Abstract
Epigenetics increasingly occupies a pivotal position in our understanding of inheritance, natural selection and, perhaps, even evolution. A survey of the PubMed database, however, reveals that the great majority (>93%) of epigenetic papers have an intra-, rather than an inter-generational focus, primarily on mechanisms and disease. Approximately ~1% of epigenetic papers even mention the nexus of epigenetics, natural selection and evolution. Yet, when environments are dynamic (e.g., climate change effects), there may be an “epigenetic advantage” to phenotypic switching by epigenetic inheritance, rather than by gene mutation. An epigenetically-inherited trait can arise simultaneously in many individuals, as opposed to a single individual with a gene mutation. Moreover, a transient epigenetically-modified phenotype can be quickly “sunsetted”, with individuals reverting to the original phenotype. Thus, epigenetic phenotype switching is dynamic and temporary and can help bridge periods of environmental stress. Epigenetic inheritance likely contributes to evolution both directly and indirectly. While there is as yet incomplete evidence of direct permanent incorporation of a complex epigenetic phenotype into the genome, doubtlessly, the presence of epigenetic markers and the phenotypes they create (which may sort quite separately from the genotype within a population) will influence natural selection and, so, drive the collective genotype of a population.
1. Introduction
Epigenetics is rapidly earning its place as a major phenomenon in modern life sciences. Indeed, according to the PubMed database, nearly 50,000 papers referencing “epigenetics” have been published to date, with >19,000 published just since 2013. The areas of the life sciences where epigenetics is influencing disciplinary thought are widespread, but not necessarily evenly spread, between the various disciplines and taxa. Indeed, as will be shown in this essay, there is a deep and prevailing focus on mechanisms within epigenetic study, with seemingly less focus on what the mechanisms actually do at the organismal phenotypic level, especially across generations. As will be argued, epigenetics has a potentially profound role in our understanding of inheritance, natural selection and evolution and deserves more attention.
2. What Is “Epigenetics”?
The term “epigenetics” was coined by Waddington C.H. in his 1942 paper entitled “The Epigenotype” [1]. This was, of course, before DNA was discovered and before the complexity of gene expression was appreciated. Against this backdrop, the field of epigenetics at first slowly developed, gaining speed in recent years. For an introduction to the plethora of both reviews and research papers on the topic, see [2,3,4,5,6,7,8,9]. Since Waddington penned his 1942 article, many researchers have attempted, to varying degrees of success, to define, redefine or simply explain the term epigenetics. Indeed, the NIH PubMed database (see more on this below) includes well in excess of 300 papers that contain the phrases “definition of epigenetics” and/or “define epigenetics” and, so, are presumably in some way weighing in on defining epigenetic mechanisms and phenotypes. Authors persist in offering up their own definition of what comprises “epigenetics” and the criteria for scoring epigenetic phenomena; an appreciation for this complex semantic landscape can be gleaned from [2,7,10,11,12,13,14,15,16,17,18,19]. Unfortunately, progress in epigenetic research is being hampered by a lack of clarity as to what is epigenetics. As Deans and Maggert [11] have so aptly summed up, the current epigenetic semantics is as follows:
“Some employ epigenetics to explain changes in gene expression, others use it to refer to transgenerational effects and/or inherited expression states. This disagreement on a clear definition has made communication difficult, synthesis of epigenetic research across fields nearly impossible, and has in many ways biased methodologies and interpretations.”
The great science philosopher Karl Popper opined that “…..we should altogether avoid, like the plague, discussing the meaning of words.”[20] With all due respect to Popper, and to underscore the concern expressed by Deans and Maggert [11], those of us interested in epigenetics do have the responsibility of coming together around a common set of comprehensive definitions, as complex as necessary and as simple as possible, of epigenetic phenomena. Having made this observation (and likely to the relief of the reader aware of the semantic confusion in the definition of epigenetics and epigenetic phenomena), it is not the purpose of this article to attempt to enforce either a new or an existing favored semantic framework on a vibrant yet often chaotic field. To this author, at least, all truly epigenetic phenomena are grist to the mill as we sort out the role of epigenetics in inheritance, natural section and evolution, though some phenomena have far more relevance to evolutionary biology than others.
This essay, then, will skirt the issue of specific definitions of epigenetics and will employ a simple (perhaps simplistic) conceptual framework that differentiates between the use of “epigenetics” to describe intragenerational phenomena and its use to describe transgenerational phenomena. This straightforward dichotomy allows us to leave somewhat in the background the voluminous literature on the intragenerational changes in gene expression within an individual during its lifetime and to focus more acutely on the inheritance of characteristics. Consequently, for the purposes of the rest of this discussion, consider the following division:
Intragenerational epigenetics: modification of gene expression through epigenetic marks (e.g., DNA methylation, covalent histone modification, microRNA action) that results in a modified phenotype, often considered at the molecular/cellular level, within an individual’s lifespan. Those who focus on this aspect of epigenetics will be familiar with phrases, such as “…. the epigenetics of cancer…” or “…. the epigenetics of cardiovascular disease”.
Transgenerational epigenetics: the inheritance of a modified phenotype from the parental generation without changes in genes or gene sequence. The same epigenetic markers mentioned above may be responsible, but a focus in this category is on the act of inheritance. Note that a few authors have used the more restrictive term “cross-generational” to identify inherited traits resulting from the direct exposure of gametes of the F1 and F2 generations while within the body of the P0 generation, as well as maternal effects, such as provisioning [2,21,22,23,24,25]. While from a mechanistic point of view, it is quite important to distinguish these different forms of epigenetic inheritance, this essay simply uses the Latin root “trans” in creating the broad category of “transgenerational” epigenetics, without attempting to link the inheritance to a specific mechanism. This may disappoint more mechanistically-focused researchers in epigenetics, but at this immature stage of exploring the role of transgenerational epigenetics in evolution and evolutionary process, the phenomena themselves may actually be as important as the specific mechanisms. Finally, it is important to point out that these two foci or categories of epigenetics (I will not call them “definitions”)—intragenerational and transgenerational—are not mutually exclusive. A change in gene expression and, thus, in phenotype in the adult P0 generation caused by DNA methylation, for example, can also be carried over into the F1 generation or beyond.
One might reasonably ask, what is the primary focus of workers in the field of epigenetics: intragenerational or transgenerational? This is not an easy question to answer, and even a thorough reading of a few select papers on the topic with their carefully chosen references is unlikely to provide further illumination. Thus, a survey of the literature was performed to determine the relative emphases on the two broad foci of epigenetics outlined above before moving on to explore the role of epigenetics in evolutionary biology.
3. A Survey of Epigenetic Research Papers: Methodology
As indicated above, various researchers use the word “epigenetics” and its derivatives in a wide variety of ways. The use of the word “epigenetic(s)” was probed by analyzing published papers in the PubMed database. Maintained by the National Center for Biotechnology Information of the U.S. National Institutes of Health (http://www.ncbi.nlm.nih.gov/pubmed/), this database catalogues >25,000,000 journal articles in the life sciences, broadly defined. Incidentally, this represents nearly half of all journal articles in all fields estimated to have ever been published [26].
In the current survey, searches were conducted to determine the total number of papers that contained carefully selected key search terms. The invariant, primary search term was “epigenetic”, which identified articles whose title or abstract contained the noun “epigenetics”, as well as the adjective “epigenetic”. Papers within this category are hereafter referred to as “epigenetic papers”. The variable secondary search term was based on a more specific probe of the epigenetic papers. Searches were conducted using a Boolean search methodology to assemble the total number of papers containing, for example, the keyword “physiology”. Secondary search terms also considered related adjectives, e.g., “physiological”, to minimize the creation of duplicate entries. Words with broad, multiple meanings, e.g., “generation”, were avoided because of the potential dilution of the search with papers that used non-pertinent meanings of a word (e.g., “generation” as in creation, as opposed to the description of offspring and parents: P0, F1). Secondary search terms were grouped around common themes. Thus, for example, the general category of “epigenetics” + “disease” actually included secondary search terms of not only “disease”, but also “illness”, “death”, “pathology”, “cancer”, “clinic”, “hospital”, etc.
There are multiple caveats to this survey, some obvious, some subtle, that need to be laid out before considering the results.
- Searches of the PubMed database only identified papers whose title or abstract contained a search term; that is, the body of the paper was not part of the search. Thus, it is possible that papers discussing epigenetics in some form or other have been overlooked. However, in a sense, this eliminated trivial references to epigenetics and focused the survey on those papers that actually thought epigenetics was important enough to include in the abstract, if not the title itself.
- The choice of search terms reflects the author’s view of biology. Thus, for example, “evo-devo” may arguably not be a “discipline” within the life sciences, per se, but it does reflect an actively growing area of biology.
- This survey were carried out in February of 2016. The PubMed database is expanded daily, if not hourly. Indeed, estimates are that an average of ~1 paper a minute is added to this database, but in batches. Therefore, this survey is unique to that specific month and year and in this sense is not exactly replicable by the reader using the current on-line database. However, the incremental changes are unlikely to affect the outcomes of the survey or its conclusions, at least in the short term, given PubMed’s huge article database.
- Importantly, any survey is affected by the source of its data, and this one is no different. While PubMed contains an enormous number of scientific articles, as implied by its name, the database does primarily focus on biomedical research. Certainly, there are journals in the area of evolution and ecology that are not indexed by PubMed (though plant biology, for example, generally is well represented). Thus, there may be a bias towards underrepresentation of evolution papers in this database. On the other hand, as the survey goes on to reveal, the near complete absence of evolutionary papers mentioning epigenetics compared to the overall number of epigenetic papers speaks volumes about the failure to link epigenetics with biology, when considering the many life science journals in animal, plant and microbial science that are indexed by PubMed.
- No distinction was made between primary research papers and review papers in this survey.
- Collectively, the caveats in this survey do point out some sources of inaccuracy in the survey. For example, the secondary search terms are somewhat subjective choices made by the author. Moreover, in some limited instances, papers are undoubtedly double counted, e.g., a paper containing the words “epigenetic”, “illness” and “death” contributes to the paper count for the overall category of “disease”. Important to emphasize is that the goal of this survey was simply to seek out general patterns, not to provide an accurate statistical analysis that would characterize a true meta-analysis. However, given the enormous number of papers in the PubMed data base, nearly 50,000 of which reference epigenetics in some form or other, it is assumed that meta-patterns will emerge despite these inherent limitations of the analytical approach. To emphasize the general nature of these findings, data on percentages are reported to the nearest whole number (but even rounding to the nearest 10% would not obscure the emergent patterns, as is evident below).
4. The Reach of Epigenetic Research in the Life Sciences
4.1. Intragenerational and Transgenerational Epigenetics
The first phase of the survey determined the relative frequency of the occurrence of intragenerational vs. transgenerational epigenetic papers. The actual use of these two specific terms as the secondary search term was not productive: only slightly above 1% of all epigenetic papers actually contained either or both of these adjectives. Subsequently, five major categories of epigenetic papers were formed for this survey. Epigenetic papers including the terms “mechanism”, “disease” and “development and ageing” (and their related topics) were considered to be more representative of an intragenerational perspective, while epigenetic studies, including the terms “evolution” and “inheritance”, were considered to be more representative of epigenetic papers with a transgenerational component to them (however little that might be) Of course, there are transgenerational epigenetic papers that discuss the mechanism of inheritance, and these would be represented in both the “mechanism” and “evolution” category. However, as is evident from Figure 1, the majority of the focus of epigenetic studies was on mechanism and disease states in approximately equal measure. Indeed, <7% of papers referencing epigenetics also mentioned either evolution and/or inheritance. Noteworthy is that while transgenerational epigenetics studies have revealed many instances of epigenetic inheritance of disease/pathologies (e.g., [3,27,28]), the epigenetic inheritance of mal-adaptive modified phenotypes receives little attention compared to the “here and now” of diseases that develop in an individual’s life span. These findings are not surprising, as even a quick examination of a sample of papers comprising the epigenetic literature reveals intensive discussion of mechanisms of epigenetic phenomena, especially as they relate to human health and disease. Additionally, to no one’s surprise, funding follows disease and its prevention and cure, which has greatly enabled the growth of epigenetic studies.
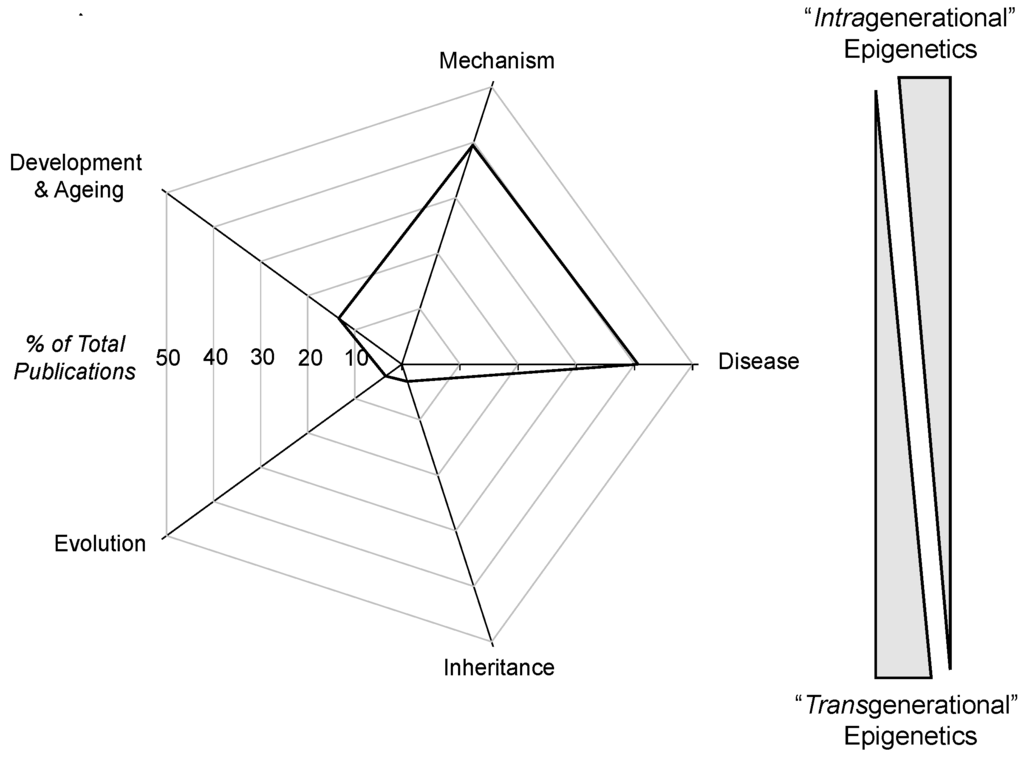
Figure 1.
Radar diagram showing the relative distribution of publications drawn from the PubMed database (http://www.ncbi.nlm.nih.gov/pubmed/) that contain the search terms “epigenetic(s)” and one of five focus areas. The graphic to the right indicates a gradient between intragenerational and transgenerational epigenetics based on the percentage of epigenetic papers emerging from each area of study indicated in the radar diagram. Thus, epigenetic papers with the terms “development and ageing” or “disease” are assumed to be more likely to be addressing intragenerational issues, such as evolution, while epigenetic papers mentioning “evolution” or “inheritance” are viewed as more likely to be focusing on transgenerational epigenetic events. See the text for an additional discussion.
4.2. Epigenetics and Taxon
The survey next explored the taxonomic distribution of epigenetic papers using the secondary search terms (and their adjectives) of “animals”, “plants”, “fungi”, “protists”, “bacteria”, “archaebacteria” and “viruses” [29]. Approximately 60% of epigenetic papers contained the search term “animal(s)”; ~10% contained “plant(s)”; and near negligible numbers of epigenetic papers specifically mentioned any of the other major taxa (Figure 2A).
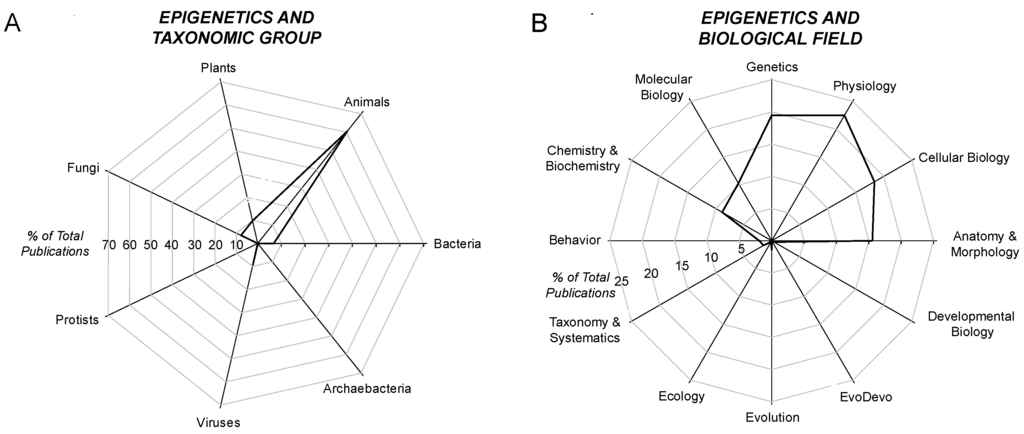
Figure 2.
Radar diagram showing the relative distribution of publications on epigenetics drawn from the PubMed database (http://www.ncbi.nlm.nih.gov/pubmed/). (A) distribution of publications that contain the search terms “epigenetic(s)” and one of seven biological taxa; (B) distribution of publications that contain the search terms “epigenetic(s)” and one of 12 biological fields.
4.3. Epigenetics and the Biological Field
The survey next considered epigenetic papers that included one of 12 major biological fields (Figure 2B). The vast majority (~95%) of epigenetic papers that even mentioned, if not actually discussed, a particular biological field was clustered in just six areas: chemistry/biochemistry, molecular biology, genetics, physiology, cellular biology or anatomy/morphology. Occurring at a very low frequency in the epigenetic literature were the fields of behavior (~2% of papers), taxonomy/systematics (1%–2%), evolution (a little above 1%) and, all being less than 0.5% of the epigenetic papers, development, ecology and evo-devo.
Combing the survey on biological fields and taxa reveals how some areas of epigenetics are almost completely unexplored. For example, combining “epigenetics” + “plant” + “evo-devo” yielded only two papers among the ~50,000 epigenetics papers warehoused in PubMed. Similarly, “epigenetics” + “virus” + “ecology” yielded just three papers. Yet, as we will now turn to, the role of epigenetics in the biology of all of these taxa may be profound.
5. Investigating Epigenetic Inheritance: The Challenges
Epigenetic papers characterized as predominantly focusing on transgenerational issues (see Section 4.1 above) account for only 5%–7% of all epigenetic papers, so it comes as no surprise that a similarly small proportion of epigenetic papers revealed in this analysis specifically consider transgenerational inheritance of a modified phenotype. Why is there so little explicit focus on epigenetic inheritance, as opposed to intragenerational epigenetics, typically associated with disease states? In partial answer to this question, and speaking pragmatically, there are numerous hurdles to well-conducted studies of transgenerational epigenetic inheritance, as will now be explored.
5.1. The Resource Cost of Epigenetic Studies
It is relatively easy to do thought experiments in epigenetic inheritance, but quite another thing to carry out actual experiments, for several reasons. First, there is an extended investment of time not found in intragenerational studies; put simply, one does not leap to study epigenetic inheritance in elephants, with their life span of up to 70 years. The study of transgenerational epigenetic inheritance by definition requires the maintenance and breeding of more than one generation of organisms and the significant costs in time, money, space, etc., that are required (even for microorganisms). Not surprisingly, then, studies of basic questions in epigenetic inheritance often turn to include animal (Drosophila, Caenorhabditis) and plant (Arabidopsis) models with relatively short generation times (e.g., [30,31,32,33,34,35,36,37,38,39]).
5.2. Elusive Epigenetic Mechanisms
A second factor that may complicate the study of epigenetic inheritance is difficulty in teasing out underlying mechanisms (which, as is evident from Figure 1, is a major driver of epigenetic research). Some researchers differentiate epigenetic inheritance due to parental effects or provisioning of eggs with hormones and other substances (so-called context-dependent epigenetic inheritance, which in some cases can span a few generations) from epigenetic inheritance due to the experiences of the egg cells and/or sperm cells and/or stem cells that produce them (so-called “germ-line dependent” epigenetic inheritance). The latter can span F1 and F2 generations at most [2,25,27]. Indeed, some researchers go to great lengths to argue what is and is not epigenetic inheritance, but likely, Waddington C.H. would not be impressed. In the end, both types of epigenetic transfer result in a modified phenotype in the F1 generation. The relevance of the mechanism (and thus, the effort to identify it) depends in part on the nature of the questions being asked. If one asks “How do I prevent this epigenetic disease?”, then knowing the mechanism is all-important. If, however, one asks “Does epigenetic inheritance drive the evolution of a population or species?”, the actual mechanism may or may not be pivotal to the answer.
5.3. The Complexities of Epigenetic “Dynamics”
A third reason why studies of transgenerational inheritance are difficult to conduct and interpret is that it increasingly appears that a complex, yet often subtle or even hidden, set of transgenerational epigenetic “dynamics” can prevail [40,41]. Essentially, most researchers in the field appear to consider an epigenetically-inherited trait as digital: it is either present/on or absent/off. Thus, experiments are typically conducted where the P0 generation is subjected to some form of stressor, and the F1 generation (in fact, often just the neonates of the F1 generation) are scored as to whether they have the trait or lack it. Yet, evidence is accruing that epigenetically-inherited traits in some organisms may take a few generations to actually “wash in”, even being completely absent in the initial F1 generation [42]. Similarly, traits may “wash out” over several generations [42]. Overall, however, the assessment of the presence and extent of the epigenetic inheritance of a trait may well pivot on whether multiple generations are observed and on the threshold level for the detection of a trait within each generation [41]. Interestingly, the trait of reduced body mass in the F1 generations resulting from parental P0 hypoxia exposure dissipates (washes out) both over time as individuals grow, but also washes out over the course of the three successive F1 broods produced by the mother [31], though the complexities introduced by germline exposure (Section 5.2) require further experimentation to clearly identify what mechanism is at play.
Epigenetic dynamics not only is evident across multiple generations, but also across multiple broods within generations. For example, exposure of adult Daphnia magna to hypoxia results in the birth of offspring with initially reduced body mass. However, this effect is only evident for the first 3–5 days after birth and only occurs in the first two broods of the mother, being absent in her third brood [31]. Thus, this epigenetically-induced phenotype of reduced body mass “washes out” over the course of development in the F1 generation and over multiple reproductive events in the P0 mother. Some researchers in epigenetic inheritance are seemingly unaware of these dynamics and fail to incorporate them into their experimental design. This may account for many “false negatives” and the underreporting of transgenerational epigenetic inheritance.
5.4. Keep Calm and Carry on with Transgenerational Epigenetic Research
As is evident from the above assessment, epigenetic transgenerational research is neither easy nor straightforward. Yet, the obstacles are not insurmountable. Tractable animal models with short generation times are available. These same models can be used in experimental designs of sufficient complexity to delineate mechanisms. Experimental designs can also be tweaked to reveal the epigenetic dynamics of the wash in/out of epigenetically-inherited traits.
6. Epigenetics in a Dynamic Environment: Consequences of Rapid and Widely-Distributed Phenotype Switching
6.1. Epigenetically-Inherited Phenotypes: Neutral, Advantageous or Disadvantageous?
The consequences of an epigenetically-inherited phenotype depend in large part on what the effect of that phenotype is on the overall fitness of the individual bearing it. Just like genetically-inherited phenotypes, epigenetically-inherited phenotypes can be neutral, advantageous or disadvantageous. In the field of medicine, most focus is on disadvantageous epigenetically-inherited phenotypes that can lead to disease states. Advantageous epigenetically-inherited phenotypes have received less attention in human health, yet certainly exist [43]. Indeed, if they can be controlled and managed, advantageous phenotypes arising by transgenerational epigenetic inheritance may have potentially large impacts on medicine [44,45] and on agriculture [10,46,47,48,49,50]. To highlight a concrete example of beneficial epigenetic inheritance, consider the transgenerational epigenetic inheritance of hypoxia resistance in zebrafish [7]. As a result of parental (P0) exposure to 2, 3 or 4 weeks of hypoxia (15%), F1 larvae had greater hypoxic resistance than controls whose parents had not experienced hypoxia. Importantly, it was not just an individual larvae or two that were hypoxic resistant, but rather statistically, the entire F1 population had elevated hypoxic resistance. Similarly, we have recently observed that F1 of zebrafish parents exposed to polycyclic aromatic hydrocarbons (PAHs) showed enhanced resistance to these toxicants when compared to control larvae whose parents were not exposed (Martinez-Bautista N. and Burggren, W. unpublished data [51]). Again, most of the population, not just a few individuals, had resistance greater than the controls whose parents were not exposed. These two examples highlight that more experimentation of potentially advantageous epigenetically-inherited phenotypes is warranted.
6.2. Comparing the Time Courses of Genetic and Epigenetic Inheritance
One of the basic tenets of evolution is that natural selection shapes populations and species over evolutionary time. Natural selection acts on organisms with enhanced or diminished fitness, derived from the accumulation of mutations. The resulting phenotypic modifications are enhanced (or not) by these mutations, but the phenotypic switch at the population level and beyond typically occurs over hundreds or thousands of generations as the genotype leading to a modified phenotype of greater fitness slowly inserts itself into the general population or, alternatively, a genotype leading to lesser fitness is eliminated from the population [52]. Klironomos et al. [53] have provided a simple, but informative model of how increases in fitness in a population can derive from either epigenetic or genetic changes in a population over tens of thousands of generations. However, the effect of epigenetic inheritance may not only be potentially broad and sweeping, but may also be felt immediately in a population [52,53,54,55,56]. To underscore this point, consider a phenotype that is advantageous in an environment when a specific stressor that occurs intermittently. Unlike an advantageous gene mutation that affects an individual and then, perhaps, spreads slowly through the population and beyond over many generations, epigenetic inheritance can simultaneously affect many (if not most or all) of a single generation of an entire population. Why? While there is certainly some variation in epigenetic markers between individuals in a population (see below), whether they result in an advantageous or disadvantageous phenotype, epigenetic markers will arise in response to an environmental stressor far more broadly and quickly within a single generation of a population than will a single point mutation occurring in a single individual. Assuming that all individuals in a population of a species presumably experience an environmental stressor at the same time and to a similar extent and that many of the individuals in that population will as a consequence possess the same epigenetic markers, then an epigenetically-switched phenotype should affect many if not most individuals in the population.
The scenario described above is depicted in Figure 3, which compares changes in a population of individuals with an advantageous phenotype arising by either epigenetic inheritance or by mutation. This scenario assumes firstly that the switched phenotype (either from genetic or epigenetic inheritance) is advantageous only in the presence of a deleterious environment, which persists over several generations (specifically, four generations in this scenario) before returning to normal, favorable environmental conditions. Second, this scenario assumes that upon return to the previous normal environment, the newly-switched phenotype is now disadvantageous and possibly lethal. Third, this scenario revolves around only a simple point mutation and, thus, ignores the complexities of pleiotropy, including antagonistic pleiotropy. Fourth, the scenario assumes that an epigenetically-inherited phenotype may persist over more than one generation. Indeed, abundant evidence now exists of epigenetically-inherited phenotypes persisting over multiple generations (e.g., [6,15,42,57,58,59]) before either suddenly disappearing or more slowly “washing out” [41].
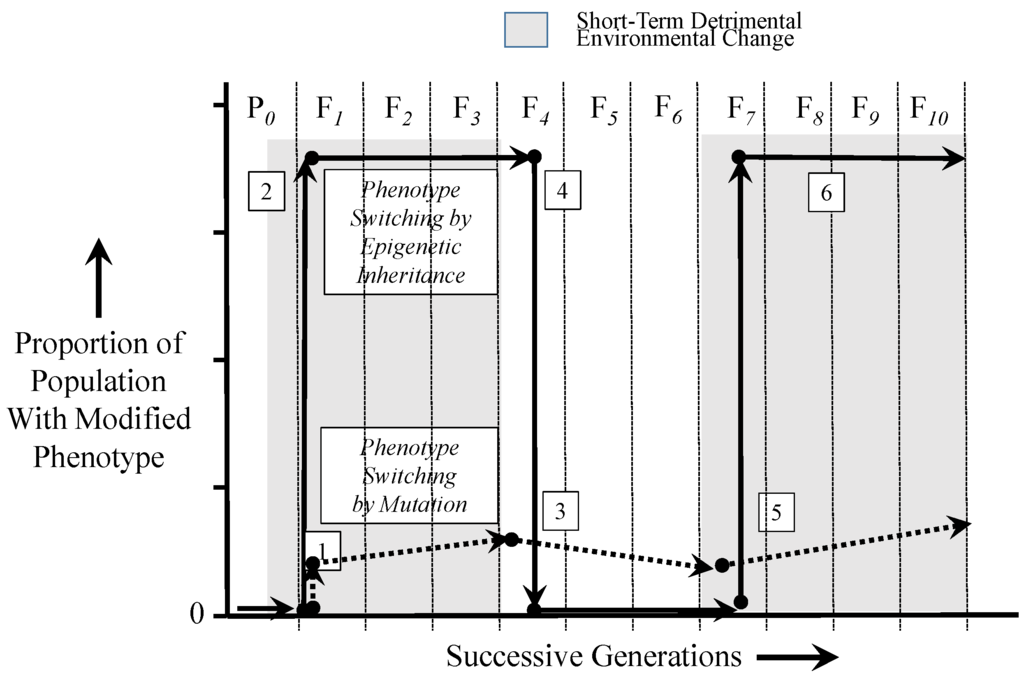
Figure 3.
A comparison of phenotype switching in a population occurring by inherited point mutation vs. inheritance through the effects of epigenetic markers. Events 1, 3 and 5 indicate proportional changes in a hypothetical population resulting from phenotype switching by point mutation that are advantageous during environmental stress, but otherwise disadvantageous (or at least energetically costly). Events 2, 4 and 6 indicate proportional changes in the population resulting from epigenetic phenotype switching. See the text for an additional explanation.
As Figure 3 illustrates, a mutation may result in an advantageous phenotype in only a single individual in a population (Event 1). Advantageous mutations occur at low frequency, become difficult to establish in the population and additionally may be easily lost to genetic drift [60,61]. Thus, this advantageous mutation (Event 1) is only slowly amplified by natural selection over numerous generations, at best. In contrast, an epigenetic phenotypic switch brought on by a deleterious environment can immediately aid in the survival of a potentially large proportion of a population (Event 2), since even allowing for the heterogeneity of epigenetic markers in a population, many in that population may have the epigenetic markers resulting in the modified phenotype. With dissipation of the deleterious environment, however, the individual(s) with the original mutation must cope with the newly disadvantageous phenotype, which cannot be eliminated from the gene pool, except by death of the individual or an unlikely second mutation back to the original gene form (Event 3). In contrast, however, the epigenetically-switched phenotype, now newly disadvantageous in the face of the return to the original environmental condition, is immediately lost by reversion to the original phenotype (Event 4). With a return of the deleterious environment after several generations, the mutant genotype and its phenotype (if they even survive the intervening return to the previous normal environment) will increase only slowly once again in the population at a rate enabled by natural selection (Event 5). Again in contrast, the epigenetically-inherited advantageous phenotype can result in the rapid re-appearance of the advantageous switched phenotype appearing in a large proportion of a population’s individuals (Event 6).
Important to acknowledge is that the scenario depicted in Figure 3 takes an “either-or” approach for epigenetic or genetic inheritance. That is, that populations are shown in this figure to either persist by epigenetic inheritance or by genetic inheritance of an advantageous phenotype, but not necessarily both. We know this approach to be an oversimplification, because presumably, there are also genetic changes that occur in populations changing by epigenetic modification. In fact, it is difficult to separate out such simultaneous phenotypic changes caused by this duality [25,53,57,62,63,64].
7. An “Epigenetic Advantage” in Changing Environments
7.1. Epigenetics in a Temporally-Complex Environment
The epigenetically-enabled survival described above and illustrated in Figure 3 becomes all the more relevant in non-stable, non-equilibrium environments subject to frequent, major changes; e.g., [5,52,53,65,66,67,68,69]. Epigenetic modifications are increasingly being viewed as providing a rapid change in phenotype that simply cannot occur quickly enough through genetic mutation [52,53,54,55,56,70,71,72,73]. Such epigenetic patterns and the phenotypes they have affected can be inherited over multiple generations, as alluded to earlier.
Of key importance is the fact that a phenotypic switch occurring broadly throughout a population caused by epigenetic inheritance is rapidly reversible (Figure 3). This differs from a mutation that requires an unlikely reversion to the pre-mutation genotype. Importantly, these characteristics of transgenerational epigenetic inheritance can allow the survival of organisms with temporarily low fitness resulting from a temporary environment change population [40,74,75,76]. Notably, however, the modified phenotype that was advantageous for ameliorating the effects of the stressor may be at best of no benefit to the individuals in the population; rather, this phenotype may actually be disadvantageous once the stressor wanes or disappears. Phenotype switching arising from mutation thus leaves surviving individuals “stuck” with that modified phenotype, which may now be, for example, more energetically expensive to maintain, after the environmental stressor disappears [40]. In stark contrast, an epigenetically-inherited phenotype may only persist as long as the environmental stressor is evident, but then, the modified phenotype (and its possible energetic costs) are “sunsetted” as the environmental stressor disappears. Consider a hypothetical population of freshwater fishes in a small, drought-plagued pond encountering severe hypoxia over several generations. A mutation resulting in thinner gill membranes might assist oxygen loading at the gills in hypoxic conditions. These thinner gills also may result in more energetically expensive osmotic uptake of water, but it is a reasonable trade-off, since the higher cost of osmoregulation is offset by the fact that the fish can continue to acquire the oxygen needed to survive in a hypoxic environment. However, when rains return, water levels rise, and water returns to full oxygen saturation; these thinner gills now carry a critical disadvantage (enhanced cost of osmoregulation) with no additional advantage to gas exchange when in well-aerated water. In this situation, fishes epigenetically inheriting thinner gills as a result of parental hypoxic exposure (presumably a high proportion of the population) revert to the original phenotype (thicker gills with reduced osmotic water uptake) when normoxic conditions return (in practice, few examples exist in fishes of transgenerational epigenetic inheritance of advantageous modifications at the level of organ structure or function, but epigenetic inheritance of hypoxia tolerance has been demonstrated in the zebrafish as described above [77]).
The “epigenetic advantage” in a seasonally- or annually-dynamic environment where environmentally-related stressors may last only a few generations is graphically expanded upon in Figure 4, through several specific scenarios. In Scenario 1, a species facing a detrimental environment over several generations may fail to thrive if no advantageous modification of phenotype occurs. Indeed, the environmental stressor could take on the trappings of an extinction event. However, in Scenario 2, a population could survive, if not thrive, through the occurrence in an individual or individuals in that population of a spontaneously-arising mutation that is advantageous in the face of the new environmental stressor. However, when the environmental stressor disappears, the population is “stuck” with the phenotypic modification, which may be disadvantageous or, at least, costly. In contrast, Scenario 3 depicts how an epigenetically-inherited advantageous phenotype can be important to the survival of the population during the period of environmental stress, but can “sunset” when the stressor disappears. This epigenetic inheritance allows the population to revert back to the original phenotype that was advantageous in the original environment. Scenario 4 and 5 are versions on this theme, showing how transient (in evolutionary terms) advantageous phenotypes acquired by epigenetic inheritance can “wash in” (4) or “wash out” (5), essentially reflecting a lag-time to epigenetic inheritance across generations.
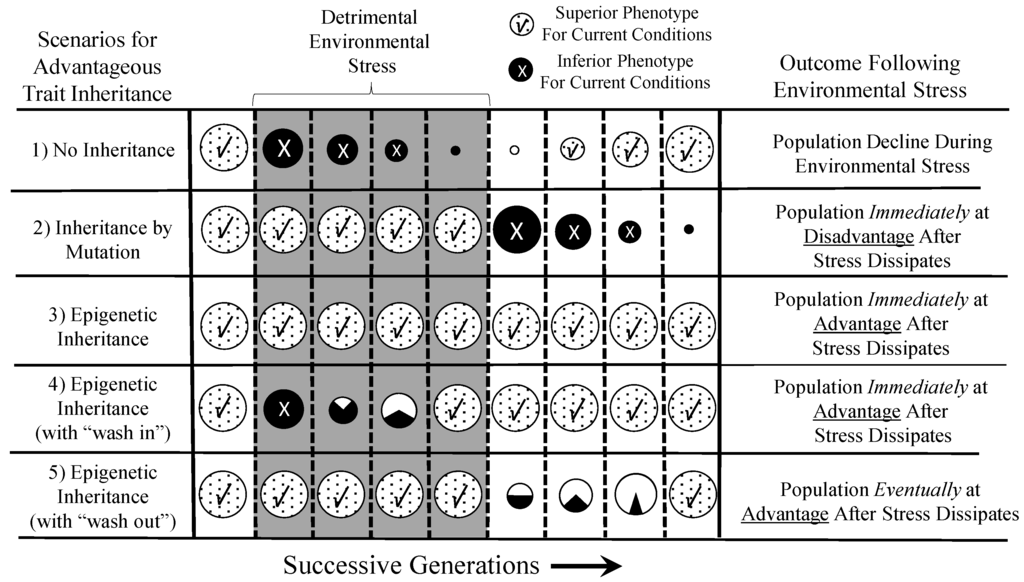
Figure 4.
The “epigenetic advantage” of epigenetic inheritance in a population living in an environment with periods of detrimental conditions. Each circle represents a population in transition over many generations, with the size of the circle reflecting the size of the population. All populations are assumed to have an advantageous phenotype in the non-detrimental environment at the beginning of the graph on the left. Upon the appearance of a critical environmental stressor, successive generations of each population either thrive with a phenotype, acquired by mutation or epigenetic inheritance, or they go into decline. Similarly, as the detrimental environment stressor wanes or disappears, the subsequent generations either have a phenotype advantageous or disadvantageous with respect to the original stressor-free environment. See the text for an additional explanation and discussion.
Table 1 summarizes, compares and contrasts the characteristics and effects of gene mutation and transgenerational epigenetic inheritance. Essentially, the epigenetic advantage in a dynamic environment derives from the immediate effect potentially in much of a population [52], the potential for an advantageous phenotype to help “bridge” a period of general low fitness by the population [40,74,75,76] and the ability for the switched phenotype to be “sunsetted” when the environmental stressor dissipates [2].

Table 1.
A comparison/contrast of the timing and reach of genetic and epigenetic inheritance.
7.2. Epigenetics in a Spatially-Complex Environment and the Exploitation of New Ecological Niches
Epigenetic transgenerational inheritance can not only be advantageous in a temporal sense, as discussed in Section 7.1, but additionally may assist spatial expansion of a species into new niches. Niche theory indicates that a given species will persist in its niche as a result of the cumulative impact of all of the factors that affect its persistence [78], and this should correctly include the potential for epigenetic phenotype switching. For example, environmentally-induced DNA methylation polymorphisms allow flower-dwelling yeasts to exploit a much broader range of niches than would otherwise be the case [78,79]. Northward range extensions in the common dandelion (Taraxacum officinale) are similarly linked to between-population variations in DNA methylation [80]. While the above examples are sufficiently compelling to warrant the expansion of this line of research, to date, the interleaving of phenotypic plasticity, epigenetic inheritance and ecological theory has been scant, at best. To return to the survey, <1% of epigenetic papers mention ecology in the title or abstract. In summary, the emerging field of “ecological epigenetics” is still in its infancy (for an introduction, see [16,81,82,83,84]), but shows great promise in understanding niche exploitation in a spatially- (and temporally-) complex environment.
8. A Role for Epigenetics in Evolution
To this point, this essay has discussed substantial qualitative and quantitative differences that emerge between genetic and epigenetic inheritance of new phenotypes. Moreover, these differences —essentially the magnitude of the effect in a population and the rapidity of the onset and dissipation of the modified phenotype—persist even when taking into account substantial genetic and epigenetic variation within a population. Inheritance is necessary for evolution, but the phenomenon of inheritance does not constitute evolution by itself. We now turn to the consideration of what the consequences of epigenetic inheritance are, if any, for evolutionary processes.
8.1. Is Epigenetics Even Relevant to Evolution?
The survey of the PubMed database reveals that the word “epigenetic” appears in <1% of all indexed “evolution papers”, and <4% of all epigenetic papers contain the word “evolution” (Figure 2B). Why so little cross-reference? One answer is that researchers may have concluded that there is no role of epigenetic inheritance in evolutionary biology. Indeed, there is a significant group of researchers that view epigenetics as uninvolved, or at least unimportant, in the process of evolution. For an introduction to this debate, see [84,85,86,87,88,89]. Yet, a key question remains largely unanswered: “To what extent, if any, do epigenetically-inherited phenotypes subsequently become fixed in the genome?” Put differently, “Do epigenetic phenomena affect evolution or does epigenetic inheritance simply co-exist independently alongside evolutionary processes, having no influence on the genotypic mix of a population”? Such questions are pivotal in the arguments for the inclusion of epigenetic inheritance into the so-called modern synthesis in evolution (see Section 8.3).
Therefore, why not assume that epigenetically-inherited characters can be fixed in the genome? Objections to inserting epigenetics into the established evolutionary paradigm often revolve around reasonable concerns involving (1) the perceived lack of substantial and compelling experimental evidence of the permanent fixation of epigenetically-inherited traits into the genome, occurring broadly across taxa and (2) no clear delineation of mechanisms by which this might occur. Thus, most discussions of the role of epigenetics and evolution appear to be based on an abundance of theory and theoretical constructs and relatively little unambiguous data. While not all evolutionary biologists immediately (or might ever) endorse a role for epigenetics in evolution, the current author concludes on largely (but not entirely) theoretical grounds that epigenetics most certainly does influence the evolution of a population, at least indirectly, though not necessarily in the way that many in the debate might think, as will now be considered.
8.2. How Epigenetic Inheritance Influences Evolution
8.2.1. Beyond the Modern Evolutionary Synthesis
Many evolutionary biologists have embraced the notion that epigenetics plays a role in evolution, while many have yet to make that conclusion. Consequently, a swirling evolutionary discourse (in fact, a controversy) has developed about modern evolutionary theory and the need for new, extended or rehashed evolutionary synthesis. A key part of that discourse is how transgenerational epigenetics factors into inheritance and evolution, with emerging concepts and terms, such as “soft inheritance”, “soft genome”, “cell memory”, “epigenetic marks” and “cultural transmission” being inserted into more traditional evolutionary theory, as represented by the modern synthesis [18,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109].
An in-depth immersion in the as-yet-unresolved debate about the role of epigenetics in shaping evolutionary theory is beyond the scope of this essay. Noteworthy in passing is that, perhaps because of the classic biological training paradigm that closely links inheritance with evolution, there is a tendency by some to assume that evolution has occurred, or is occurring, if a phenotype apparently becomes “fixed” in the population, as ascertained by its continual appearance over several consecutive generations. This simple litmus test for evolution can erroneously lead to the conclusion that an epigenetically-inherited phenotype has become incorporated into the genome when, in fact, the persistence of an epigenetically-inherited phenotype can result from continued inheritance of epigenetic markers over numerous generations. Consider the zebrafish exposed to polycyclic aromatic hydrocarbons, such as benzo-a-pyrene. The morphological effects (pericardial edema) may not disappear until the F3 of F4 generation (and may not even appear in the F1 generation) [42]. In a few more extreme, yet illustrative examples, epigenetically-inherited phenotypes caused by RNA interference persist across as many as 50 generations in Caenorhabditis elegans [34,58,110,111]. In the plant toadflax, epigenetically-inherited phenotypes are said to persist across possibly hundreds of generations [59]. Thus, the mere persistence of a modified phenotype across as many generations as most investigators have the patience to wait for is in and of itself insufficient evidence for the fixation of epigenetically-modified traits into the genome. Yet, there is evidence and theoretical arguments for both the direct and indirect involvement of epigenetics altering the genome, as will now be considered.
8.2.2. Direct Incorporation of an Epigenetically-Inherited Phenotype into the Genome
Numerous papers have been published over the last several decades that have analyzed how genomic DNA methylation has changed (or remained stable) over evolutionary time scales. Typically, these studies reveal the functional specificity of species-specific DNA methylation, correlate of evolutionary DNA methylation signatures, document extensive phylogenetic conservation of DNA methylation mechanisms and show paramutation through RNA interference. The phenomenon of paramutation—where directed allelic interactions create heritable changes in the state of an allele—is being intensively studied in plants [50,112,113] for its implications to crop productivity, for example.
Collectively, then, compelling evidence points to epigenetic influences on speciation with both short- and long-term evolutionary implications [19,50,54,112,114,115,116,117,118]. The fascinating topic of the influence of epigenetics in speciation is beyond the scope of this essay; for a comprehensive review of that literature, see [107].
Confounding the determination of the role of epigenetics in evolution and speciation is the key question of whether epigenetically-acquired phenotypic traits themselves can actually become fixed intact, i.e. permanently added to the genome, as opposed to having the markers that generate them have indirect influence on the subsequent genome through genome destabilization, mutation, natural selection or other mechanisms. Mechanisms by which this might occur are emerging, but evidence remains somewhat circumstantial [4,19,27,119]. A considerable amount of attention is focused on methylated CpG islands, which are considered mutational “hotspots”, because the methylated cytosine can undergo spontaneous conversion to thymine, while methylated guanine converts to uracil in these islands. These mutations, not necessarily recognized as damaged DNA, are not subject to excision or correction by DNA repair mechanisms [19,120]. Consequently, the mutations become fixed in the genome. While interspecific differences in amino acid sequence have been attributed to CpG hypermutability in primates [121,122,123], it remains unclear as yet, however, whether CpG mutation is actually linked to the creation of phenotypes that actually alter fitness and, so, contribute to evolution and speciation.
8.2.3. Indirect Epigenetic Effects on Evolution: Altered Gene Stability and Mutation Rates
Beyond the direct incorporation of an epigenetically-inherited phenotype into the genome lie multiple indirect effects of environmentally-induced epigenetic markers upon gene stability and mutation rates. Elevated levels of cytosine methylation increase mutation frequencies as a result of elevated levels of cytosine-to-thymine transitions and other DNA characteristics [4,19,106,107,124,125,126]. Additionally, DNA methylation can interfere with both DNA damage repair genes [127,128] and apoptotic pathways that help maintain gene integrity [129]. While these last two phenomena have been examined largely in the intragenerational context of human disease, they do lead to permanent genetic change. As such, these changes along with altered mutation rates from cytosine methylation dynamics can all contribute to evolutionary changes through their indirect effects on gene stability.
Important to emphasize is that these epigenetic effects on evolution are categorized as indirect because the action of the epigenetic markers generated by environmental stressors is not necessarily related (and most likely unrelated) to the actual phenotype that the markers created.
8.2.4. Indirect Epigenetic Effects on Evolution: Altered Natural Selection
A second indirect effect of epigenetic inheritance is the creation of a phenotype that leads to differential natural selection of genotypes [6,53,54,56,114,115,130,131,132]. Such a process is, of course, evolutionary in nature and leads to permanent shifts in the genetic makeup of a population. A classic example of this is the so-called “Baldwin effect”, in which an animal’s ability to learn new behaviors (which we know can be epigenetically inherited) may affect its success in reproducing (e.g., [19,133].
To further develop the notion of indirect influences on evolution by epigenetically-driven natural selection, consider the following “knowns”:
- (1)
- Phenotypes can be epigenetically inherited. Abundant evidence of epigenetic inheritance exists in numerous organisms, as already discussed.
- (2)
- Epigenetically-inherited phenotypes can be neutral, adaptive or maladaptive. Again, this has been discussed above in the context of epigenetically-inherited disease, for example.
- (3)
- Variation in genotype exists between members of a population. This is, of course, a basic tenet of Mendelian inheritance, although fitness is increasingly viewed as a complex combination of population density, genotype and genotype frequencies [98].
- (4)
- Variation in epigenetic markers exists between members of a population. There is increasing appreciation that there is variation between individuals within a population (and in an individual as a function of age) in the DNA methylation or post-translational acetylation, methylation, phosphorylation or ubiquitination of histones associated with the nucleosome cores and the phenotypes they ultimately produce; e.g., [53,55,64,78,81,106,125,132,134,135,136,137,138,139,140,141,142,143,144]. There are also spontaneous changes in DNA methylation patterns [89,145]. Thus, both the epigenome and genome of a population show heterogeneity to a greater or lesser extent.
Perhaps most critically, consider that:
- (5)
- Within a population, a particular set of epigenetic markers may be distributed completely independently from a particular genotype. That is, a particular genotype in a population may or may not be associated with a particular set of epigenetic markers and the associated phenotype.
If all points above hold, then epigenetics unequivocally affects evolution through its indirect action on the process of the natural selection of genotypes. Importantly, it is not required for the epigenetically-inherited phenotype to become fixed into the genotype to affect the evolution of traits. Rather, what drives the changing genotype of a population is the differential survival of individuals due to unevenly-distributed epigenetic markers creating advantageous or disadvantageous phenotypes that are subject to natural selection. Figure 5 illustrates this point. In Scenario 1, the parental generation is exposed to an environmental stressor, resulting in epigenetic inheritance of a maladaptive epigenetic marker (X) in the F1 generation. The F1 bearing this marker, which creates an inferior phenotype, happens in Scenario 1 to be evenly distributed between individuals of Genotype 1 (G1) and Genotype 2 (G2). Selection against the F1 occurs evenly because of the presence of the epigenetic marker in both genotypes, so the proportion of the G1 and G2 genotypes is not changed across time in the population. In Scenario 2, more G1 individuals happen to have the disadvantageous epigenetic marker and its associated inferior phenotype than G1s. This results in a selective disadvantage for G1, which then decreases in the overall F2 population as a result. In Scenario 3, the situation is reversed, with initially more G2 individuals bearing the disadvantageous epigenetic marker X. Consequently, G2 is selected against and declines in the overall F2 population through natural selection on disadvantaged individuals.
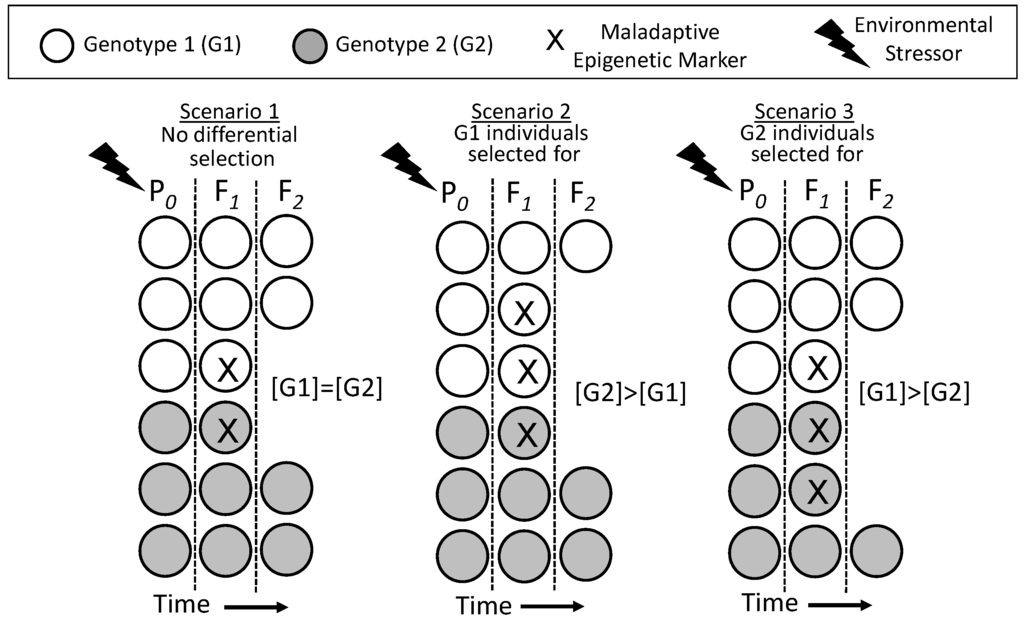
Figure 5.
Changes in the distribution of genotype within a population as a result of differential selection against individuals with a maladaptive epigenetically-inherited phenotype (X) arising in the F1 generation. An environmental stressor experienced by the P0 generation induces maladaptive epigenetic markers in the F1 generation, and the individuals with the maladaptive phenotype are selected against. Importantly, the distribution of epigenetic markers in the F1 in Scenarios 2 and 3 is both variable and independent of genotype. See the text for an additional discussion.
8.3. Traditional Genetic Inheritance of Mechanisms for Epigenetic Inheritance
Inherited traits are often thought of as discrete structures, processes, behaviors, etc. Yet, the ability for phenotypic switching is also a heritable trait. For example, well documented is the inheritance of phenotypic plasticity, including developmental phenotypic plasticity and critical windows (e.g., [73,146,147,148,149]). What about the inheritance of an “epigenetic potential”? Somewhat ironically, especially given the continuing discourse involving the Modern and extended evolutionary syntheses discussed in Section 4, traditional genetics must be involved in the process of the creation and persistence in populations as a result of epigenetic inheritance. Indeed, several authors have opined that the mechanisms by which epigenetic inheritance and phenotypic switching occurs are themselves selected for and evolve over time [74,89,114,140,150,151,152]. A natural extension of this conjecture is that some populations of organisms may carry in their genes a lesser or greater ability to inherit a phenotype by epigenetic transgenerational transfer. Numerous studies have shown population differences in, for example, the degree of DNA methylation between populations of the same species in microbes [78,81], plants [136,153] and animals (including humans) [137,142,143]. While there is undoubtedly a stochastic component to such variation (e.g., [154,155,156]), variation in epigenetic markers between populations may be both evidence of and a mechanism for natural selection acting on the capability of organisms to show epigenetic phenotypic switching. How do these emerging phenomena play into a rethinking of the Hardy–Weinberg law, which states that allele and genotype frequencies remain constant in a population over generations in the absence of other evolutionary influences? Epigenetic studies on inheritance, natural selection and evolution and the phenomenon of paramutation collectively indicate that transgenerational epigenetic inheritance needs to be factored into the venerable Hardy–Weinberg law.
9. Conclusions and A Possible Future for Epigenetic Research
The epigenetic juggernaut is gaining momentum and is likely to touch most if not all aspects of biology in the coming years. While the importance of epigenetics in human disease is being intensively studied (especially intragenerational aspects), the role that epigenetics plays in inheritance and beyond to evolution is receiving much less attention, even though the implications may be far reaching. The importance of epigenetic inheritance in natural selection and evolution is not yet fully understood, and few definitive experimental studies have explored actual fitness enhancements over time in individuals and populations with epigenetically-inherited modified phenotypes, which by their nature are transient. Theoretical considerations, some of which have been explored in this essay, argue for the importance of thoroughly integrating epigenetic phenomena into the process of evolution. Whether this requires another theoretical synthesis is perhaps less important than aggressively moving forward to add actual experimental data to the few existing studies operating at the nexus of epigenetics, natural selection and evolution.
Acknowledgments
I thank John Torday for the invitation to participate in this special issue of Biology. Ben Dubansky, Keith Maggert and Pamella Padilla provided many useful comments on the manuscript. Support for this manuscript was provided for in part by NSF Operating Grant IOS-1543301.
Conflicts of Interest
The author declares no conflict of interest.
References and Notes
- Waddington, C.H. The epigenotype. Endeavour 1942, 1, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Burggren, W.W.; Crews, D. Epigenetics in comparative biology: Why we should pay attention. Integr. Comp. Biol. 2014, 54, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.E.; Skinner, M.K. Environmentally induced epigenetic transgenerational inheritance of reproductive disease. Biol. Reprod. 2015, 93. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K.; Guerrero-Bosagna, C.; Haque, M.M. Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations. Epigenetics 2015, 10, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K. Role of epigenetics in developmental biology and transgenerational inheritance. Birth. Defects. Res. C. Embryo Today 2011, 93, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K. Environmental Epigenetics and a Unified Theory of the Molecular Aspects of Evolution: A Neo-Lamarckian Concept that Facilitates Neo-Darwinian Evolution. Genome Biol. Evol. 2015, 7, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.H.; Burggren, W.W. Epigenetics and transgenerational transfer: A physiological perspective. J. Exp. Biol. 2010, 213, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Crews, D.; Gillette, R.; Miller-Crews, I.; Gore, A.C.; Skinner, M.K. Nature, nurture and epigenetics. Mol. Cell. Endocrinol. 2014, 398, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Relton, L.R.; Smith, G.D. Is epidemiology ready for epigenetics? Int. J. Epidemiol. 2012, 41, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Recio, O.; Toro, M.A.; Bach, A. Past, present, and future of epigenetics applied to livestock breeding. Front. Genet. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Deans, C.; Maggert, K.A. What do you mean, "epigenetic"? Genetics 2015, 199, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Mann, J.R. Epigenetics and memigenetics. Cell Mol. Life Sci. 2014, 71, 1117–1122. [Google Scholar] [CrossRef] [PubMed]
- Tammen, S.A.; Friso, S.; Choi, S.W. Epigenetics: The link between nature and nurture. Mol. Aspects Med. 2013, 34, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, mechanisms and clinical perspective. Semin. Reprod. Med. 2009, 27, 351–357. [Google Scholar]
- Guerrero-Bosagna, C.M.; Skinner, M.K. Epigenetic transgenerational effects of endocrine disruptors on male reproduction. Semin. Reprod. Med. 2009, 27, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, K.J.; von Holdt, B.M.; Sork, V.L. Epigenetics in ecology and evolution: What we know and what we need to know. Mol. Ecol. 2016. [Google Scholar] [CrossRef]
- Martos, S.N.; Tang, W.Y.; Wang, Z. Elusive inheritance: Transgenerational effects and epigenetic inheritance in human environmental disease. Prog. Biophys. Mol. Biol. 2015, 118, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Gissis, S.; Jablonka, E. Trarnsformations of Lamarckism; MIT Press: Cambridge, MA, USA, 2011; pp. 1–457. [Google Scholar]
- Bateson, P. The impact of the organism on its descendants. Genet. Res. Int. 2012. [Google Scholar] [CrossRef] [PubMed]
- Popper, K. Objective Knowledge: An evolutionary approach; Oxford University Press: Oxford, UK, 1979. [Google Scholar]
- Nelson, T.C.; Groth, K.D.; Sotherland, P.R. Maternal investment and nutrient use affect phenotype of American alligator and domestic chicken hatchlings. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2010, 157, 19–27. [Google Scholar]
- Reed, W.L.; Clark, M.E. Beyond maternal effects in birds: Responses of the embryo to the environment. Integr. Comp. Biol. 2011, 51, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.H. Transgenerational epigenetics: The role of maternal effects in cardiovascular development. Integr. Comp. Biol. 2014, 54, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Seemann, F.; Peterson, D.R.; Witten, P.E.; Guo, B.S.; Shanthanagouda, A.H.; Ye, R.R.; Zhang, G.; Au, D.W. Insight into the transgenerational effect of benzo[a]pyrene on bone formation in a teleost fish (Oryzias latipes). Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 2015, 178, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Clarke, H.J.; Vieux, K.F. Epigenetic inheritance through the female germ-line: The known, the unknown, and the possible. Semin. Cell Dev. Biol. 2015, 43, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Jinha, A.E. Article 50 million: An estimate of the number of scholarly articles in existince. Learn. Publ. 2010, 23, 258–263. [Google Scholar] [CrossRef]
- Prokopuk, L.; Western, P.S.; Stringer, J.M. Transgenerational epigenetic inheritance: Adaptation through the germline epigenome? Epigenomics 2015, 7, 829–846. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Chillaron, J.C.; Ramon-Krauel, M.; Ribo, S.; Diaz, R. Transgenerational epigenetic inheritance of diabetes risk as a consequence of early nutritional imbalances. Proc. Nutr. Soc. 2016, 75, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Not all would agree with these taxonomic choices of search terms, but, again, the purpose of this meta-analysis is to determine relative frequencies and patterns of various taxa in epigenetic studies, not to provide a comprehensive quantitative analysis.
- Ronsseray, S. Paramutation phenomena in non-vertebrate animals. Semin. Cell Dev. Biol. 2015, 44, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Andrewartha, S.J.; Burggren, W.W. Transgenerational variation in metabolism and life-history traits induced by maternal hypoxia in Daphnia magna. Physiol. Biochem. Zool. 2012, 85, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Twyman, R.M.; Vilcinskas, A. Insects as models to study the epigenetic basis of disease. Prog. Biophys. Mol. Biol. 2015, 118, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Schatten, H.; Sun, Q.Y. Environmental epigenetic inheritance through gametes and implications for human reproduction. Hum. Reprod. Update 2015, 21, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Padilla, P.A.; Garcia, A.M.; Ladage, M.L.; Toni, L.S. Caenorhabditis elegans: An old genetic model can learn new epigenetic tricks. Integr. Comp. Biol. 2014, 54, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Perfus-Barbeoch, L.; Castagnone-Sereno, P.; Reichelt, M.; Fneich, S.; Roquis, D.; Pratx, L.; Cosseau, C.; Grunau, C.; Abad, P. Elucidating the molecular bases of epigenetic inheritance in non-model invertebrates: The case of the root-knot nematode Meloidogyne incognita. Front. Physiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, S.; Thompson, M.B.; Seebacher, F. Energetic cost of a meal in a frequent feeding lizard. Comp. Biochem. Physiol. 2003, 135, 377–382. [Google Scholar] [CrossRef]
- Iglesias, F.M.; Cerdan, P.D. Maintaining Epigenetic Inheritance During DNA Replication in Plants. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Sun, B.; Looi, L.; Xu, Y.; Gan, E.S.; Huang, J.; Ito, T. Co-ordination of Flower Development Through Epigenetic Regulation in Two Model Species: Rice and Arabidopsis. Plant Cell Physiol. 2015, 56, 830–842. [Google Scholar] [CrossRef] [PubMed]
- Jobson, M.A.; Jordan, J.M.; Sandrof, M.A.; Hibshman, J.D.; Lennox, A.L.; Baugh, L.R. Transgenerational Effects of Early Life Starvation on Growth, Reproduction, and Stress Resistance in Caenorhabditis elegans. Genetics 2015, 201, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Burggren, W.W. Epigenetics as a source of variation in comparative animal physiology—or—Lamarck is lookin’ pretty good these days. J. Exp. Biol. 2014, 217, 682–689. [Google Scholar] [CrossRef] [PubMed]
- Burggren, W.W. Dynamics of epigenetic phenomena: Intergenerational and intragenerational phenotype 'washout'. J. Exp. Biol. 2015, 218, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Corrales, J.; Thornton, C.; White, M.; Willett, K.L. Multigenerational effects of benzo[a]pyrene exposure on survival and developmental deformities in zebrafish larvae. Aquat. Toxicol. 2014, 148, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Saab, B.J.; Mansuy, I.M. Neurobiological disease etiology and inheritance: An epigenetic perspective. J. Exp. Biol. 2014, 217, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Vaiserman, A.M. Epigenetic engineering and its possible role in anti-aging intervention. Rejuvenation Res. 2008, 11, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Felling, R.J.; Song, H. Epigenetic mechanisms of neuroplasticity and the implications for stroke recovery. Exp. Neurol. 2015, 268, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Goddard, M.E.; Whitelaw, E. The use of epigenetic phenomena for the improvement of sheep and cattle. Front Genet. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Bilichak, A.; Kovalchuk, I. Transgenerational response to stress in plants and its application for breeding. J. Exp. Bot. 2016, 67, 2081–2092. [Google Scholar] [CrossRef] [PubMed]
- López-Arredondo, D.; González-Morales, S.I.; Bello-Bello, E.; Alejo-Jacuinde, G.; Herrera, L. Engineering food crops to grow in harsh environments. F1000Res. 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sun, Y.; Shen, K.; Sun, S.; Wang, J.; Jiang, T.; Cao, S.; Josiah, S.M.; Pang, J.; Lin, X.; et al. Immediate Genetic and Epigenetic Changes in F1 Hybrids Parented by Species with Divergent Genomes in the Rice Genus (Oryza). PLoS ONE 2015, 10, e0132911. [Google Scholar] [CrossRef] [PubMed]
- Springer, N.M.; McGinnis, K.M. Paramutation in evolution, population genetics and breeding. Semin. Cell Dev. Biol. 2015, 44, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Martinez Bautista, N.; Burggren, W.W.; University of North Texas, Denton, TX, USA. Unpublished data. 2016.
- Rando, O.J.; Verstrepen, K.J. Timescales of genetic and epigenetic inheritance. Cell 2007, 128, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Klironomos, F.D.; Berg, J.; Collins, S. How epigenetic mutations can affect genetic evolution: Model and mechanism. Bioessays 2013, 35, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.A.; Martin, M.D.; Nguyen, M.; Mendelson, T.C. Epigenetic divergence as a potential first step in darter speciation. Mol. Ecol. 2016, 25, 1883–1894. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C.M.; Medrano, M.; Bazaga, P. Comparative spatial genetics and epigenetics of plant populations: Heuristic value and a proof of concept. Mol. Ecol. 2016, 25, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Flatscher, R.; Frajman, B.; Schönswetter, P.; Paun, O. Environmental heterogeneity and phenotypic divergence: Can heritable epigenetic variation aid speciation? Genet. Res. Int. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Eichten, S.R.; Briskine, R.; Song, J.; Li, Q.; Swanson-Wagner, R.; Hermanson, P.J.; Waters, A.J.; Starr, E.; West, P.T.; Tiffin, F.; et al. Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 2013, 25, 2783–2797. [Google Scholar] [CrossRef] [PubMed]
- Vastenhouw, N.L.; Brunschwig, K.; Okihara, K.L.; Müller, F.; Tijsterman, M.; Plasterk, R.H. Gene expression: Long-term gene silencing by RNAi. Nature 2006, 442. [Google Scholar] [CrossRef] [PubMed]
- Cubas, P.; Vincent, C.; Coen, E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 1999, 401, 157–161. [Google Scholar] [PubMed]
- Richards, C.L.; Bossdorf, O.; Pigliucci, M. What role does heritable epigenetic variation play in phenotypic evolution? BioScience 2010, 60, 232–237. [Google Scholar] [CrossRef]
- Hartl, D.L.; Clark, A.G. Principles of Population Genetics; Sinauer Associates: Sunderland, MA, USA, 2007. [Google Scholar]
- Dubin, M.J.; Zhang, P.; Meng, D.; Remigereau, M.S.; Osborne, E.J.; Paolo, C.F.; Drewe, P.; Kahles, A.; Jean, G.; Vilhjálmsson, B.; et al. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. Elife 2015, 4, e05255. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Arcelus, M.; Lappalainen, T.; Montgomery, S.B.; Buil, A.; Ongen, H.; Yurovsky, A.; Bryois, J.; Giger, T.; Romano, L.; Planchon, A.; et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife 2013, 2, e00523. [Google Scholar] [PubMed]
- Schmitz, R.J.; Schultz, M.D.; Urich, M.A.; Nery, J.R.; Pelizzola, M.; Libiger, O.; Alix, A.; McCosh, R.B.; Chen, H.; Schork, N.J.; et al. Patterns of population epigenomic diversity. Nature 2013, 495, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Brandvain, Y.; Wright, S.I. The Limits of Natural Selection in a Nonequilibrium World. Trends Genet. 2016, 32, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Taute, K.M.; Gude, S.; Nghe, P.; Tans, S.J. Evolutionary constraints in variable environments, from proteins to networks. Trends Genet. 2014, 30, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Bell, G. Fluctuating selection: The perpetual renewal of adaptation in variable environments. Philos. Trans. R. Soc. Lond. B. 2010, 365, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Angers, B.; Castonguay, E.; Massicotte, R. Environmentally induced phenotypes and DNA methylation: How to deal with unpredictable conditions until the next generation and after. Mol. Ecol. 2010, 19, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Bonduriansky, R.; Crean, A.J.; Day, T. The implications of nongenetic inheritance for evolution in changing environments. Evol. Appl. 2012, 5, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Dolinoy, D.C.; Huang, D.; Jirtle, R.L. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. USA 2007, 104, 13056–13061. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Geng, Y.; Li, B.; Chen, J.; Yang, J. Genome-wide DNA methylation alterations of Alternanthera philoxeroides in natural and manipulated habitats: Implications for epigenetic regulation of rapid responses to environmental fluctuation and phenotypic variation. Plant Cell Environ. 2010, 33, 1820–1827. [Google Scholar] [CrossRef] [PubMed]
- Soubry, A. Epigenetic inheritance and evolution: A paternal perspective on dietary influences. Prog. Biophys. Mol. Biol. 2015, 118, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Jablonka, E. Epigenetic inheritance and plasticity: The responsive germline. Prog. Biophys. Mol. Biol. 2013, 111, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Badyaev, A.V. Epigenetic resolution of the 'curse of complexity' in adaptive evolution of complex traits. J. Physiol. 2014, 592, 2251–2260. [Google Scholar] [CrossRef] [PubMed]
- Roux, F.; Colomé-Tatché, M.; Edelist, C.; Wardenaar, R.; Guerche, P.; Hospital, F.; Colot, V.; Jansen, R.C.; Johannes, F. Genome-wide epigenetic perturbation jump-starts patterns of heritable variation found in nature. Genetics 2011, 188, 1015–1017. [Google Scholar] [CrossRef] [PubMed]
- De Vos, M.G.; Poelwijk, F.J.; Battich, N.; Ndika, J.D.; Tans, S.J. Environmental dependence of genetic constraint. PLoS Genet. 2013, 9, e1003580. [Google Scholar] [PubMed]
- Ho, D.H.; Burggren, W.W. Parental hypoxic exposure confers offspring hypoxia resistance in zebrafish (Danio rerio). J. Exp. Biol. 2012, 215, 4208–4216. [Google Scholar] [CrossRef] [PubMed]
- Schrey, A.W.; Richards, C.L. Within-genotype epigenetic variation enables broad niche width in a flower living yeast. Mol. Ecol. 2012, 21, 2559–2561. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C.M.; Pozo, M.I.; Bazaga, P. Jack of all nectars, master of most: DNA methylation and the epigenetic basis of niche width in a flower-living yeast. Mol. Ecol. 2012, 21, 2602–2616. [Google Scholar] [CrossRef] [PubMed]
- Preite, V.; Snoek, L.B.; Oplaat, C.; Biere, A.; van der Putten, W.H.; Verhoeven, K.J.F. The epigenetic footprint of poleward range-expanding plants in apomictic dandelions. Mol. Ecol. 2015, 24, 4406–4418. [Google Scholar] [CrossRef] [PubMed]
- Kilvitis, H.J.; Alvarez, M.; Foust, C.M.; Schrey, A.W.; Robertson, M.; Richards, C.L. Ecological epigenetics. Adv. Exp. Med. Biol. 2014, 781, 191–210. [Google Scholar] [PubMed]
- Robertson, M.; Richards, C. Opportunities and challenges of next-generation sequencing applications in ecological epigenetics. Mol. Ecol. 2015, 24, 3799–3801. [Google Scholar] [CrossRef] [PubMed]
- Richards, E.J. Population epigenetics. Curr. Opin. Genet. Dev. 2008, 18, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Bossdorf, O.; Richards, C.L.; Pigliucci, M. Epigenetics for ecologists. Ecol. Lett. 2008, 11, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, J. The epigenetics heretic. Science 2014, 343, 361–363. [Google Scholar] [CrossRef] [PubMed]
- Heard, E.; Martienssen, R.A. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 2014, 157, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Jablonka, E.; Lamb, M.J.; Zeligowski, A. Evolution in Four Dimensions: Genetics, Epigenetic, Behavioral, and Symbolic Variation in the History of Life; MIT Press: Cambridge, MA, USA, 2006; pp. 1–472. [Google Scholar]
- Smith, G.D. Epidemiology, epigenetics and the 'Gloomy Prospect': Embracing randomness in population health research and practice. Int. J. Epidemiol. 2011, 40, 537–562. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Colot, V. Epialleles in plant evolution. Genome Biol. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Stotz, K. Extended evolutionary psychology: The importance of transgenerational developmental plasticity. Front. Psychol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Dickins, T.E.; Rahman, Q. The extended evolutionary synthesis and the role of soft inheritance in evolution. Proc. Biol. Sci. 2012, 279, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Danchin, E.; Charmantier, A.; Champagne, F.A.; Mesoudi, A.; Pujol, B.; Blanchet, S. Beyond DNA: Integrating inclusive inheritance into an extended theory of evolution. Nat. Rev. Genet. 2011, 12, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Danchin, E.; Pocheville, A. Inheritance is where physiology meets evolution. J. Physiol. 2014, 592, 2307–2317. [Google Scholar] [CrossRef] [PubMed]
- Laland, K.N.; Uller, T.; Feldman, M.W.; Sterelny, K.; Müller, G.B.; Moczek, A.; Jablonka, E.; John Odling-Smee, J. The extended evolutionary synthesis: Its structure, assumptions and predictions. Proc. Biol. Sci. 2015, 282. [Google Scholar] [CrossRef] [PubMed]
- Boero, F. From Darwin's Origin of Species toward a theory of natural history. F1000Prime Rep. 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Noble, D. Evolution beyond neo-Darwinism: A new conceptual framework. J. Exp. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Tricker, P.J. Transgenerational inheritance or resetting of stress-induced epigenetic modifications: Two sides of the same coin. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Reznick, D. Hard and Soft Selection Revisited: How Evolution by Natural Selection Works in the Real World. J. Hered. 2016, 107, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Pigliucci, M. An extended synthesis for evolutionary biology. Ann. N. Y. Acad. Sci. 2009, 1168, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Holland, M.L.; Rakyan, V.K. Transgenerational inheritance of non-genetically determined phenotypes. Biochem. Soc. Trans. 2013, 41, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Anava, S.; Posner, R.; Rechavi, O. The soft genome. Worm 2014, 3, e989798. [Google Scholar] [CrossRef] [PubMed]
- Moya, C.; Boyd, R.; Henrich, J. Reasoning About Cultural and Genetic Transmission: Developmental and Cross-Cultural Evidence From Peru, Fiji, and the United States on How People Make Inferences About Trait Transmission. Top. Cogn. Sci. 2015, 7, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Torday, J.S. On the evolution of development. Trends Dev. Biol. 2014, 8, 17–37. [Google Scholar] [PubMed]
- Laland, K.; Uller, T.; Feldman, M.; Sterelny, K.; Müller, G.B.; Moczek, A.; Jablonka, E.; Odling-Smee, J.; Wray, G.A.; Hoekstra, H.E.; et al. Does evolutionary theory need a rethink? Nature 2014, 514, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Day, T.; Bonduriansky, R. A unified approach to the evolutionary consequences of genetic and nongenetic inheritance. Am. Nat. 2011, 178, E18–E36. [Google Scholar] [CrossRef] [PubMed]
- Hauser, M.T.; Aufsatz, W.; Jonak, C.; Luschnig, C. Transgenerational epigenetic inheritance in plants. Biochim. Biophys. Acta 2011, 1809, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Mendizabal, I.; Keller, T.E.; Zeng, J.; Yi, S.V. Epigenetics and evolution. Integr. Comp. Biol. 2014, 54, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S. The decline of soft inheritance. In Transformations of Lamarckism; Gissis, S., Jablonka, E., Eds.; MIT Press: Cambridge, MA, USA, 2011; pp. 121–126. [Google Scholar]
- Wilkins, A. Why did the Modern Synthesis give short shrift to "Soft Inheritance"? In Transformations of Lamarckism; Gissis, S., Jablonka, E., Eds.; MIT Press: Cambridge, MA, USA, 2011; pp. 127–132. [Google Scholar]
- Katz, D.J.; Edwards, T.M.; Reinke, V.; Kelly, W.G. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 2009, 137, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Rechavi, O.; Houri-Ze'evi, L.; Anava, S.; Goh, W.S.; Kerk, S.Y.; Hannon, G.J.; Hobert, O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 2014, 158, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Pilu, R. Paramutation phenomena in plants. Semin. Cell Dev. Biol. 2015, 44, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell. Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Boffelli, D.; Martin, D.I. Epigenetic inheritance: A contributor to species differentiation? DNA Cell Biol. 2012, 31, S11–S16. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.D.; O'Neill, R.J. Chromosomes, conflict, and epigenetics: Chromosomal speciation revisited. Annu. Rev. Genomics Hum. Genet. 2010, 11, 291–316. [Google Scholar] [CrossRef] [PubMed]
- Rebollo, R.; Horard, B.; Hubert, B.; Vieira, C. Jumping genes and epigenetics: Towards new species. Gene 2010, 454, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R. The inheritance of epigenetic defects. Science 1987, 238, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Bonduriansky, R.; Day, T. Nongenetic inheritance and the evolution of costly female preference. J. Evol. Biol. 2013, 26, 76–87. [Google Scholar] [PubMed]
- Rehan, V.K.; Liu, J.; Naeem, E.; Tian, J.; Sakurai, R.; Kwong, K.; Akbari, O.; Torday, J.S. Perinatal nicotine exposure induces asthma in second generation offspring. BMC Med. 2012, 10. [Google Scholar] [CrossRef] [PubMed]
- Schorderet, D.F.; Gartler, S.M. Analysis of CpG suppression in methylated and nonmethylated species. Proc. Natl. Acad. Sci. USA 1992, 89, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Misawa, K.; Kamatani, N.; Kikuno, R.F. The universal trend of amino acid gain-loss is caused by CpG hypermutability. J. Mol. Evol. 2008, 67, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Mugal, C.F.; Arndt, P.F.; Holm, L.; Ellegren, H. Evolutionary consequences of DNA methylation on the GC content in vertebrate genomes. G3 (Bethesda) 2015, 5, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Borstnik, B.; Pumpernik, D. The apparent enhancement of CpG transversions in primate lineage is a consequence of multiple replacements. J. Bioinform. Comput. Biol. 2014, 12. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980, 8, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Dubrovina, A.S.; Kiselev, K.V. Age-associated alterations in the somatic mutation and DNA methylation levels in plants. Plant Biol. (Stuttg) 2016, 18, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumari, R.; Sharma, V.; Sharma, V. Roles, and establishment, maintenance and erasing of the epigenetic cytosine methylation marks in plants. J. Genet. 2013, 92, 629–666. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.T.; Kim, D.K.; Cockayne, D.A.; Belousov, A.; Bitter, H.; Cho, M.H.; Duvoix, A.; Edwards, L.D.; Lomas, D.A.; Miller, B.E.; et al. Systemic soluble receptor for advanced glycation endproducts is a biomarker of emphysema and associated with AGER genetic variants in patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2013, 188, 948–957. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Cao, Y.; Qin, J.; Song, X.; Zhang, Q.; Shi, Y.; Cao, L. DNA methylation, its mediators and genome integrity. Int. J. Biol. Sci. 2015, 11, 604–617. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.X.; Hackett, J.A.; Nestor, C.; Dunican, D.S.; Madej, M.; Reddington, J.P.; Pennings, S.; Harrison, D.J.; Meehan, R.R. Apoptosis and DNA methylation. Cancers 2011, 3, 1798–1820. [Google Scholar] [CrossRef] [PubMed]
- Putiri, E.L.; Robertson, K.D. Epigenetic mechanisms and genome stability. Clin. Epigenetics 2011, 2, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Zhu, B.; Li, M.; Wang, L.; Xu, L.; Zhang, H.; Zheng, S.; Qi, B.; Han, F.; Liu, B. Extensive and heritable epigenetic remodeling and genetic stability accompany allohexaploidization of wheat. Genetics 2011, 188, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Fischer, M.; Colot, V.; Bossdorf, O. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol. 2013, 197, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Sznajder, B.; Sabelis, M.W.; Egas, M. How Adaptive Learning Affects Evolution: Reviewing Theory on the Baldwin Effect. Evol. Biol. 2012, 39, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Sun, K.; Jiang, T.; Feng, J. Natural epigenetic variation in bats and its role in evolution. J. Exp. Biol. 2015, 218, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Tabansky, I.; Stern, J.N.; Pfaff, D.W. Implications of Epigenetic Variability within a Cell Population for "Cell Type" Classification. Front Behav. Neurosci. 2015, 9. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.; Pérez, R.; Bazaga, P.; Medrano, M.; Herrera, C.M. Individual variation in size and fecundity is correlated with differences in global DNA cytosine methylation in the perennial herb Helleborus foetidus (Ranunculaceae). Am. J. Bot. 2014, 101, 1309–1313. [Google Scholar] [CrossRef] [PubMed]
- Oey, H.; Isbel, L.; Hickey, P.; Ebaid, B.; Whitelaw, E. Genetic and epigenetic variation among inbred mouse littermates: Identification of inter-individual differentially methylated regions. Epigenetics Chromatin. 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Lévesque, M.L.; Casey, K.F.; Szyf, M.; Ismaylova, E.; Ly, V.; Verner, M.P.; Suderman, M.; Brendgen, M.; Vitaro, F.; Dionne, G.; et al. Genome-wide DNA methylation variability in adolescent monozygotic twins followed since birth. Epigenetics 2014, 9, 1410–1421. [Google Scholar] [CrossRef] [PubMed]
- Lemmers, R.J.; Goeman, J.J.; van der Vliet, P.J.; van Nieuwenhuizen, M.P.; Balog, J.; Vos-Versteeg, M.; Camano, P.; Ramos Arroyo, M.A.; Jerico, I.; Rogers, M.T.; et al. Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum. Mol. Genet. 2015, 24, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Mathers, J.C. Session 2: Personalised nutrition. Epigenomics: A basis for understanding individual differences? Proc. Nutr. Soc. 2008, 67, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.B.; Abdallah, B.Y.; Horne, S.D.; Liu, G.; Bremer, S.W.; Heng, H.H. Genetic and Epigenetic heterogeneity in Cancer; John Wiley and Sons Ltd.: Chichester, UK, 2011. [Google Scholar]
- Heyn, H.; Moran, S.; Hernando-Herraez, I.; Sergi Sayols, S.; Gomez, A.; Sandoval, J.; Monk, D.; Hata, K.; Marques-Bonet, T.; Wang, L.; et al. DNA methylation contributes to natural human variation. Genome Res. 2013, 23, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Whitelaw, N.C.; Whitelaw, E. How lifetimes shape epigenotype within and across generations. Hum. Mol. Genet. 2006, 15, R131–R137. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.J.; Tricker, P.J. Epigenomic plasticity within populations: Its evolutionary significance and potential. Heredity (Edinb) 2010, 105, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Van der Graaf, A.; Wardenaar, R.; Neumann, D.A.; Taudt, A.; Shaw, R.G.; Jansen, R.C.; Schmitz, R.J.; Colomé-Tatché, M.; Johannes, F. Rate, spectrum, and evolutionary dynamics of spontaneous epimutations. Proc. Natl. Acad. Sci. USA 2015, 112, 6676–6681. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, T.W.; Frankenhuis, W.E. Adaptive explanations for sensitive windows in development. Front Zool. 2015, 12. [Google Scholar] [CrossRef] [PubMed]
- Carretero, A.; Ditrich, H.; Pérez-Aparicio, F.J.; Splechtna, H.; Ruberte, J. Development and degeneration of the arterial system in the mesonephros and metanephros of chicken embryos. Anat. Rec. 1995, 243, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Kopp, M.; Matuszewski, S. Rapid evolution of quantitative traits: Theoretical perspectives. Evol. Appl. 2014, 7, 169–191. [Google Scholar] [CrossRef] [PubMed]
- Diaz, S.A.; Viney, M. The evolution of plasticity of dauer larva developmental arrest in the nematode Caenorhabditis elegans. Ecol. Evol. 2015, 5, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, S.; Baumberger, R.; Grossniklaus, U. Epigenetic variation, inheritance, and selection in plant populations. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Herraez, I.; Garcia-Perez, R.; Sharp, A.J.; Marques-Bonet, T. DNA Methylation: Insights into Human Evolution. PLoS Genet. 2015, 11, e1005661. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X. Comparative epigenomics: A powerful tool to understand the evolution of DNA methylation. New Phytol. 2016, 210, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Kooke, R.; Keurentjes, J.J. Epigenetic variation contributes to environmental adaptation of Arabidopsis thaliana. Plant Signal Behav. 2015, 10, e1057368. [Google Scholar] [CrossRef] [PubMed]
- Bintu, L.; Yong, J.; Antebi1, Y.E.; McCue, K.; Kazuki, Y.; Uno, N.; Oshimura, M.; Elowitz, M.B. Dynamics of epigenetic regulation at the single-cell level. Science 2016, 351, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Massah, S.; Beischlag, T.V.; Prefontaine, G.G. Epigenetic events regulating monoallelic gene expression. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 337–358. [Google Scholar] [CrossRef] [PubMed]
- Vogt, G. Stochastic developmental variation, an epigenetic source of phenotypic diversity with far-reaching biological consequences. J. Biosci. 2015, 40, 159–204. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).