The Function of Fish Cytokines
Abstract
:1. Introduction
2. β-Trefoil Cytokines
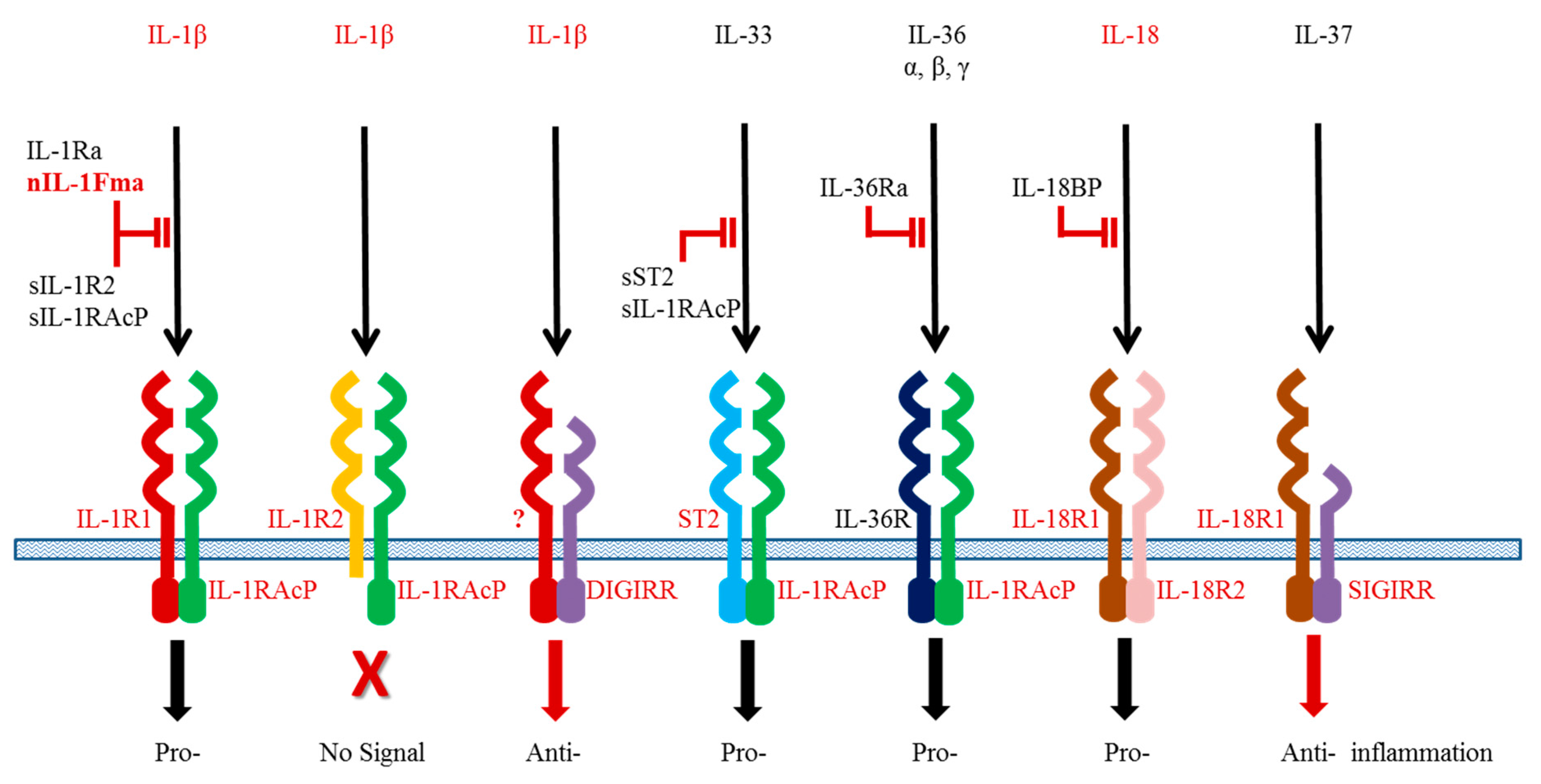
2.1. IL-1β
2.2. IL-18
2.3. Fish Specific Novel IL-1 Family Member
3. B-Jellyroll Cytokines
4. Cysteine Knot Cytokines
4.1. Interleukin-17
4.2. Transforming Growth Factor-β
5. Type I α Helical Cytokines
5.1. IL-2 Subfamily
5.2. Beta Chain Cytokines
5.3. IL-6 Subfamily
5.4. IL-12 Subfamily
5.5. Colony Stimulating Factors
6. Type II α Helical Cytokines
6.1. Type I IFN
6.2. Group I IFNs
6.3. Group II IFNs
6.4. Type II IFNs
6.5. IFN-γrel
6.6. IL-10
6.7. Other Members of the Type II α-Helical Cytokine Family
7. Conclusions
Conflicts of Interest
Abbreviations
| CCK | CC chemokine |
| CD | cluster of differentiation |
| CISH | cytokine inducible Src homology 2 (SH2)-containing protein |
| CNTF | ciliary neurotrophic factor |
| COX-2 | cyclooxygenase-2 |
| CRFB | class II cytokine receptor family member |
| CSF | colony stimulating factor |
| DAMP | danger associated molecular pattern |
| DC-SIGN | dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin |
| DIGIRR | double immunoglobulin IL-1R related molecule |
| γIP | gamma interferon inducible protein |
| GBP | guanylate binding protein |
| G-CSF | granulocyte colony stimulating factor |
| GM-CSF | granulocyte-macrophage colony-stimulating factor |
| HK | head kidney |
| HSC | haematopoietic stem cell |
| IGFBP | insulin growth factor binding protein |
| ICE | IL-1β converting enzyme |
| IEC | intestinal epithelial cell |
| IFN | interferon |
| IFN-γrel | interferon gamma related molecule |
| Ig | immunoglobulin |
| IL | interleukin |
| IL-1F | interleukin-1 family member |
| IL-1Ra | interleukin 1 receptor antagonist |
| IL-1RAcP | interleukin-1 receptor accessory protein |
| IL-36Ra | interleukin-36 receptor antagonist |
| ILC | innate lymphoid cell |
| iNOS | inducible nitric oxide synthase |
| IRAK | interleukin 1 receptor associated kinase |
| IRF | interferon regulatory factor |
| ISG | interferon stimulated gene |
| JAK | Janus kinase |
| LPS | lipopolysaccharide |
| LT | lymphotoxin |
| MAPK | mitogen-activated protein kinase |
| M-CSFR | macrophage colony stimulating factor receptor |
| MyD88 | myeloid differentiation factor 88 |
| NF-κB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| nIL-1Fm | novel interleukin-1 family member |
| PBL | peripheral blood leucocyte |
| PAMP | pathogen associated molecular pattern |
| poly(I:C) | Polyinosinic-polycytidylic acid |
| PRR | pattern recognition receptor |
| ROS | reactive oxidative species |
| SIGIRR | single domain interleukin-1 receptor related molecule |
| SOCS | suppressors of cytokine signalling |
| STAT | signal transducers and activators of transcription |
| TACE | TNF-α-converting enzyme |
| TGF | transforming growth factor |
| TIR | Toll/IL-1 receptor |
| TNF | tumour necrosis factor |
| TNFSF | tumour necrosis factor superfamily member |
| TNFSFR | tumour necrosis factor superfamily member receptor |
| TRAF | tumour necrosis factor receptor associated factor |
| Treg | regulatory T cell |
| Tyk | tyrosine kinase |
| WGD | whole genome duplication |
References
- Secombes, C.J.; Wang, T.H.; Bird, S. The interleukins of fish. Dev. Comp. Immunol. 2011, 35, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Secombes, C.J. The cytokine networks of adaptive immunity in fish. Fish Shellfish Immunol. 2013, 35, 1703–1718. [Google Scholar] [CrossRef] [PubMed]
- Secombes, C.J.; Zou, J.; Bird, S. Cytokines of cartilaginous fish. In Immunobiology of the Shark; Smith, S.L., Sim, R.B., Flajnik, M.F., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 123–142. [Google Scholar]
- Secombes, C.J.; Wang, T.; Bird, S. Vertebrate cytokines and their evolution. In The Evolution of the Immune System: Conservation and Diversification; Malagoli, D., Ed.; Academic Press Inc.: Cambridge, MA, USA, 2016; Chapter 5; pp. 87–151. [Google Scholar]
- Bird, S.; Tafalla, C. Teleost chemokines and their receptors. Biology 2015, 4, 756–784. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, J.; Owyang, A.; Oldham, E.; Song, Y.; Murphy, E.; McClanahan, T.K.; Zurawski, G.; Moshrefi, M.; Qin, J.; Li, X.; et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity 2005, 23, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Sims, J.E.; Nicklin, M.J.; Bazan, J.F.; Barton, J.L.; Busfield, S.J.; Ford, J.E.; Kastelein, R.A.; Kumar, S.; Lin, H.; Mulero, J.J.; et al. A new nomenclature for IL-1-family genes. Trends Immunol. 2001, 22, 536–537. [Google Scholar] [CrossRef]
- Bird, S.; Wang, T.; Zou, J.; Cunningham, C.; Secombes, C.J. The first cytokine sequence within cartilaginous fish: IL-1β in the small spotted catshark (Scyliorhinus canicula). J. Immunol. 2002, 168, 3329–3340. [Google Scholar] [CrossRef] [PubMed]
- Engelsma, M.Y.; Stet, R.J.; Schipper, H.; Verburg-van Kemenade, B.M. Regulation of interleukin 1 beta RNA expression in the common carp, Cyprinus carpio L. Dev. Comp. Immunol. 2001, 25, 195–203. [Google Scholar] [CrossRef]
- Fujiki, K.; Shin, D.H.; Nakao, M.; Yano, T. Molecular cloning and expression analysis of carp (Cyprinus carpio) interleukin-1 beta, high affinity immunoglobulin E Fc receptor gamma subunit and serum amyloid A. Fish Shellfish Immunol. 2000, 10, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos, O.; Zou, J.; Cunningham, C.; Secombes, C.J. Cloning, sequencing, and analysis of expression of a second IL-1beta gene in rainbow trout (Oncorhynchus mykiss). Immunogenetics 2000, 51, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Scapigliati, G.; Buonocore, F.; Bird, S.; Zou, J.; Pelegrin, P.; Falasca, C.; Prugnoli, D.; Secombes, C.J. Phylogeny of cytokines: Molecular cloning and expression analysis of sea bass Dicentrarchus labrax interleukin-1beta. Fish Shellfish Immunol. 2001, 11, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Kwang, J. Carp interleukin-1 beta in the role of an immuno-adjuvant. Fish Shellfish Immunol. 2000, 10, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Grabowski, P.S.; Cunningham, C.; Secombes, C.J. Molecular cloning of interleukin 1beta from rainbow trout Oncorhynchus mykiss reveals no evidence of an ice cut site. Cytokine 1999, 11, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Cunningham, C.; Secombes, C.J. The rainbow trout Oncorhynchus mykiss interleukin-1 beta gene has a differ organization to mammals and undergoes incomplete splicing. Eur. J. Biochem. 1999, 259, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Huising, M.O.; Stet, R.J.; Savelkoul, H.F.; Verburg-van Kemenade, B.M. The molecular evolution of the interleukin-1 family of cytokines; IL-18 in teleost fish. Dev. Comp. Immunol. 2004, 28, 395–413. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Bird, S.; Koussounadis, A.; Holland, J.W.; Carrington, A.; Zou, J.; Secombes, C.J. Identification of a novel IL-1 cytokine family member in teleost fish. J. Immunol. 2009, 183, 962–974. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Bird, S.; Truckle, J.; Bols, N.; Horne, M.; Secombes, C. Identification and expression analysis of an IL-18 homologue and its alternatively spliced form in rainbow trout (Oncorhynchus mykiss). Eur. J. Biochem. 2004, 271, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Engelsma, M.Y.; Stet, R.J.; Saeij, J.P.; Verburg-van Kemenade, B.M. Differential expression and haplotypic variation of two interleukin-1beta genes in the common carp (Cyprinus carpio L.). Cytokine 2003, 22, 21–32. [Google Scholar] [CrossRef]
- Husain, M.; Bird, S.; van Zwieten, R.; Secombes, C.J.; Wang, T. Cloning of the IL-1beta3 gene and IL-1beta4 pseudogene in salmonids uncovers a second type of IL-1beta gene in teleost fish. Dev. Comp. Immunol. 2012, 38, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Hong, S.H.; Lee, H.J.; Jun, L.J.; Chung, J.K.; Kim, K.H.; Jeong, H.D. Molecular cDNA cloning and analysis of the organization and expression of the IL-1beta gene in the Nile tilapia, Oreochromis niloticus. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2006, 143, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Ogryzko, N.V.; Renshaw, S.A.; Wilson, H.L. The IL-1 family in fish: Swimming through the muddy waters of inflammasome evolution. Dev. Comp. Immunol. 2014, 46, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Overgard, A.C.; Nepstad, I.; Nerland, A.H.; Patel, S. Characterisation and expression analysis of the Atlantic halibut (Hippoglossus hippoglossus L.) cytokines: IL-1β, IL-6, IL-11, IL-12β and IFNγ. Mol. Biol. Rep. 2012, 39, 2201–2213. [Google Scholar] [CrossRef] [PubMed]
- Taechavasonyoo, A.; Kondo, H.; Nozaki, R.; Suzuki, Y.; Hirono, I. Identification of novel interleukin 1 beta family genes in Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2013, 34, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Baoprasertkul, P.; Peatman, E.; Liu, Z. Genomic organization, gene duplication, and expression analysis of interleukin-1beta in channel catfish (Ictalurus punctatus). Mol. Immunol. 2006, 43, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Angosto, D.; Lopez-Castejon, G.; Lopez-Munoz, A.; Sepulcre, M.P.; Arizcun, M.; Meseguer, J.; Mulero, V. Evolution of inflammasome functions in vertebrates: Inflammasome and caspase-1 trigger fish macrophage cell death but are dispensable for the processing of IL-1β. Innate Immun. 2012, 18, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Pozo, E.; Pelegrin, P.; Garcia-Castillo, J.; Garcia-Ayala, A.; Mulero, V.; Meseguer, J. Acidophilic granulocytes of the marine fish gilthead seabream (Sparus aurata L.) produce interleukin-1beta following infection with Vibrio anguillarum. Cell Tissue Res. 2004, 316, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Pelegrin, P.; Chaves-Pozo, E.; Mulero, V.; Meseguer, J. Production and mechanism of secretion of interleukin-1beta from the marine fish gilthead seabream. Dev. Comp. Immunol. 2004, 28, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Vojtech, L.N.; Scharping, N.; Woodson, J.C.; Hansen, J.D. Roles of inflammatory caspases during processing of zebrafish interleukin-1beta in Francisella noatunensis infection. Infect. Immun. 2012, 80, 2878–2885. [Google Scholar] [CrossRef] [PubMed]
- Garlanda, C.; Dinarello, C.A.; Mantovani, A. The interleukin-1 family: Back to the future. Immunity 2013, 39, 1003–1018. [Google Scholar] [CrossRef] [PubMed]
- Afonina, I.S.; Muller, C.; Martin, S.J.; Beyaert, R. Proteolytic processing of interleukin-1 family cytokines: Variations on a common theme. Immunity 2015, 42, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Bo, Y.X.; Song, X.H.; Wu, K.; Hu, B.; Sun, B.Y.; Liu, Z.J.; Fu, J.G. Characterization of interleukin-1beta as a proinflammatory cytokine in grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 2015, 46, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Buonocore, F.; Mazzini, M.; Forlenza, M.; Randelli, E.; Secombes, C.J.; Zou, J.; Scapigliati, G. Expression in Escherchia coli and purification of sea bass (Dicentrarchus labrax) interleukin 1beta, a possible immunoadjuvant in aquaculture. Mar. Biotechnol. 2004, 6, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Zou, J.; Crampe, M.; Peddie, S.; Scapigliati, G.; Bols, N.; Cunningham, C.; Secombes, C.J. The production and bioactivity of rainbow trout (Oncorhynchus mykiss) recombinant IL-1 beta. Vet. Immunol. Immunopathol. 2001, 81, 1–14. [Google Scholar] [CrossRef]
- Yu, Y.; Zhong, Q.; Li, C.; Jiang, L.; Sun, Y.; Wang, X.; Wang, Z.; Zhang, Q. Molecular cloning and characterization of interleukin-1beta in half-smooth tongue sole Cynoglossus semilaevis. Vet. Immunol. Immunopathol. 2012, 146, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Latz, E.; Xiao, T.S.; Stutz, A. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 2013, 13, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Reis, M.I.; do Vale, A.; Pereira, P.J.; Azevedo, J.E.; Dos Santos, N.M. Caspase-1 and IL-1β processing in a teleost fish. PLoS ONE 2012, 7, e50450. [Google Scholar] [CrossRef] [PubMed]
- Reis, M.I.; Costa-Ramos, C.; do Vale, A.; dos Santos, N.M. Molecular cloning of sea bass (Dicentrarchus labrax L.) caspase-8 gene and its involvement in Photobacterium damselae ssp. piscicida triggered apoptosis. Fish Shellfish Immunol. 2010, 29, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Mathew, J.A.; Guo, Y.X.; Goh, K.P.; Chan, J.; Verburg-van Kemenade, B.M.; Kwang, J. Characterisation of a monoclonal antibody to carp IL-1beta and the development of a sensitive capture ELISA. Fish Shellfish Immunol. 2002, 13, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Zou, J.; Collet, B.; Bols, N.C.; Secombes, C.J. Analysis and characterisation of IL-1beta processing in rainbow trout, Oncorhynchus mykiss. Fish Shellfish Immunol. 2004, 16, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Aachoui, Y.; Leaf, I.A.; Hagar, J.A.; Fontana, M.F.; Campos, C.G.; Zak, D.E.; Tan, M.H.; Cotter, P.A.; Vance, R.E.; Aderem, A.; et al. Caspase-11 protects against bacteria that escape the vacuole. Science 2013, 339, 975–978. [Google Scholar] [CrossRef] [PubMed]
- Maelfait, J.; Vercammen, E.; Janssens, S.; Schotte, P.; Haegman, M.; Magez, S.; Beyaert, R. Stimulation of Toll-like receptor 3 and 4 induces interleukin-1beta maturation by caspase-8. J. Exp. Med. 2008, 205, 1967–1973. [Google Scholar] [CrossRef] [PubMed]
- Rojas, V.; Camus-Guerra, H.; Guzman, F.; Mercado, L. Pro-inflammatory caspase-1 activation during the immune response in cells from rainbow trout Oncorhynchus mykiss (Walbaum 1792) challenged with pathogen-associated molecular patterns. J. Fish Dis. 2015, 38, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Sakata, S.; Yan, Y.; Satou, Y.; Momoi, A.; Ngo-Hazelett, P.; Nozaki, M.; Furutani-Seiki, M.; Postlethwait, J.H.; Yonehara, S.; Sakamaki, K. Conserved function of caspase-8 in apoptosis during bony fish evolution. Gene 2007, 396, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Ge, X.; Zhu, J.; Zhang, W.; Zhang, Q. Molecular cloning, immunohistochemical localization, characterization and expression analysis of caspase-8 from the blunt snout bream (Megalobrama amblycephala) exposed to ammonia. Fish Shellfish Immunol. 2015, 47, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Alfaidi, M.; Wilson, H.; Daigneault, M.; Burnett, A.; Ridger, V.; Chamberlain, J.; Francis, S. Neutrophil elastase promotes interleukin-1beta secretion from human coronary endothelium. J. Biol. Chem. 2015, 290, 24067–24078. [Google Scholar] [CrossRef] [PubMed]
- Terada, K.; Yamada, J.; Hayashi, Y.; Wu, Z.; Uchiyama, Y.; Peters, C.; Nakanishi, H. Involvement of cathepsin B in the processing and secretion of interleukin-1beta in chromogranin A-stimulated microglia. Glia 2010, 58, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Peddie, S.; Campos-Perez, J.J.; Zou, J.; Secombes, C.J. The effect of intraperitoneally administered recombinant IL-1beta on immune parameters and resistance to Aeromonas salmonicida in the rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 2003, 27, 801–812. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, D.; Li, J.; Liu, Z. Molecular characterization, recombinant expression and bioactivity analysis of the interleukin-1 beta from the yellowfin sea bream, Acanthopagrus latus (Houttuyn). Fish Shellfish Immunol. 2008, 24, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Kono, T.; Fujiki, K.; Nakao, M.; Yano, T.; Endo, M.; Sakai, M. The immune responses of common carp, Cyprinus carpio L., injected with carp interleukin-1beta gene. J. Interferon Cytokine Res. 2002, 22, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.Q.; Bei, J.X.; Feng, L.N.; Zhang, Y.; Liu, X.C.; Wang, L.; Chen, J.L.; Lin, H.R. Interleukin-1beta gene in orange-spotted grouper, Epinephelus coioides: Molecular cloning, expression, biological activities and signal transduction. Mol. Immunol. 2008, 45, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Taechavasonyoo, A.; Hirono, I.; Kondo, H. The immune-adjuvant effect of Japanese flounder Paralichthys olivaceus IL-1β. Dev. Comp. Immunol. 2013, 41, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Kono, T.; Korenaga, H.; Sakai, M. Genomics of fish IL-17 ligand and receptors: A review. Fish Shellfish Immunol. 2011, 31, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, C.; Thongda, W.; Luo, Y.; Beck, B.; Peatman, E. Characterization and mucosal responses of interleukin 17 family ligand and receptor genes in channel catfish Ictalurus punctatus. Fish Shellfish Immunol. 2014, 38, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Martin, S.A.; Secombes, C.J. Two interleukin-17C-like genes exist in rainbow trout Oncorhynchus mykiss that are differentially expressed and modulated. Dev. Comp. Immunol. 2010, 34, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Jiang, Y.; Wang, A.; Husain, M.; Xu, Q.; Secombes, C.J. Identification of the salmonid IL-17A/F1a/b, IL-17A/F2b, IL-17A/F3 and IL-17N genes and analysis of their expression following in vitro stimulation and infection. Immunogenetics 2015, 67, 395–412. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A.; Zou, J.; Houlihan, D.F.; Secombes, C.J. Directional responses following recombinant cytokine stimulation of rainbow trout (Oncorhynchus mykiss) RTS-11 macrophage cells as revealed by transcriptome profiling. BMC Genomics 2007. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Li, R.; Xu, Q.; Secombes, C.J.; Wang, T. Two types of TNF-α exist in teleost fish: Phylogeny, expression, and bioactivity analysis of type-II TNF-α3 in rainbow trout Oncorhynchus mykiss. J. Immunol. 2013, 191, 5959–5972. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.; Zou, J.; Secombes, C.J.; Martin, S.A. Cortisol modulates the induction of inflammatory gene expression in a rainbow trout macrophage cell line. Fish Shellfish Immunol. 2011, 30, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Kono, T.; Monte, M.M.; Kuse, H.; Costa, M.M.; Korenaga, H.; Maehr, T.; Husain, M.; Sakai, M.; Secombes, C.J. Identification of IL-34 in teleost fish: Differential expression of rainbow trout IL-34, MCSF1 and MCSF2, ligands of the MCSF receptor. Mol. Immunol. 2013, 53, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Maehr, T.; Vecino, J.L.; Wadsworth, S.; Wang, T.; Secombes, C.J. Four CISH paralogues are present in rainbow trout Oncorhynchus mykiss: Differential expression and modulation during immune responses and development. Mol. Immunol. 2014, 62, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Maehr, T.; Costa, M.M.; Vecino, J.L.; Wadsworth, S.; Martin, S.A.; Wang, T.; Secombes, C.J. Transforming growth factor-β1b: A second TGF-β1 paralogue in the rainbow trout (Oncorhynchus mykiss) that has a lower constitutive expression but is more responsive to immune stimulation. Fish Shellfish Immunol. 2013, 34, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wei, H.; Qin, S.; Zhang, S.; Wang, X.; Zhang, A.; Du, L. Reciprocal interactions between fish TGF-β1 and IL-1β is responsible for restraining IL-1β signalling activity in grass carp head kidney leukocytes. Dev. Comp. Immunol. 2014, 47, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Peddie, S.; Zou, J.; Cunningham, C.; Secombes, C.J. Rainbow trout (Oncorhynchus mykiss) recombinant IL-1beta and derived peptides induce migration of head-kidney leucocytes in vitro. Fish Shellfish Immunol. 2001, 11, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, S.; Randelli, E.; Buonocore, F.; Zou, J.; Secombes, C.J.; Scapigliati, G. Evolution of cytokine responses: IL-1beta directly affects intracellular Ca2+ concentration of teleost fish leukocytes through a receptor-mediated mechanism. Cytokine 2006, 34, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Li, R.; Monte, M.M.; Jiang, Y.; Nie, P.; Holland, J.W.; Secombes, C.J.; Wang, T. Sequence and expression analysis of rainbow trout CXCR2, CXCR3a and CXCR3b aids interpretation of lineage-specific conversion, loss and expansion of these receptors during vertebrate evolution. Dev. Comp. Immunol. 2014, 45, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, Q.; Wang, T.; Collet, B.; Corripio-Miyar, Y.; Bird, S.; Xie, P.; Nie, P.; Secombes, C.J.; Zou, J. Phylogenetic analysis of vertebrate CXC chemokines reveals novel lineage specific groups in teleost fish. Dev. Comp. Immunol. 2013, 41, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Pooley, N.J.; Tacchi, L.; Secombes, C.J.; Martin, S.A. Inflammatory responses in primary muscle cell cultures in Atlantic salmon (Salmo salar). BMC Genomics 2013. [Google Scholar] [CrossRef] [PubMed]
- Heidari, Z.; Bickerdike, R.; Tinsley, J.; Zou, J.; Wang, T.Y.; Chen, T.Y.; Martin, S.A. Regulatory factors controlling muscle mass: Competition between innate immune function and anabolic signals in regulation of atrogin-1 in Atlantic salmon. Mol. Immunol. 2015, 67, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Costa, I.A.; Hein, T.W.; Secombes, C.J.; Gamperl, A.K. Recombinant interleukin-1beta dilates steelhead trout coronary microvessels: Effect of temperature and role of the endothelium, nitric oxide and prostaglandins. J. Exp. Biol. 2015, 218, 2269–2278. [Google Scholar] [CrossRef] [PubMed]
- Palomo, J.; Dietrich, D.; Martin, P.; Palmer, G.; Gabay, C. The interleukin (IL)-1 cytokine family—Balance between agonists and antagonists in inflammatory diseases. Cytokine 2015, 76, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, S.; Stansberg, C.; Olsen, L.; Zou, J.; Secombes, C.J.; Cunningham, C. Cloning of a Salmo salar interleukin-1 receptor-like cDNA. Dev. Comp. Immunol. 2002, 26, 415–431. [Google Scholar] [CrossRef]
- Stansberg, C.; Subramaniam, S.; Collet, B.; Secombes, C.J.; Cunningham, C. Cloning of the Atlantic salmon (Salmo salar) IL-1 receptor associated protein. Fish Shellfish Immunol. 2005, 19, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.Q.; Yao, M.; Yi, S.B.; Li, Y.W.; Liu, X.C.; Zhang, Y.; Lin, H.R. Soluble interleukin-1 receptor, a potential negative regulator of orange-spotted grouper Epinephelus coioides interleukin-1 system. J. Fish Biol. 2013, 83, 642–658. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Castejon, G.; Sepulcre, M.P.; Roca, F.J.; Castellana, B.; Planas, J.V.; Meseguer, J.; Mulero, V. The type II interleukin-1 receptor (IL-1RII) of the bony fish gilthead seabream Sparus aurata is strongly induced after infection and tightly regulated at transcriptional and post-transcriptional levels. Mol. Immunol. 2007, 44, 2772–2780. [Google Scholar] [CrossRef] [PubMed]
- Morrison, R.N.; Young, N.D.; Nowak, B.F. Description of an Atlantic salmon (Salmo salar L.) type II interleukin-1 receptor cDNA and analysis of interleukin-1 receptor expression in amoebic gill disease-affected fish. Fish Shellfish Immunol. 2012, 32, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Sangrador-Vegas, A.; Martin, S.A.; O’Dea, P.G.; Smith, T.J. Cloning and characterization of the rainbow trout (Oncorhynchus mykiss) type II interleukin-1 receptor cDNA. Eur. J. Biochem. 2000, 267, 7031–7037. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, S.; Du, L.; Yang, K.; Wang, X.; Zhang, A.; Zhou, H. Molecular and functional characterization of IL-1 receptor type 2 in grass carp: A potent inhibitor of IL-1β signaling in head kidney leukocytes. Dev. Comp. Immunol. 2013, 41, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Li, S.; Qi, J.; Zeng, L.; Zhong, Q.; Zhang, Q. Cloning and characterization of type II interleukin-1 receptor cDNA from Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2010, 157, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Sigh, J.; Lindenstrom, T.; Buchmann, K. Expression of pro-inflammatory cytokines in rainbow trout (Oncorhynchus mykiss) during an infection with Ichthyophthirius multifiliis. Fish Shellfish Immunol. 2004, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.F.; Fang, Y.; Jin, Y.; Dong, W.R.; Xiang, L.X.; Shao, J.Z. Discovery of the DIGIRR gene from teleost fish: A novel Toll-IL-1 receptor family member serving as a negative regulator of IL-1 signaling. J. Immunol. 2011, 187, 2514–2530. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. IL-18: A TH1-inducing, proinflammatory cytokine and new member of the IL-1 family. J. Allergy Clin. Immunol. 1999, 103, 11–24. [Google Scholar] [CrossRef]
- Okamura, H.; Kashiwamura, S.; Tsutsui, H.; Yoshimoto, T.; Nakanishi, K. Regulation of interferon-gamma production by IL-12 and IL-18. Curr. Opin. Immunol. 1998, 10, 259–264. [Google Scholar] [CrossRef]
- Perez-Cordon, G.; Estensoro, I.; Benedito-Palos, L.; Calduch-Giner, J.A.; Sitja-Bobadilla, A.; Perez-Sanchez, J. Interleukin gene expression is strongly modulated at the local level in a fish-parasite model. Fish Shellfish Immunol. 2014, 37, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Pereiro, P.; Balseiro, P.; Romero, A.; Dios, S.; Forn-Cuni, G.; Fuste, B.; Planas, J.V.; Beltran, S.; Novoa, B.; Figueras, A. High-throughput sequence analysis of turbot (Scophthalmus maximus) transcriptome using 454-pyrosequencing for the discovery of antiviral immune genes. PLoS ONE 2012, 7, e35369. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, B.; Lee, A.P.; Ravi, V.; Maurya, A.K.; Lian, M.M.; Swann, J.B.; Ohta, Y.; Flajnik, M.F.; Sutoh, Y.; Kasahara, M.; et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature 2014, 505, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Meijer, A.H.; Gabby Krens, S.F.; Medina Rodriguez, I.A.; He, S.; Bitter, W.; Ewa Snaar-Jagalska, B.; Spaink, H.P. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol. Immunol. 2004, 40, 773–783. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Yang, X.; Wang, X.; Wei, H.; Zhang, A.; Zhou, H. Molecular and functional characterization of an IL-1beta receptor antagonist in grass carp (Ctenopharyngodon idella). Dev. Comp. Immunol. 2015, 49, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B. Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 2003, 3, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Androlewicz, M.J.; Browning, J.L.; Ware, C.F. Lymphotoxin is expressed as a heteromeric complex with a distinct 33-kDa glycoprotein on the surface of an activated human T cell hybridoma. J. Biol. Chem. 1992, 267, 2542–2547. [Google Scholar] [PubMed]
- Horiuchi, T.; Mitoma, H.; Harashima, S.; Tsukamoto, H.; Shimoda, T. Transmembrane TNF-alpha: Structure, function and interaction with anti-TNF agents. Rheumatology 2010, 49, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Young, J.; Yu, X.; Wolslegel, K.; Nguyen, A.; Kung, C.; Chiang, E.; Kolumam, G.; Wei, N.; Wong, W.L.; DeForge, L.; et al. Lymphotoxin-alphabeta heterotrimers are cleaved by metalloproteinases and contribute to synovitis in rheumatoid arthritis. Cytokine 2010, 51, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Bobe, J.; Goetz, F.W. Molecular cloning and expression of a TNF receptor and two TNF ligands in the fish ovary. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2001, 129, 475–481. [Google Scholar] [CrossRef]
- Covello, J.M.; Bird, S.; Morrison, R.N.; Battaglene, S.C.; Secombes, C.J.; Nowak, B.F. Cloning and expression analysis of three striped trumpeter (Latris lineata) pro-inflammatory cytokines, TNF-alpha, IL-1beta and IL-8, in response to infection by the ectoparasitic, Chondracanthus goldsmidi. Fish Shellfish Immunol. 2009, 26, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Glenney, G.W.; Wiens, G.D. Early diversification of the TNF superfamily in teleosts: Genomic characterization and expression analysis. J. Immunol. 2007, 178, 7955–7973. [Google Scholar] [CrossRef] [PubMed]
- Hirono, I.; Nam, B.H.; Kurobe, T.; Aoki, T. Molecular cloning, characterization, and expression of TNF cDNA and gene from Japanese flounder Paralichthys olivaceus. J. Immunol. 2000, 165, 4423–4427. [Google Scholar] [CrossRef] [PubMed]
- Laing, K.J.; Wang, T.; Zou, J.; Holland, J.; Hong, S.; Bols, N.; Hirono, I.; Aoki, T.; Secombes, C.J. Cloning and expression analysis of rainbow trout Oncorhynchus mykiss tumour necrosis factor-alpha. Eur. J. Biochem. 2001, 268, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, D.S.; Pereira, P.J.; Reis, M.I.; do Vale, A.; Zou, J.; Silva, M.T.; Secombes, C.J.; dos Santos, N.M. Molecular cloning and expression analysis of sea bass (Dicentrarchus labrax L.) tumor necrosis factor-alpha (TNF-alpha). Fish Shellfish Immunol. 2007, 23, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Pleic, I.L.; Secombes, C.J.; Bird, S.; Mladineo, I. Characterization of three pro-inflammatory cytokines, TNFalpha1, TNFalpha2 and IL-1beta, in cage-reared Atlantic bluefin tuna Thunnus thynnus. Fish Shellfish Immunol. 2014, 36, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Savan, R.; Kono, T.; Igawa, D.; Sakai, M. A novel tumor necrosis factor (TNF) gene present in tandem with the TNF-alpha gene on the same chromosome in teleosts. Immunogenetics 2005, 57, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Uenobe, M.; Kohchi, C.; Yoshioka, N.; Yuasa, A.; Inagawa, H.; Morii, K.; Nishizawa, T.; Takahashi, Y.; Soma, G. Cloning and characterization of a TNF-like protein of Plecoglossus altivelis (ayu fish). Mol. Immunol. 2007, 44, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhou, Z.C.; Chen, C.; Huo, W.L.; Yin, Z.X.; Weng, S.P.; Chan, S.M.; Yu, X.Q.; He, J.G. Tumor necrosis factor-alpha gene from mandarin fish, Siniperca chuatsi: Molecular cloning, cytotoxicity analysis and expression profile. Mol. Immunol. 2007, 44, 3615–3622. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Chen, D.; Wei, H.; Du, L.; Zhao, T.; Wang, X.; Zhou, H. Functional characterization of TNF-α in grass carp head kidney leukocytes: Induction and involvement in the regulation of NF-κB signaling. Fish Shellfish Immunol. 2012, 33, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Wang, T.; Hirono, I.; Aoki, T.; Inagawa, H.; Honda, T.; Soma, G.I.; Ototake, M.; Nakanishi, T.; Ellis, A.E.; et al. Differential expression of two tumor necrosis factor genes in rainbow trout, Oncorhynchus mykiss. Dev. Comp. Immunol. 2002, 26, 161–172. [Google Scholar] [CrossRef]
- Zou, J.; Peddie, S.; Scapigliati, G.; Zhang, Y.; Bols, N.C.; Ellis, A.E.; Secombes, C.J. Functional characterisation of the recombinant tumor necrosis factors in rainbow trout, Oncorhynchus mykiss. Dev. Comp. Immunol. 2003, 27, 813–822. [Google Scholar] [CrossRef]
- Goetz, F.W.; Planas, J.V.; MacKenzie, S. Tumor necrosis factors. Dev. Comp. Immunol. 2004, 28, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Wiens, G.D.; Glenney, G.W. Origin and evolution of TNF and TNF receptor superfamilies. Dev. Comp. Immunol. 2011, 35, 1324–1335. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, S.; Biswas, G.; Kono, T.; Hikima, J.; Sakai, M. Presence of two tumor necrosis factor (tnf)-alpha homologs on different chromosomes of zebrafish (Danio rerio) and medaka (Oryzias latipes). Mar. Genomics 2014, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kono, T.; Zou, J.; Bird, S.; Savan, R.; Sakai, M.; Secombes, C.J. Identification and expression analysis of lymphotoxin-beta like homologues in rainbow trout Oncorhynchus mykiss. Mol. Immunol. 2006, 43, 1390–1401. [Google Scholar] [CrossRef] [PubMed]
- Grayfer, L.; Walsh, J.G.; Belosevic, M. Characterization and functional analysis of goldfish (Carassius auratus L.) tumor necrosis factor-alpha. Dev. Comp. Immunol. 2008, 32, 532–543. [Google Scholar] [CrossRef] [PubMed]
- Morrison, R.N.; Zou, J.; Secombes, C.J.; Scapigliati, G.; Adams, M.B.; Nowak, B.F. Molecular cloning and expression analysis of tumour necrosis factor-alpha in amoebic gill disease (AGD)-affected Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 2007, 23, 1015–1031. [Google Scholar] [CrossRef] [PubMed]
- Saeij, J.P.; Stet, R.J.; de Vries, B.J.; van Muiswinkel, W.B.; Wiegertjes, G.F. Molecular and functional characterization of carp TNF: A link between TNF polymorphism and trypanotolerance? Dev. Comp. Immunol. 2003, 27, 29–41. [Google Scholar] [CrossRef]
- Savan, R.; Sakai, M. Presence of multiple isoforms of TNF alpha in carp (Cyprinus carpio L.): Genomic and expression analysis. Fish Shellfish Immunol. 2004, 17, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Benedicenti, O.; Collins, C.; Wang, T.; McCarthy, U.; Secombes, C.J. Which Th pathway is involved during late stage amoebic gill disease? Fish Shellfish Immunol. 2015, 46, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Roher, N.; Callol, A.; Planas, J.V.; Goetz, F.W.; MacKenzie, S.A. Endotoxin recognition in fish results in inflammatory cytokine secretion not gene expression. Innate Immun. 2011, 17, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.J.; Chen, Q.; Yang, G.J.; Chen, J. The TNFα converting enzyme (TACE) from ayu (Plecoglossus altivelis) exhibits TNFα shedding activity. Mol. Immunol. 2015, 63, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Castillo, J.; Chaves-Pozo, E.; Olivares, P.; Pelegrin, P.; Meseguer, J.; Mulero, V. The tumor necrosis factor alpha of the bony fish seabream exhibits the in vivo proinflammatory and proliferative activities of its mammalian counterparts, yet it functions in a species-specific manner. Cell Mol. Life Sci. 2004, 61, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Secombes, C.J.; Long, S.; Miller, N.; Clem, L.W.; Chinchar, V.G. Molecular identification and expression analysis of tumor necrosis factor in channel catfish (Ictalurus punctatus). Dev. Comp. Immunol. 2003, 27, 845–858. [Google Scholar] [CrossRef]
- Kim, M.S.; Hwang, Y.J.; Yoon, K.J.; Zenke, K.; Nam, Y.K.; Kim, S.K.; Kim, K.H. Molecular cloning of rock bream (Oplegnathus fasciatus) tumor necrosis factor-alpha and its effect on the respiratory burst activity of phagocytes. Fish Shellfish Immunol. 2009, 27, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.L.; Kim, M.S.; Kim, K.H. Molecular cloning of rock bream’s (Oplegnathus fasciatus) tumor necrosis factor receptor-associated factor 2 and its role in NF-κB activiation. Fish Shellfish Immunol. 2011, 30, 1178–1183. [Google Scholar] [CrossRef] [PubMed]
- Clay, H.; Volkman, H.E.; Ramakrishnan, L. Tumor necrosis factor signaling mediates resistance to mycobacteria by inhibiting bacterial growth and macrophage death. Immunity 2008, 29, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Roca, F.J.; Ramakrishnan, L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell 2013, 153, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Hino, K.; Nakamura, O.; Yoshiura, Y.; Suetake, H.; Suzuki, Y.; Watanabe, T. TNF induces the growth of thymocytes in rainbow trout. Dev. Comp. Immunol. 2006, 30, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Roca, F.J.; Mulero, I.; Lopez-Munoz, A.; Sepulcre, M.P.; Renshaw, S.A.; Meseguer, J.; Mulero, V. Evolution of the inflammatory response in vertebrates: Fish TNF-alpha is a powerful activator of endothelial cells but hardly activates phagocytes. J. Immunol. 2008, 181, 5071–5081. [Google Scholar] [CrossRef] [PubMed]
- Forlenza, M.; Magez, S.; Scharsack, J.P.; Westphal, A.; Savelkoul, H.F.; Wiegertjes, G.F. Receptor-mediated and lectin-like activities of carp (Cyprinus carpio) TNF-alpha. J. Immunol. 2009, 183, 5319–5332. [Google Scholar] [CrossRef] [PubMed]
- Boecke, A.; Sieger, D.; Neacsu, C.D.; Kashkar, H.; Kronke, M. Factor associated with neutral sphingomyelinase activity mediates navigational capacity of leukocytes responding to wounds and infection: Live imaging studies in zebrafish larvae. J. Immunol. 2012, 189, 1559–1566. [Google Scholar] [CrossRef] [PubMed]
- Praveen, K.; Evans, D.L.; Jaso-Friedmann, L. Constitutive expression of tumor necrosis factor-alpha in cytotoxic cells of teleosts and its role in regulation of cell-mediated cytotoxicity. Mol. Immunol. 2006, 43, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Guo, T.C.; Mutoloki, S.; Haugland, O.; Evensen, O. Gene expression studies of host response to Salmonid alphavirus subtype 3 experimental infections in Atlantic salmon. Vet. Res. 2012. [Google Scholar] [CrossRef] [PubMed]
- Ronza, P.; Bermudez, R.; Losada, A.P.; Sitja-Bobadilla, A.; Pardo, B.G.; Quiroga, M.I. Immunohistochemical detection and gene expression of TNFalpha in turbot (Scophthalmus maximus) enteromyxosis. Fish Shellfish Immunol. 2015, 47, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Marjoram, L.; Alvers, A.; Deerhake, M.E.; Bagwell, J.; Mankiewicz, J.; Cocchiaro, J.L.; Beerman, R.W.; Willer, J.; Sumigray, K.D.; Katsanis, N.; et al. Epigenetic control of intestinal barrier function and inflammation in zebrafish. Proc. Natl. Acad. Sci. USA 2015, 112, 2770–2775. [Google Scholar] [CrossRef] [PubMed]
- Crespo, D.; Bonnet, E.; Roher, N.; MacKenzie, S.A.; Krasnov, A.; Goetz, F.W.; Bobe, J.; Planas, J.V. Cellular and molecular evidence for a role of tumor necrosis factor alpha in the ovulatory mechanism of trout. Reprod. Biol. Endocrinol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Crespo, D.; Mananos, E.L.; Roher, N.; MacKenzie, S.A.; Planas, J.V. Tumor necrosis factor alpha may act as an intraovarian mediator of luteinizing hormone-induced oocyte maturation in trout. Biol. Reprod. 2012, 86, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Song, J.; Yang, H.; Gao, W.; Liu, N.A.; Zhang, B.; Lin, S. Mmp23b promotes liver development and hepatocyte proliferation through the tumor necrosis factor pathway in zebrafish. Hepatology 2010, 52, 2158–2166. [Google Scholar] [CrossRef] [PubMed]
- Hymowitz, S.G.; Filvaroff, E.H.; Yin, J.P.; Lee, J.; Cai, L.; Risser, P.; Maruoka, M.; Mao, W.; Foster, J.; Kelley, R.F.; et al. IL-17s adopt a cystine knot fold: Structure and activity of a novel cytokine, IL-17F, and implications for receptor binding. EMBO J. 2001, 20, 5332–5341. [Google Scholar] [CrossRef] [PubMed]
- Parthier, C.; Stelter, M.; Ursel, C.; Fandrich, U.; Lilie, H.; Breithaupt, C.; Stubbs, M.T. Structure of the Toll-Spatzle complex, a molecular hub in Drosophila development and innate immunity. Proc. Natl. Acad. Sci. USA 2014, 111, 6281–6286. [Google Scholar] [CrossRef] [PubMed]
- Gunimaladevi, I.; Savan, R.; Sakai, M. Identification, cloning and characterization of interleukin-17 and its family from zebrafish. Fish Shellfish Immunol. 2006, 21, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Feng, S.; Yin, L.; Wang, X.; Zhang, A.; Yang, K.; Zhou, H. Identification and functional characterization of grass carp IL-17A/F1: An evaluation of the immunoregulatory role of teleost IL-17A/F1. Dev. Comp. Immunol. 2015, 51, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Monte, M.M.; Wang, T.; Holland, J.W.; Zou, J.; Secombes, C.J. Cloning and characterisation of rainbow trout interleukin-17A/F2 (IL-17A/F2) and IL-17 receptor A: Expression during infection and bioactivity of recombinant IL-17A/F2. Infect. Immun. 2013, 81, 340–353. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Qin, L.; Wang, X.; Zhang, A.; Wei, H.; Zhou, H. Characterization of grass carp (Ctenopharyngodon idella) IL-17D: Molecular cloning, functional implication and signal transduction. Dev. Comp. Immunol. 2014, 42, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Mitra, S.; Wyse, C.; Alnabulsi, A.; Zou, J.; Weerdenburg, E.M.; van der Sar, A.M.; Wang, D.; Secombes, C.J.; Bird, S. First demonstration of antigen induced cytokine expression by CD4-1+ lymphocytes in a poikilotherm: Studies in zebrafish (Danio rerio). PLoS ONE 2015, 10, e0126378. [Google Scholar] [CrossRef] [PubMed]
- Quintana, F.J.; Iglesias, A.H.; Farez, M.F.; Caccamo, M.; Burns, E.J.; Kassam, N.; Oukka, M.; Weiner, H.L. Adaptive autoimmunity and Foxp3-based immunoregulation in zebrafish. PLoS ONE 2010, 5, e9478. [Google Scholar] [CrossRef] [PubMed]
- Funkenstein, B.; Olekh, E.; Jakowlew, S.B. Identification of a novel transforming growth factor-beta (TGF-beta6) gene in fish: Regulation in skeletal muscle by nutritional state. BMC Mol. Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Haddad, G.; Hanington, P.C.; Wilson, E.C.; Grayfer, L.; Belosevic, M. Molecular and functional characterization of goldfish (Carassius auratus L.) transforming growth factor beta. Dev. Comp. Immunol. 2008, 32, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yin, L.; Feng, S.; Wang, X.; Yang, K.; Zhang, A.; Zhou, H. Dual-parallel inhibition of IL-10 and TGF-β1 controls LPS-induced inflammatory response via NF-κB signalling in grass carp monocytes/macrophages. Fish Shellfish Immunol. 2015, 44, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Wang, X.; Chen, D.; Wang, Y.; Zhang, A.; Zhou, H. TGF-β1 exerts opposing effects on grass carp leukocytes: Implications in teleost immunity, receptor signaling and potential self-regulatory mechanisms. PLoS ONE 2012, 7, e35011. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhou, H. Grass carp transforming growth factor-β1 (TGF-β1): Molecular cloning, tissue distribution and immunobiological activity in teleost peripheral blood lymphocytes. Mol. Immunol. 2008, 45, 1792–1798. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Cai, C.; Hu, X.; Liu, Y.; Guo, Y.; Hu, P.; Chen, Z.; Peng, S.; Zhang, D.; Jiang, S.; et al. Evolutionary suppression of erythropoiesis via the modulation of TGF-β signalling in an Antarctic icefish. Mol. Ecol. 2015, 24, 4664–4678. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.L.; Ma, T.Y.; Wu, J.Y.; Yi, L.Y.; Wang, J.Y.; Gao, X.K. Cloning and primary immunological study of TGF-β1 and its receptors TβR I/TβR II in tilapia (Oreochromis niloticus). Dev. Comp. Immunol. 2015, 51, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Vosshenrich, C.A.J.; Di Santo, J.P. Interleukin signalling. Curr. Biol. 2002, 12, 760–763. [Google Scholar] [CrossRef]
- Ohtani, M.; Hayashi, N.; Hashimoto, K.; Nakanishi, T.; Dijkstra, J.M. Comprehensive clarification of two paralogous interleukin 4/13 loci in teleost fish. Immunogenetics 2008, 60, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Secombes, C.J. The evolution of IL-4 and IL-13 and their receptor subunits. Cytokine 2015, 75, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Rosales, P.; Bird, S.; Wang, T.H.; Fujiki, K.; Davidson, W.S.; Zou, J.; Secombes, C.J. Rainbow trout interleukin-2: Cloning, expression and bioactivity analysis. Fish Shellfish Immunol. 2009, 27, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Johansson, P.; Abós, B.; Holt, A.; Tafalla, C.; Jiang, Y.; Wang, A.; Xu, Q.; Qi, Z.; Huang, W.; et al. First in-depth analysis of the novel Th2-type cytokines in salmonid fish reveals distinct patterns of expression and modulation but overlapping bioactivities. Oncotarget 2016, 7, 10917–10946. [Google Scholar] [PubMed]
- Corripio-Miyar, Y.; Secombes, C.J.; Zou, J. Long-term stimulation of trout head kidney cells with the cytokines MCSF, IL-2 and IL-6: Gene expression dynamics. Fish Shellfish Immunol. 2012, 32, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.F.; Xiang, L.X.; Wang, Q.L.; Dong, W.R.; Gong, Y.F.; Shao, J.Z. The DC-SIGN of zebrafish: Insights into the existence of a CD209 homologue in a lower vertebrate and its involvement in adaptive immunity. J. Immunol. 2009, 183, 7398–7410. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.L.; Xiang, L.X.; Shao, J.Z. Identification and characterization of a novel immunoglobulin Z isotype in zebrafish: Implications for a distinct B cell receptor in lower vertebreates. Mol. Immunol. 2010, 47, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.Y.; Pan, P.P.; Fang, W.; Shao, J.Z.; Xiang, L.X. Essential role of IL-4 and IL-4Rα in interaction in adaptive immunity of zebrafish: Insight into the origin of Th2-like regulatory mechanisms in ancient vertebrates. J. Immunol. 2012, 188, 5571–5584. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Miyata, S.; Katakura, F.; Nagasawa, T.; Shibasaki, Y.; Yabu, T.; Fischer, U.; Nakayasu, C.; Nakanishi, T.; Moritomo, T. Recombinant carp IL-4/13B stimulates in vitro proliferation of carp IgM+ B cells. Fish Shellfish Immunol. 2016, 49, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Kono, T.; Bird, S.; Sonoda, K.; Savan, R.; Secombes, C.J.; Sakai, M. Characterization and expression analysis of an interleukin-7 homologue in the Japanese pufferfish, Takifugu rubripes. FEBS J. 2008, 275, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Iwanami, N.; Mateos, F.; Hess, I.; Riffel, N.; Soza-Ried, C.; Schorpp, M.; Boehm, T. Genetic evidence for an evolutionarily conserved role of IL-7 signaling in T cell development in zebrafish. J. Immunol. 2011, 186, 7060–7066. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Holland, J.W.; Carrington, A.; Zou, J.; Secombes, C.J. Molecular and functional characterisation of interleukin-15 in rainbow trout Oncorhynchus mykiss: A potent inducer of interferon-gamma expression in spleen leucocytes. J. Immunol. 2007, 179, 1475–1488. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.H.; Husain, M.; Hong, S.; Holland, J.W. Differential expression, modulation and bioactivity of distinct fish IL-12 isoforms: Implication towards the evolution of Th1-like immune responses. Eur. J. Immunol. 2014, 44, 1541–1551. [Google Scholar] [CrossRef] [PubMed]
- Bei, J.X.; Suetake, H.; Araki, K.; Kikuchi, K.; Yoshiura, Y.; Lin, H.R.; Suzuki, Y. Two interleukin (IL)-15 homologues in fish from two distinct origins. Mol. Immunol. 2006, 43, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Xiang, L.X.; Shao, J.Z.; Wen, Y.; Chen, S.Y. Identification and characterization of an interleukin-15 homologue from Tetraodon nigroviridis. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2006, 143, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.M.; Takizawa, F.; Fischer, U.; Friedrich, M.; Soto-Lampe, V.; Lefèvre, C.; Lenk, M.; Karger, A.; Matsui, T.; Hashimoto, K. Identification of a gene for an ancient cytokine, interleukin 15-like, in mammals; interleukins 2 and 15 co-evolved with this third family member, all sharing binding motifs for IL-15Rα. Immunogenetics 2014, 66, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Diaz-Rosales, P.; Costa, M.M.; Campbell, S.; Snow, M.; Collet, B.; Martin, S.A.M.; Secombes, C.J. Functional characterisation of a nonmammalian IL-21: Rainbow trout Oncorhynchus mykiss IL-21 upregulates the expression of the Th cell signature cytokines IFN-g, IL-10 and IL-22. J. Immunol. 2011, 186, 708–721. [Google Scholar] [CrossRef] [PubMed]
- Sertori, R.; Liongue, C.; Basheer, F.; Lewis, K.L.; Rasighaemi, P.; de Coninck, D.; Traver, D.; Ward, A.C. Conserved IL-2Rγc signaling mediates lymphopoiesis in zebrafish. J. Immunol. 2016, 196, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Huang, W.; Costa, M.M.; Secombes, C.J. The gamma-chain cytokine/receptor system in fish: More ligands and receptors. Fish Shellfish Immunol. 2011, 31, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Fang, W.; Xiang, L.X.; Pan, R.L.; Shao, J.Z. Identification of Treg-like cells in Tetraodon: Insight into the origin of regulatory T subsets during early vertebrate evolution. Cell. Mol. Life Sci. 2011, 68, 2615–2626. [Google Scholar] [CrossRef] [PubMed]
- Liongue, C.; Ward, A.C. Evolution of class I cytokine receptors. BMC Evol. Biol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.M. Th2 and Treg candidate genes in elephant shark. Nature 2014, 511, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Takizawa, F.; Fischer, U.; Dijkstra, J.M. Along the axis between type I and type 2 immunity; Principles conserved in evolution from fish to mammals. Biology 2015, 4, 814–859. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, K.; Nakao, M.; Dixon, B. Molecular cloning and characterisation of a carp (Cyprinus carpio) cytokine-like cDNA that shares sequence similarity withIL-6 subfamily cytokines CNTF, OSM and LIF. Dev. Comp. Immunol. 2003, 27, 127–136. [Google Scholar] [CrossRef]
- Wang, T.; Secombes, C.J. Identification and expression analysis of two fish-specific IL-6 cytokine family members, the ciliary neurotrophic factor (CNTF)-like and M17 genes, in rainbow trout Oncorhynchus mykiss. Mol. Immunol. 2009, 46, 2290–2298. [Google Scholar] [CrossRef] [PubMed]
- Huising, M.O.; Kruiswijk, C.P.; van Schijndel, J.E.; Savelkoul, H.F.; Flik, G.; Verburg-van Kemenade, B.M. Multiple and highly divergent IL-11 genes in teleost fish. Immunogenetics 2005, 57, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.D.; Yasuike, M.; Kondo, H.; Hirono, I.; Aoki, T. Teleostean IL11b exhibits complementing function to IL11a and expansive involvement in antibacterial and antiviral responses. Mol. Immunol. 2008, 45, 3494–3501. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.M.; Maehr, T.; Diaz-Rosales, P.; Secombes, C.J.; Wang, T. Bioactivity studies of rainbow trout (Oncorhynchus mykiss) interleukin-6: Effects on macrophage growth and antimicrobial peptide gene expression. Mol. Immunol. 2011, 48, 1903–1916. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.; Lin, H.T.; Foung, Y.F.; Lin, J.H. The bioactivity of teleost IL-6: IL-6 protein in orange-spotted grouper (Epinephelus coioides) induces Th2 cell differentiation and antibody production. Dev. Comp. Immunol. 2012, 38, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, M.; Odaka, T.; Suetake, H.; Tahara, D.; Miyadai, T. Teleost IL-6 promotes antibody production through STAT3 signaling via IL-6R and gp130. Dev. Comp. Immunol. 2012, 38, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Hanington, P.C.; Belosevic, M. Interleukin-6 family cytokine M17 induces differentiation and nitric oxide response of goldfish (Carassius auratus L.) macrophages. Dev. Comp. Immunol. 2007, 31, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Ogai, K.; Kuwana, A.; Hisano, S.; Nagashima, M.; Koriyama, Y.; Sugitani, K.; Mawatari, K.; Nakashima, H.; Kato, S. Upregulation of leukemia inhibitory factor (LIF) during the early stage of optic nerve regeneration in zebrafish. PLoS ONE 2014, 9, e106010. [Google Scholar] [CrossRef] [PubMed]
- Collison, L.W.; Delgoffe, G.M.; Guy, C.S.; Vignali, K.M.; Chaturvedi, V.; Fairweather, D.; Satoskar, A.R.; Garcia, K.C.; Hunter, C.A.; Drake, C.G.; et al. The composition and signalling of the IL-35 receptor are unconventional. Nat. Immunol. 2012, 13, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Heinzel, F.P.; Hujer, A.M.; Ahmed, F.N.; Rerko, R.M. Vivo production and function of IL-12 p40 homodimers. J. Immunol. 1997, 158, 4381–4388. [Google Scholar] [PubMed]
- Zhang, L.; Zhang, B.C.; Hu, Y.H. Rock bream (Oplegnathus fasciatus) IL-12p40: Identification, expression, and effect on bacterial infection. Fish Shellfish Immunol. 2014, 39, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.L.; Priya, T.A.J.; Hu, K.Y.; Yan, H.Y.; Shen, S.T.; Song, Y.L. Grouper interleukin-12, linked by an ancient disulphide-bond architecture, exhibits cytokine and chemokine activities. Fish Shellfish Immunol. 2014, 36, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Segaliny, A.I.; Brion, R.; Brulin, B.; Maillasson, M.; Charrier, C.; Teletchea, S.; Heymann, D. IL-34 and M-CSF form a novel heteromeric cytokine and regulate M-CSF receptor activation and localization. Cytokine 2015, 76, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Hanington, P.C.; Wang, T.; Secombes, C.J.; Belosevic, M. Growth factors of lower vertebrates: Characterisation of goldfish (Carassius auratus L.) macrophage colony stimulating factor-1. J. Biol. Chem. 2007, 282, 31865–31872. [Google Scholar] [CrossRef] [PubMed]
- Barreda, D.R.; Hanington, P.C.; Stafford, J.L.; Belosevic, M. A novel soluble form of the CSF-1 receptor inhibits proliferation of self-renewing macrophages of goldfish (Carassius auratus L.). Dev. Comp. Immunol. 2005, 29, 879–894. [Google Scholar] [CrossRef] [PubMed]
- Grayfer, L.; Hanington, P.C.; Belosevic, M. Macrophage colony-stimulating factor (CSF-1) induces pro-inflammatory gene expression and enhances antimicrobial responses of goldfish (Carassius auratus L.) macrophages. Fish Shellfish Immunol. 2009, 26, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Hanington, P.C.; Hitchen, S.J.; Beamish, L.A.; Belosevic, M. Macrophage colony stimulating factor (CSF-1) is a central growth factor of goldfish macrophages. Fish Shellfish Immunol. 2009, 26, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hanington, P.C.; Belosevic, M.; Secombes, C.J. Two macrophage colony-stimulating factor genes exist in fish that differ in gene organisation and are differentially expressed. J. Immunol. 2008, 181, 3310–3322. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Kono, T.; Monte, M.M.; Kuse, H.; Costa, M.M.; Korenaga, H.; Maehr, T.; Husain, M.; Sakai, M.; Secombes, C.J. Identification of IL-34 in teleost fish: Differential expression of rainbow trout IL-34, MCSF1 and MCSF2, ligands of the MCSF receptor. Mol. Immunol. 2013, 53, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.Q.; Li, Y.W.; Zhou, L.; Li, A.X.; Luo, X.C.; Dan, X.M. Grouper (Epinephelus coiodes) IL-34/MCSF2 and MSCFR1/MCSFR2 were involved in mononuclear phagocytes activation against Cryptocaryon irritans infection. Fish Shellfish Immunol. 2015, 43, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Szretter, K.J.; Vermi, W.; Gilfillan, S.; Rossini, C.; Cella, M.; Barrow, A.D.; Diamond, M.S.; Colonna, M. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 2012, 13, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Liongue, C.; Hall, C.J.; O’Connell, B.A.; Crosier, P.; Ward, A.C. Zebrafish granulocyte colony-stimulating factor receptor signalling promotes myelopoiesis and myeloid cell migration. Blood 2009, 113, 2535–2546. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, O.; Stachura, D.L.; Machoňová, O.; Pajer, P.; Brynda, J.; Zon, L.I.; Traver, D.; Bartůněk, P. Dissection of vertebrate hematopoiesis using zebrafish thrombopoietin. Blood 2014, 124, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Stachura, D.L.; Svoboda, O.; Campbell, C.A.; Espin-Palazón, R.; Lau, R.P.; Zon, L.I.; Bartůněk, P.; Traver, D. The zebrafish granulocyte colony-stimulating factors (Gcsfs): 2 paralogous cytokines and their roles in hematopoietic development and maintenance. Blood 2013, 122, 3918–3928. [Google Scholar] [CrossRef] [PubMed]
- Galdames, J.A.; Zuñiga-Traslaviña, C.; Reyes, A.E.; Feijóo, C.G. Gcsf-Chr19 promotes neutrophil migration to damaged tissue through blood vessels in zebrafish. J. Immunol. 2014, 193, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Niethammer, P.; Grabher, C.; Look, A.T.; Mitchison, T.J. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 2009, 459, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Lutfalla, G.; Roest Crollius, H.; Stange-Thomann, N.; Jaillon, O.; Mogensen, K.; Monneron, D. Comparative genomic analysis reveals independent expansion of a lineage-specific gene family in vertebrates: The class II cytokine receptors and their ligands in mammals and fish. BMC Genomics 2003. [Google Scholar] [CrossRef] [PubMed]
- Altmann, S.M.; Mellon, M.T.; Distel, D.L.; Kim, C.H. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J. Virol. 2003, 77, 1992–2002. [Google Scholar] [CrossRef] [PubMed]
- Boudinot, P.; Zou, J.; Ota, T.; Buonocore, F.; Scapigliati, G.; Canapa, A.; Cannon, J.; Litman, G.; Hansen, J.D. A tetrapod-like repertoire of innate immune receptors and effectors for coelacanths. J. Exp. Zool. B. Mol. Dev. Evol. 2014, 322, 415–437. [Google Scholar] [CrossRef] [PubMed]
- Langevin, C.; Aleksejeva, E.; Passoni, G.; Palha, N.; Levraud, J.P.; Boudinot, P. The antiviral innate immune response in fish: Evolution and conservation of the IFN system. J. Mol. Biol. 2013, 425, 4904–4920. [Google Scholar] [CrossRef] [PubMed]
- Robertsen, B. The interferon system of teleost fish. Fish Shellfish Immunol. 2006, 20, 172–191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.B.; Gui, J.F. Molecular regulation of interferon antiviral response in fish. Dev. Comp. Immunol. 2012, 38, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Secombes, C.J. Teleost fish interferons and their role in immunity. Dev. Comp. Immunol. 2011, 35, 1376–1387. [Google Scholar] [CrossRef] [PubMed]
- Aggad, D.; Mazel, M.; Boudinot, P.; Mogensen, K.E.; Hamming, O.J.; Hartmann, R.; Kotenko, S.; Herbomel, P.; Lutfalla, G.; Levraud, J.P. The two groups of zebrafish virus-induced interferons signal via distinct receptors with specific and shared chains. J. Immunol. 2009, 183, 3924–3931. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Nie, P.; Collet, B.; Secombes, C.J.; Zou, J. Identification of an additional two-cysteine containing type I interferon in rainbow trout Oncorhynchus mykiss provides evidence of a major gene duplication event within this gene family in teleosts. Immunogenetics 2009, 61, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Robertsen, B.; Wang, Z.; Liu, B. Identification of an Atlantic salmon IFN multigene cluster encoding three IFN subtypes with very different expression properties. Dev. Comp. Immunol. 2009, 33, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Tafalla, C.; Truckle, J.; Secombes, C.J. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J. Immunol. 2007, 179, 3859–3871. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Gorgoglione, B.; Taylor, N.G.; Summathed, T.; Lee, P.T.; Panigrahi, A.; Genet, C.; Chen, Y.M.; Chen, T.Y.; Ul Hassan, M.; et al. Salmonids have an extraordinary complex type I IFN system: Characterization of the IFN locus in rainbow trout Oncorhynchus mykiss reveals two novel IFN subgroups. J. Immunol. 2014, 193, 2273–2286. [Google Scholar] [CrossRef] [PubMed]
- Hamming, O.J.; Lutfalla, G.; Levraud, J.P.; Hartmann, R. Crystal structure of zebrafish interferons I and II reveals conservation of type I interferon structure in vertebrates. J. Virol. 2011, 85, 8181–8187. [Google Scholar] [CrossRef] [PubMed]
- Casani, D.; Randelli, E.; Costantini, S.; Facchiano, A.M.; Zou, J.; Martin, S.; Secombes, C.J.; Scapigliati, G.; Buonocore, F. Molecular characterisation and structural analysis of an interferon homologue in sea bass (Dicentrarchus labrax L.). Mol. Immunol. 2009, 46, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Kuo, C.E.; Chen, G.R.; Kao, Y.T.; Zou, J.; Secombes, C.J.; Chen, T.Y. Functional analysis of an orange-spotted grouper (Epinephelus coioides) interferon gene and characterisation of its expression in response to nodavirus infection. Dev. Comp. Immunol. 2014, 46, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Hikima, J.; Hwang, S.D.; Morita, T.; Suzuki, Y.; Kato, G.; Kondo, H.; Hirono, I.; Jung, T.S.; Aoki, T. Transcriptional regulation of type I interferon gene expression by interferon regulatory factor-3 in Japanese flounder, Paralichthys olivaceus. Dev. Comp. Immunol. 2012, 36, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; Milev-Milovanovic, I.; Wilson, M.; Bengten, E.; Clem, L.W.; Miller, N.W.; Chinchar, V.G. Identification and expression analysis of cDNAs encoding channel catfish type I interferons. Fish Shellfish Immunol. 2006, 21, 42–59. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; Wilson, M.; Bengten, E.; Bryan, L.; Clem, L.W.; Miller, N.W.; Chinchar, V.G. Identification of a cDNA encoding channel catfish interferon. Dev. Comp. Immunol. 2004, 28, 97–111. [Google Scholar] [CrossRef]
- Robertsen, B.; Bergan, V.; Rokenes, T.; Larsen, R.; Albuquerque, A. Atlantic salmon interferon genes: Cloning, sequence analysis, expression, and biological activity. J. Interferon Cytokine Res. 2003, 23, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, S.; Chiang, Y.A.; Hikima, J.; Sakai, M.; Lo, C.F.; Wang, H.C.; Aoki, T. Expression and biological activity of two types of interferon genes in medaka (Oryzias latipes). Fish Shellfish Immunol. 2016, 48, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Pereiro, P.; Costa, M.M.; Diaz-Rosales, P.; Dios, S.; Figueras, A.; Novoa, B. The first characterization of two type I interferons in turbot (Scophthalmus maximus) reveals their differential role, expression pattern and gene induction. Dev. Comp. Immunol. 2014, 45, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Svingerud, T.; Solstad, T.; Sun, B.; Nyrud, M.L.; Kileng, O.; Greiner-Tollersrud, L.; Robertsen, B. Atlantic salmon type I IFN subtypes show differences in antiviral activity and cell-dependent expression: Evidence for high IFNb/IFNc-producing cells in fish lymphoid tissues. J. Immunol. 2012, 189, 5912–5923. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.B.; Li, Q.; Gui, J.F. Differential expression of two Carassius auratus Mx genes in cultured CAB cells induced by grass carp hemorrhage virus and interferon. Immunogenetics 2004, 56, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Robertsen, C.; Sun, B.; Robertsen, B. Protection of Atlantic salmon against virus infection by intramuscular injection of IFNc expression plasmid. Vaccine 2014, 32, 4695–4702. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lu, L.F.; Feng, H.; Wu, N.; Chen, D.D.; Zhang, Y.B.; Gui, J.F.; Nie, P.; Zhang, Y.A. IFN regulatory factor 10 is a negative regulator of the IFN responses in fish. J. Immunol. 2014, 193, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhang, Y.B.; Liu, T.K.; Gan, L.; Yu, F.F.; Liu, Y.; Gui, J.F. Characterization of fish IRF3 as an IFN-inducible protein reveals evolving regulation of IFN response in vertebrates. J. Immunol. 2010, 185, 7573–7582. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhang, Y.B.; Jiang, J.; Wang, B.; Chen, C.; Zhang, J.; Gui, J.F. Gig1, a novel antiviral effector involved in fish interferon response. Virology 2014, 448, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Collet, B.; Nie, P.; Lester, K.; Campbell, S.; Secombes, C.J.; Zou, J. Expression and functional characterization of the RIG-I-like receptors MDA5 and LGP2 in rainbow trout (Oncorhynchus mykiss). J. Virol. 2011, 85, 8403–8412. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A.; Taggart, J.B.; Seear, P.; Bron, J.E.; Talbot, R.; Teale, A.J.; Sweeney, G.E.; Hoyheim, B.; Houlihan, D.F.; Tocher, D.R.; et al. Interferon type I and type II responses in an Atlantic salmon (Salmo salar) SHK-1 cell line by the salmon TRAITS/SGP microarray. Physiol. Genomics 2007, 32, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Evensen, O.; Munang'andu, H.M. De novo assembly and transcriptome analysis of Atlantic salmon macrophage/dendritic-like TO cells following type I IFN treatment and Salmonid alphavirus subtype-3 infection. BMC Genomics 2015. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, S.; Liu, J.; Xiao, J.; Yan, J.; Feng, H. IFNa of black carp is an antiviral cytokine modified with N-linked glycosylation. Fish Shellfish Immunol. 2015, 46, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Kileng, O.; Brundtland, M.I.; Robertsen, B. Infectious salmon anemia virus is a powerful inducer of key genes of the type I interferon system of Atlantic salmon, but is not inhibited by interferon. Fish Shellfish Immunol. 2007, 23, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Skjaeveland, I.; Svingerud, T.; Zou, J.; Jorgensen, J.; Robertsen, B. Antiviral activity of salmonid gamma interferon against infectious pancreatic necrosis virus and salmonid alphavirus and its dependency on type I interferon. J. Virol. 2011, 85, 9188–9198. [Google Scholar] [CrossRef] [PubMed]
- Svingerud, T.; Holand, J.K.; Robertsen, B. Infectious salmon anemia virus (ISAV) replication is transiently inhibited by Atlantic salmon type I interferon in cell culture. Virus Res. 2013, 177, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Guo, T.C.; Mutoloki, S.; Haugland, O.; Marjara, I.S.; Evensen, O. Alpha interferon and not gamma interferon inhibits salmonid alphavirus subtype 3 replication in vitro. J. Virol. 2010, 84, 8903–8912. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Munoz, A.; Roca, F.J.; Meseguer, J.; Mulero, V. New insights into the evolution of IFNs: Zebrafish group II IFNs induce a rapid and transient expression of IFN-dependent genes and display powerful antiviral activities. J. Immunol. 2009, 182, 3440–3449. [Google Scholar] [CrossRef] [PubMed]
- Bergan, V.; Steinsvik, S.; Xu, H.; Kileng, O.; Robertsen, B. Promoters of type I interferon genes from Atlantic salmon contain two main regulatory regions. FEBS J. 2006, 273, 3893–3906. [Google Scholar] [CrossRef] [PubMed]
- Purcell, M.K.; Laing, K.J.; Woodson, J.C.; Thorgaard, G.H.; Hansen, J.D. Characterization of the interferon genes in homozygous rainbow trout reveals two novel genes, alternate splicing and differential regulation of duplicated genes. Fish Shellfish Immunol. 2009, 26, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.X.; Zou, J.; Nie, P.; Huang, B.; Yu, Z.; Collet, B.; Secombes, C.J. Intracellular interferons in fish: A unique means to combat viral infection. PLoS Pathog. 2013. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.P.; Chung, C.L.; Hung, Y.F.; Lai, Y.S.; Chiou, P.P.; Lu, M.W.; Kong, Z.L. Comparison of the responses of different recombinant fish type I interferons against betanodavirus infection in grouper. Fish Shellfish Immunol. 2016, 49, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Greiner-Tollersrud, L.; Koop, B.F.; Robertsen, B. Atlantic salmon possesses two clusters of type I interferon receptor genes on different chromosomes, which allows for a larger repertoire of interferon receptors than in zebrafish and mammals. Dev. Comp. Immunol. 2014, 47, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.X.; Zheng, Z.; Chen, X.; Perrimon, N. The Jak/STAT pathway in model organisms: Emerging roles in cell movement. Dev. Cell 2002, 3, 765–778. [Google Scholar] [CrossRef]
- Stein, C.; Caccamo, M.; Laird, G.; Leptin, M. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.C.; Ward, A.C.; Liang, R.; Leung, A.Y. The role of jak2a in zebrafish hematopoiesis. Blood 2007, 110, 1824–1830. [Google Scholar] [CrossRef] [PubMed]
- Sobhkhez, M.; Hansen, T.; Iliev, D.B.; Skjesol, A.; Jorgensen, J.B. The Atlantic salmon protein tyrosine kinase Tyk2: Molecular cloning, modulation of expression and function. Dev. Comp. Immunol. 2013, 41, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Sobhkhez, M.; Skjesol, A.; Thomassen, E.; Tollersrud, L.G.; Iliev, D.B.; Sun, B.; Robertsen, B.; Jorgensen, J.B. Structural and functional characterization of salmon STAT1, STAT2 and IRF9 homologs sheds light on interferon signaling in teleosts. FEBS Open Bio. 2014, 4, 858–871. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Xiong, R.; Zhang, Y.S.; Zhu, L.Y.; Shao, J.Z.; Xiang, L.X. Conserved inhibitory role of teleost SOCS-1s in IFN signaling pathways. Dev. Comp. Immunol. 2014, 43, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Skjesol, A.; Liebe, T.; Iliev, D.B.; Thomassen, E.I.; Tollersrud, L.G.; Sobhkhez, M.; Lindenskov Joensen, L.; Secombes, C.J.; Jorgensen, J.B. Functional conservation of suppressors of cytokine signaling proteins between teleosts and mammals: Atlantic salmon SOCS1 binds to JAK/STAT family members and suppresses type I and II IFN signaling. Dev. Comp. Immunol. 2014, 45, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Xiong, R.; Nie, L.; Xiang, L.X.; Shao, J.Z. Characterization of a PIAS4 homologue from zebrafish: Insights into its conserved negative regulatory mechanism in the TRIF, MAVS, and IFN signaling pathways during vertebrate evolution. J. Immunol. 2012, 188, 2653–2668. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Q.; Xu, Q.Q.; Chang, M.X.; Zou, J.; Secombes, C.J.; Peng, K.M.; Nie, P. Molecular characterization and expression analysis of the IFN-gamma related gene (IFN-gammarel) in grass carp Ctenopharyngodon idella. Vet. Immunol. Immunopathol. 2010, 134, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Igawa, D.; Sakai, M.; Savan, R. An unexpected discovery of two interferon gamma-like genes along with interleukin (IL)-22 and -26 from teleost: IL-22 and -26 genes have been described for the first time outside mammals. Mol. Immunol. 2006, 43, 999–1009. [Google Scholar] [CrossRef] [PubMed]
- Swain, B.; Basu, M.; Lenka, S.S.; Das, S.; Jayasankar, P.; Samanta, M. Characterization and inductive expression analysis of interferon gamma-related gene in the Indian major carp, rohu (Labeo rohita). DNA Cell Biol. 2015, 34, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Yabu, T.; Toda, H.; Shibasaki, Y.; Araki, K.; Yamashita, M.; Anzai, H.; Mano, N.; Masuhiro, Y.; Hanazawa, S.; Shiba, H.; et al. Antiviral protection mechanisms mediated by ginbuna crucian carp interferon gamma isoforms 1 and 2 through two distinct interferon gamma-receptors. J. Biochem. 2011, 150, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Yoshiura, Y.; Dijkstra, J.M.; Sakai, M.; Ototake, M.; Secombes, C. Identification of an interferon gamma homologue in Fugu, Takifugu rubripes. Fish Shellfish Immunol. 2004, 17, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Carrington, A.; Collet, B.; Dijkstra, J.M.; Yoshiura, Y.; Bols, N.; Secombes, C. Identification and bioactivities of IFN-gamma in rainbow trout Oncorhynchus mykiss: The first Th1-type cytokine characterized functionally in fish. J. Immunol. 2005, 175, 2484–2494. [Google Scholar] [CrossRef] [PubMed]
- Grayfer, L.; Garcia, E.G.; Belosevic, M. Comparison of macrophage antimicrobial responses induced by type II interferons of the goldfish (Carassius auratus L.). J. Biol. Chem. 2010, 285, 23537–23547. [Google Scholar] [CrossRef] [PubMed]
- Pijanowski, L.; Scheer, M.; Verburg-van Kemenade, B.M.; Chadzinska, M. Production of inflammatory mediators and extracellular traps by carp macrophages and neutrophils in response to lipopolysaccharide and/or interferon-gamma2. Fish Shellfish Immunol. 2015, 42, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Zhang, S.; Chen, D.; Zhang, A.; Wang, X.; Zhou, H. IFN-gamma-activated lymphocytes boost nitric oxide production in grass carp monocytes/macrophages. Fish Shellfish Immunol. 2013, 35, 1635–1641. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Diaz-Rosales, P.; Martin, S.A.; Secombes, C.J. Cloning of a novel interleukin (IL)-20-like gene in rainbow trout Oncorhynchus mykiss gives an insight into the evolution of the IL-10 family. Dev. Comp. Immunol. 2010, 34, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Arts, J.A.; Tijhaar, E.J.; Chadzinska, M.; Savelkoul, H.F.; Verburg-van Kemenade, B.M. Functional analysis of carp interferon-gamma: Evolutionary conservation of classical phagocyte activation. Fish Shellfish Immunol. 2010, 29, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.Y.; Hikima, J.; Ohtani, M.; Jang, H.B.; del Castillo, C.S.; Nho, S.W.; Cha, I.S.; Park, S.B.; Aoki, T.; Jung, T.S. Recombinant interferon-gamma activates immune responses against Edwardsiella tarda infection in the olive flounder, Paralichthys olivaceus. Fish Shellfish Immunol. 2012, 33, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, Y.; Yabu, T.; Araki, K.; Mano, N.; Shiba, H.; Moritomo, T.; Nakanishi, T. Peculiar monomeric interferon gammas, IFNγrel 1 and IFNγrel 2, in ginbuna crucian carp. FEBS J. 2014, 281, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Stolte, E.H.; Savelkoul, H.F.; Wiegertjes, G.; Flik, G.; Lidy Verburg-van Kemenade, B.M. Differential expression of two interferon-gamma genes in common carp (Cyprinus carpio L.). Dev. Comp. Immunol. 2008, 32, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Aggad, D.; Stein, C.; Sieger, D.; Mazel, M.; Boudinot, P.; Herbomel, P.; Levraud, J.P.; Lutfalla, G.; Leptin, M. In vivo analysis of Ifn-gamma1 and Ifn-gamma2 signaling in zebrafish. J. Immunol. 2010, 185, 6774–6782. [Google Scholar] [CrossRef] [PubMed]
- Biswas, G.; Nagamine, R.; Hikima, J.; Sakai, M.; Kono, T. Inductive immune responses in the Japanese pufferfish (Takifugu rubripes) treated with recombinant IFN-gamma, IFN-gammarel, IL-4/13A and IL-4/13B. Int. Immunopharmacol. 2016, 31, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Grayfer, L.; Hodgkinson, J.W.; Hitchen, S.J.; Belosevic, M. Characterization and functional analysis of goldfish (Carassius auratus L.) interleukin-10. Mol. Immunol. 2011, 48, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Harun, N.O.; Costa, M.M.; Secombes, C.J.; Wang, T. Sequencing of a second interleukin-10 gene in rainbow trout Oncorhynchus mykiss and comparative investigation of the expression and modulation of the paralogues in vitro and in vivo. Fish Shellfish Immunol. 2011, 31, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Kamota, S.; Ito, K.; Yoshiura, Y.; Ototake, M.; Moritomo, T.; Nakanishi, T. Molecular cloning and expression analysis of rainbow trout (Oncorhynchus mykiss) interleukin-10 cDNAs. Fish Shellfish Immunol. 2005, 18, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Pinto, R.D.; Nascimento, D.S.; Reis, M.I.; do Vale, A.; Dos Santos, N.M. Molecular characterization, 3D modelling and expression analysis of sea bass (Dicentrarchus labrax L.) interleukin-10. Mol. Immunol. 2007, 44, 2056–2065. [Google Scholar] [CrossRef] [PubMed]
- Savan, R.; Igawa, D.; Sakai, M. Cloning, characterization and expression analysis of interleukin-10 from the common carp, Cyprinus carpio L. Eur. J. Biochem. 2003, 270, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Seppola, M.; Larsen, A.N.; Steiro, K.; Robertsen, B.; Jensen, I. Characterisation and expression analysis of the interleukin genes, IL-1beta, IL-8 and IL-10, in Atlantic cod (Gadus morhua L.). Mol. Immunol. 2008, 45, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yang, M.; Zhao, T.; Wang, X.; Zhou, H. Functional expression and characterization of grass carp IL-10: An essential mediator of TGF-beta1 immune regulation in peripheral blood lymphocytes. Mol. Immunol. 2013, 53, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Clark, M.S.; Secombes, C.J. Characterisation, expression and promoter analysis of an interleukin 10 homologue in the puffer fish, Fugu rubripes. Immunogenetics 2003, 55, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Piazzon, M.C.; Wentzel, A.S.; Tijhaar, E.J.; Rakus, K.L.; Vanderplasschen, A.; Wiegertjes, G.F.; Forlenza, M. Cyprinid herpesvirus 3 IL10 inhibits inflammatory activities of carp macrophages and promotes proliferation of IgM+ B cells and memory T cells in a manner similar to carp IL10. J. Immunol. 2015, 195, 3694–3704. [Google Scholar] [CrossRef] [PubMed]
- Sunarto, A.; Liongue, C.; McColl, K.A.; Adams, M.M.; Bulach, D.; Crane, M.S.; Schat, K.A.; Slobedman, B.; Barnes, A.C.; Ward, A.C.; et al. Koi herpesvirus encodes and expresses a functional interleukin-10. J. Virol. 2012, 86, 11512–11520. [Google Scholar] [CrossRef] [PubMed]
- Van Beurden, S.J.; Forlenza, M.; Westphal, A.H.; Wiegertjes, G.F.; Haenen, O.L.; Engelsma, M.Y. The alloherpesviral counterparts of interleukin 10 in European eel and common carp. Fish Shellfish Immunol. 2011, 31, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Piazzon, M.C.; Savelkoul, H.S.; Pietretti, D.; Wiegertjes, G.F.; Forlenza, M. Carp IL10 has anti-inflammatory activities on phagocytes, promotes proliferation of memory T cells, and regulates B cell differentiation and antibody secretion. J. Immunol. 2015, 194, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Monte, M.M.; Zou, J.; Wang, T.; Carrington, A.; Secombes, C.J. Cloning, expression analysis and bioactivity studies of rainbow trout (Oncorhynchus mykiss) interleukin-22. Cytokine 2011, 55, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Zhang, Q.; Wang, Z.; Zhao, W.; Chen, S.; Gao, Q. Molecular cloning, expression analysis and functional characterization of interleukin-22 in So-iny mullet, Liza haematocheila. Mol. Immunol. 2015, 63, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Corripio-Miyar, Y.; Zou, J.; Richmond, H.; Secombes, C.J. Identification of interleukin-22 in gadoids and examination of its expression level in vaccinated fish. Mol. Immunol. 2009, 46, 2098–2106. [Google Scholar] [CrossRef] [PubMed]
| Cytokine | Human homologue | Receptors | Known immune functions | references |
|---|---|---|---|---|
| IL-10, viral IL-10 | IL-10 | IL-10R1/CRFB12, IL-10R2/CRFB4 | Suppress immune responses, inhibit inflammation, promote T cell proliferation, memory B cells, and IgM production | [265,273,276] |
| IL-20L | IL19, IL-20, IL-24 | Not characterised | Not characterised | [200,258] |
| IL-22 | IL-22 | Not characterised | Activate antimicrobial peptide genes and antibacterial immunity | [277,278] |
| IL-26 | IL-26 | Not characterised | Not characterised | [242,250] |
| Group I IFN (IFN-a, d, e) | Type I IFN | IFNAR1/CRFB5, IFNAR2/CRFB1 | Induce expression of the antiviral effectors, promote apoptosis, regulate inflammation | [201,207,210,221,234] |
| Group II IFN (IFN-b, c, f) | Type I IFN | IFNAR1/CRFB5, IFNAR2/CRFB2 | Induce expression of the antiviral effectors, promote apoptosis | [207,210,221,223] |
| IFN-γ | Type II IFN | IFN-γR1/CRFB13, IFN-γR2/CRFB6 | Activate phagocytes, enhance antigen presentation, promote Th1 cytokine expression | [254,255,256,257] |
| IFN-γrel | Type II IFN | CRFB17, IFN-γR2/CRFB6 | Regulate anti-bacterial and antiviral immunity | [255,261] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, J.; Secombes, C.J. The Function of Fish Cytokines. Biology 2016, 5, 23. https://doi.org/10.3390/biology5020023
Zou J, Secombes CJ. The Function of Fish Cytokines. Biology. 2016; 5(2):23. https://doi.org/10.3390/biology5020023
Chicago/Turabian StyleZou, Jun, and Christopher J. Secombes. 2016. "The Function of Fish Cytokines" Biology 5, no. 2: 23. https://doi.org/10.3390/biology5020023
APA StyleZou, J., & Secombes, C. J. (2016). The Function of Fish Cytokines. Biology, 5(2), 23. https://doi.org/10.3390/biology5020023





