected quality of the resulting models, as estimated by Global Model Quality Estimate (GMQE) and Quaternary Structure Quality Estimate (QSQE). For each single domain antibody, the top-ranked template was chosen and the homology structure was built based on that template [25,26,27]. For sdAbEA1, clone G10, the top hit was 5F10 with a GMQE score of 0.78, and for sdAbEA1, clone A1, the top ranked template was 6GLW with a GMQE score of 0.77. The models were downloaded and the 3D protein structure visualizations and alignments were done with the open source protein structure visualization program Pymol [28]. Since neither of the top-ranked templates was a structure with bound antigen, we searched further in the template lists with lower GMQE scores to look for sdAb structures with bound antigen. Two protein structures were found: a fusion protein of two domains, Rpn8 and Rpn11, of the 26S proteasome’s deubiquitylation module bound to an sdAb (pdb bank ID 4OCN) [29] and Shiga toxin (stx2e) binding to a neutralizing sdAb (pdb bank ID 4P2C) [30] (Figure S2).
2.5. Statistical Analysis
The data were analyzed using one-way Analysis of Variance and Tukey’s HSD (honest significant difference) posthoc test for between-group differences (significant at p ≤ 0.05). The graphical comparisons in Figures 2 and 3 were derived from a study comparing diatoms transformed with generation 1 and generation 2, i.e., pDDV1 and 2 pDDV2, expression vectors- and a reference group comprising untransformed native diatoms.
3. Results
3.1. Re-Designing the Fusion Constructs
Previous reports that used Silaffin 3 (Sil3) [6] or its truncated derivative Sil3T8 [15] for in vivo tethering of fusion proteins to T. pseudonana diatom biosilica, located the silaffin domain at the N-terminus of the protein. When tethering an sdAb to the diatom biosilica in this fashion, we hypothesized based on structural considerations that the antigen binding loops would be oriented inward and adjacent to the biosilica rather than being exposed and facing outward away from the biosilica. In order to rotate the binding loops of our sdAbs away from the biosilica surface, and thereby increase antigen accessibility, the Sil3T8 tether needed to become a C-terminal fusion to the sdAb. But, because the Sil3T8 peptide contains an endoplasmic reticulum (ER) trafficking sequence [15,16], which needed to remain at the N-terminus of the fusion protein sequence, simply swapping the order of sdAbEA1 and Sil3T8 in our modular Gateway (Invitrogen) cloning system was not a feasible solution. Thus, re-designing the biosilica-targeting fusion constructs for retention of the ER trafficking sequence at the N-terminus of the peptide sequence was of primary concern.
Two changes were made to the ER trafficking sequence in relation to our previous Sil3T8 fusion constructs: the ER trafficking sequence was (1) lengthened and (2) relocated closer to the N-terminus of the fusion protein (Figure 1b). Analysis of the T. pseudonana Sil3 peptide sequence predicted that the original trafficking sequence of 17 amino acids ended at the cleavage site, and that the cleavage site may be more complex than previously assumed. Therefore, we increased the length of the trafficking sequence to include the first 21 amino acids of the Sil3 gene. By extending this peptide sequence we hoped to facilitate trafficking by allowing more efficient cleavage of the signal sequence.
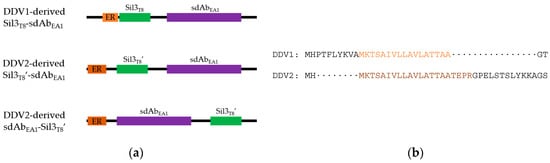
Figure 1.
Diagrams of fusion proteins produced for this study. (a) Order of peptides in fusion proteins. Black lines—residual cloning scars; orange—ER trafficking sequence (light orange, 17 amino acids; dark orange, 21 amino acids); green—Sil3T8 biosilica-targeting peptide (Sil3T8, original composite peptide with ER trafficking sequence and silica targeting peptide; Sil3T8′, uncoupled silica-targeting peptide); purple—sdAbEA1 (can be either clone A1 or G10, exact identity is not specified here). Diagrams are for illustration purposes, and are not drawn to scale. (b) Alignment of ER trafficking sequences in each diatom expression vector. The sequence shown is from the N-terminus of the fusion protein through the second peptide feature of the protein. Colors are the same as in (a).
Further, the Gateway (Invitrogen) modular cloning system leaves substantial scars between units. Our original design intended for the promoter to be one of the modular units. By leaving a scar after the promoter (which also contained the start codon for the fusion protein), the ER trafficking signal began 10 amino acids downstream from the N-terminus of the protein (Figure 1b, top). In order to continue to employ the modular Gateway (Invitrogen) cloning of our fusion partners, and use the scars as flexible linkers between fusion components, a new diatom-specific destination vector (pDDV2) was created with a static fcp promoter [22] that was immediately followed by the ER trafficking sequence. The Gateway (Invitrogen) scar would then be located downstream of the cleaved ER trafficking sequence (Figure 1b, bottom). Not only would this rearrangement of cloning elements allow the ER trafficking sequence to move closer to the N-terminus of the fusion protein (potentially allowing more efficient recognition), but it also uncoupled the remaining 37 amino acids of the Sil3T8 biosilica targeting peptide from the trafficking sequence so that the silica targeting peptide could be placed on either side of its fusion partner (Figure 1a).
In summary, the first generation diatom expression vector (pDDV1, Ref. [18]) was used to transform T. pseudonana resulting in the in vivo self-assembly of fusion proteins having a presumably suboptimal ER trafficking sequence (both in length and placement) and an N-terminal trafficking + targeting peptide/C-terminal sdAb orientation (Figure 1a, top). The second generation diatom expression vector (pDDV2) was used to transform T. pseudonana, resulting in the in vivo self-assembly of fusion proteins with the putatively improved ER trafficking sequence (both in length and placement), and the uncoupled targeting peptide Sil3T8′ located either on the N- (Sil3T8′-sdAbEA1; Figure 1a, middle) or C- (sdAbEA1-Sil3T8′; Figure 1a, bottom) terminus of the processed fusion protein.
3.2. Comparision of Two Generations of Fusion Constructs
Our earlier efforts to create an environmental biosensor for B. anthracis focused on the in vivo assembly of two sdAbs in diatom biosilica: sdAbEA1 clone G10, which binds intact spores, and sdAbEA1 clone A1, which binds lysed spores [20]. Given our previously inconsistent results in functionalizing diatom biosilica with these two sdAbs, we hoped that redesigning the expression vector would increase functionalization for both sdAbs. To that end, EA1-EGFP binding of three versions of each sdAbEA1 clone A1 (sdAbEA1/A1)–functionalized and clone G10 (sdAbEA1/G10)–functionalized diatom biosilica were compared (Figure 2 and Figure 3, respectively).
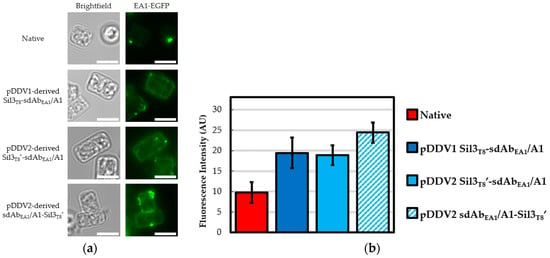
Figure 2.
sdAbEA1/A1 in vivo functionalized biosilica. Diatom biosilica was isolated from native T. pseudonana and cell lines transformed with expression constructs containing various Sil3T8-sdAbEA1/A1 fusions. Presence of functional, in vivo assembled, biosilica-tethered sdAbEA1/A1 was confirmed by ability to bind EA1-EGFP protein. (a) Epifluorescence images of representative frustules showing different EA1-EGFP binding potentials. Given that S-layer proteins like extractable antigen 1 (EA1) (and, by extension, the EA1-EGFP used here) are self-assembling [31], the punctate fluorescence observed in all four panels is expected and contributes to the fluorescent signal observed for the native frustules. All calibration bars are 5 μm. (b) Average measured fluorescence intensities in arbitrary units (AU). Error bars are 95% confidence intervals (±t × 1 SEM). For native and pDDV1-derived Sil3T8-sdAbEA1/A1 biosilica frustules, n = 25 (df = 24, t = 2.06); pDDV2-derived Sil3T8′-sdAbEA1/A1 and sdAbEA1/A1-Sil3T8′ biosilica frustules, n = 100 (df = 99, t = 1.984).
Figure 3.
sdAbEA1/G10 in vivo functionalized biosilica. Diatom biosilica was isolated from native T. pseudonana and cell lines transformed with constructs containing various Sil3T8-sdAbEA1/G10 fusions. Presence of functional, in vivo assembled, biosilica-tethered sdAbEA1/G10 was confirmed by ability to bind EA1-EGFP protein. As all six single domain antibody (sdAb) fusions were tested in a single experiment, the native diatom biosilica results are identical to those presented in the previous figure. (a) Epifluorescence images of representative frustules showing different EA1-EGFP binding potentials. Given that S-layer proteins like EA1 (and, by extension, the EA1-EGFP used here) are self-assembling [31], the punctate fluorescence observed in all four panels is expected and contributes to the fluorescent signal observed for the native frustules. All calibration bars are 5 μm. (b) Average measured fluorescence intensities in arbitrary units (AU). Error bars are 95% confidence intervals (±t × 1 SEM). For native and pDDV1-derived Sil3T8-sdAbEA1/G10 biosilica frustules, n = 25 (df = 24, t = 2.06); pDDV2-derived Sil3T8′-sdAbEA1/G10 and sdAbEA1/G10-Sil3T8′ biosilica frustules, n = 100 (df = 99, t = 1.984).
Considering sdAbEA1/A1 (Figure 2), the first generation pDDV1-derived Sil3T8-sdAbEA1/A1 functionalized biosilica frustules exhibited EA1-EGFP binding greater than that of native biosilica, similar to previous findings [18] in spite of the suboptimal ER trafficking sequence. Yet functionalized biosilica frustules with neither of the second generation pDDV2-derived constructs exhibited significantly improved EA1-EGFP binding in relation to the pDDV1-derived Sil3T8-sdAbEA1/A1 functionalized biosilica. These results indicated that the alterations to the ER trafficking signal sequence did not result in improved binding due to increased density of sdAbEA1/A1 in the frustule. The rotation of the sdAb in relation to the diatom biosilica also did not increase the amount of EA1-EGFP able to bind for this particular sdAb.
Considering sdAbEA1/G10 (Figure 3), the first generation pDDV1-derived Sil3T8-sdAbEA1/G10 in vivo functionalized biosilica frustules did not show EA1-EGFP fluorescence different from that of native biosilica, as expected from our previous observations. Conversely, in vivo functionalized biosilica frustules from both the second generation pDDV2-derived Sil3T8′-sdAbEA1/G10 and sdAbEA1/G10-Sil3T8′ exhibited significantly greater EA1-EGFP binding in comparison with the first generation pDDV1-derived construct. Although these findings do not exclude other factors associated with protein instability, they are consistent with results intended from the logical re-design of the constructs and pDDV2 to improve both ER trafficking and the release of steric hindrance at the antigen binding site.
3.3. Homology Modeling Suggests Differences in Antigen Binding Between sdAbEA1/A1 and sdAbEA1/G10
Based on sequence homology, the overall structures of both sdAbEA1/A1 and sdAbEA1/G10 are typical for sdAbs (Figure 4). This structure includes a heavy chain comprised of beta sheets, with N- and C-termini protruding from opposite ends of the 3-dimensional structure. The binding site, i.e., the complementarity determining region 3 (CDR3), is a loop structure (Figure 4, arrows). We predict the binding site for sdAbEA1/G10 to be located close to the N-terminus in the folded protein. Thus, N-terminal fusions with Sil3T8 would be expected to introduce steric hindrance that could prevent effective binding between the sdAb and the antigen. Antigen binding would not be disturbed if Sil3T8 was fused at the C-terminus of sdAbEA1/G10. We predict the binding site for sdAbEA1/A1, however, to be side facing, so that little steric hindrance would be introduced regardless of the orientation of the Sil3T8 peptide fusion with sdAbEA1/A1.
Figure 4.
Aligned homology models of single domain antibodies with presumed antigen binding configuration. The ribbon diagrams of the homology structures of sdAbEA1/A1 (cyan, based on pdb ID 6GLW) and sdAbEA1/G10 (green, based on pdb ID 5F10) were aligned using Pymol. The ovals show the presumed binding positions of the EA1 antigen for sdAbEA1/A1 (light blue) or sdAbEA1/G10 (light green). The N- and C-terminal regions of the sdAbs are also noted in blue and red, respectively. See Figure S2 for the individual pdb bank structures used to support the orientations of antigen binding.
These models help to explain our observations in relation to sdAbEA1/A1, whereby EA1-EGFP binding is not substantially improved by our construct re-design. A side-facing binding surface would mean that sdAbEA1/A1 binding would not be sterically hindered by a fusion partner at either terminus of the peptide.
These models also supported our hypothesis that the primary reason that we did not observe EA1 binding by our first generation Sil3T8-sdAbEA1/G10-functionalized diatom biosilica was due to steric hindrance when the fusion protein was tethered to diatom biosilica by the N-terminally-located Sil3T8 silica-targeting peptide. Tethering to the diatom biosilica using the C-terminal fusion with the uncoupled Sil3T8′ biosilica-targeting peptide would have alleviated some of that stress.
4. Discussion
Individual protein structures often dictate the order of complex fusion protein partners. Problems arise when combining peptides with incompatible fusion preferences, as was found to be the case in creating a series of diatom biosilica-targeted constructs encoding sdAbs against the B. anthracis S-layer protein EA1. In particular, both the Sil3T8 (e.g., [12,15,16,32]) and sdAbEA1 (e.g., [33,34]) peptides previously have been made only as N-terminal fusions to their partners. One reason for this order of fusion partners is the requirement of many signal peptides’ proximity to the protein terminus, rather than just their amino acid sequence. Given our results with sdAbEA1/A1, some flexibility likely does exist with this placement (i.e., “close” to the N-terminus is good enough).
Further, the three-dimensional structure of each peptide in a chimeric protein must be considered. Even though much of the sdAb structure is conserved, the structure of the binding loops by necessity will vary by antigen and epitope recognized by the sdAb. For the two sdAbs described in this work, which bind unique epitopes in the same antigen, predicted structural differences of their respective binding loops may have caused the antigen to be positioned differently in relation to the static regions of the sdAb peptide, consequently altering the functionality of the sdAbs when they were tethered in diatom biosilica. This difference in binding location supports our observations whereby sdAbEA1/G10 is more sensitive to fusion partner orientation than sdAbEA1/A1. By uncoupling the trafficking and biosilica targeting domains of the Sil3T8 peptide sequence, we have increased the flexibility of using this particular peptide as a fusion partner for proteins that are more selective in regard to partner orientation.
In conclusion, we note that while modular cloning systems for synthetic biology are attractive due to their inherent convenience, cellular biology (e.g., protein trafficking) and biochemical (e.g., protein folding) considerations ultimately must guide the design of fusion proteins. This principle was especially relevant to the construction of functionalized diatom biosilica through genetic modification with two related sdAbs for the detection of the pathogenic bacteria B. anthracis.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-7737/9/1/14/s1, Supplementary Methods, Table S1: PCR primers for cloning and analysis, Table S2: PCR primers for gateway entry clones; Table S3: Gateway entry clones, Table S4: Gateway expression clones, Figure S1: SignalP prediction of ER trafficking sequence, Figure S2: Illustration of two single domain antibodies bound to antigen, Peptide sequences of fusion constructs.
Author Contributions
Conceptualization, N.R.F., G.R. and K.A.H.; Investigation, N.R.F., Y.X. and K.A.H.; Writing—Original draft preparation, N.R.F., Y.X. and K.A.H.; Writing—Review and editing, N.R.F., Y.X., K.A.H., T.C.S., G.L.R. and G.R.; Project administration, G.R.; Funding acquisition, G.R., G.L.R. and T.C.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Defense Threat Reduction Agency (G.R. and T.C.S) and by the Office of Naval Research (G.R. and G.L.R.). The APC was funded by Kansas City University of Medicine and Biosciences.
Acknowledgments
Nils Kroger and Nicole Poulsen (CUBE Center for Molecular Bioengineering, Dresden, DE) provided expression plasmids containing the diatom-specific genes; Ellen Goldman (Naval Research Laboratory, Washington, D.C.) provided plasmids containing the sequence for the sdAbEA1 clones.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Round, F.E.; Crawford, R.M.; Mann, D.G. Diatoms: Biology and Morphology of the Genera; Cambridge University Press: New York, NY, USA, 1990; p. 760. [Google Scholar]
- Gordon, R.; Losic, D.; Tiffany, M.A.; Nagy, S.S.; Sterrenburg, F.A. The Glass Menagerie: Diatoms for novel applications in nanotechnology. Trends Biotechnol. 2009, 27, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.J.; Theriot, J.A.; Sood, P.; Marshall, W.F.; Landweber, L.F.; Fritz-Laylin, L.; Polka, J.K.; Oliferenko, S.; Gerbich, T.; Gladfelter, A.; et al. Non-model model organisms. BMC Biol. 2017, 15, 55. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhen, L.; Ren, F.; Campbell, J.; Rorrer, G.L.; Wang, A.X. Ultra-sensitive immunoassay biosensors using hybrid plasmonic-biosilica nanostructured materials. J. Biophotonics 2015, 8, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Cicco, S.R.; Vona, D.; Gristina, R.; Sardella, E.; Ragni, R.; Lo Presti, M.; Farinola, G.M. Biosilica from Living Diatoms: Investigations on Biocompatibility of Bare and Chemically Modified Thalassiosira weissflogii Silica Shells. Bioengineering 2016, 3, 35. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, N.; Berne, C.; Spain, J.; Kroger, N. Silica immobilization of an enzyme through genetic engineering of the diatom Thalassiosira pseudonana. Angew. Chem. Int. Ed. 2007, 46, 1843–1846. [Google Scholar] [CrossRef] [PubMed]
- Narayan, R.; Nayak, U.Y.; Raichur, A.M.; Garg, S. Mesoporous Silica Nanoparticles: A Comprehensive Review on Synthesis and Recent Advances. Pharmaceutics 2018, 10, 118. [Google Scholar] [CrossRef]
- Bilal, M.; Iqbal, H.M.N. Chemical, physical, and biological coordination: An interplay between materials and enzymes as potential platforms for immobilization. Coord. Chem. Rev. 2019, 388, 1–23. [Google Scholar] [CrossRef]
- von Baeckmann, C.; Guillet-Nicolas, R.; Renfer, D.; Kahlig, H.; Kleitz, F. A Toolbox for the Synthesis of Multifunctionalized Mesoporous Silica Nanoparticles for Biomedical Applications. ACS Omega 2018, 3, 17496–17510. [Google Scholar] [CrossRef]
- Shadjou, N.; Hasanzadeh, M. Bone tissue engineering using silica-based mesoporous nanobiomaterials: Recent progress. Mater. Sci. Eng. C Mater. Biol. Appl. 2015, 55, 401–409. [Google Scholar] [CrossRef]
- Gotz, W.; Tobiasch, E.; Witzleben, S.; Schulze, M. Effects of Silicon Compounds on Biomineralization, Osteogenesis, and Hard Tissue Formation. Pharmaceutics 2019, 11, 117. [Google Scholar] [CrossRef]
- Delalat, B.; Sheppard, V.C.; Rasi Ghaemi, S.; Rao, S.; Prestidge, C.A.; McPhee, G.; Rogers, M.L.; Donoghue, J.F.; Pillay, V.; Johns, T.G.; et al. Targeted drug delivery using genetically engineered diatom biosilica. Nat. Commun. 2015, 6, 8791. [Google Scholar] [CrossRef] [PubMed]
- Terracciano, M.; De Stefano, L.; Rea, I. Diatoms Green Nanotechnology for Biosilica-Based Drug Delivery Systems. Pharmaceutics 2018, 10, 242. [Google Scholar] [CrossRef] [PubMed]
- Manayil, J.C.; Lee, A.F.; Wilson, K. Functionalized Periodic Mesoporous Organosilicas: Tunable Hydrophobic Solid Acids for Biomass Conversion. Molecules 2019, 24, 239. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, V.C.; Scheffel, A.; Poulsen, N.; Kroger, N. Live diatom silica immobilization of multimeric and redox-active enzymes. Appl. Environ. Microbiol. 2012, 78, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, N.; Scheffel, A.; Sheppard, V.C.; Chesley, P.M.; Kroger, N. Pentalysine clusters mediate silica targeting of silaffins in Thalassiosira pseudonana. J. Biol. Chem. 2013, 288, 20100–20109. [Google Scholar] [CrossRef] [PubMed]
- Marshall, K.E.; Robinson, E.W.; Hengel, S.M.; Pasa-Tolic, L.; Roesijadi, G. FRET imaging of diatoms expressing a biosilica-localized ribose sensor. PLoS ONE 2012, 7, e33771. [Google Scholar] [CrossRef]
- Ford, N.R.; Hecht, K.A.; Hu, D.; Orr, G.; Xiong, Y.; Squier, T.C.; Rorrer, G.L.; Roesijadi, G. Antigen Binding and Site-Directed Labeling of Biosilica-Immobilized Fusion Proteins Expressed in Diatoms. ACS Synth. Biol. 2016, 5, 193–199. [Google Scholar] [CrossRef]
- Xiong, Y.; Ford, N.R.; Hecht, K.A.; Roesijadi, G.; Squier, T.C. Dynamic Stabilization of Expressed Proteins in Engineered Diatom Biosilica Matrices. Bioconjug. Chem. 2016, 27, 1205–1209. [Google Scholar] [CrossRef]
- Walper, S.A.; Anderson, G.P.; Brozozog Lee, P.A.; Glaven, R.H.; Liu, J.L.; Bernstein, R.D.; Zabetakis, D.; Johnson, L.; Czarnecki, J.M.; Goldman, E.R. Rugged single domain antibody detection elements for Bacillus anthracis spores and vegetative cells. PLoS ONE 2012, 7, e32801. [Google Scholar] [CrossRef]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef]
- Poulsen, N.; Chesley, P.M.; Kröger, N. Molecular genetic manipulation of the diatom thalassiosira pseudonana (bacillariophyceae)1. J. Phycol. 2006, 42, 1059–1065. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.; Engelbrecht, J.; Brunak, S.; von Heijne, G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997, 10, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Schrodinger, L.L.C. The PyMOL Molecular Graphics System, Version 1.7. 2014. Available online: https://pymol.org/2/support.html? (accessed on 30 November 2019).
- Pathare, G.R.; Nagy, I.; Sledz, P.; Anderson, D.J.; Zhou, H.J.; Pardon, E.; Steyaert, J.; Forster, F.; Bracher, A.; Baumeister, W. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc. Natl. Acad. Sci. USA 2014, 111, 2984–2989. [Google Scholar] [CrossRef]
- Lo, A.W.; Moonens, K.; De Kerpel, M.; Brys, L.; Pardon, E.; Remaut, H.; De Greve, H. The molecular mechanism of Shiga toxin Stx2e neutralization by a single-domain antibody targeting the cell receptor-binding domain. J. Biol. Chem. 2014, 289, 25374–25381. [Google Scholar] [CrossRef]
- Sára, M.; Sleytr, U.B. S-Layer proteins. J. Bacteriol. 2000, 182, 859–868. [Google Scholar] [CrossRef]
- Groger, P.; Poulsen, N.; Klemm, J.; Kroger, N.; Schlierf, M. Establishing super-resolution imaging for proteins in diatom biosilica. Sci. Rep. 2016, 6, 36824. [Google Scholar] [CrossRef]
- Walper, S.A.; Brozozog Lee, P.A.; Goldman, E.R.; Anderson, G.P. Comparison of single domain antibody immobilization strategies evaluated by surface plasmon resonance. J. Immunol. Methods 2013, 388, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.P.; Shriver-Lake, L.C.; Walper, S.A.; Ashford, L.; Zabetakis, D.; Liu, J.L.; Breger, J.C.; Brozozog Lee, P.A.; Goldman, E.R. Genetic Fusion of an Anti-BclA Single-Domain Antibody with Beta Galactosidase. Antibodies 2018, 7, 36. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).