Anaphase A: Disassembling Microtubules Move Chromosomes toward Spindle Poles
Abstract
:1. Introduction and Distinction between Anaphase “A” and “B”
2. Centromeres and Kinetochores Usually Lead Anaphase Movements While Chromosome Arms Follow
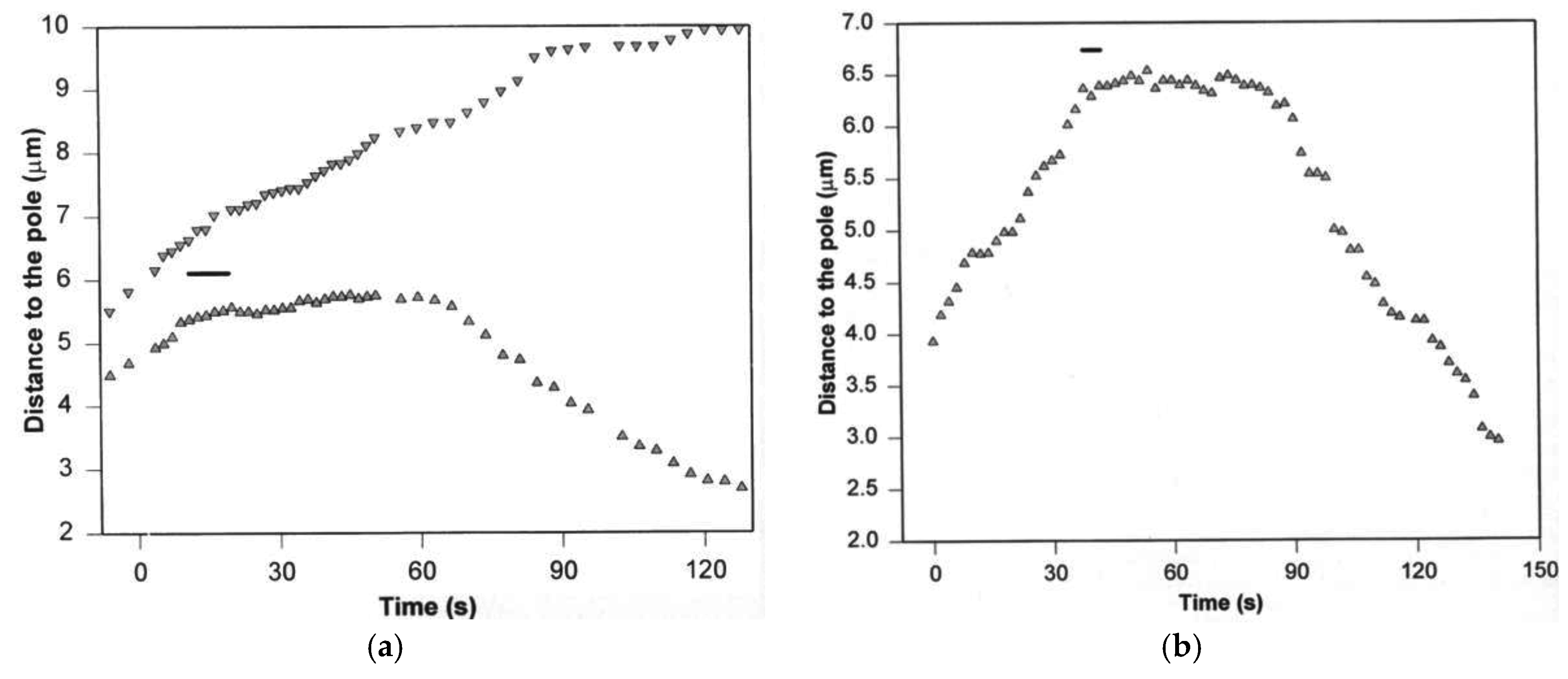
3. Poleward Movement during Anaphase A Is Mostly but Not Entirely Unidirectional
4. Poleward Chromosome Movement Is Coupled to Shortening of the Connecting Microtubules
5. Kinetochore-Attached Microtubules Can ‘Flux’ Continuously toward the Poles
6. Anaphase in Some Cell Types Does Not Conform to the Canonical View
7. Kinetochores Can Either Be Actively Pulling Poleward or Passively Slipping Anti-Poleward
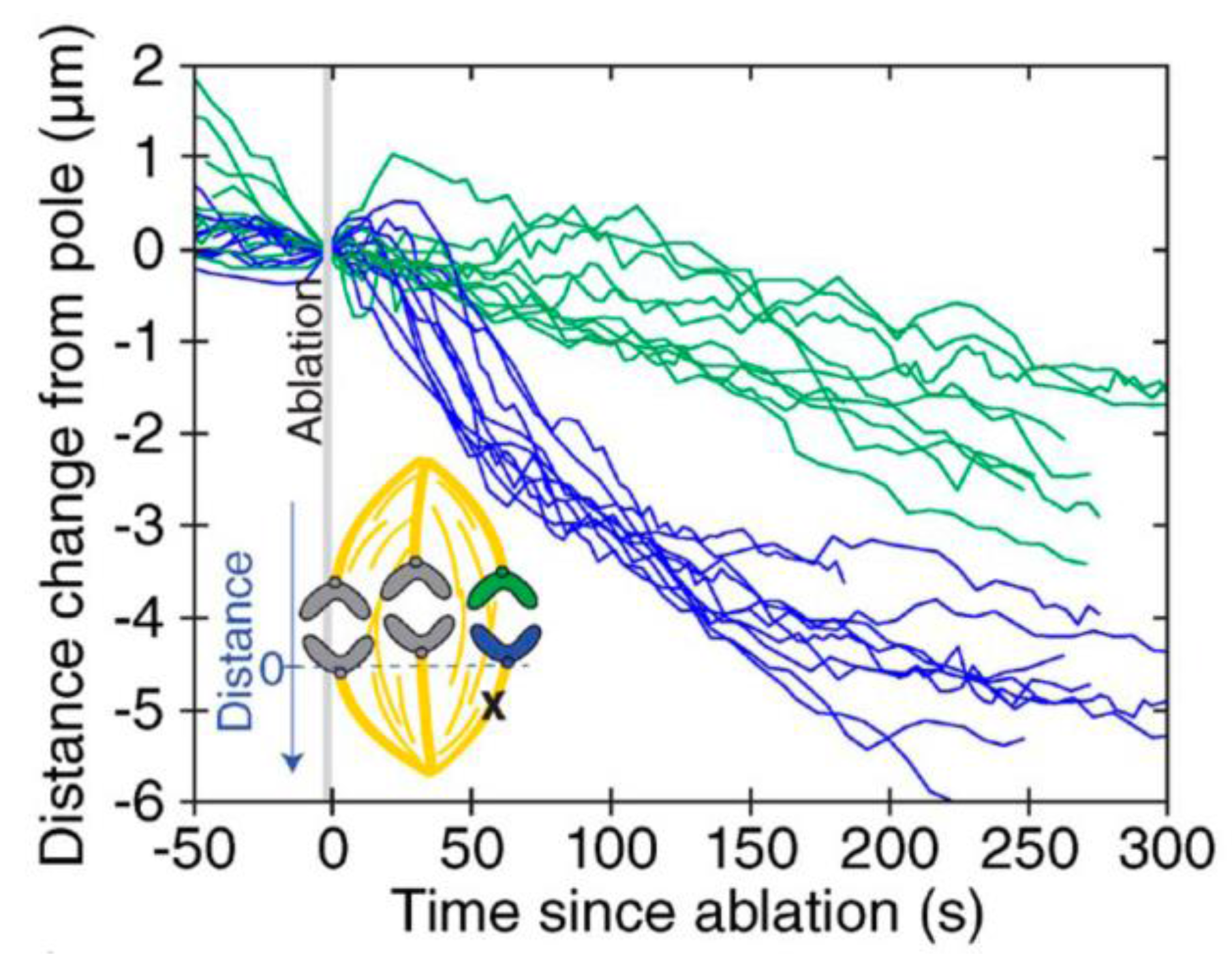
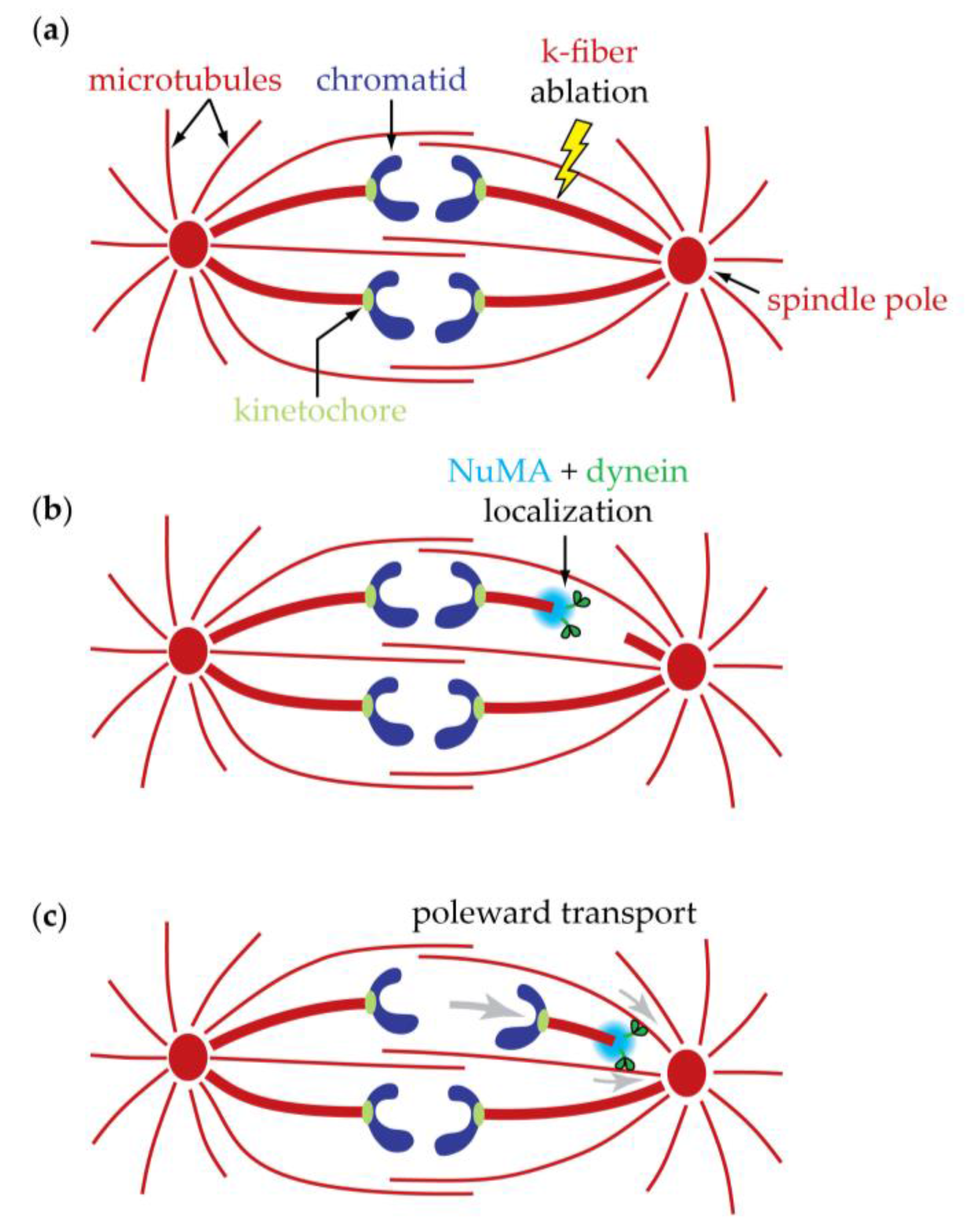
8. Anaphase Spindle Generates More Force than Needed for Anaphase Chromosome Movement
9. Why Is the Anaphase Spindle ‘Over-Engineered’ to Produce Forces so Much Higher than Needed?
10. New Techniques Are Providing Force Estimates from a Wider Variety of Cell Types
11. Tip-Coupling: One of the Most Conserved Features of Mitosis and One of the Most Puzzling
12. Conventional Motors Are Found at Kinetochores but Might Not Be the Primary Basis for Tip-Coupling
13. Kinetochores Also Contain Non-Motor Microtubule-Binding Elements
14. Toward an Integrated View of the Tip-Coupling Apparatus of the Kinetochore
15. Microtubules Could Be the Engines that Drive Poleward Chromosome Movement during Anaphase A
16. Purified Kinetochores and Sub-Complexes Are Excellent Tip-Couplers
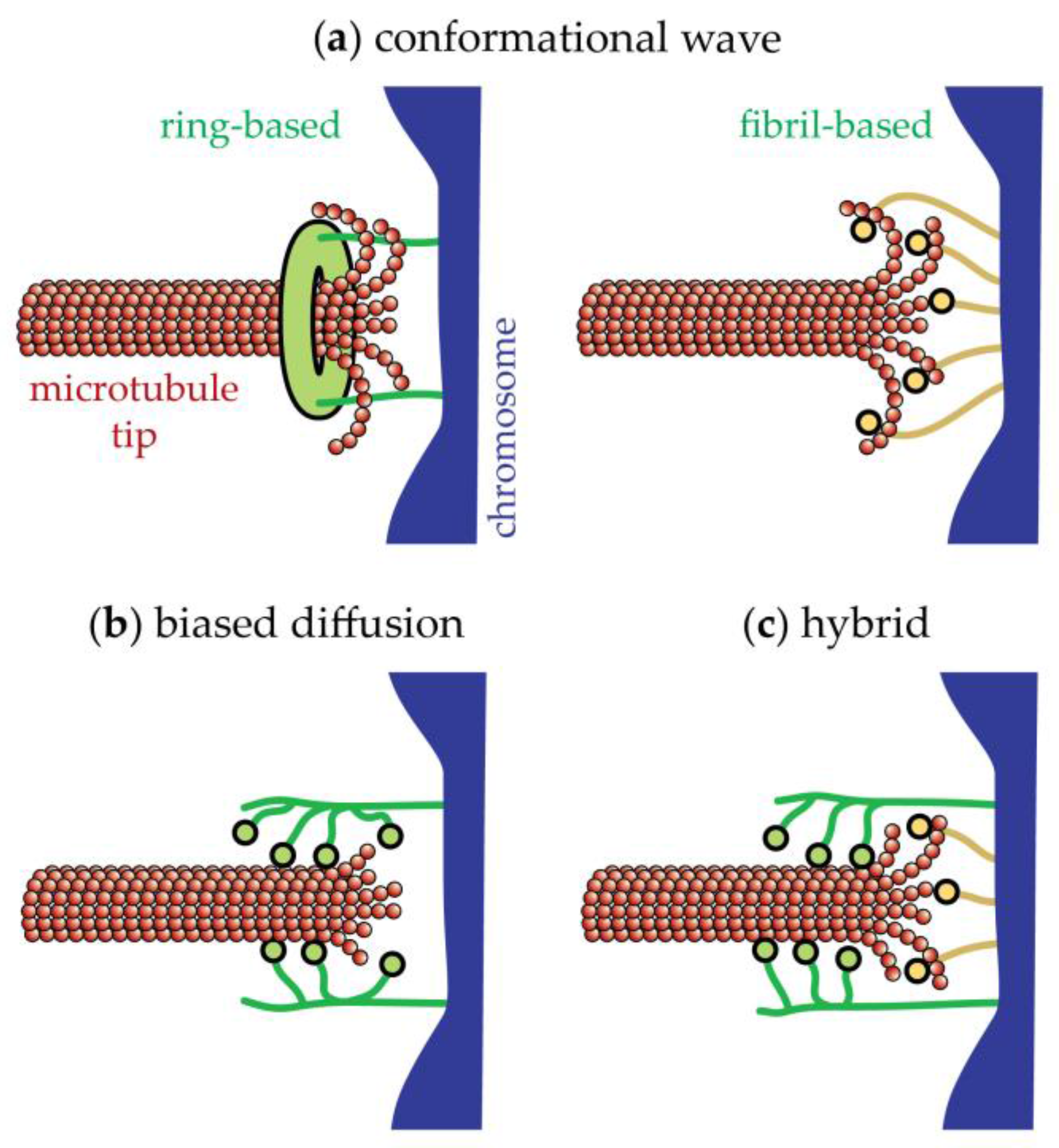
17. The Conformational Wave Model for Disassembly-Driven Movement
18. The Biased Diffusion Model for Disassembly-Driven Movement
19. Movement Coupled to Tip Assembly
20. Mechanism of Poleward Flux Might Differ for Kinetochore-Attached Versus Non-Kinetochore Microtubules
21. Potential Biophysical Mechanisms for Kinetochore-Microtubule Flux
22. Loss of Tension by Itself Might Be Sufficient to Trigger Anaphase Chromosome-to-Pole Movement
23. Phosphoregulatory Changes at the Metaphase-to-Anaphase Transition
24. Conclusions
Acknowledgments
Conflicts of Interest
References
- Mazia, D. Mitosis and the physiology of cell division. In The Cell: Biochemistry, Physiology, Morphology, Vol III; Brachet, J., Mirsky, A.E., Eds.; Academic Press: London, UK, 1961; pp. 77–412. [Google Scholar]
- Strasburger, E. Die Controversen der indirecten Kerntheilung. Arch. Mikrosk. Anat. 1884, 23, 246–304. [Google Scholar] [CrossRef]
- Scholey, J.M.; Civelekoglu-Scholey, G.; Brust-Mascher, I. Anaphase B. Biology 2016, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Ris, H. The anaphase movement of chromosomes in the spermatocytes of the grasshopper. Biol. Bull. 1949, 96, 90–106. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.R.; Hays, T. A Brief History of Research on Mitotic Mechanisms. Biology 2016, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Sharp, L.W. Introduction to Cytology; McGraw-Hill: New York, NY, USA; London, UK, 1934. [Google Scholar]
- Rieder, C.L.; Davison, E.A.; Jensen, L.C.; Cassimeris, L.; Salmon, E.D. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 1986, 103, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Straight, A.F.; Marshall, W.F.; Sedat, J.W.; Murray, A.W. Mitosis in living budding yeast: Anaphase A but no metaphase plate. Science 1997, 277, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Khodjakov, A.; Cole, R.W.; Bajer, A.S.; Rieder, C.L. The force for poleward chromosome motion in Haemanthus cells acts along the length of the chromosome during metaphase but only at the kinetochore during anaphase. J. Cell Biol. 1996, 132, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- LaFountain, J.R., Jr.; Oldenbourg, R.; Cole, R.W.; Rieder, C.L. Microtubule flux mediates poleward motion of acentric chromosome fragments during meiosis in insect spermatocytes. Mol. Biol. Cell 2001, 12, 4054–4065. [Google Scholar] [CrossRef] [PubMed]
- Rieder, C.L.; Salmon, E.D. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol. 1994, 124, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Skibbens, R.V.; Skeen, V.P.; Salmon, E.D. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: A push-pull mechanism. J. Cell Biol. 1993, 122, 859–875. [Google Scholar] [CrossRef]
- Mitchison, T.; Kirschner, M. Dynamic instability of microtubule growth. Nature 1984, 312, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Walker, R.A.; O’Brien, E.T.; Pryer, N.K.; Soboeiro, M.F.; Voter, W.A.; Erickson, H.P.; Salmon, E.D. Dynamic instability of individual microtubules analyzed by video light microscopy: Rate constants and transition frequencies. J. Cell Biol. 1988, 107, 1437–1448. [Google Scholar] [CrossRef] [PubMed]
- Stumpff, J.; von Dassow, G.; Wagenbach, M.; Asbury, C.; Wordeman, L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell 2008, 14, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Cimini, D.; Moree, B.; Canman, J.C.; Salmon, E.D. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 2003, 116, 4213–4225. [Google Scholar] [CrossRef]
- Grishchuk, E.L.; Lampson, M. Mechanisms to avoid and correct erroneous kinetochore-microtubule attachments. Biology 2017, 6, 1. [Google Scholar] [CrossRef]
- Uretz, R.B.; Bloom, W.; Zirkle, R.E. Irradiation of parts of individual cells. II. Effects of an ultraviolet microbeam focused on parts of chromosomes. Science 1954, 120, 197–199. [Google Scholar] [CrossRef] [PubMed]
- McNeill, P.A.; Berns, M.W. Chromosome behavior after laser microirradiation of a single kinetochore in mitotic PtK2 cells. J. Cell Biol. 1981, 88, 543–553. [Google Scholar] [CrossRef] [PubMed]
- McDonald, K.L.; O’Toole, E.T.; Mastronarde, D.N.; McIntosh, J.R. Kinetochore microtubules in PTK cells. J. Cell Biol. 1992, 118, 369–383. [Google Scholar] [CrossRef]
- Winey, M.; Mamay, C.L.; O’Toole, E.T.; Mastronarde, D.N.; Giddings, T.H., Jr.; McDonald, K.L.; McIntosh, J.R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 1995, 129, 1601–1615. [Google Scholar] [CrossRef] [PubMed]
- Gorbsky, G.J.; Sammak, P.J.; Borisy, G.G. Chromosomes move poleward in anaphase along stationary microtubules that coordinately disassemble from their kinetochore ends. J. Cell Biol. 1987, 104, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.; Evans, L.; Schulze, E.; Kirschner, M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 1986, 45, 515–527. [Google Scholar] [CrossRef]
- Shelden, E.; Wadsworth, P. Microinjection of biotin-tubulin into anaphase cells induces transient elongation of kinetochore microtubules and reversal of chromosome-to-pole motion. J. Cell Biol. 1992, 116, 1409–1420. [Google Scholar] [CrossRef] [PubMed]
- Cimini, D.; Cameron, L.A.; Salmon, E.D. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 2004, 14, 2149–2155. [Google Scholar] [CrossRef] [PubMed]
- Maddox, P.S.; Bloom, K.S.; Salmon, E.D. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2000, 2, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Khodjakov, A.; La Terra, S.; Chang, F. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr. Biol. 2004, 14, 1330–1340. [Google Scholar] [CrossRef] [PubMed]
- Sagolla, M.J.; Uzawa, S.; Cande, W.Z. Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J. Cell Sci. 2003, 116, 4891–4903. [Google Scholar] [CrossRef] [PubMed]
- Mallavarapu, A.; Sawin, K.; Mitchison, T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr. Biol. 1999, 9, 1423–1426. [Google Scholar] [CrossRef]
- Ganem, N.J.; Upton, K.; Compton, D.A. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 2005, 15, 1827–1832. [Google Scholar] [CrossRef] [PubMed]
- LaFountain, J.R., Jr.; Cohan, C.S.; Siegel, A.J.; LaFountain, D.J. Direct visualization of microtubule flux during metaphase and anaphase in crane-fly spermatocytes. Mol. Biol. Cell 2004, 15, 5724–5732. [Google Scholar] [CrossRef] [PubMed]
- Maddox, P.; Straight, A.; Coughlin, P.; Mitchison, T.J.; Salmon, E.D. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: Implications for spindle mechanics. J. Cell Biol. 2003, 162, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.J.; Salmon, E.D. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 1992, 119, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Kronebusch, P.J.; Borisy, G.G. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol. 1995, 131, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Maddox, P.; Desai, A.; Oegema, K.; Mitchison, T.J.; Salmon, E.D. Poleward microtubule flux is a major component of spindle dynamics and anaphase a in mitotic Drosophila embryos. Curr. Biol. 2002, 12, 1670–1674. [Google Scholar] [CrossRef]
- Rogers, G.C.; Rogers, S.L.; Schwimmer, T.A.; Ems-McClung, S.C.; Walczak, C.E.; Vale, R.D.; Scholey, J.M.; Sharp, D.J. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 2004, 427, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Dhonukshe, P.; Vischer, N.; Gadella, T.W., Jr. Contribution of microtubule growth polarity and flux to spindle assembly and functioning in plant cells. J. Cell Sci. 2006, 119, 3193–3205. [Google Scholar] [CrossRef] [PubMed]
- Hamaguchi, Y.; Toriyama, M.; Sakai, H.; Hiramoto, Y. Redistribution of fluorescently labeled tubulin in the mitotic apparatus of sand dollar eggs and the effects of taxol. Cell Struct. Funct. 1987, 12, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.C.; Mitchison, T.J.; Rieder, C.L.; Salmon, E.D. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell 1996, 7, 1547–1558. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Maddox, P.S.; Mitchison, T.J.; Salmon, E.D. Anaphase A chromosome movement and poleward spindle microtubule flux occur At similar rates in Xenopus extract spindles. J. Cell Biol. 1998, 141, 703–713. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Asthana, S.; Sorger, P.K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 2000, 101, 763–775. [Google Scholar] [CrossRef]
- Pearson, C.G.; Maddox, P.S.; Salmon, E.D.; Bloom, K. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 2001, 152, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Brust-Mascher, I.; Scholey, J.M. Microtubule flux and sliding in mitotic spindles of Drosophila embryos. Mol. Biol. Cell 2002, 13, 3967–3975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Rogers, G.C.; Buster, D.W.; Sharp, D.J. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 2007, 177, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Rath, U.; Rogers, G.C.; Tan, D.; Gomez-Ferreria, M.A.; Buster, D.W.; Sosa, H.J.; Sharp, D.J. The Drosophila kinesin-13, KLP59D, impacts Pacman- and Flux-based chromosome movement. Mol. Biol. Cell 2009, 20, 4696–4705. [Google Scholar] [CrossRef] [PubMed]
- Matos, I.; Pereira, A.J.; Lince-Faria, M.; Cameron, L.A.; Salmon, E.D.; Maiato, H. Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J. Cell Biol. 2009, 186, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Savoian, M.S. Using Photobleaching to Measure Spindle Microtubule Dynamics in Primary Cultures of Dividing Drosophila Meiotic Spermatocytes. J. Biomol. Tech. 2015, 26, 66–73. [Google Scholar] [CrossRef] [PubMed]
- LaFountain, J.R., Jr.; Cohan, C.S.; Oldenbourg, R. Pac-man motility of kinetochores unleashed by laser microsurgery. Mol. Biol. Cell 2012, 23, 3133–3142. [Google Scholar] [CrossRef]
- LaFountain, J.R., Jr.; Cohan, C.S.; Oldenbourg, R. Functional states of kinetochores revealed by laser microsurgery and fluorescent speckle microscopy. Mol. Biol. Cell 2011, 22, 4801–4808. [Google Scholar] [CrossRef] [PubMed]
- Maffini, S.; Maia, A.R.; Manning, A.L.; Maliga, Z.; Pereira, A.L.; Junqueira, M.; Shevchenko, A.; Hyman, A.; Yates, J.R., 3rd; Galjart, N.; et al. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr. Biol. 2009, 19, 1566–1572. [Google Scholar] [CrossRef] [PubMed]
- Wandke, C.; Barisic, M.; Sigl, R.; Rauch, V.; Wolf, F.; Amaro, A.C.; Tan, C.H.; Pereira, A.J.; Kutay, U.; Maiato, H.; et al. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J. Cell Biol. 2012, 198, 847–863. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Diaz Valencia, J.D.; Stewman, S.; Metz, J.; Monnier, S.; Rath, U.; Asenjo, A.B.; Charafeddine, R.A.; Sosa, H.J.; Ross, J.L.; et al. Human Fidgetin is a microtubule severing the enzyme and minus-end depolymerase that regulates mitosis. Cell Cycle 2012, 11, 2359–2366. [Google Scholar] [CrossRef] [PubMed]
- Wordeman, L.; Wagenbach, M.; von Dassow, G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J. Cell Biol. 2007, 179, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Dumont, J.; Oegema, K.; Desai, A. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 2010, 12, 894–901. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Skibbens, R.V.; Rieder, C.L.; Salmon, E.D. Kinetochore motility after severing between sister centromeres using laser microsurgery: Evidence that kinetochore directional instability and position is regulated by tension. J. Cell Sci. 1995, 108 Pt 7, 2537–2548. [Google Scholar] [PubMed]
- Khodjakov, A.; Rieder, C.L. Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J. Cell Biol. 1996, 135, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Akiyoshi, B.; Sarangapani, K.K.; Powers, A.F.; Nelson, C.R.; Reichow, S.L.; Arellano-Santoyo, H.; Gonen, T.; Ranish, J.A.; Asbury, C.L.; Biggins, S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 2010, 468, 576–579. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B. Measurements of the force produced by the mitotic spindle in anaphase. J. Cell Biol. 1983, 97, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Elting, M.W.; Hueschen, C.L.; Udy, D.B.; Dumont, S. Force on spindle microtubule minus ends moves chromosomes. J. Cell Biol. 2014, 206, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Sikirzhytski, V.; Magidson, V.; Steinman, J.B.; He, J.; Le Berre, M.; Tikhonenko, I.; Ault, J.G.; McEwen, B.F.; Chen, J.K.; Sui, H.; et al. Direct kinetochore-spindle pole connections are not required for chromosome segregation. J. Cell Biol. 2014, 206, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B. Chromosome Velocity during Mitosis as a Function of Chromosome Size and Position. J. Cell Biol. 1965, 25, 119–135. [Google Scholar] [CrossRef]
- Marshall, W.F.; Marko, J.F.; Agard, D.A.; Sedat, J.W. Chromosome elasticity and mitotic polar ejection force measured in living Drosophila embryos by four-dimensional microscopy-based motion analysis. Curr. Biol. 2001, 11, 569–578. [Google Scholar] [CrossRef]
- Sarangapani, K.K.; Asbury, C.L. Catch and release: How do kinetochores hook the right microtubules during mitosis? Trends Genet. TIG 2014, 30, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B. How cells get the right chromosomes. Science 1997, 275, 632–637. [Google Scholar] [CrossRef]
- Nicklas, R.B.; Koch, C.A. Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J. Cell Biol. 1969, 43, 40–50. [Google Scholar] [CrossRef]
- Joglekar, A.P. A Cell Biological Perspective on Past, Present and Future Investigations of the Spindle Assembly Checkpoint. Biology 2016, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B. The forces that move chromosomes in mitosis. Annu. Rev. Biophys. Biophys. Chem. 1988, 17, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Chacon, J.M.; Mukherjee, S.; Schuster, B.M.; Clarke, D.J.; Gardner, M.K. Pericentromere tension is self-regulated by spindle structure in metaphase. J. Cell Biol. 2014, 205, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Ye, A.A.; Cane, S.; Maresca, T.J. Chromosome biorientation produces hundreds of piconewtons at a metazoan kinetochore. Nat. Commun. 2016, 7, 13221. [Google Scholar] [CrossRef] [PubMed]
- Greenleaf, W.J.; Woodside, M.T.; Block, S.M. High-resolution, single-molecule measurements of biomolecular motion. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 171–190. [Google Scholar] [CrossRef] [PubMed]
- Leidel, C.; Longoria, R.A.; Gutierrez, F.M.; Shubeita, G.T. Measuring molecular motor forces in vivo: Implications for tug-of-war models of bidirectional transport. Biophys. J. 2012, 103, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Sims, P.A.; Xie, X.S. Probing dynein and kinesin stepping with mechanical manipulation in a living cell. Chemphyschem 2009, 10, 1511–1516. [Google Scholar] [CrossRef]
- Hendricks, A.G.; Holzbaur, E.L.; Goldman, Y.E. Force measurements on cargoes in living cells reveal collective dynamics of microtubule motors. Proc. Natl. Acad. Sci. USA 2012, 109, 18447–18452. [Google Scholar] [CrossRef] [PubMed]
- Shubeita, G.T.; Tran, S.L.; Xu, J.; Vershinin, M.; Cermelli, S.; Cotton, S.L.; Welte, M.A.; Gross, S.P. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell 2008, 135, 1098–1107. [Google Scholar] [CrossRef] [PubMed]
- Jun, Y.; Tripathy, S.K.; Narayanareddy, B.R.; Mattson-Hoss, M.K.; Gross, S.P. Calibration of optical tweezers for in vivo force measurements: How do different approaches compare? Biophys. J. 2014, 107, 1474–1484. [Google Scholar] [CrossRef] [PubMed]
- Neuman, K.C.; Chadd, E.H.; Liou, G.F.; Bergman, K.; Block, S.M. Characterization of photodamage to Escherichia coli in optical traps. Biophys. J. 1999, 77, 2856–2863. [Google Scholar] [CrossRef]
- Gross, S.P. Application of optical traps in vivo. Methods Enzymol. 2003, 361, 162–174. [Google Scholar] [PubMed]
- Ferraro-Gideon, J.; Sheykhani, R.; Zhu, Q.; Duquette, M.L.; Berns, M.W.; Forer, A. Measurements of forces produced by the mitotic spindle using optical tweezers. Mol. Biol. Cell 2013, 24, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Steuer, E.R.; Wordeman, L.; Schroer, T.A.; Sheetz, M.P. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature 1990, 345, 266–268. [Google Scholar] [CrossRef] [PubMed]
- Yen, T.J.; Li, G.; Schaar, B.T.; Szilak, I.; Cleveland, D.W. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature 1992, 359, 536–539. [Google Scholar] [CrossRef]
- Pfarr, C.M.; Coue, M.; Grissom, P.M.; Hays, T.S.; Porter, M.E.; McIntosh, J.R. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature 1990, 345, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Earnshaw, W.C.; Migeon, B.R. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma 1985, 92, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Savoian, M.S.; Goldberg, M.L.; Rieder, C.L. The rate of poleward chromosome motion is attenuated in Drosophila zw10 and rod mutants. Nat. Cell Biol. 2000, 2, 948–952. [Google Scholar] [PubMed]
- Sharp, D.J.; Rogers, G.C.; Scholey, J.M. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2000, 2, 922–930. [Google Scholar] [PubMed]
- Howell, B.J.; McEwen, B.F.; Canman, J.C.; Hoffman, D.B.; Farrar, E.M.; Rieder, C.L.; Salmon, E.D. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001, 155, 1159–1172. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, T.M. Spindle Assembly. Biology 2017, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Goshima, G.; Yamada, M. Mitotic spindle assembly in land plants: Molecules and mechanisms. Biology 2017, 6, 6. [Google Scholar] [CrossRef]
- Grishchuk, E.L.; McIntosh, J.R. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. Embo J. 2006, 25, 4888–4896. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Kitamura, E.; Kitamura, Y.; Tanaka, T.U. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 2007, 178, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Tytell, J.D.; Sorger, P.K. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 2006, 172, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Weaver, B.A.; Bonday, Z.Q.; Putkey, F.R.; Kops, G.J.; Silk, A.D.; Cleveland, D.W. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 2003, 162, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, T.M.; Lampson, M.A.; Hergert, P.; Cameron, L.; Cimini, D.; Salmon, E.D.; McEwen, B.F.; Khodjakov, A. Chromosomes can congress to the metaphase plate before biorientation. Science 2006, 311, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Tulu, U.S.; Wadsworth, P.; Rieder, C.L. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 2007, 17, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A.; Desai, A. Kinetochore assembly, structure, and function. Biology 2017, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.R.; Al-Bassam, J.; Harrison, S.C. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 2007, 14, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.R.; Schnell, J.R.; Larsen, N.A.; Sorger, P.K.; Chou, J.J.; Harrison, S.C. Structure of a central component of the yeast kinetochore: The Spc24p/Spc25p globular domain. Structure 2006, 14, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.R.; Sorger, P.K.; Harrison, S.C. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl. Acad. Sci. USA 2005, 102, 5363–5367. [Google Scholar] [CrossRef]
- Ciferri, C.; Pasqualato, S.; Screpanti, E.; Varetti, G.; Santaguida, S.; Dos Reis, G.; Maiolica, A.; Polka, J.; De Luca, J.G.; De Wulf, P.; et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 2008, 133, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Alushin, G.M.; Ramey, V.H.; Pasqualato, S.; Ball, D.A.; Grigorieff, N.; Musacchio, A.; Nogales, E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature 2010, 467, 805–810. [Google Scholar] [CrossRef] [PubMed]
- DeLuca, J.G.; Dong, Y.; Hergert, P.; Strauss, J.; Hickey, J.M.; Salmon, E.D.; McEwen, B.F. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell 2005, 16, 519–531. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Rines, D.R.; Espelin, C.W.; Sorger, P.K. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 2001, 106, 195–206. [Google Scholar] [CrossRef]
- McCleland, M.L.; Gardner, R.D.; Kallio, M.J.; Daum, J.R.; Gorbsky, G.J.; Burke, D.J.; Stukenberg, P.T. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003, 17, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Long, S.; Ciferri, C.; Westermann, S.; Drubin, D.; Barnes, G.; Nogales, E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J. Mol. Biol. 2008, 383, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.P.; Bloom, K.; Salmon, E.D. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 2009, 19, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Tien, J.F.; Umbreit, N.T.; Zelter, A.; Riffle, M.; Hoopmann, M.R.; Johnson, R.S.; Fonslow, B.R.; Yates, J.R., 3rd; MacCoss, M.J.; Moritz, R.L.; et al. Kinetochore Biorientation in Saccharomyces cerevisiae Requires a Tightly Folded Conformation of the Ndc80 Complex. Genetics 2014, 198, 1483–1493. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.P.; Bouck, D.C.; Molk, J.N.; Bloom, K.S.; Salmon, E.D. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 2006, 8, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Lawrimore, J.; Bloom, K.S.; Salmon, E.D. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 2011, 195, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Emanuele, M.J.; McCleland, M.L.; Satinover, D.L.; Stukenberg, P.T. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol. Biol. Cell 2005, 16, 4882–4892. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.P.; Bouck, D.; Finley, K.; Liu, X.; Wan, Y.; Berman, J.; He, X.; Salmon, E.D.; Bloom, K.S. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 2008, 181, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Badger, B.L.; Salmon, E.D. A quantitative description of Ndc80 complex linkage to human kinetochores. Nat. Commun. 2015, 6, 8161. [Google Scholar] [CrossRef] [PubMed]
- Zinkowski, R.P.; Meyne, J.; Brinkley, B.R. The centromere-kinetochore complex: A repeat subunit model. J. Cell Biol. 1991, 113, 1091–1110. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Brew, C.; Wolyniak, M.; Desai, A.; Anderson, S.; Muster, N.; Yates, J.R.; Huffaker, T.C.; Drubin, D.G.; Barnes, G. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 2001, 155, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Enquist-Newman, M.; Muller-Reichert, T.; Drubin, D.G.; Barnes, G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 2001, 152, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, C.; Cheeseman, I.M.; Goode, B.L.; McDonald, K.L.; Barnes, G.; Drubin, D.G. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 1998, 143, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Janke, C.; Ortiz, J.; Tanaka, T.U.; Lechner, J.; Schiebel, E. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. Embo J. 2002, 21, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Miranda, J.J.; De Wulf, P.; Sorger, P.K.; Harrison, S.C. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 2005, 12, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Westermann, S.; Avila-Sakar, A.; Wang, H.W.; Niederstrasser, H.; Wong, J.; Drubin, D.G.; Nogales, E.; Barnes, G. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol. Cell 2005, 17, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Asbury, C.L.; Gestaut, D.R.; Powers, A.F.; Franck, A.D.; Davis, T.N. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA 2006, 103, 9873–9878. [Google Scholar] [CrossRef] [PubMed]
- Westermann, S.; Wang, H.W.; Avila-Sakar, A.; Drubin, D.G.; Nogales, E.; Barnes, G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature 2006, 440, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Welburn, J.P.; Grishchuk, E.L.; Backer, C.B.; Wilson-Kubalek, E.M.; Yates, J.R., 3rd; Cheeseman, I.M. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell 2009, 16, 374–385. [Google Scholar] [CrossRef] [PubMed]
- Jeyaprakash, A.A.; Santamaria, A.; Jayachandran, U.; Chan, Y.W.; Benda, C.; Nigg, E.A.; Conti, E. Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol. Cell 2012, 46, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Coue, M.; Lombillo, V.A.; McIntosh, J.R. Microtubule depolymerization promotes particle and chromosome movement in vitro. J. Cell Biol. 1991, 112, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Lombillo, V.A.; Coue, M.; McIntosh, J.R. In vitro motility assays using microtubules tethered to Tetrahymena pellicles. Methods Cell Biol. 1993, 39, 149–165. [Google Scholar] [PubMed]
- Lombillo, V.A.; Nislow, C.; Yen, T.J.; Gelfand, V.I.; McIntosh, J.R. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J. Cell Biol. 1995, 128, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Lombillo, V.A.; Stewart, R.J.; McIntosh, J.R. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature 1995, 373, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Gudimchuk, N.; Vitre, B.; Kim, Y.; Kiyatkin, A.; Cleveland, D.W.; Ataullakhanov, F.I.; Grishchuk, E.L. Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat. Cell Biol. 2013, 15, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Gard, D.L.; Kirschner, M.W. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol. 1987, 105, 2203–2215. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.S.; Toda, T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr. Biol. 2011, 21, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Kalantzaki, M.; Kitamura, E.; Zhang, T.; Mino, A.; Novak, B.; Tanaka, T.U. Kinetochore-microtubule error correction is driven by differentially regulated interaction modes. Nat. Cell Biol. 2015, 17, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Ohkura, H.; Adachi, Y.; Kinoshita, N.; Niwa, O.; Toda, T.; Yanagida, M. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 1988, 7, 1465–1473. [Google Scholar] [PubMed]
- Tanaka, K.; Mukae, N.; Dewar, H.; van Breugel, M.; James, E.K.; Prescott, A.R.; Antony, C.; Tanaka, T.U. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 2005, 434, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.H.; Takada, H.; Hsu, K.S.; Toda, T. The internal loop of fission yeast Ndc80 binds Alp7/TACC-Alp14/TOG and ensures proper chromosome attachment. Mol. Biol. Cell 2013, 24, 1122–1133. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.J.; Huffaker, T.C. Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 1997, 139, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.P.; Asbury, C.L.; Biggins, S. A TOG Protein Confers Tension Sensitivity to Kinetochore-Microtubule Attachments. Cell 2016, 165, 1428–1439. [Google Scholar] [CrossRef] [PubMed]
- Trushko, A.; Schaffer, E.; Howard, J. The growth speed of microtubules with XMAP215-coated beads coupled to their ends is increased by tensile force. Proc. Natl. Acad. Sci. USA 2013, 110, 14670–14675. [Google Scholar] [CrossRef] [PubMed]
- Gonen, S.; Akiyoshi, B.; Iadanza, M.G.; Shi, D.; Duggan, N.; Biggins, S.; Gonen, T. The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nat. Struct. Mol. Biol. 2012, 19, 925–929. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.R.; Grishchuk, E.L.; Morphew, M.K.; Efremov, A.K.; Zhudenkov, K.; Volkov, V.A.; Cheeseman, I.M.; Desai, A.; Mastronarde, D.N.; Ataullakhanov, F.I. Fibrils connect microtubule tips with kinetochores: A mechanism to couple tubulin dynamics to chromosome motion. Cell 2008, 135, 322–333. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.R.; O’Toole, E.; Zhudenkov, K.; Morphew, M.; Schwartz, C.; Ataullakhanov, F.I.; Grishchuk, E.L. Conserved and divergent features of kinetochores and spindle microtubule ends from five species. J. Cell Biol. 2013, 200, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Salmon, E.D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell 1995, 6, 1619–1640. [Google Scholar] [CrossRef] [PubMed]
- Koshland, D.E.; Mitchison, T.J.; Kirschner, M.W. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature 1988, 331, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Amos, L.; Klug, A. Arrangement of subunits in flagellar microtubules. J. Cell Sci. 1974, 14, 523–549. [Google Scholar] [PubMed]
- Desai, A.; Mitchison, T.J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83–117. [Google Scholar] [CrossRef] [PubMed]
- Mandelkow, E.M.; Mandelkow, E.; Milligan, R.A. Microtubule dynamics and microtubule caps: A time-resolved cryo-electron microscopy study. J. Cell Biol. 1991, 114, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Muller-Reichert, T.; Chretien, D.; Severin, F.; Hyman, A.A. Structural changes at microtubule ends accompanying GTP hydrolysis: Information from a slowly hydrolyzable analogue of GTP, guanylyl (alpha,beta)methylenediphosphonate. Proc. Natl. Acad. Sci. USA 1998, 95, 3661–3666. [Google Scholar] [CrossRef] [PubMed]
- Rice, L.M.; Montabana, E.A.; Agard, D.A. The lattice as allosteric effector: Structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc. Natl. Acad. Sci. USA 2008, 105, 5378–5383. [Google Scholar] [CrossRef] [PubMed]
- Caplow, M.; Ruhlen, R.L.; Shanks, J. The free energy for hydrolysis of a microtubule-bound nucleotide triphosphate is near zero: All of the free energy for hydrolysis is stored in the microtubule lattice. J. Cell Biol. 1994, 127, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Caplow, M.; Shanks, J. Evidence that a single monolayer tubulin-GTP cap is both necessary and sufficient to stabilize microtubules. Mol. Biol. Cell 1996, 7, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Walker, R.A.; Inoue, S.; Salmon, E.D. Asymmetric behavior of severed microtubule ends after ultraviolet-microbeam irradiation of individual microtubules in vitro. J. Cell Biol. 1989, 108, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Powers, A.F.; Franck, A.D.; Gestaut, D.R.; Cooper, J.; Gracyzk, B.; Wei, R.R.; Wordeman, L.; Davis, T.N.; Asbury, C.L. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 2009, 136, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Gestaut, D.R.; Graczyk, B.; Cooper, J.; Widlund, P.O.; Zelter, A.; Wordeman, L.; Asbury, C.L.; Davis, T.N. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 2008, 10, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Grishchuk, E.L.; Spiridonov, I.S.; Volkov, V.A.; Efremov, A.; Westermann, S.; Drubin, D.; Barnes, G.; Ataullakhanov, F.I.; McIntosh, J.R. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc. Natl. Acad. Sci. USA 2008, 105, 6918–6923. [Google Scholar] [CrossRef] [PubMed]
- Kudalkar, E.M.; Scarborough, E.A.; Umbreit, N.T.; Zelter, A.; Gestaut, D.R.; Riffle, M.; Johnson, R.S.; MacCoss, M.J.; Asbury, C.L.; Davis, T.N. Regulation of outer kinetochore Ndc80 complex-based microtubule attachments by the central kinetochore Mis12/MIND complex. Proc. Natl. Acad. Sci. USA 2015, 112, E5583–E5589. [Google Scholar] [CrossRef] [PubMed]
- Tien, J.F.; Umbreit, N.T.; Gestaut, D.R.; Franck, A.D.; Cooper, J.; Wordeman, L.; Gonen, T.; Asbury, C.L.; Davis, T.N. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 2010, 189, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Volkov, V.A.; Zaytsev, A.V.; Gudimchuk, N.; Grissom, P.M.; Gintsburg, A.L.; Ataullakhanov, F.I.; McIntosh, J.R.; Grishchuk, E.L. Long tethers provide high-force coupling of the Dam1 ring to shortening microtubules. Proc. Natl. Acad. Sci. USA 2013, 110, 7708–7713. [Google Scholar] [CrossRef] [PubMed]
- Weir, J.R.; Faesen, A.C.; Klare, K.; Petrovic, A.; Basilico, F.; Fischbock, J.; Pentakota, S.; Keller, J.; Pesenti, M.E.; Pan, D.; et al. Insights from biochemical reconstitution into the architecture of human kinetochores. Nature 2016, 537, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, A.; Keller, J.; Liu, Y.; Overlack, K.; John, J.; Dimitrova, Y.N.; Jenni, S.; van Gerwen, S.; Stege, P.; Wohlgemuth, S.; et al. Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores. Cell 2016, 167, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, Y.N.; Jenni, S.; Valverde, R.; Khin, Y.; Harrison, S.C. Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly. Cell 2016, 167, 1014–1027. [Google Scholar] [CrossRef] [PubMed]
- Asbury, C.L.; Tien, J.F.; Davis, T.N. Kinetochores’ gripping feat: Conformational wave or biased diffusion? Trends Cell Biol. 2011, 21, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Efremov, A.; Grishchuk, E.L.; McIntosh, J.R.; Ataullakhanov, F.I. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proc. Natl. Acad. Sci. USA 2007, 104, 19017–19022. [Google Scholar] [CrossRef] [PubMed]
- Salmon, E.D. Microtubules: A ring for the depolymerization motor. Curr. Biol. 2005, 15, R299–R302. [Google Scholar] [CrossRef] [PubMed]
- Umbreit, N.T.; Miller, M.P.; Tien, J.F.; Cattin Ortola, J.; Gui, L.; Lee, K.K.; Biggins, S.; Asbury, C.L.; Davis, T.N. Kinetochores require oligomerization of Dam1 complex to maintain microtubule attachments against tension and promote biorientation. Nat. Commun. 2014, 5, 4951. [Google Scholar] [CrossRef] [PubMed]
- Lampert, F.; Hornung, P.; Westermann, S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 2010, 189, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Miranda, J.J.; King, D.S.; Harrison, S.C. Protein arms in the kinetochore-microtubule interface of the yeast DASH complex. Mol. Biol. Cell 2007, 18, 2503–2510. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, G.J.; Deluca, J.G. Connecting with Ska, a key complex at the kinetochore-microtubule interface. EMBO J. 2009, 28, 1375–1377. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.C.; Arthanari, H.; Boeszoermenyi, A.; Dashkevich, N.M.; Wilson-Kubalek, E.M.; Monnier, N.; Markus, M.; Oberer, M.; Milligan, R.A.; Bathe, M.; et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell 2012, 23, 968–980. [Google Scholar] [CrossRef] [PubMed]
- VandenBeldt, K.J.; Barnard, R.M.; Hergert, P.J.; Meng, X.; Maiato, H.; McEwen, B.F. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr. Biol. 2006, 16, 1217–1223. [Google Scholar] [CrossRef] [PubMed]
- Volkov, V.A.; Grissom, P.M.; Arzhanik, V.K.; Zaytsev, A.V.; Renganathan, K.; McClure-Begley, T.; Old, W.M.; Ahn, N.; McIntosh, J.R. Centromere protein F includes two sites that couple efficiently to depolymerizing microtubules. J. Cell Biol. 2015, 209, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Hill, T.L. Theoretical problems related to the attachment of microtubules to kinetochores. Proc. Natl. Acad. Sci. USA 1985, 82, 4404–4408. [Google Scholar] [CrossRef] [PubMed]
- Musinipally, V.; Howes, S.; Alushin, G.M.; Nogales, E. The microtubule binding properties of CENP-E’s C-terminus and CENP-F. J. Mol. Biol. 2013, 425, 4427–4441. [Google Scholar] [CrossRef] [PubMed]
- Sarangapani, K.K.; Akiyoshi, B.; Duggan, N.M.; Biggins, S.; Asbury, C.L. Phosphoregulation promotes release of kinetochores from dynamic microtubules via multiple mechanisms. Proc. Natl. Acad. Sci. USA 2013, 110, 7282–7287. [Google Scholar] [CrossRef] [PubMed]
- Sarangapani, K.K.; Duro, E.; Deng, Y.; Alves Fde, L.; Ye, Q.; Opoku, K.N.; Ceto, S.; Rappsilber, J.; Corbett, K.D.; Biggins, S.; et al. Sister kinetochores are mechanically fused during meiosis I in yeast. Science 2014, 346, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Hoog, J.L.; Huisman, S.M.; Sebo-Lemke, Z.; Sandblad, L.; McIntosh, J.R.; Antony, C.; Brunner, D. Electron tomography reveals a flared morphology on growing microtubule ends. J. Cell Sci. 2011, 124, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Chretien, D.; Fuller, S.D.; Karsenti, E. Structure of growing microtubule ends: Two-dimensional sheets close into tubes at variable rates. J. Cell Biol. 1995, 129, 1311–1328. [Google Scholar] [CrossRef] [PubMed]
- Arnal, I.; Karsenti, E.; Hyman, A.A. Structural transitions at microtubule ends correlate with their dynamic properties in Xenopus egg extracts. J. Cell Biol. 2000, 149, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Guesdon, A.; Bazile, F.; Buey, R.M.; Mohan, R.; Monier, S.; Garcia, R.R.; Angevin, M.; Heichette, C.; Wieneke, R.; Tampe, R.; et al. EB1 interacts with outwardly curved and straight regions of the microtubule lattice. Nat. Cell Biol. 2016, 18, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Kapitein, L.C.; Peterman, E.J.; Kwok, B.H.; Kim, J.H.; Kapoor, T.M.; Schmidt, C.F. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 2005, 435, 114–118. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.R.; Euteneuer, U. Tubulin hooks as probes for microtubule polarity: An analysis of the method and an evaluation of data on microtubule polarity in the mitotic spindle. J. Cell Biol. 1984, 98, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Cameron, L.A.; Yang, G.; Cimini, D.; Canman, J.C.; Kisurina-Evgenieva, O.; Khodjakov, A.; Danuser, G.; Salmon, E.D. Kinesin 5-independent poleward flux of kinetochore microtubules in PtK1 cells. J. Cell Biol. 2006, 173, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, D.T.; Perlman, Z.E.; Burbank, K.S.; Groen, A.C.; Mitchison, T.J. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J. Cell Biol. 2004, 167, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Shirasu-Hiza, M.; Perlman, Z.E.; Wittmann, T.; Karsenti, E.; Mitchison, T.J. Eg5 causes elongation of meiotic spindles when flux-associated microtubule depolymerization is blocked. Curr. Biol. 2004, 14, 1941–1945. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, D. Kinetochore fibre dynamics outside the context of the spindle during anaphase. Nat. Cell Biol. 2004, 6, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Verma, S.; Mitchison, T.J.; Walczak, C.E. Kin I kinesins are microtubule-destabilizing enzymes. Cell 1999, 96, 69–78. [Google Scholar] [CrossRef]
- Cooper, J.R.; Wagenbach, M.; Asbury, C.L.; Wordeman, L. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat. Struct. Mol. Biol. 2010, 17, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Hunter, A.W.; Caplow, M.; Coy, D.L.; Hancock, W.O.; Diez, S.; Wordeman, L.; Howard, J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell 2003, 11, 445–457. [Google Scholar] [CrossRef]
- Ganem, N.J.; Compton, D.A. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 2004, 166, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Manning, A.L.; Ganem, N.J.; Bakhoum, S.F.; Wagenbach, M.; Wordeman, L.; Compton, D.A. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol. Biol. Cell 2007, 18, 2970–2979. [Google Scholar] [CrossRef] [PubMed]
- Wilbur, J.D.; Heald, R. Mitotic spindle scaling during Xenopus development by kif2a and importin alpha. eLife 2013, 2, e00290. [Google Scholar] [CrossRef] [PubMed]
- Sharp, D.J.; Ross, J.L. Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 2012, 125, 2561–2569. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.; Asenjo, A.B.; Mennella, V.; Sharp, D.J.; Sosa, H. Kinesin-13s form rings around microtubules. J. Cell Biol. 2006, 175, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Asenjo, A.B.; Greenbaum, M.; Xie, L.; Sharp, D.J.; Sosa, H. A second tubulin binding site on the kinesin-13 motor head domain is important during mitosis. PLoS ONE 2013, 8, e73075. [Google Scholar] [CrossRef] [PubMed]
- Wordeman, L.; Mitchison, T.J. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 1995, 128, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Oguchi, Y.; Uchimura, S.; Ohki, T.; Mikhailenko, S.V.; Ishiwata, S. The bidirectional depolymerizer MCAK generates force by disassembling both microtubule ends. Nat. Cell Biol. 2011, 13, 846–852. [Google Scholar] [CrossRef] [PubMed]
- Verde, F.; Dogterom, M.; Stelzer, E.; Karsenti, E.; Leibler, S. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J. Cell Biol. 1992, 118, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Merdes, A.; Ramyar, K.; Vechio, J.D.; Cleveland, D.W. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 1996, 87, 447–458. [Google Scholar] [CrossRef]
- Maiato, H.; Rieder, C.L.; Khodjakov, A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 2004, 167, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Khodjakov, A.; Cole, R.W.; Oakley, B.R.; Rieder, C.L. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 2000, 10, 59–67. [Google Scholar] [CrossRef]
- Gaglio, T.; Dionne, M.A.; Compton, D.A. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J. Cell Biol. 1997, 138, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Surrey, T.; Nedelec, F.; Leibler, S.; Karsenti, E. Physical properties determining self-organization of motors and microtubules. Science 2001, 292, 1167–1171. [Google Scholar] [CrossRef] [PubMed]
- Nedelec, F.; Surrey, T.; Karsenti, E. Self-organisation and forces in the microtubule cytoskeleton. Curr. Opin. Cell Biol. 2003, 15, 118–124. [Google Scholar] [CrossRef]
- Maiato, H.; Khodjakov, A.; Rieder, C.L. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 2005, 7, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Östergren, G. Luzula and the mechanism of chromosome movements. Hereditas 1949, 35, 445–468. [Google Scholar] [CrossRef]
- Ostergren, G.; Mole-Bajer, J.; Bajer, A. An interpretation of transport phenomena at mitosis. Ann. N. Y. Acad. Sci. 1960, 90, 381–408. [Google Scholar] [CrossRef] [PubMed]
- Franck, A.D.; Powers, A.F.; Gestaut, D.R.; Gonen, T.; Davis, T.N.; Asbury, C.L. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat. Cell Biol. 2007, 9, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Llauró, A.; Asbury, C.L.; University of Washington, Seattle, WA, USA. unpublished observations. 2016.
- Higuchi, T.; Uhlmann, F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature 2005, 433, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Brust-Mascher, I.; Scholey, J.M. Mitotic motors and chromosome segregation: The mechanism of anaphase B. Biochem. Soc. Trans. 2011, 39, 1149–1153. [Google Scholar] [CrossRef] [PubMed]
- Su, K.C.; Barry, Z.; Schweizer, N.; Maiato, H.; Bathe, M.; Cheeseman, I.M. A Regulatory Switch Alters Chromosome Motions at the Metaphase-to-Anaphase Transition. Cell Rep. 2016, 17, 1728–1738. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, R.B.; Staehly, C.A. Chromosome micromanipulation. I. The mechanics of chromosome attachment to the spindle. Chromosoma 1967, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lampson, M.A.; Renduchitala, K.; Khodjakov, A.; Kapoor, T.M. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 2004, 6, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Umbreit, N.T.; Gestaut, D.R.; Tien, J.F.; Vollmar, B.S.; Gonen, T.; Asbury, C.L.; Davis, T.N. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc. Natl. Acad. Sci. USA 2012, 109, 16113–16118. [Google Scholar] [CrossRef] [PubMed]
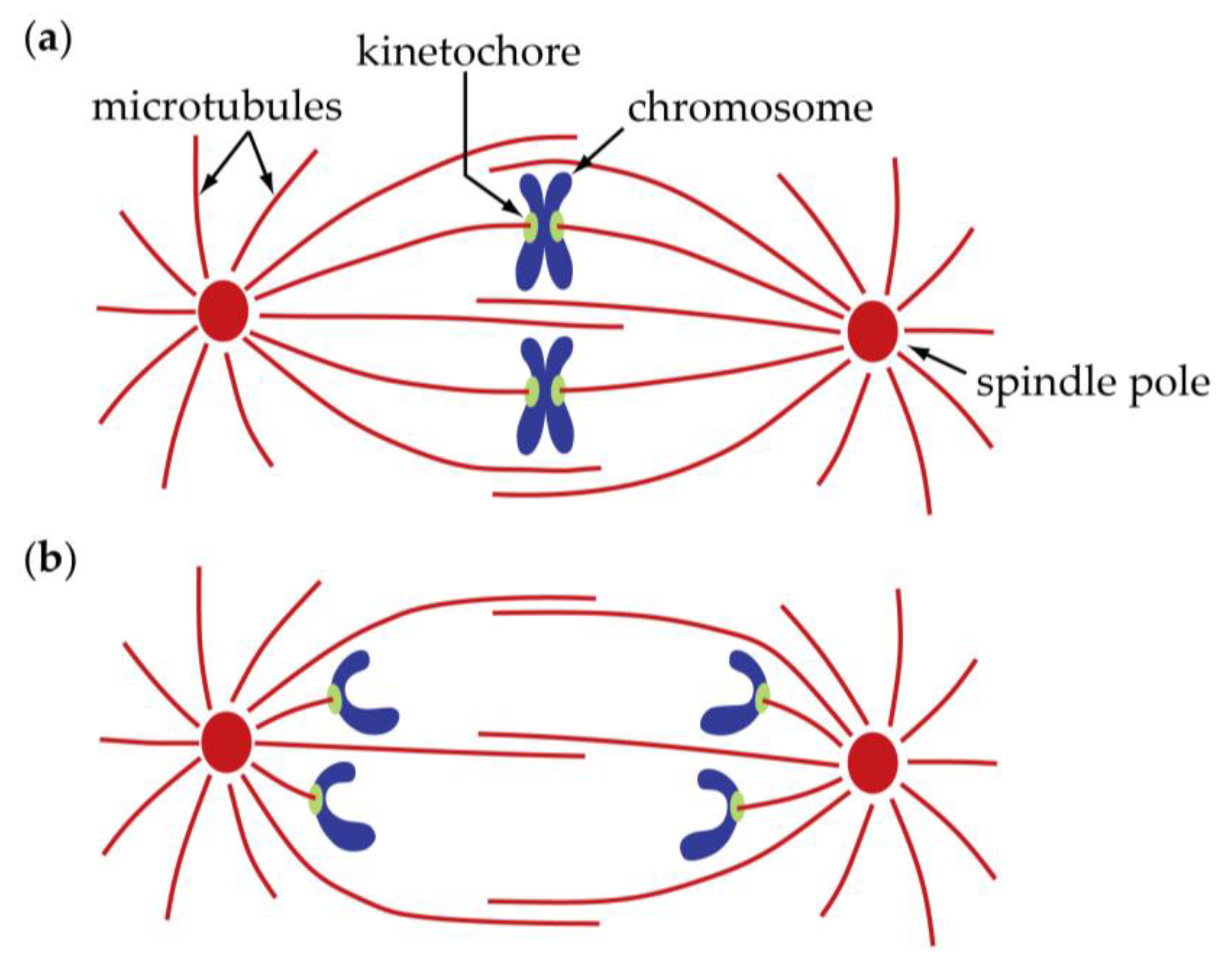
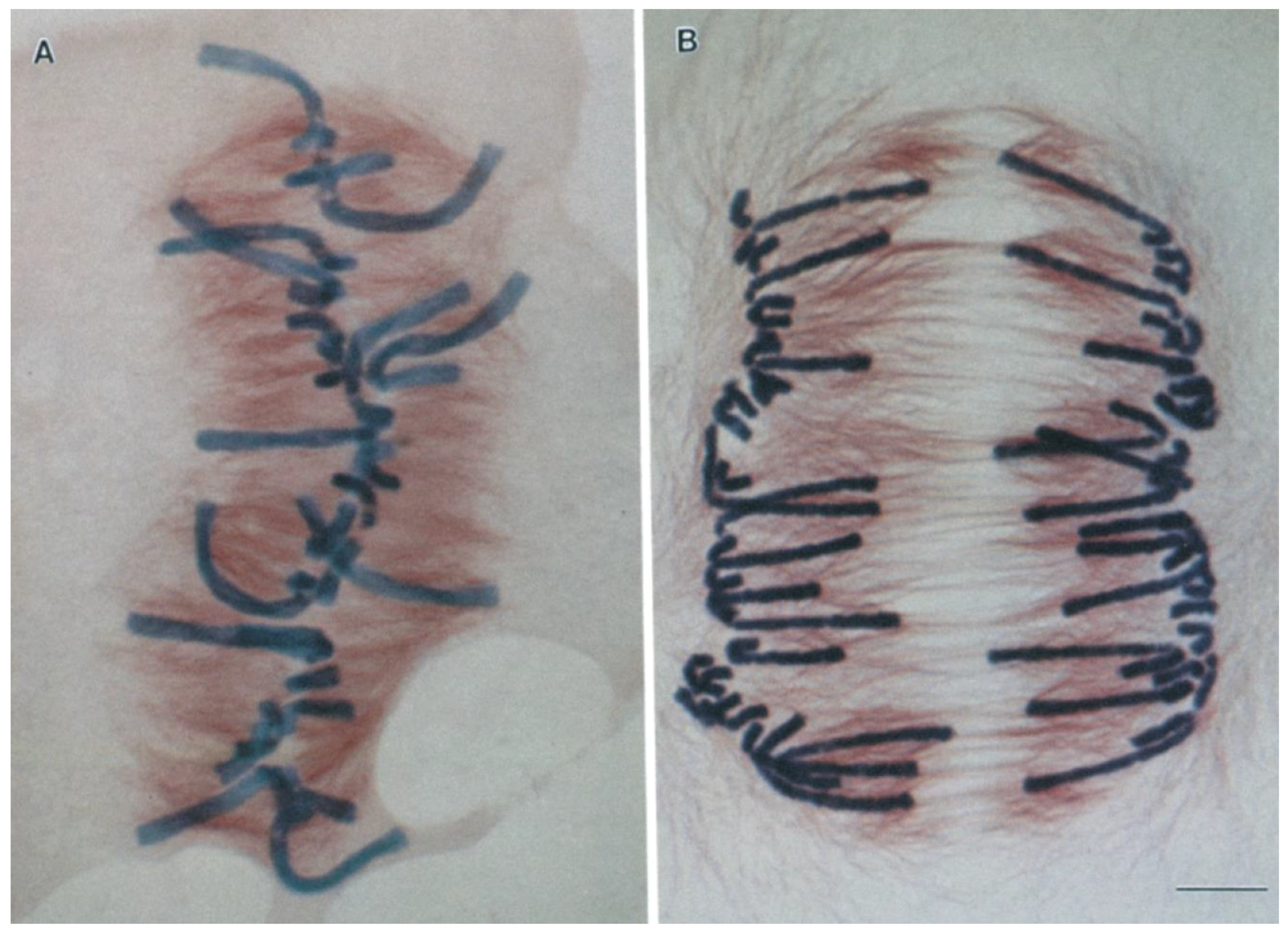
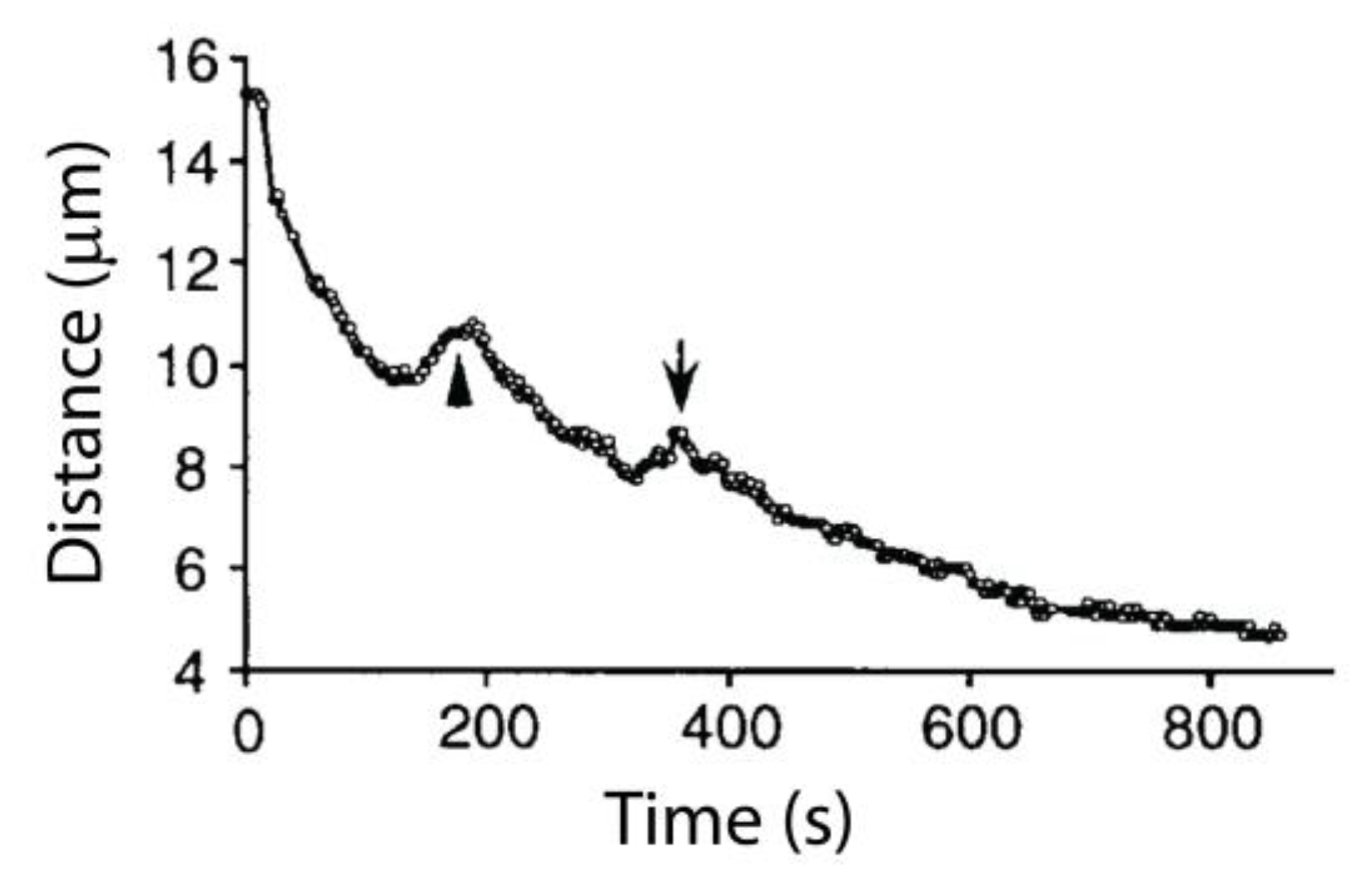
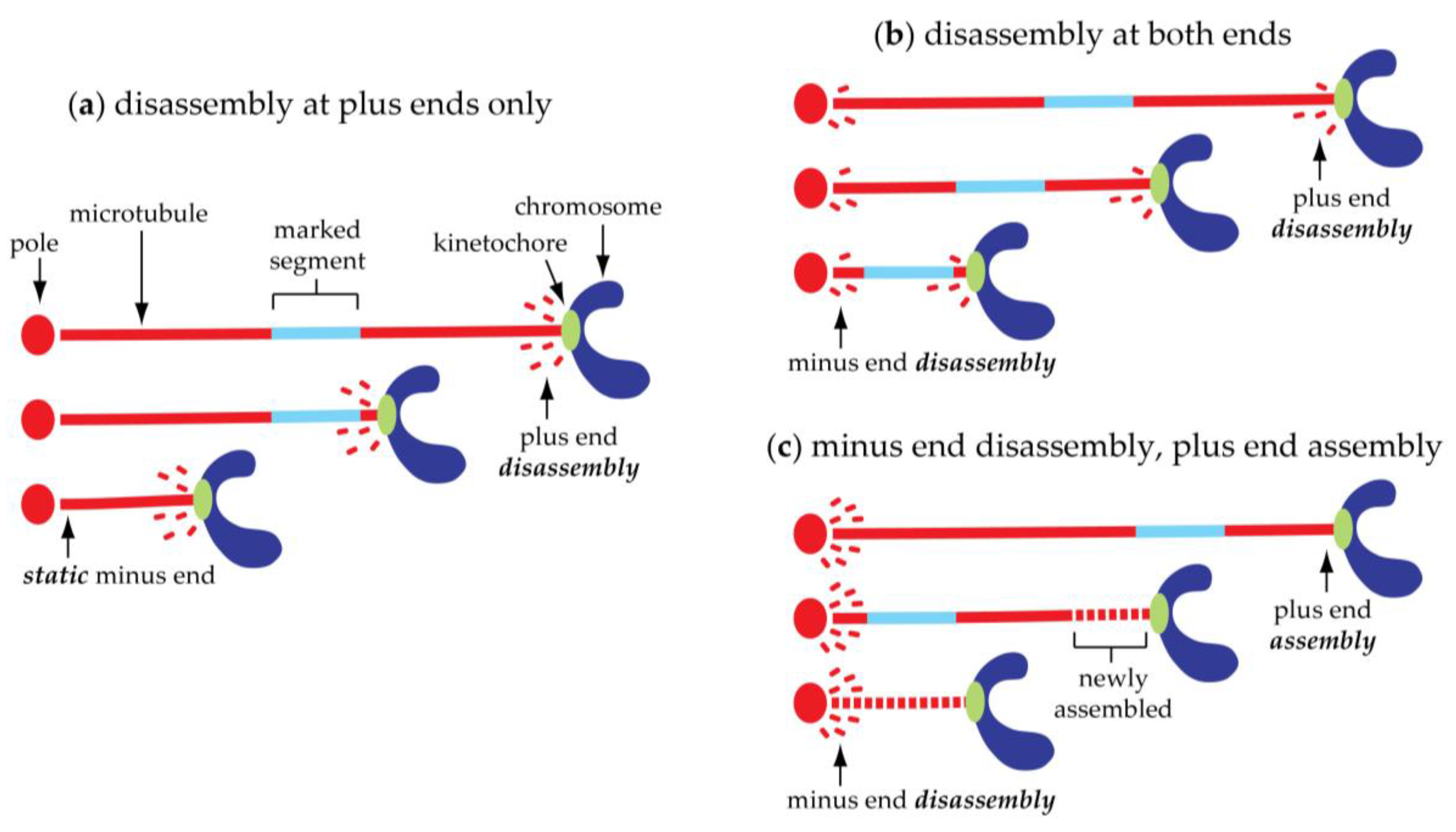


| Spindle/Cell Type | Chromosome-to-Pole Speed (μm/min) | Speed Measured in Anaphase A? | Microtubule-to-Pole Flux Speed (μm/min) | Technique for Flux Measurement | Flux Measured in Anaphase A? | Experimental Condition | Fraction of Anaphase A Speed Due to Flux (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Sand dollar embryos | 1 | yes | 1.8 | photobleaching | yes | control | 180 | [38] |
| Newt lung cells | 1.7 | yes | 0.44 | photoactivation | yes | early anaphase | 26 | [33] |
| 0.54 | yes | 0.18 | photoactivation | yes | late anaphase | 33 | ||
| Newt lung cells | 0.2 | yes | 0.2 | photoactivation | yes | 10 uM taxol, late anaphase | 100 | [39] |
| Pig kidney (LLC-PK) and rat kangaroo (PtK1) cells | 1.2 | yes | 0.2 | photoactivation | yes | early anaphase | 17 | [34] |
| Xenopus (meiotic) extract spindles | 2 | yes | 2 | photoactivation | yes | control | 100 | [40] |
| 0.2 | yes | 0.2 | photoactivation | yes | 1.5 mM AMPPNP | 100 | ||
| 2 | yes | 2 | photoactivation | no (metaphase) | 1 uM taxol | 100 | ||
| Xenopus (meiotic) extract spindles | 2.8 | yes | 1.6 | speckle | yes | plus end depol. | 57 | [32] |
| 0.7 | yes | 1.6 | speckle | yes | plus end polym. | 229 | ||
| Budding yeast | 1.3 | yes | - | - | - | CEN dots (14 kb LacO array) | - | [8] |
| Budding yeast | - | - | 0 | photobleaching | no (anaphase B) | ipMTs (not kMTs) | - | [26] |
| Budding yeast | 0.3 | yes | - | - | - | CEN dots (2–11 kb LacO arrays) | - | [41] |
| Budding yeast | 0.3 | yes | - | - | - | CEN dots (2 kb LacO array) | - | [42] |
| Fission yeast | - | - | 0 | photobleaching | no (anaphase B) | ipMTs (not kMTs) | - | [29] |
| Fission yeast | - | - | 0 | speckle | no (anaphase B) | ipMTs (not kMTs) | - | [28] |
| Fission yeast | - | - | 0 | photobleaching | no (anaphase B) | ipMTs (not kMTs) | - | [27] |
| Drosophila embryos | 3.6 | yes | 3.2 | speckle | yes | control, 18 °C | 89 | [35] |
| Drosophila embryos | 6.4 | yes | 1.9 | speckle | yes | control | 30 | [43] |
| Drosophila embryos | 5.6 | yes | 2.2 | speckle | yes | control | 39 | [36] |
| 3.4 | yes | 3.4 | speckle | yes | anti-KLP59C | 100 | ||
| 3.2 | yes | 0 | speckle | no (metaphase) | anti-KLP10A | 0 | ||
| Drosophila (S2) cells | 1.2 | yes | 0.6 | photobleaching | yes | control | 50 | [44] |
| 0.6 | yes | 0.5 | photobleaching | yes | katanin RNAi | 83 | ||
| 0.8 | yes | 0.2 | photobleaching | yes | spastin RNAi | 25 | ||
| 0.7 | yes | 0.1 | photobleaching | yes | fidgetin RNAi | 14 | ||
| Drosophila (S2) cells | 1.7 | yes | 0.9 | photobleaching | yes | control | 53 | [45] |
| 0.7 | yes | 0.5 | photobleaching | yes | KLP59D RNAi | 71 | ||
| 1.7 | yes | 0.9 | photobleaching | yes | KLP59C RNAi | 53 | ||
| 0.8 | yes | 0.3 | photobleaching | yes | KLP10A RNAi | 38 | ||
| Drosophila (S2) cells | 0.8 | yes | 0.4 | speckle | yes | control | 50 | [46] |
| 0.7 | yes | 0.2 | speckle | yes | CLASP & KLP10A RNAi | 28 | ||
| Drosophila spermatocytes (meiosis) | - | - | 0.6 | photobleaching | no (metaphase) | metaphase | - | [47] |
| 1.7 | yes | 0.6 | photobleaching | yes | disjoining | 35 | ||
| 2.7 | yes | 1 | photobleaching | yes | separated | 37 | ||
| Crane-fly spermatocytes (meiosis) | 0.5 | yes | 0.9 | speckle | yes | autosomal half-bivalents | 180 | [31] |
| Crane-fly spermatocytes (meiosis) | 1.3 | yes | 0.8 | speckle | yes | sex univalent, bipolar link cut | 62 | [48] |
| Crane-fly spermatocytes (meiosis) | 0.7 | no (metaphase) | 0.7 | speckle | no (metaphase) | autosomal, cut K-fragment | 100 | [49] |
| Human (U20S) cells | 1.5 | yes | 0.5 | photoactivation | no (metaphase) | control | 33 | [30] |
| 1.2 | yes | 0 | photoactivation | no (metaphase) | MCAK & Kif2a RNAi | 0 | ||
| Human (U20S) cells | - | - | 0.5 | photoactivation | no (metaphase) | control | - | [50] |
| - | - | 0.3 | photoactivation | no (metaphase) | CLASPs RNAi | - | ||
| - | - | 0.3 | photoactivation | no (metaphase) | Cenp-E RNAi | - | ||
| Human (U20S) cells | - | - | 0.6 | photoconversion | no (metaphase) | control | - | [51] |
| - | - | 0.3 | photoconversion | no (metaphase) | Kif4A RNAi | - | ||
| Human (U20S) cells | 0.44 | yes | 0.9 | photoactivation | no (metaphase) | control | - | [52] |
| 0.3 | yes | 0.6 | photoactivation | no (metaphase) | fidgetin siRNA | - | ||
| Human (HeLa) cells | - | - | 0.4 | photoactivation | no (metaphase) | control | - | [53] |
| - | - | 0.2 | photoactivation | no (metaphase) | ectopic MCAK at CEN | - | ||
| Human (HeLa) cells | 1.7 | yes | - | - | - | control | - | [15] |
| 0.9 | yes | - | - | - | Kif18A overexpress. | - | ||
| 2.8 | yes | - | - | - | Kif18A siRNA | - | ||
| Tobacco (BY-2) cells | 2.1 | yes | 2 | photobleaching | yes | control | 95 | [37] |
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asbury, C.L. Anaphase A: Disassembling Microtubules Move Chromosomes toward Spindle Poles. Biology 2017, 6, 15. https://doi.org/10.3390/biology6010015
Asbury CL. Anaphase A: Disassembling Microtubules Move Chromosomes toward Spindle Poles. Biology. 2017; 6(1):15. https://doi.org/10.3390/biology6010015
Chicago/Turabian StyleAsbury, Charles L. 2017. "Anaphase A: Disassembling Microtubules Move Chromosomes toward Spindle Poles" Biology 6, no. 1: 15. https://doi.org/10.3390/biology6010015
APA StyleAsbury, C. L. (2017). Anaphase A: Disassembling Microtubules Move Chromosomes toward Spindle Poles. Biology, 6(1), 15. https://doi.org/10.3390/biology6010015





