Easy and Rapid Binding Assay for Functional Analysis of Disulfide-Containing Peptides by a Pull-Down Method Using a Puromycin-Linker and a Cell-Free Translation System
Abstract
:1. Introduction
2. Experimental Section
2.1. Pull-Down Method for Disulfide-Containing Peptides
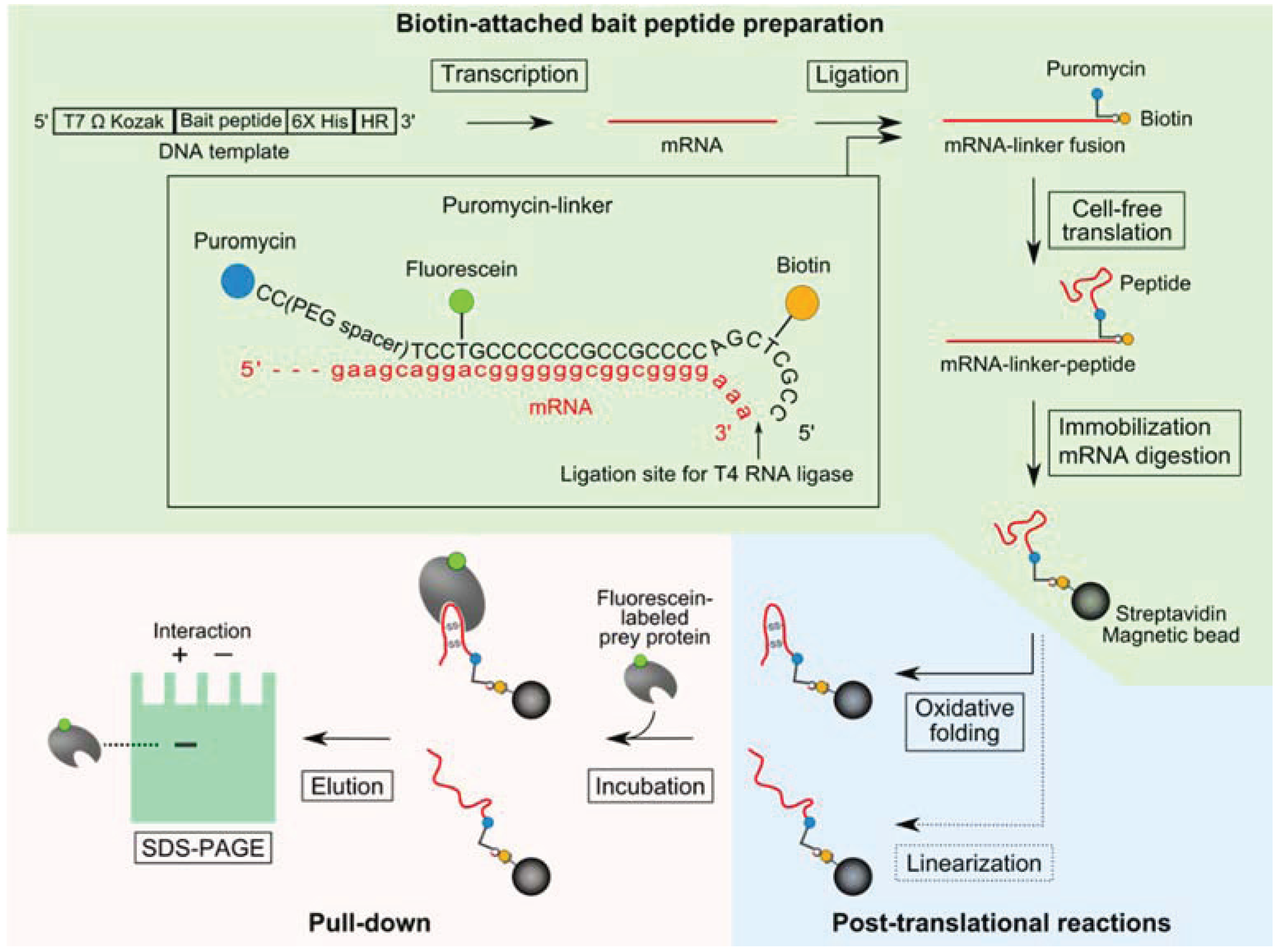
2.2. Introduction of a Chemical Cross-Linker into a Disulfide-Containing Peptide
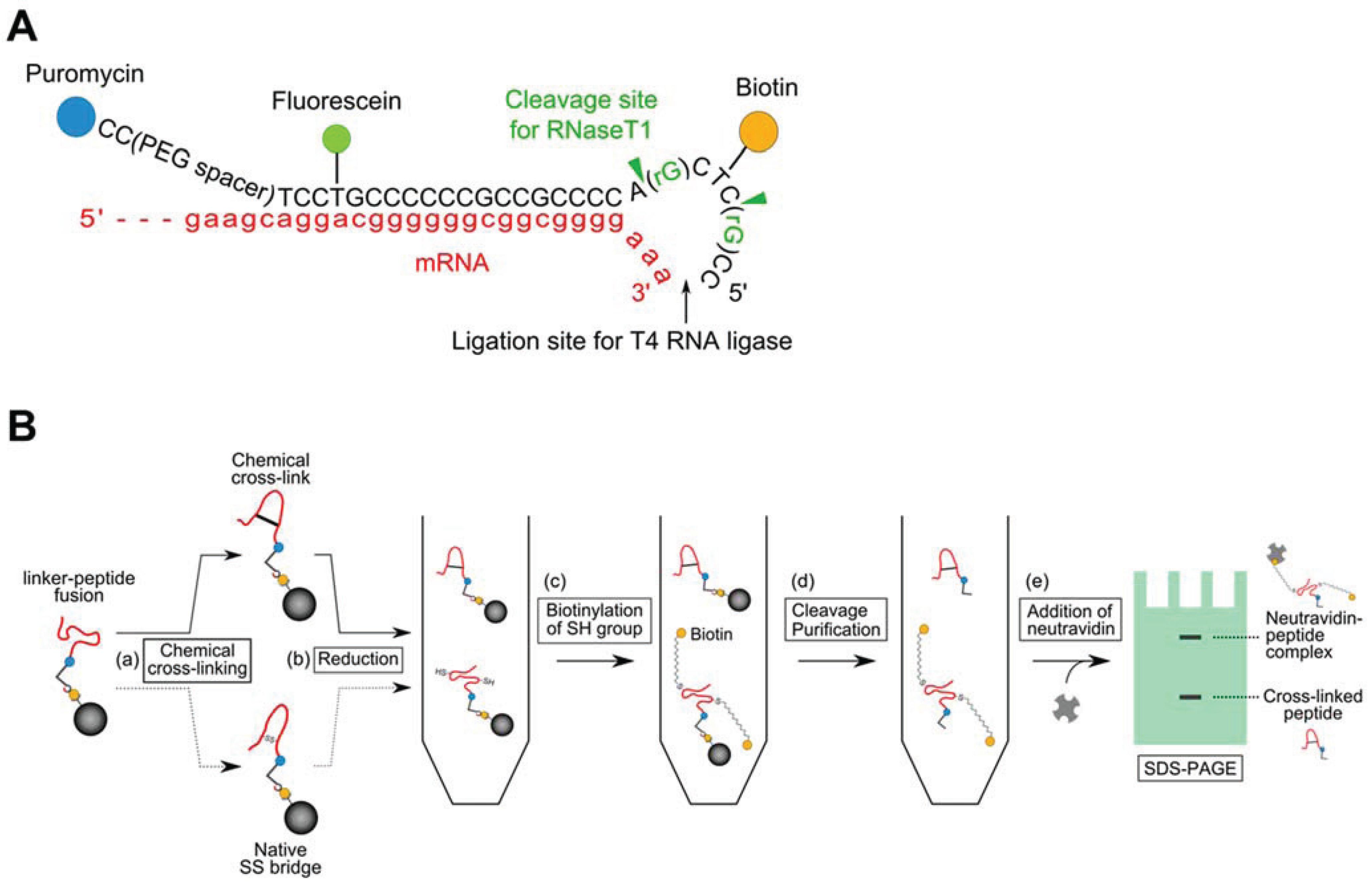
3. Results and Discussion
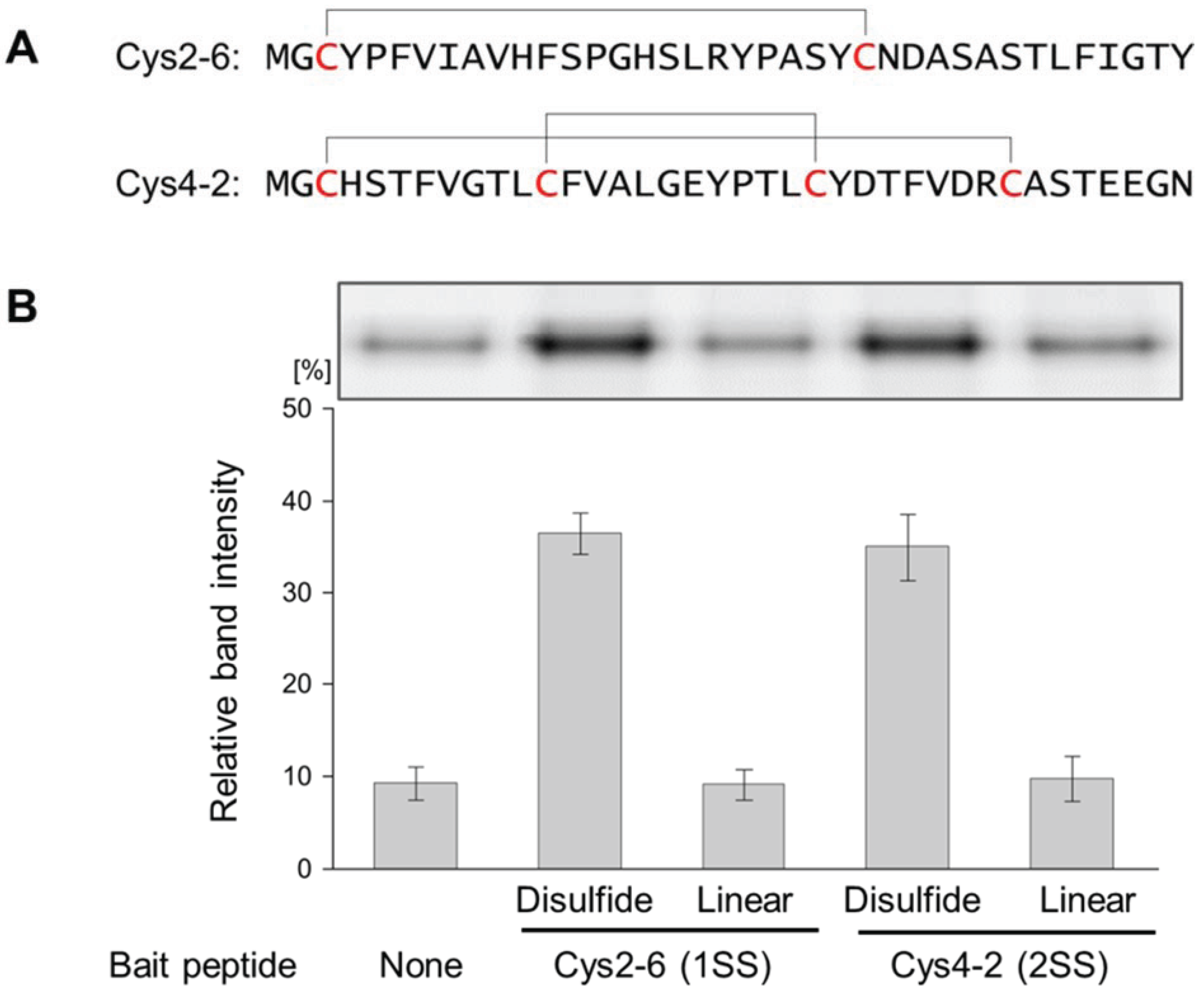
3.1. Evaluation of the Interaction between the Disulfide-Containing Peptides and the Target Protein
3.2. Evaluation of the Interaction between the Chemically cross-Linked Peptide and the Target Protein


4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013, 81, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Bockus, A.T.; McEwen, C.M.; Lokey, R.S. Form and function in cyclic peptide natural products: A pharmacokinetic perspective. Curr. Top. Med. Chem. 2013, 13, 821–836. [Google Scholar] [CrossRef] [PubMed]
- Daly, N.L.; Craik, D. Bioactive cystine knot proteins. Curr. Opin. Chem. Biol. 2011, 15, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.M.; Gonçalves, S.; Santos, N.C. Defensins: Anti-fungal lessons from eukaryotes. Front. Microbiol. 2014, 5, 97. [Google Scholar] [PubMed]
- Kolmar, H. Biological diversity and therapeutic potential of natural and engineered cystine knot miniproteins. Curr. Opin. Pharmacol. 2009, 9, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Benítez, M.G.; Puche, J.T.; Albericio, F. Multifaceted roles of disulfide bonds. Peptides as Therapeutics. Chem. Rev. 2014, 114, 901–926. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, J.R.; Lewis, R.J.; Dutertre, S. Towards an integrated venomics approach for accelerated conopeptide discovery. Toxicon 2012, 60, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Hernández, V.Q.; Ortiz, E.; Anaya, M.R.; Schwartz, E.F.; Becerril, B.; Corzo, G.; Possani, L.D. Scorpion and spider venom peptides: Gene cloning and peptide expression. Toxicon 2011, 58, 644–663. [Google Scholar] [CrossRef] [PubMed]
- Quimbar, P.; Malik, U.; Sommerhoff, C.P.; Kaas, Q.; Chan, L.Y.; Huang, Y.H.; Grundhuber, M.; Dunse, K.; Craik, D.J.; Anderson, M.A.; et al. High-affinity cyclic peptide matriptase inhibitors. J. Biol. Chem. 2013, 288, 13885–13896. [Google Scholar] [CrossRef] [PubMed]
- Gunasekera, S.; Foley, F.M.; Clark, R.J.; Sando, L.; Fabri, L.J.; Craik, D.J.; Daly, N.L. Engineering stabilized vascular endothelial growth factor-A antagonists: Synthesis, structural characterization, and bioactivity of grafted analogues of cyclotides. J. Med. Chem. 2008, 51, 7697–7704. [Google Scholar] [CrossRef] [PubMed]
- Krause, S.; Schmoldt, H.U.; Wentzel, A.; Ballmaier, M.; Friedrich, K.; Kolmar, H. Grafting of thrombopoietin-mimetic peptides into cystine knot miniproteins yields high-affinity thrombopoietin antagonists and agonists. FEBS J. 2007, 274, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Aboye, T.L.; Ha, H.; Majumder, S.; Christ, F.; Debyser, Z.; Shekhtman, A.; Neamati, N.; Camarero, J.A. Design of a novel cyclotide-based CXCR4 antagonist with anti-human immunodeficiency virus (HIV)-1 activity. J. Med. Chem. 2012, 55, 10729–10734. [Google Scholar] [CrossRef] [PubMed]
- Zoller, F.; Haberkorn, U.; Mier, W. Miniproteins as phage display-scaffolds for clinical applications. Molecules 2011, 16, 2467–2485. [Google Scholar] [CrossRef] [PubMed]
- Zoller, F.; Markert, A.; Barthe, P.; Hebling, U.; Altmann, A.; Lindner, T.; Mier, W.; Haberkorn, U. A disulfide-constrained miniprotein with striking tumor-binding specificity developed by ribosome display. Angew. Chem. Int. Ed. 2013, 52, 11760–11764. [Google Scholar] [CrossRef]
- Nemoto, N.; Miyamoto-Sato, E.; Husimi, Y.; Yanagawa, H. In vitro virus: Bonding of mRNA bearing puromycin at the 3'-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 1997, 414, 405–408. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.W.; Szostak, J.W. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA 1997, 94, 12297–12302. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, J.; Naimuddin, M.; Biyani, M.; Sasaki, T.; Machida, M.; Kubo, T.; Funatsu, T.; Husimi, Y.; Nemoto, N. cDNA display: A novel screening method for functional disulfide-rich peptides by solid-phase synthesis and stabilization of mRNA-protein fusions. Nucl. Acids Res. 2009, 37, e108. [Google Scholar] [CrossRef] [PubMed]
- Funamoto, S.; Sasaki, T.; Ishihara, S.; Nobuhara, M.; Nakano, M.; Watanabe-Takahashi, M.; Saito, T.; Kakuda, N.; Miyasaka, T.; Nishikawa, K.; et al. Substrate ectodomain is critical for substrate preference and inhibition of γ-secretase. Nat. Commun. 2013, 4, 2529. [Google Scholar] [CrossRef] [PubMed]
- Arolas, J.L.; Aviles, F.X.; Chang, J.Y.; Ventura, S. Folding of small disulfide-rich proteins: Clarifying the puzzle. Trends Biochem. Sci. 2006, 31, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Akondi, K.B.; Muttenthaler, M.; Dutertre, S.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Discovery, synthesis, and structure-activity relationships of conotoxins. Chem. Rev. 2014, 114, 5815–5847. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, Y.; Kohno, F.; Nishigaki, K.; Nemoto, N. A pull-down method with a biotinylated bait protein prepared by cell-free translation using a puromycin linker. Anal. Biochem. 2013, 434, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Nell, K.H.; Meldal, M. Maintaining biological activity by using triazoles as disulfide bond mimetics. Angew. Chem. Int. Ed. 2011, 50, 5204–5206. [Google Scholar] [CrossRef]
- Bondebjerg, J.; Grunnet, M.; Jespersen, T.; Meldal, M. Solid-phase synthesis and biological activity of a thioether analogue of conotoxin G1. ChemBioChem 2003, 4, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Gyanda, R.; Banerjee, J.; Chang, Y.P.; Phillips, A.M.; Toll, L.; Armishaw, C.J. Oxidative folding and preparation of α-conotoxins for use in high-throughput structure-activity relationship studies. J. Pept. Sci. 2013, 19, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Gee, S.H.; Sekely, S.A.; Lombardo, C.; Kurakin, A.; Froehner, S.C.; Kay, B.K. Cyclic peptides as non-carboxyl-terminal ligands of syntrophin PDZ domains. J. Chem. Biol. 1998, 273, 21980–21987. [Google Scholar] [CrossRef]
- Mochizuki, Y.; Biyani, M.; Tsuji-Ueno, S.; Suzuki, M.; Nishigaki, K.; Husimi, Y.; Nemoto, N. One-pot preparation of mRNA/cDNA display by a novel and versatile puromycin-linker DNA. ACS Comb. Sci. 2011, 13, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Buczek, O.; Bulaj, G.; Olivera, B.M. Conotoxins and the posttranslational modification of secreted gene products. Cell. Mol. Life Sci. 2005, 62, 3067–3079. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Passioura, T.; Suga, H. Technologies for the synthesis of mRNA-encoding libraries and discovery of bioactive natural product-inspired non-traditional macrocyclic peptides. Molecules 2013, 18, 3502–3528. [Google Scholar] [CrossRef] [PubMed]
- Menegatti, S.; Hussain, M.; Naik, A.D.; Carbonell, R.G.; Rao, B.M. mRNA display selection and solid-phase synthesis of Fc-binding cyclic peptide affinity ligands. Biotechnol. Bioeng. 2013, 110, 857–870. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanemura, Y.; Mochizuki, Y.; Kumachi, S.; Nemoto, N. Easy and Rapid Binding Assay for Functional Analysis of Disulfide-Containing Peptides by a Pull-Down Method Using a Puromycin-Linker and a Cell-Free Translation System. Biology 2015, 4, 161-172. https://doi.org/10.3390/biology4010161
Tanemura Y, Mochizuki Y, Kumachi S, Nemoto N. Easy and Rapid Binding Assay for Functional Analysis of Disulfide-Containing Peptides by a Pull-Down Method Using a Puromycin-Linker and a Cell-Free Translation System. Biology. 2015; 4(1):161-172. https://doi.org/10.3390/biology4010161
Chicago/Turabian StyleTanemura, Yutaro, Yuki Mochizuki, Shigefumi Kumachi, and Naoto Nemoto. 2015. "Easy and Rapid Binding Assay for Functional Analysis of Disulfide-Containing Peptides by a Pull-Down Method Using a Puromycin-Linker and a Cell-Free Translation System" Biology 4, no. 1: 161-172. https://doi.org/10.3390/biology4010161
APA StyleTanemura, Y., Mochizuki, Y., Kumachi, S., & Nemoto, N. (2015). Easy and Rapid Binding Assay for Functional Analysis of Disulfide-Containing Peptides by a Pull-Down Method Using a Puromycin-Linker and a Cell-Free Translation System. Biology, 4(1), 161-172. https://doi.org/10.3390/biology4010161





