Three Complete Mitochondrial Genomes of Ocellarnaca (Orthoptera, Gryllacrididae) and Their Phylogenies
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Acquisition and DNA Extraction
2.2. Next-Generation Sequencing, Genomic Assembly and Analysis
2.3. Phylogenetic Analysis
3. Results and Discussion
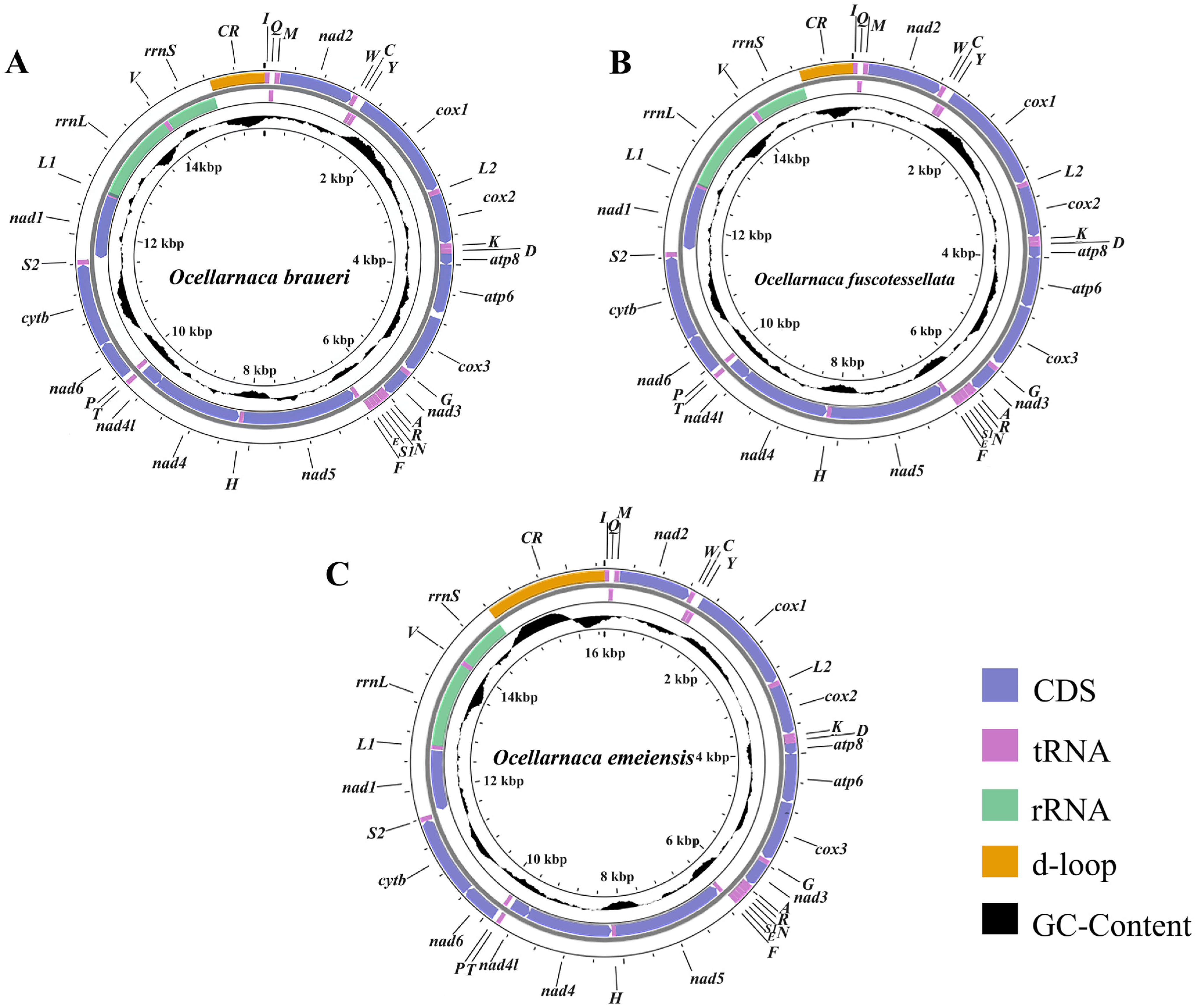
3.1. Genome Structure and Composition of the Three Raspy Cricket Species
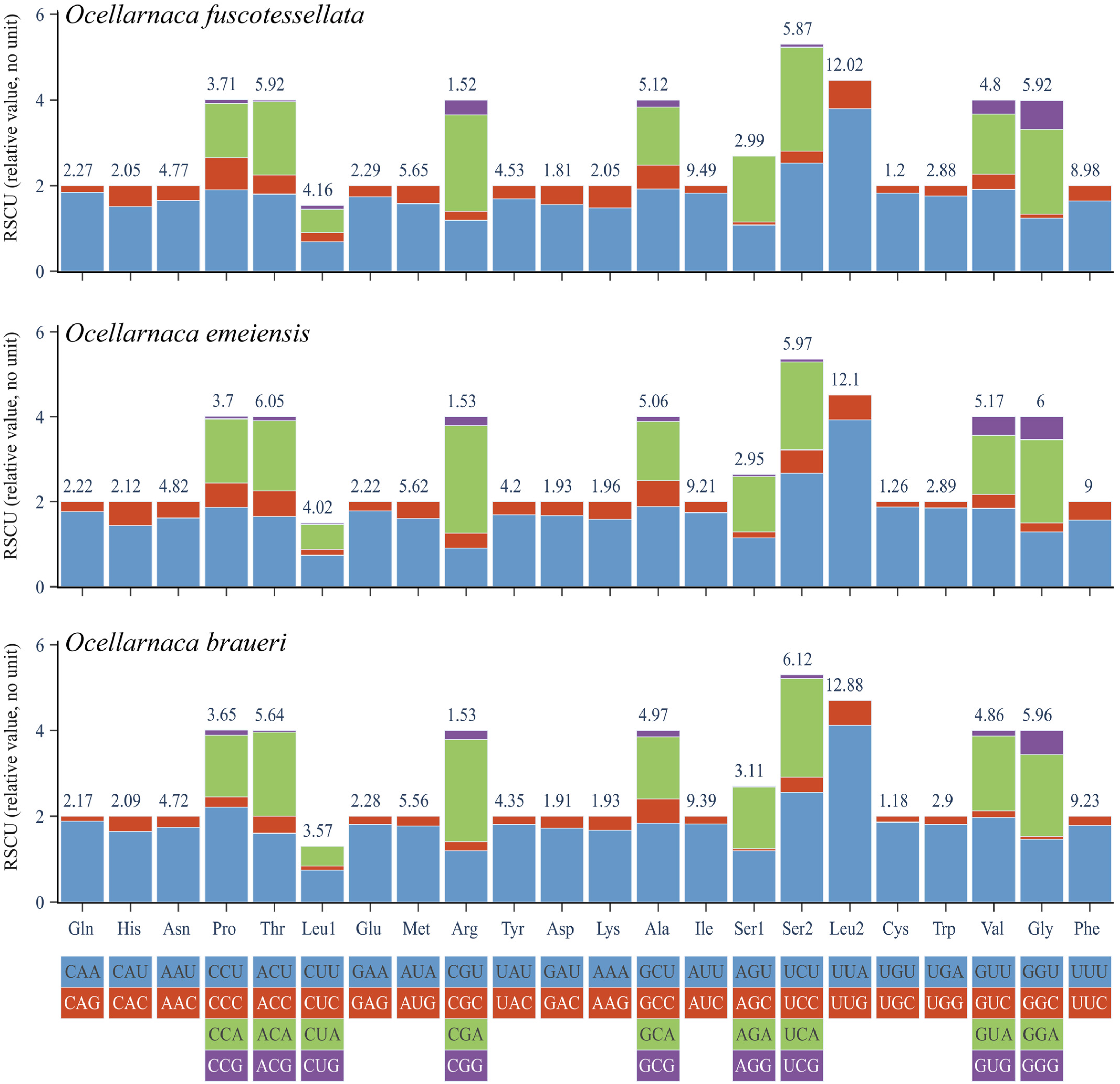
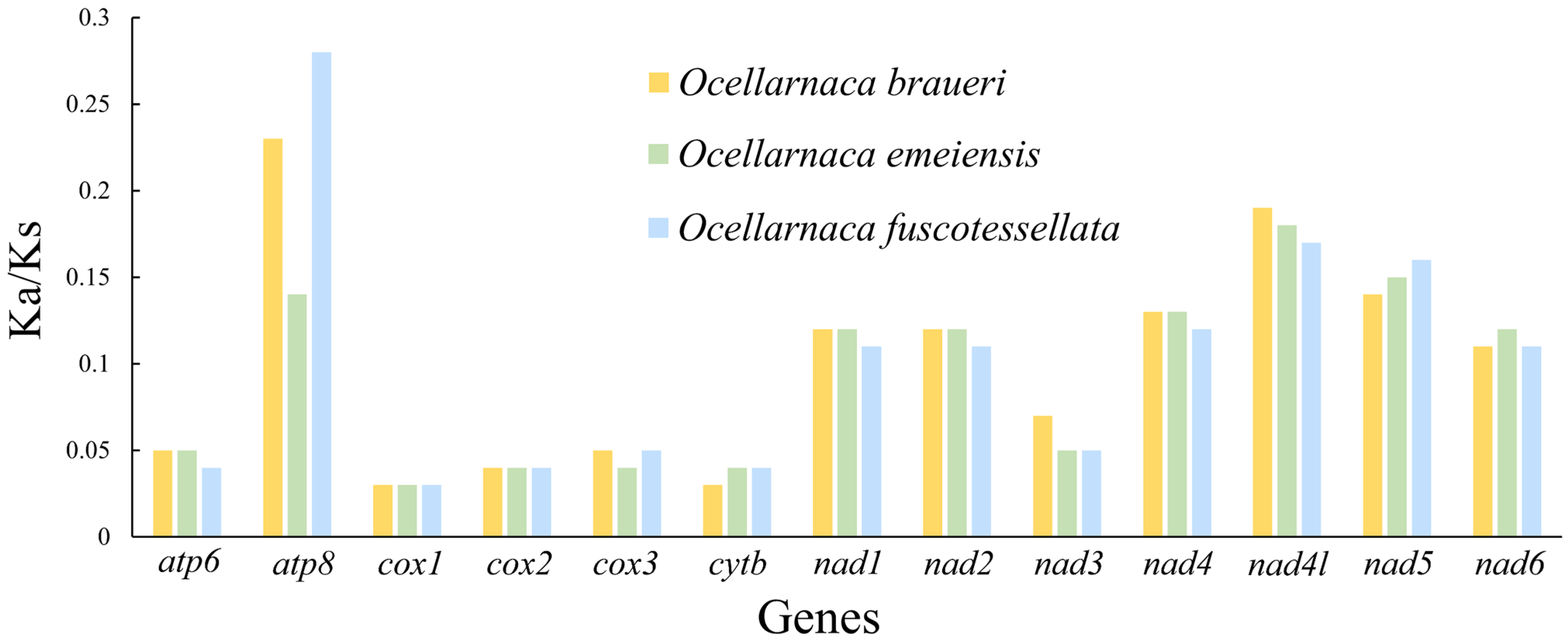
3.2. Protein-Coding Genes and Evolutionary Rates
3.3. RNAs and Control Regions
3.4. Mitochondrial Microsatellites (mtSSRs)
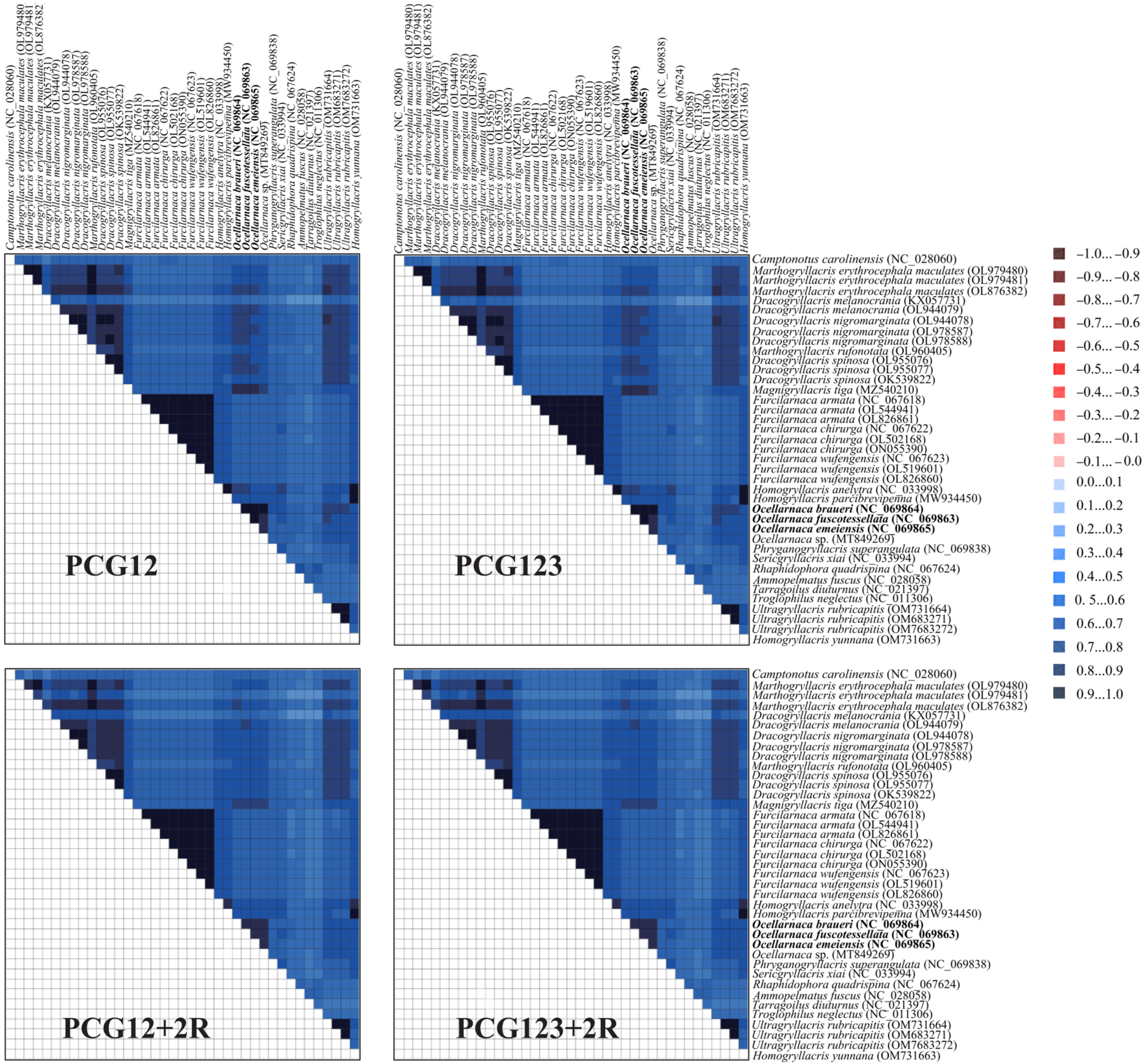
3.5. Substitution Saturation and Heterogeneity Analysis
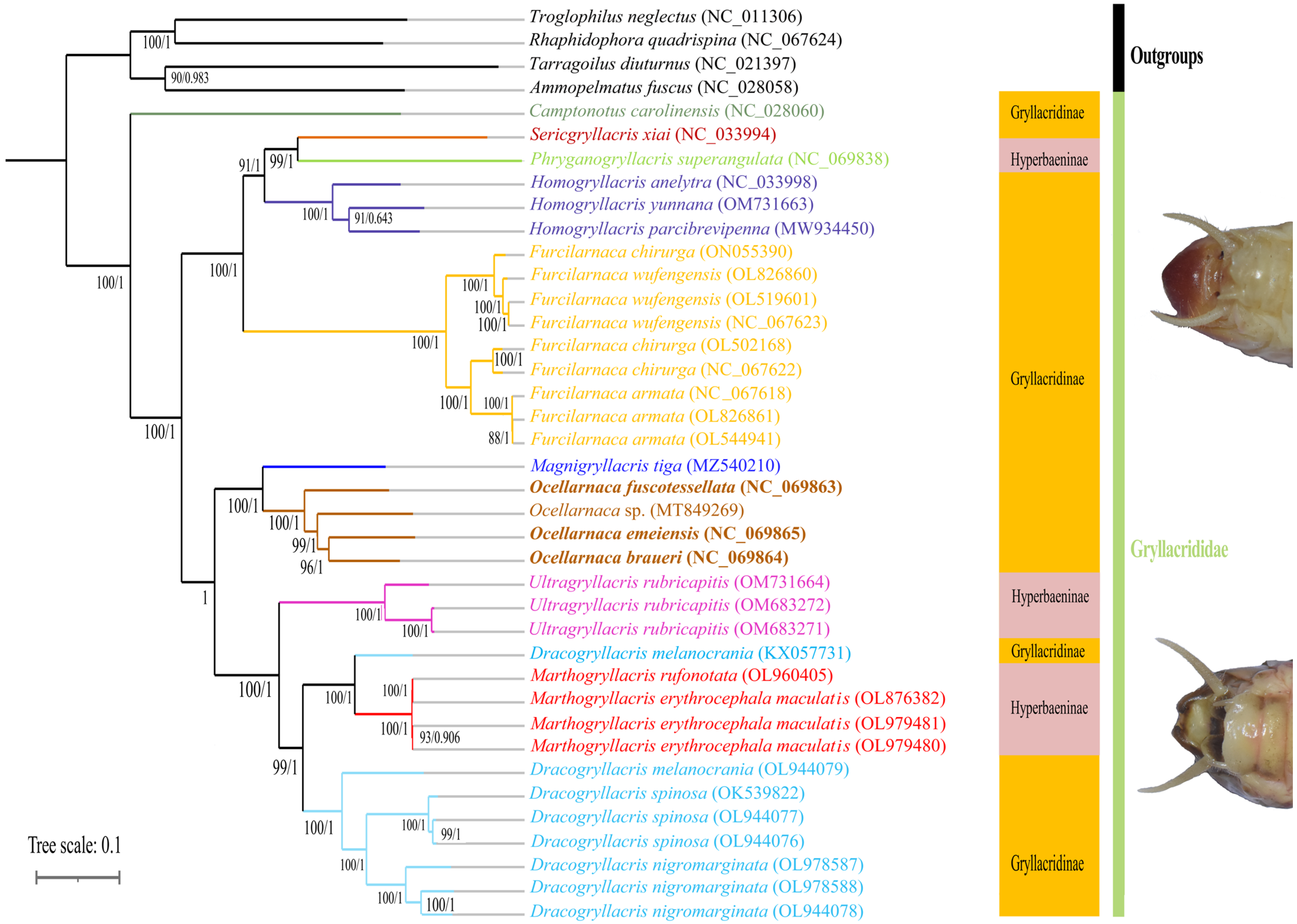
3.6. Phylogenetic Analysis
3.7. Implications for Conservation and Monitoring
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, S.Y.; Liu, Y.J.; Xu, J.Y.; Yin, Z.X.; He, Z.Q. Molecular phylogeny of Chinese raspy crickets (Orthoptera: Gryllacrididae) reveals incongruences in current classification. Zool. J. Linn. Soc.-Lond. 2024, 201, zlae051. [Google Scholar] [CrossRef]
- Cigliano, M.M.; Holger, B.; David, C.E.; Daniel, O.; Orthoptera Species File. Version 5.0/5.0. Available online: http://Orthoptera.SpeciesFile.org (accessed on 25 July 2025).
- Bian, X. Contribution to the knowledge of Chinese Gryllacrididae (Orthoptera: Ensifera: Stenopelmatoidea) XXIV: Update on the Raspy Cricket species checklist with some comments. Zootaxa 2023, 5383, 585–593. [Google Scholar] [CrossRef]
- Robinson, D.J.; Hall, M.J. Sound signalling in Orthoptera. Adv. Insect Physiol. 2002, 29, 151–278. [Google Scholar] [CrossRef]
- Andrew, A.W.; Sarah, W.; Jeffrey, S.C.; David, J.M.; Stephen, T.M.; Tara, D.S. Silk from Crickets: A New Twist on Spinning. PLoS ONE 2012, 7, e30408. [Google Scholar] [CrossRef]
- Brunner von Wattenwyl, C. Monographie der Stenopelmatiden und Gryllacriden. Verhandlungen Kais. Zool.–Bot. Ges. Wien 1888, 38, 247–394. [Google Scholar]
- Jena, F.S. Zoologische und Anthropologische Ergebnisse Einer Forschungsreise im Westlichen und Zentralen Sudafrika, Ausgefuhrt in den Jahren 1903–1905; G. Fischer: Berlin, Germany, 1909; pp. 1–52. [Google Scholar]
- Karny, H.H. Beiträge zur malayischen Orthopterenfauna VI. Die Gryllacriden des Buitenzorger Museums. Treubia 1924, 5, 19–105. [Google Scholar]
- Vandergast, A.G.; Weissman, D.B.; Wood, D.A.; Rentz, D.C.F.; Bazelet, C.S.; Ueshima, N. Tackling an intractable problem: Can greater taxon sampling help resolve relationships within the Stenopelmatoidea (Orthoptera: Ensifera)? Zootaxa 2017, 4291, 1–33. [Google Scholar] [CrossRef]
- Zhou, Z.J.; Zhao, L.; Liu, N.; Guo, H.F.; Guan, B.; Di, J.X.; Shi, F.M. Towards a higher–level Ensifera phylogeny inferred from mitogenome sequences. Mol. Phylogenet. Evol. 2017, 108, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Ingrisch, S. New taxa and records of Gryllacrididae (Orthoptera, Stenopelmatoidea) from South East Asia and New Guinea with a key to the genera. Zootaxa 2018, 4510, 1–278. [Google Scholar] [CrossRef]
- Cadena-Castañeda, O.J. A proposal towards classification of the Raspy Crickets (Orthoptera: Stenopelmatoidea: Gryllacrididae) with zoogeographical comments: An initial contribution to the higher classification of the Gryllacridines. Zootaxa 2019, 4605, 1–100. [Google Scholar] [CrossRef]
- Gorochov, A.V. The Families Stenopelmatidae and Anostostomatidae (Orthoptera). 1. Higher Classification, New and Little Known Taxa. Entomol. Rev. 2020, 100, 1106–1151. [Google Scholar] [CrossRef]
- Behar, D.M.; van Oven, M.; Rosset, S.; Metspalu, M.; Loogväli, E.L.; Silva, N.M.; Kivisild, T.; Torroni, A.; Villems, R.A. “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 2012, 90, 675–684. [Google Scholar] [CrossRef]
- Guo, W.H.; Grewe, F.; Fan, W.S.; Young, G.J.; Knoop, V.; Palmer, J.D.; Mower, J.P. Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol. Bio Evol. 2016, 33, 1448–1460. [Google Scholar] [CrossRef]
- Lv, F.H.; Peng, W.F.; Yang, J.; Zhao, Y.X.; Li, W.R.; Liu, M.J.; Ma, Y.H.; Zhao, Q.J.; Yang, G.L.; Wang, F.; et al. Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep. Mol. Biol. Evol. 2015, 32, 2515–2533. [Google Scholar] [CrossRef]
- Li, H.; Leavengood, J.M.; Chapman, E.G.; Burkhardt, D.; Song, F.; Jiang, P.; Liu, J.P.; Zhou, X.G.; Cai, W.Z. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc. Biol. Sci. 2017, 284, 20171223. [Google Scholar] [CrossRef]
- Kinkar, L.; Laurimäe, T.; Sharbatkhori, M.; Mirhendi, H.; Kia, E.B.; Ponce-Gordo, F.; Andresiuk, V.; Simsek, S.; Lavikainen, A.; Irshadullah, M.; et al. New mitogenome and nuclear evidence on the phylogeny and taxonomy of the highly zoonotic tapeworm Echinococcus granulosus sensu stricto. Infect. Genet. Evol. 2017, 52, 52–58. [Google Scholar] [CrossRef]
- Chen, C.; Li, Q.; Fu, R.T.; Wang, J.; Deng, G.M.; Chen, X.J.; Lu, D.H. Comparative mitochondrial genome analysis reveals intron dynamics and gene rearrangements in two Trametes species. Sci. Rep. 2010, 11, 2569. [Google Scholar] [CrossRef]
- Song, H.J.; Béthoux, O.; Shin, S.; Donath, A.; Letsch, H.; Liu, S.L.; McKenna, D.D.; Meng, G.L.; Misof, B.; Podsiadlowski, L.; et al. Phylogenomic analysis sheds light on the evolutionary pathways towards acoustic communication in Orthoptera. Nat. Commun. 2020, 11, 4939. [Google Scholar] [CrossRef]
- Liu, J.; Lu, X.Y.; Zhang, Q.W.; Wu, X.Y.; Yang, D.D.; Bian, X. Contribution to the knowledge of Chinese Gryllacrididae (Orthoptera) V: Further study on the Chinese Capnogryllacris and comment on the phylogenetic relationships of the Gryllacrididae. Zootaxa 2022, 5099, 1–45. [Google Scholar] [CrossRef]
- Lu, X.Y.; Luo, H.Y.; Zhang, Q.W.; Bian, X. Comparative mitogenome analysis and phylogenetic inference of the genus Ultragryllacris (Orthoptera: Gryllacrididae). Zootaxa 2023, 5230, 439–455. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI). Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. 1988. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 5 May 2025).
- de Freitas, K.E.J.; Busanello, C.; Viana, V.E.; Pegoraro, C.; de Carvalho Victoria, F.; da Maia, L.C.; Costa de Oliveira, A. An empirical analysis of mtSSRs: Could microsatellite distribution patterns explain the evolution of mitogenomes in plants? Funct. Integr. Genom. 2022, 22, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Kapil, A.; Shanker, A. MitoSatPlant: Mitochondrial microsatellites database of viridiplantae. Mitochondrion 2014, 19, 334–337. [Google Scholar] [CrossRef]
- Matvienko, M. Genomics Workbench and other products. Qiagen Bioinformatics Workshop at PAG 2015. Plant Anim. Genome 2015, 1, 1–42. [Google Scholar]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Julio, R.; Albert, F.; Juan, C.S.; Sara, G.; Pablo, L.; Sebastián, E.R.; Alejandro, S. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Dallagnol, L.C.; Cônsoli, F.L. Evolutionary and phylogenetic insights from the mitochondrial genomic analysis of Diceraeus melacanthus and D. furcatus (Hemiptera: Pentatomidae). Sci. Rep. 2024, 14, 12861. [Google Scholar] [CrossRef] [PubMed]
- Fenn, J.D.; Song, H.; Cameron, S.L.; Whiting, M.F. A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Mol. Phylogenet. Evol. 2008, 49, 59–68. [Google Scholar] [CrossRef]
- Lu, X.; Liu, J.; Huang, X.; Bian, X. Contribution to the Chinese subfamily Rhaphidophorinae Walker, 1869 (Orthoptera: Rhaphidophoridae: Rhaphidophorinae) IV: Seven new species of Rhaphidophora and one new mitogenome. Zootaxa 2022, 5087, 129–153. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Amédégnato, C.; Cigliano, M.M.; Desutter-Grandcolas, L.; Heads, S.W.; Huang, Y.; Otte, D.; Whiting, M.F. 300 million years of diversification: Elucidating the patterns of orthopteran evolution based on comprehensive taxon and gene sampling. Cladistics 2015, 31, 621–651. [Google Scholar] [CrossRef]
- Zhou, Z.; Shi, F.; Zhao, L. The first mitochondrial genome for the superfamily Hagloidea and implications for its systematic status in Ensifera. PLoS ONE 2014, 9, e86027. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kuck, P.; Meid, S.A.; Gross, C.; Wägele, J.W.; Misof, B. AliGROOVE-visualization of heterogeneous sequence divergence within multiple sequence alignments and detection of inflated branch support. BMC Bioinform. 2014, 15, 294. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Xiang, C.Y.; Gao, F.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. Imeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Xu, K.D.; Zhao, F.; Liu, W.Y.; Li, L.Z.; Hua, Z.Y.; Zhou, X. Itol.toolkit accelerates working with iTOL (Interactive Tree of Life) by an automated generation of annotation files. Bioinformatics 2023, 39, btad339. [Google Scholar] [CrossRef]
- Ma, C.; Li, J. Comparative analysis of mitochondrial genomes of the superfamily Grylloidea (Insecta, Orthoptera) reveals phylogenetic distribution of gene rearrangements. Int. J. Biol. Macromol. 2018, 120, 1048–1054. [Google Scholar] [CrossRef]
- Li, R.; Ying, X.; Deng, W.; Rong, W.; Li, X. Mitochondrial genomes of eight Scelimeninae species (Orthoptera) and their phylogenetic implications within Tetrigoidea. PeerJ 2021, 9, e10523. [Google Scholar] [CrossRef]
- Ma, Y.; Miao, Y. Mitogenomic Comparison of the Mole Crickets Gryllotalpidae with the Phylogenetic Implications (Orthoptera: Ensifera). Insects 2022, 13, 919. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Chen, S.; Cheng, S.; Zhou, W.; Ma, Q.; Chen, Z.; Fu, C.X.; Liu, X.; Zhao, Y.P.; Soltis, P.S.; et al. Natural selection and repeated patterns of molecular evolution following allopatric divergence. eLife 2019, 8, e45199. [Google Scholar] [CrossRef] [PubMed]
- Popadin, K.Y.; Nikolaev, S.I.; Junier, T.; Baranova, M.; Antonarakis, S.E. Purifying selection in mammalian mitochondrial protein-coding genes is highly effective and congruent with evolution of nuclear genes. Mol. Biol. Evol. 2012, 30, 347–355. [Google Scholar] [CrossRef]
- Cvijović, I.; Good, B.H.; Desai, M.M. The effect of strong purifying selection on genetic diversity. Genetics 2018, 209, 1235–1278. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Jühling, F.; Pütz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef]
- Pons, J.; Bover, P.; Bidegaray-Batista, L.; Arnedo, M.A. Arm-less mitochondrial tRNAs conserved for over 30 millions of years in spiders. BMC Genom. 2019, 20, 665. [Google Scholar] [CrossRef]
- Canto, C.; Auwerx, J. NAD+ as a signaling molecule modulating metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Ying, W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: Regulation and biological consequences. Antioxid. Redox Signal. 2008, 10, 179–206. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, R.S.; Handy, D.E.; Loscalzo, J. NAD (H) and NADP (H) redox couples and cellular energy metabolism. Antioxid. Redox Signal. 2018, 28, 251–272. [Google Scholar] [CrossRef]
- Keating, J.N.; Garwood, R.J.; Sansom, R.S. Phylogenetic congruence, conflict and consilience between molecular and morphological data. BMC Ecol. Evol. 2023, 23, 30. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.J. The role of morphological data in phylogeny reconstruction. Syst. Biol. 2004, 53, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.T.; Zhang, J.Z. Morphological and molecular convergences in mammalian phylogenetics. Nat. Commun. 2016, 7, 12758. [Google Scholar] [CrossRef]
- Whitfield, J.B.; Kjer, K.M. Ancient rapid radiations of insects: Challenges for phylogenetic analysis. Annu. Rev. Entomol. 2008, 53, 449–472. [Google Scholar] [CrossRef]
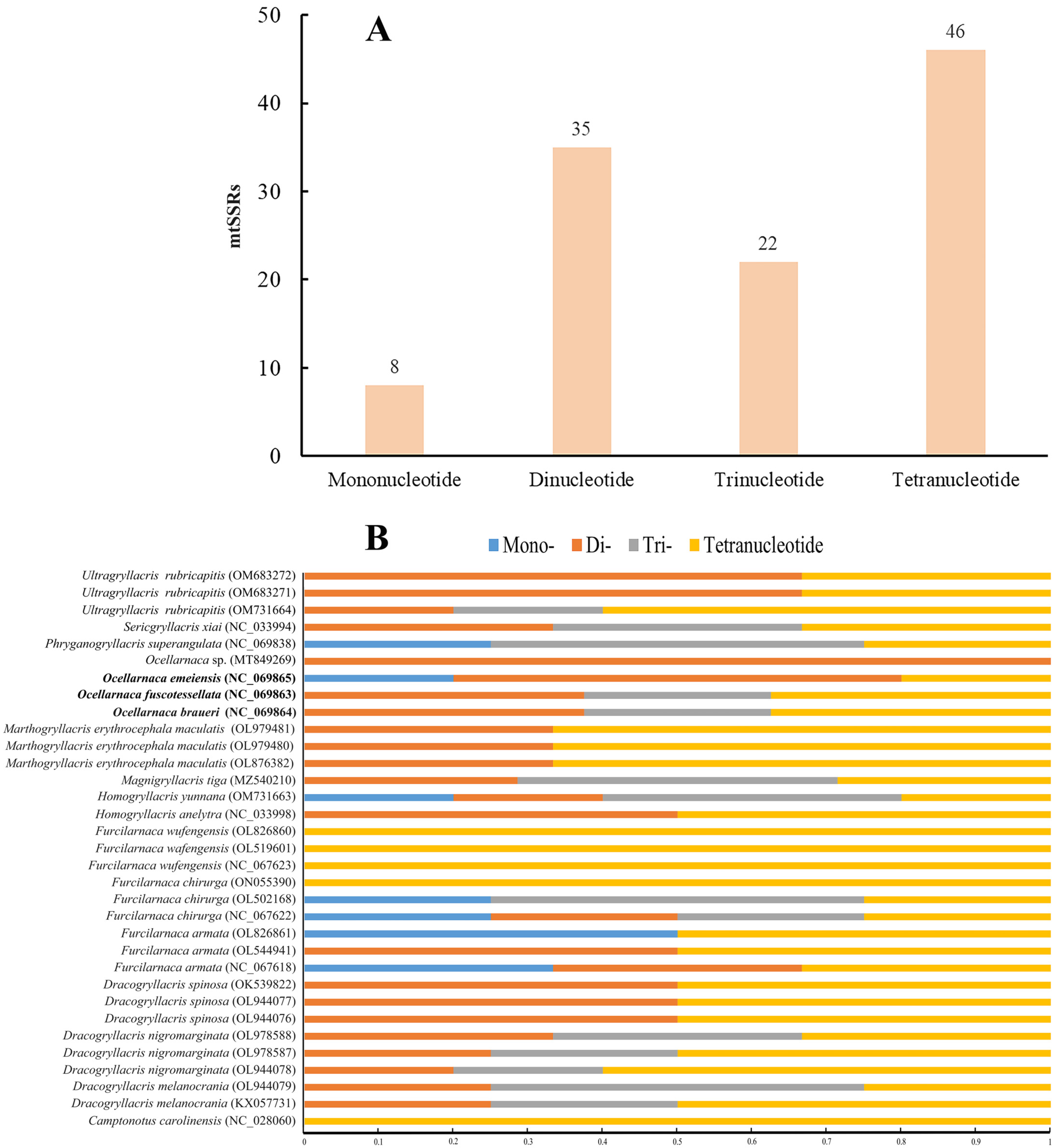

| Species | mtSSR | Repeats | Size (bp) | Start | End | Location Region |
|---|---|---|---|---|---|---|
| Ocellarnaca braueri (NC_069864) | (ATT)4 | 4 | 12 | 3876 | 3887 | IR |
| (TAA)4 | 4 | 12 | 9948 | 9959 | nad6 | |
| (TTAT)3 | 3 | 12 | 10,124 | 10,135 | nad6 | |
| (TAAA)3 | 3 | 12 | 13,847 | 13,858 | rrnL | |
| (AATA)3 | 3 | 12 | 14,577 | 14,588 | rrnS | |
| (TA)8 | 8 | 16 | 15,236 | 15,251 | CR | |
| (AT)7 | 7 | 14 | 15,476 | 15,489 | CR | |
| (AT)7 | 7 | 14 | 15,508 | 15,521 | CR | |
| Ocellarnaca emeiensis (NC_069865) | (ATTA)3 | 3 | 12 | 3830 | 3841 | trnD |
| (T)12 | 12 | 12 | 16,003 | 16,014 | CR | |
| (TA)7 | 7 | 14 | 16,103 | 16,116 | CR | |
| (AT)7 | 7 | 14 | 16,341 | 16,354 | CR | |
| (AT)6 | 6 | 12 | 16,373 | 16,384 | CR | |
| Ocellarnaca fuscotessellata (NC_069863) | (ATT)4 | 4 | 12 | 3880 | 3891 | atp8 |
| (AT)8 | 8 | 16 | 5576 | 5591 | IR | |
| (TTAT)3 | 3 | 12 | 6321 | 6332 | trnE | |
| (ATT)4 | 4 | 12 | 6407 | 6418 | nad5 | |
| (TAAA)3 | 3 | 12 | 9020 | 9031 | nad4 | |
| (TTAT)3 | 3 | 12 | 10,141 | 10,152 | nad6 | |
| (CA)7 | 7 | 14 | 11,678 | 11,691 | IR | |
| (AT)7 | 7 | 14 | 15,472 | 15,485 | CR |
| Species | Kb | Total Number of SSR | RA | Total Length of SSR (bp) | RD |
|---|---|---|---|---|---|
| Camptonotus carolinensis (NC_028060) | 15.211 | 3 | 0.197225692 | 50 | 3.287095 |
| Dracogryllacris melanocrania (KX057731) | 16.136 | 4 | 0.24789291 | 36 | 2.231036 |
| Dracogryllacris melanocrania (OL944079) | 15.711 | 4 | 0.254598689 | 24 | 1.527592 |
| Dracogryllacris nigromarginat (OL944078) | 16.639 | 5 | 0.300498828 | 36 | 2.163592 |
| Dracogryllacris nigromarginata (OL978587) | 16.218 | 4 | 0.246639536 | 50 | 3.082994 |
| Dracogryllacris nigromarginata (OL978588) | 15.776 | 3 | 0.190162272 | 12 | 0.760649 |
| Dracogryllacris spinosa (OL944076) | 15.825 | 2 | 0.126382306 | 60 | 3.791469 |
| Dracogryllacris spinosa (OL944077) | 15.817 | 2 | 0.126446229 | 104 | 6.575204 |
| Dracogryllacris spinosa (OK539822) | 16.505 | 2 | 0.121175401 | 104 | 6.301121 |
| Furcilarnaca armata (NC_067618) | 15.787 | 3 | 0.190029771 | 26 | 1.646925 |
| Furcilarnaca armata (OL544941) | 15.830 | 2 | 0.126342388 | 48 | 3.032217 |
| Furcilarnaca armata (OL826861) | 15.677 | 2 | 0.127575429 | 12 | 0.765453 |
| Furcilarnaca chirurga (NC_067622) | 15.454 | 4 | 0.258832665 | 24 | 1.552996 |
| Furcilarnaca chirurga (OL502168) | 15.441 | 4 | 0.25905058 | 12 | 0.777152 |
| Furcilarnaca chirurga (ON055390) | 15.550 | 1 | 0.064308682 | 24 | 1.543408 |
| Furcilarnaca wufengensis (NC_067623) | 15.954 | 1 | 0.062680206 | 38 | 2.381848 |
| Furcilarnaca wufengensis (OL519601) | 16.022 | 1 | 0.062414181 | 26 | 1.622769 |
| Furcilarnaca wufengensis (OL826860) | 15.478 | 1 | 0.06460783 | 26 | 1.679804 |
| Homogryllacris anelytra (NC_033998) | 15.706 | 2 | 0.12733987 | 60 | 3.820196 |
| Homogryllacris yunnana (OM731663) | 16.209 | 5 | 0.308470603 | 48 | 2.961318 |
| Magnigryllacris tiga (MZ540210) | 15.513 | 7 | 0.451234449 | 64 | 4.125572 |
| Marthogryllacris erythrocephala maculatis (OL876382) | 16.163 | 3 | 0.185609107 | 44 | 2.722267 |
| Marthogryllacris erythrocephala maculatis (OL979480) | 16.171 | 3 | 0.185517284 | 86 | 5.318162 |
| Marthogryllacris erythrocephala maculatis (OL979481) | 15.845 | 3 | 0.189334175 | 12 | 0.757337 |
| Ocellarnaca braueri (NC_069864) | 15.597 | 8 | 0.512919151 | 50 | 3.205745 |
| Ocellarnaca emeiensis (NC_069865) | 16.510 | 5 | 0.512590504 | 38 | 2.301635 |
| Ocellarnaca fuscotessellata (NC_069863) | 15.607 | 8 | 0.30284676 | 40 | 2.562953 |
| Ocellarnaca sp. (MT849269) | 16.157 | 3 | 0.185678034 | 38 | 2.351922 |
| Phryganogryllacris superangulata (NC_069838) | 15.976 | 4 | 0.250375563 | 36 | 2.253380 |
| Sericgryllacris xiai (NC_033994) | 15.876 | 3 | 0.188964475 | 38 | 2.393550 |
| Ultragryllacris rubricapitis (OM731664) | 15.558 | 5 | 0.321378069 | 66 | 4.242191 |
| Ultragryllacris rubricapitis (OM683271) | 16.625 | 3 | 0.180451128 | 60 | 3.609023 |
| Ultragryllacris rubricapitis (OM683272) | 15.766 | 3 | 0.190282887 | 12 | 0.761132 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, T.; Qin, Y.; Lu, X.; Pang, S.; Bian, X. Three Complete Mitochondrial Genomes of Ocellarnaca (Orthoptera, Gryllacrididae) and Their Phylogenies. Biology 2025, 14, 1231. https://doi.org/10.3390/biology14091231
Luo T, Qin Y, Lu X, Pang S, Bian X. Three Complete Mitochondrial Genomes of Ocellarnaca (Orthoptera, Gryllacrididae) and Their Phylogenies. Biology. 2025; 14(9):1231. https://doi.org/10.3390/biology14091231
Chicago/Turabian StyleLuo, Ting, Yanting Qin, Xiangyi Lu, Siyu Pang, and Xun Bian. 2025. "Three Complete Mitochondrial Genomes of Ocellarnaca (Orthoptera, Gryllacrididae) and Their Phylogenies" Biology 14, no. 9: 1231. https://doi.org/10.3390/biology14091231
APA StyleLuo, T., Qin, Y., Lu, X., Pang, S., & Bian, X. (2025). Three Complete Mitochondrial Genomes of Ocellarnaca (Orthoptera, Gryllacrididae) and Their Phylogenies. Biology, 14(9), 1231. https://doi.org/10.3390/biology14091231





