Characterization of the Complete Mitogenome of Polypedates braueri (Anura, Rhacophoridae, Polypedates) and Insights into the Phylogenetic Relationships of Rhacophoridae
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Specimen Collection and DNA Extraction
2.3. Primer Design and PCR Amplification
2.4. Sequence Assembly, Analysis, and Annotation
2.5. Phylogenetic Analyses
2.6. Divergence Time Estimates Focused on Rhacophoridae
2.7. Ka and Ks Analysis
3. Results
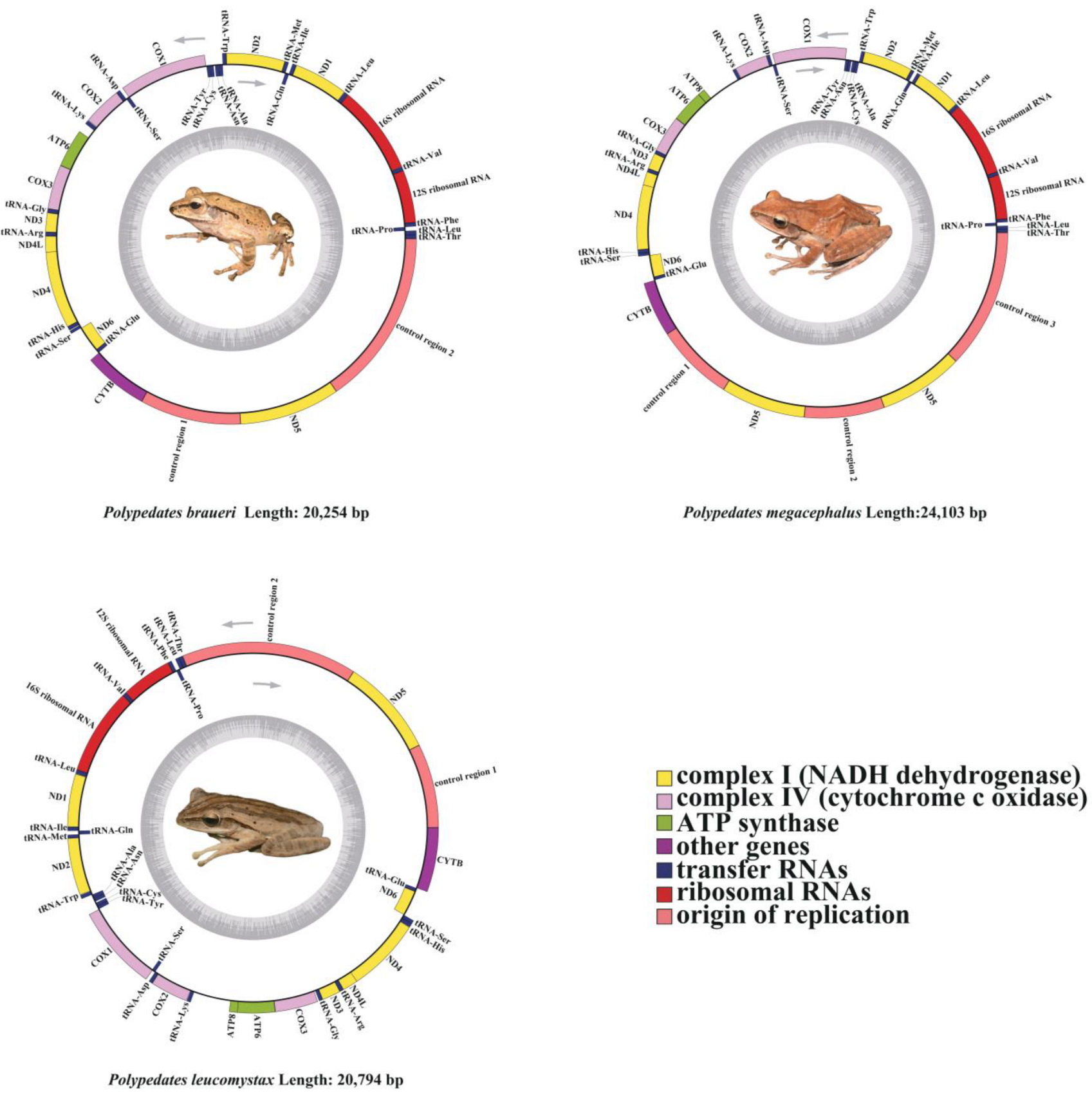
3.1. Mitochondrial Genome Structure
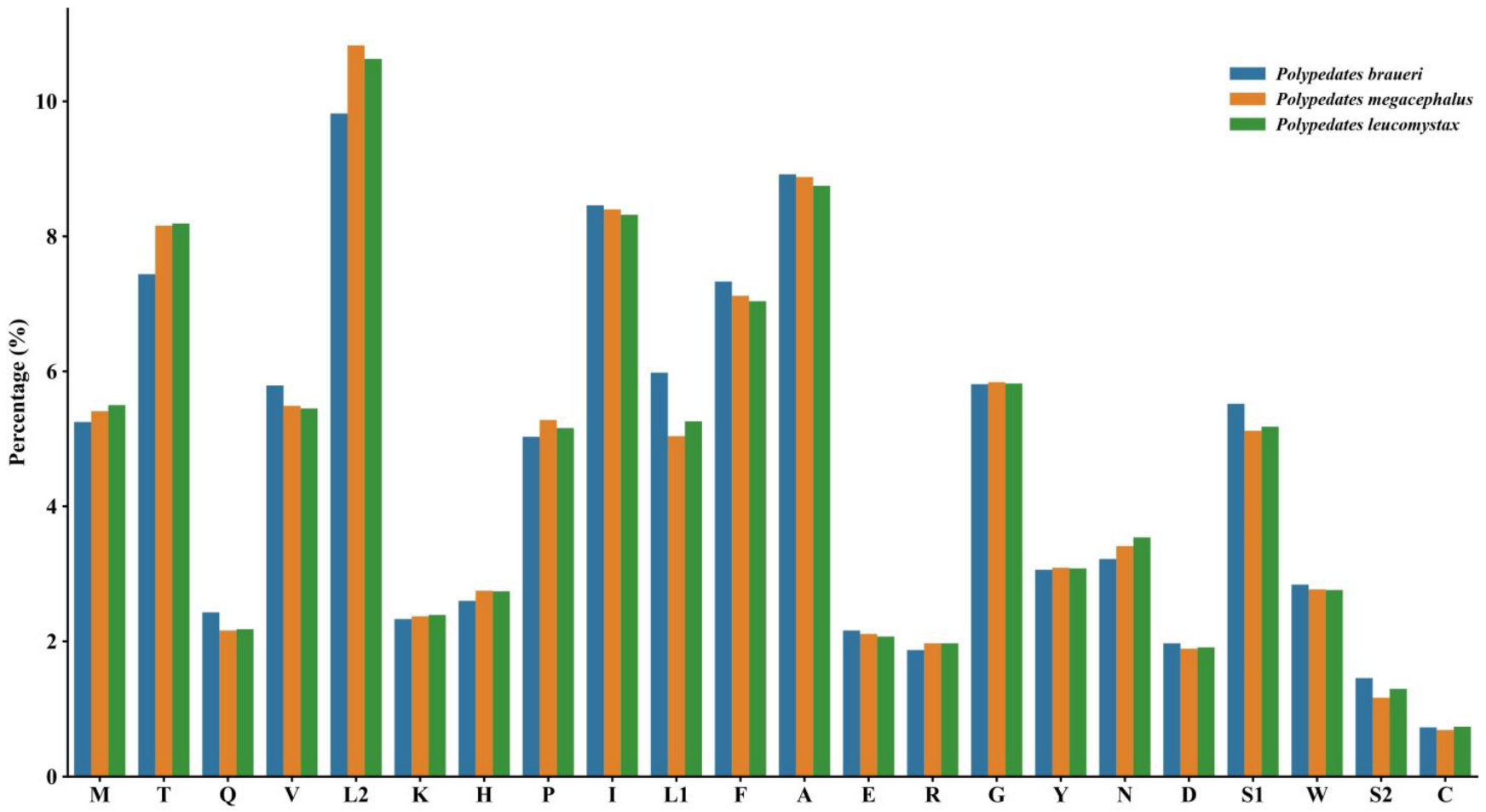
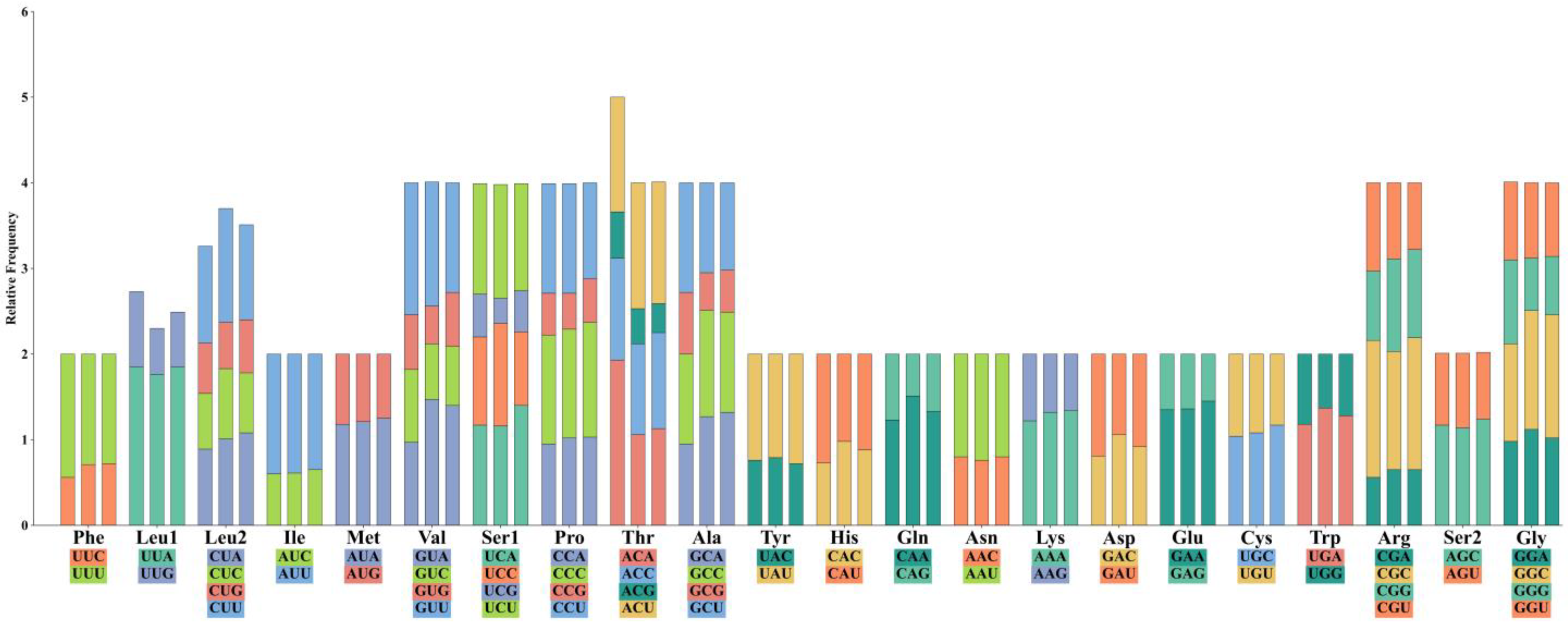
3.2. Protein-Coding Genes and Codon Usage
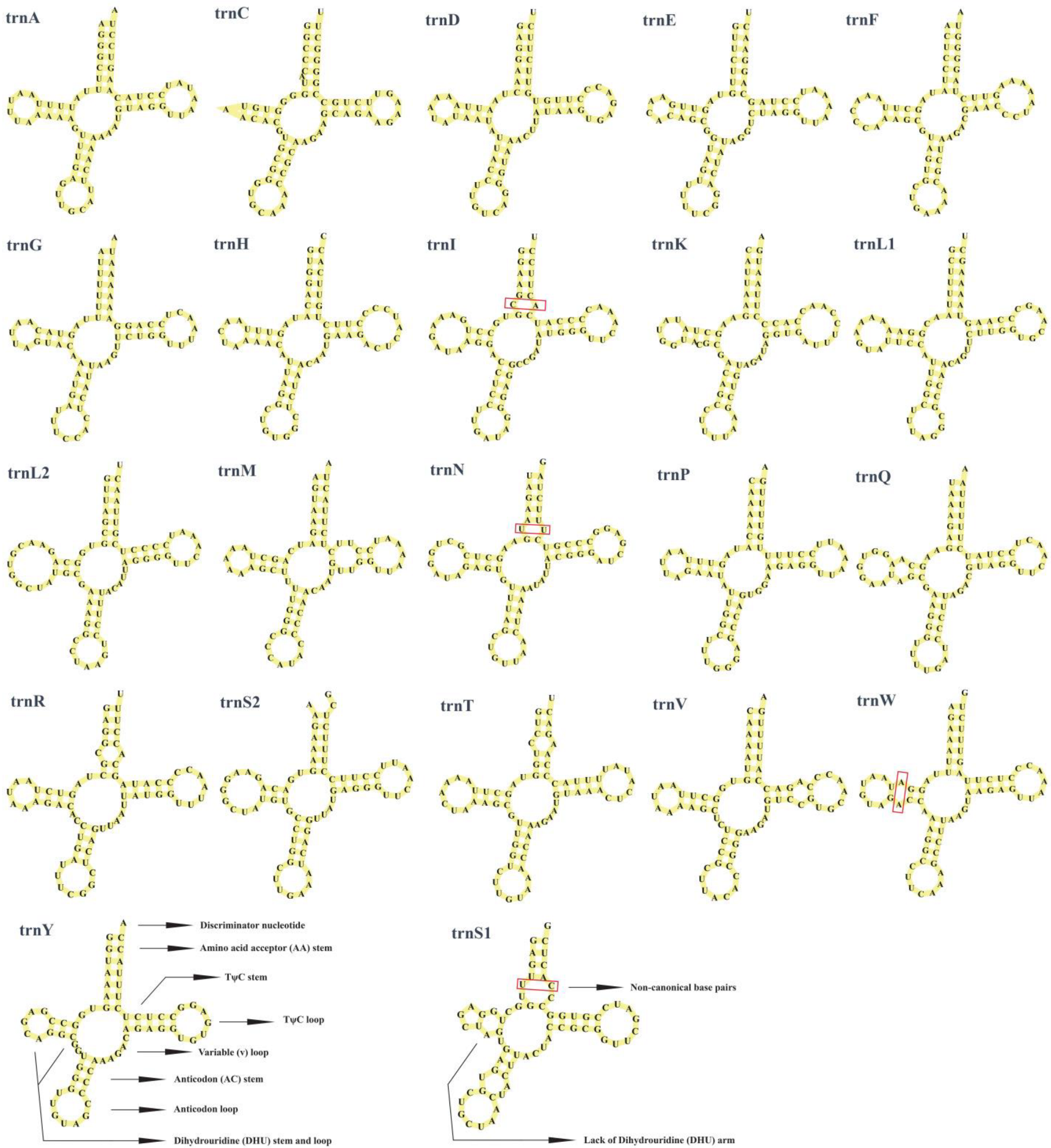
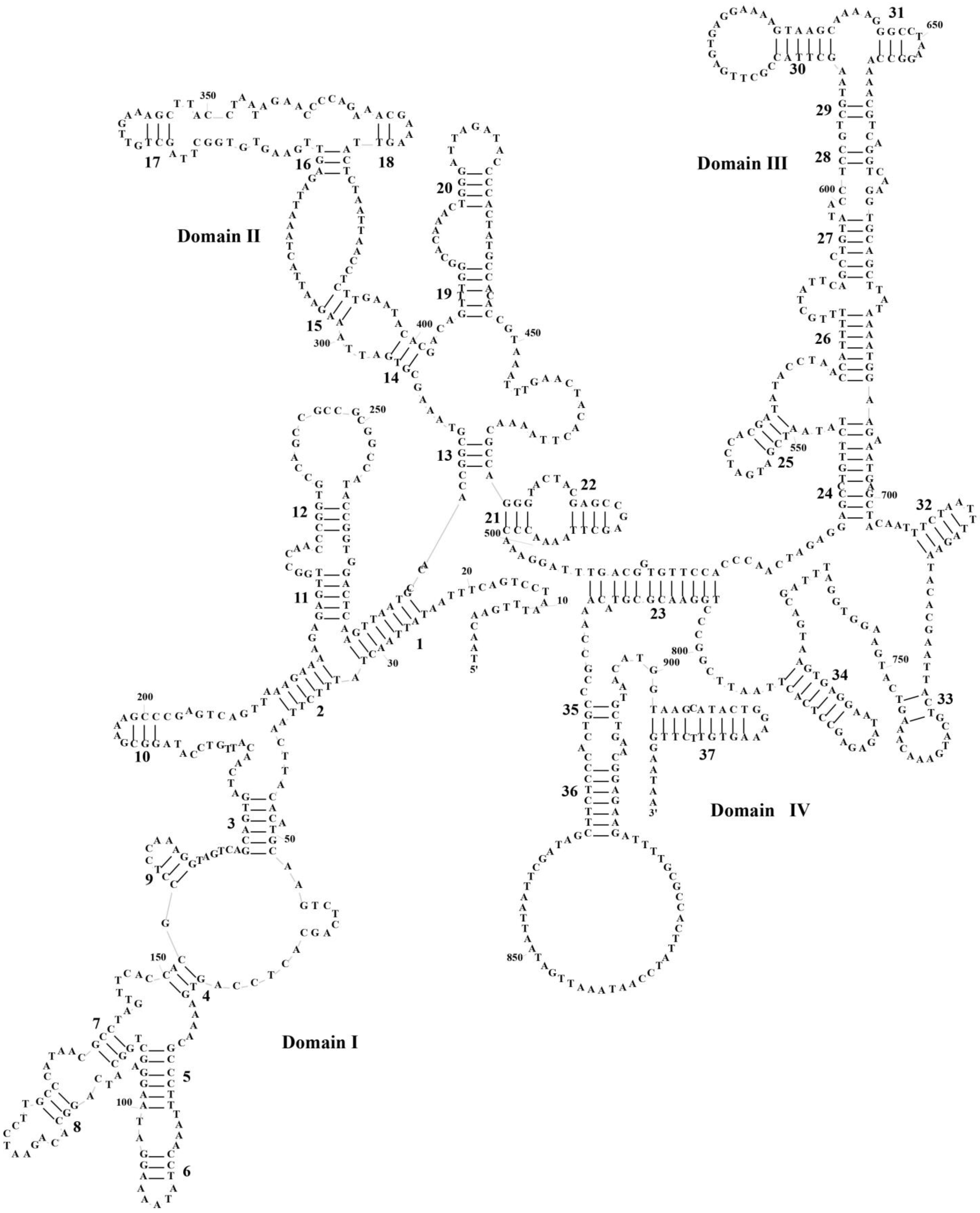
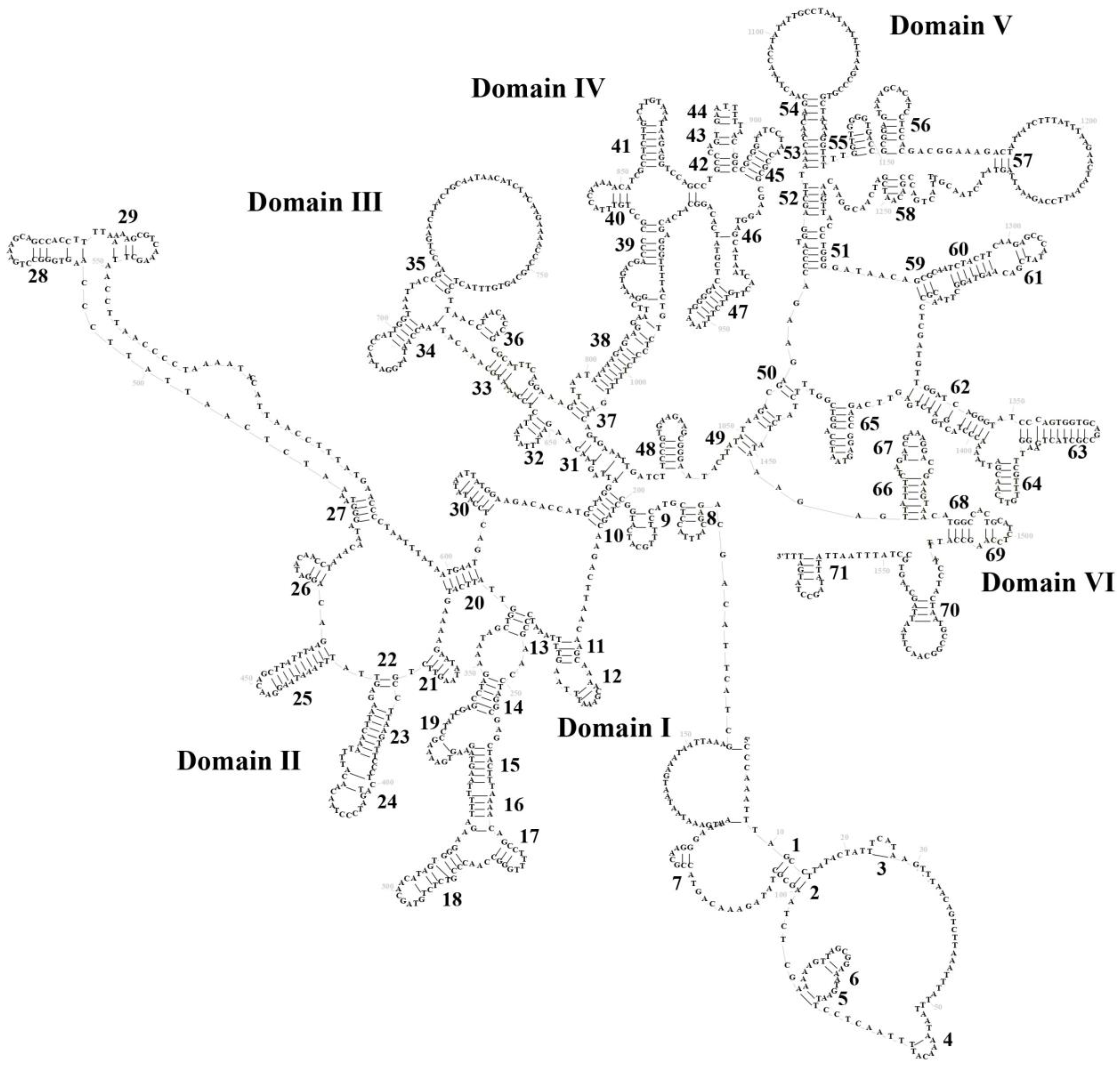
3.3. Transfer RNAs and Ribosomal RNA Genes
3.4. Control Region
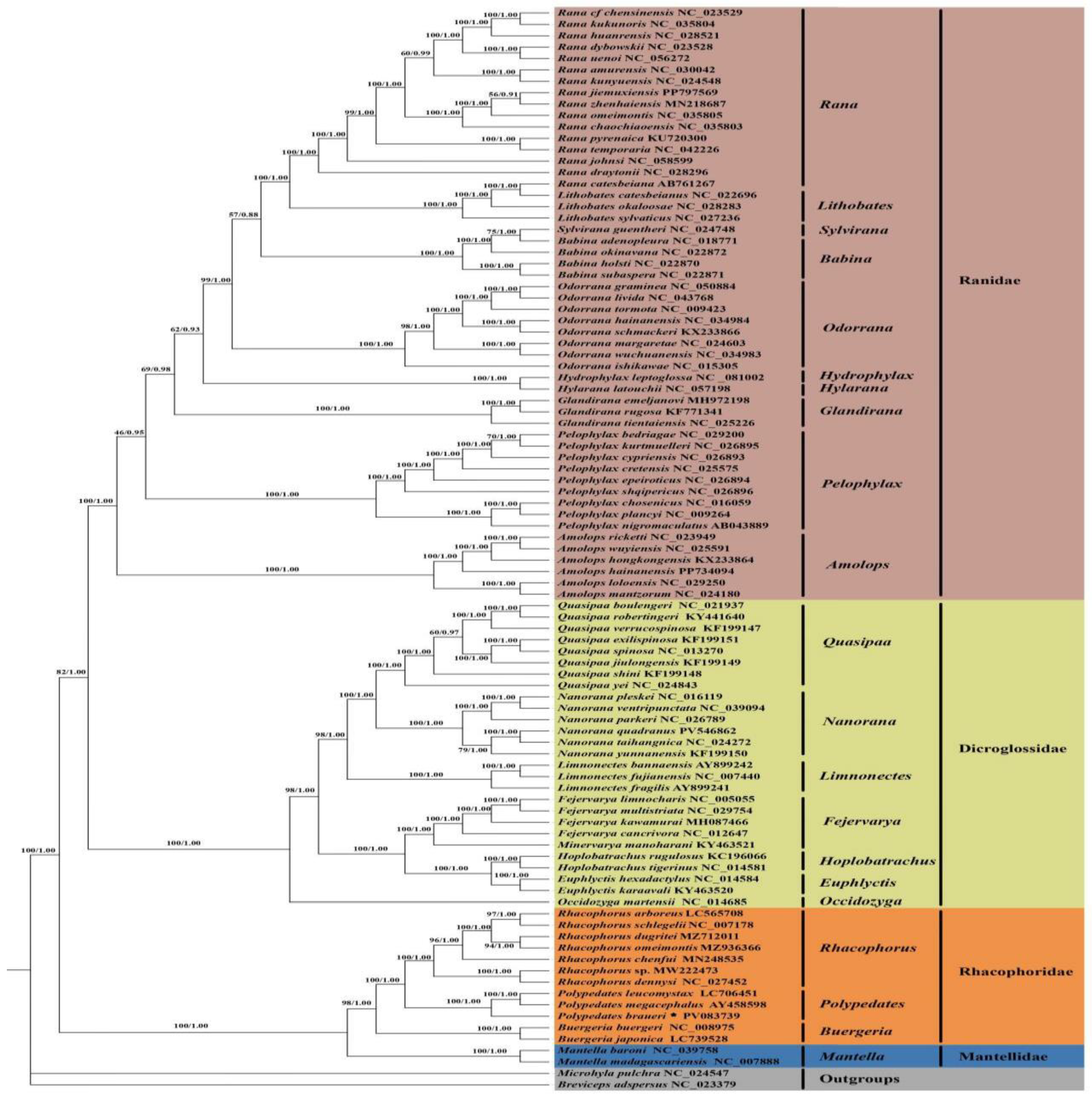
3.5. Phylogenetic Analysis
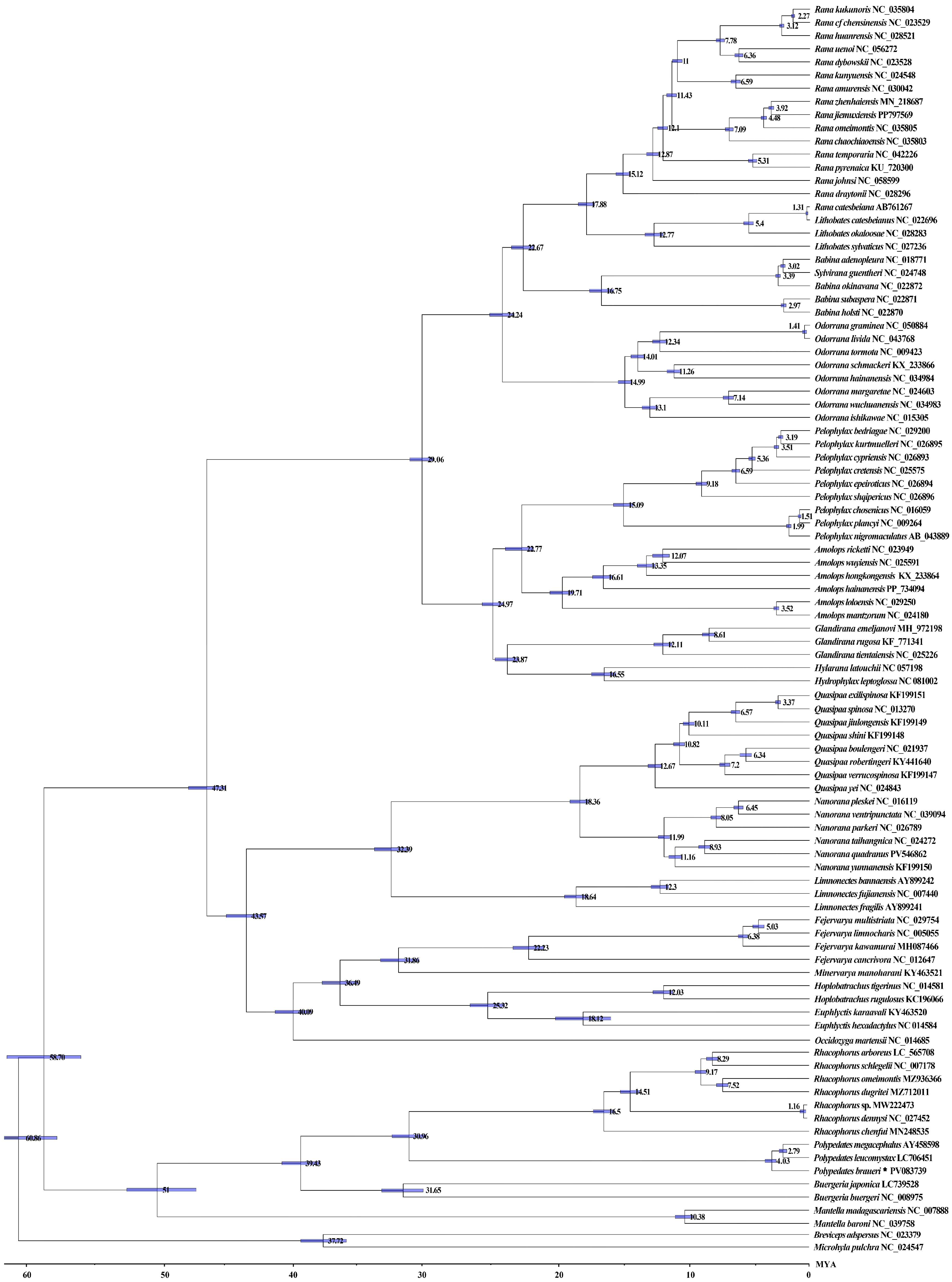
3.6. Divergence Time Estimation
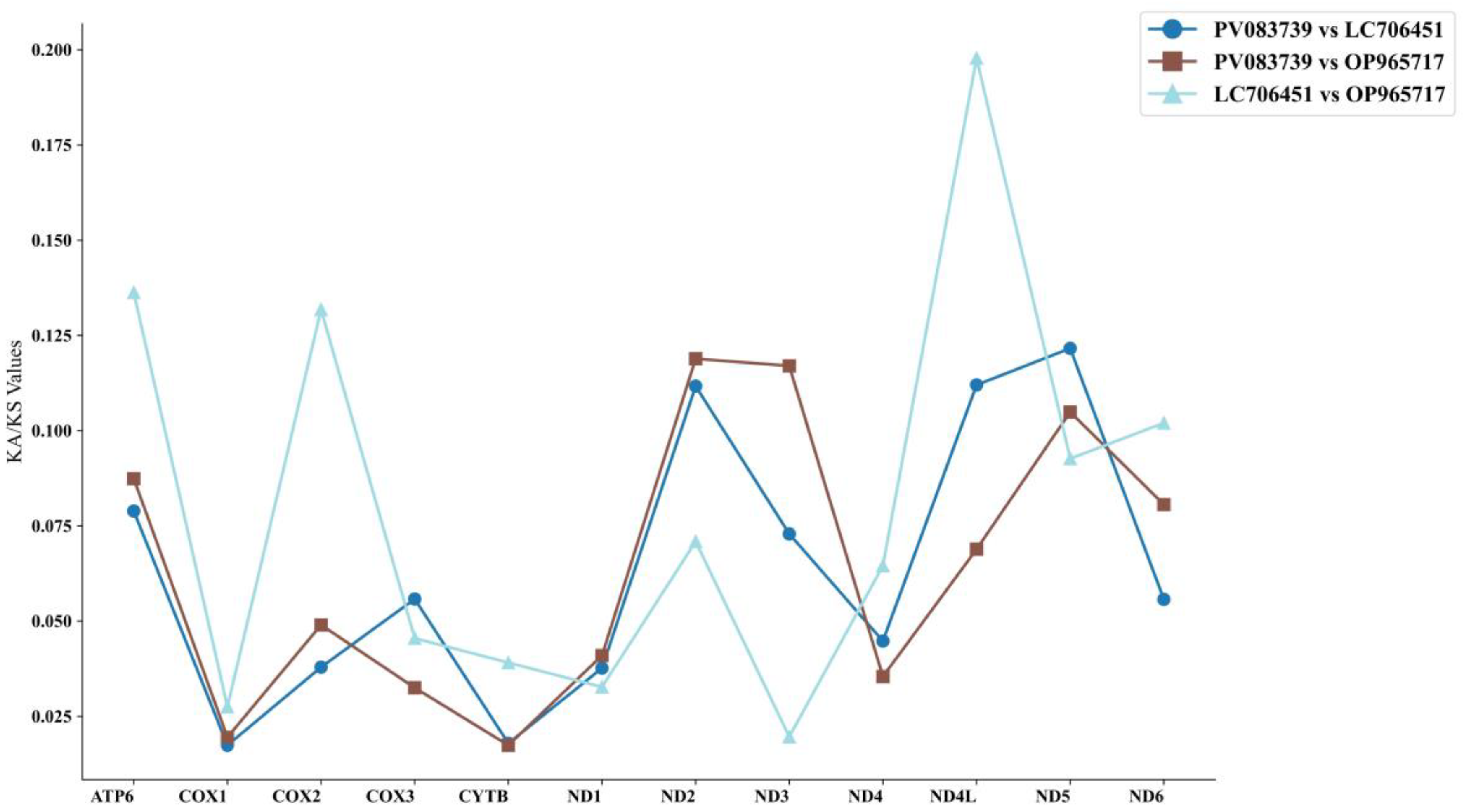
3.7. Non-Synonymous and Synonymous Substitution Rates
4. Discussion
4.1. Mitogenome Sructural Analyses of Genus Polypedates
4.2. Phylogenetic Analysis and Divergence Time Estimation
4.3. Inference into ATP8 Gene Loss in Polypedates Frogs
4.4. Selective Pressure of Genus Polypedates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cui, L.; Huang, A.; He, Z.; Ao, L.; Ge, F.; Fan, X.; Zeng, B.; Yang, M.; Yang, D.; Ni, Q.; et al. Complete Mitogenomes of Polypedates Tree Frogs Unveil Gene Rearrangement and Concerted Evolution within Rhacophoridae. Animals 2022, 12, 2449. [Google Scholar] [CrossRef]
- McDonough, K. Amphibian Species of the World: An Online Reference (Version 6). Ref. Rev. 2014, 28, 32. [Google Scholar] [CrossRef]
- Pan, S.; Dang, N.; Wang, J.; Zheng, Y.; Rao, D.; Li, J. Molecular Phylogeny Supports the Validity of Polypedates impresus Yang 2008. Asian Herpetol. Res. 2013, 4, 124–133. [Google Scholar] [CrossRef]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999, 14, 394–398. [Google Scholar] [CrossRef]
- Nass, S.; Nass, M.M. Intramitochondrial fibers with DNA characteristics. II. Enzymatic and other hydrolytic treatments. J. Cell Biol. 1963, 19, 613–629. [Google Scholar] [CrossRef]
- Keeling, P.J.; Archibald, J.M. Organelle evolution: What’s in a name? Curr. Biol. 2008, 18, 345–347. [Google Scholar] [CrossRef]
- Birky, C.W., Jr. The inheritance of genes in mitochondria and chloroplasts: Laws, mechanisms, and models. Annu. Rev. Genet. 2001, 35, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Sato, K. Maternal inheritance of mitochondrial DNA by diverse mechanisms to eliminate paternal mitochondrial DNA. Biochim. Biophys. Acta 2013, 1833, 1979–1984. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Jiang, H.; Zhu, K.; Lu, X.; Liu, L.; Liu, B.; Jiang, L.; Ye, Y.; Lü, Z. Large-scale mitochondrial gene rearrangements in the hermit crab Pagurus nigrofascia and phylogenetic analysis of the Anomura. Gene 2019, 695, 75–83. [Google Scholar] [CrossRef]
- Wu, Q.L.; Li, Q.; Gu, Y.; Shi, B.C.; van Achterberg, C.; Wei, S.J.; Chen, X.X. The complete mitochondrial genome of Taeniogonalos taihorina (Bischoff) (Hymenoptera: Trigonalyidae) reveals a novel gene rearrangement pattern in the Hymenoptera. Gene 2014, 543, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Liu, S.; Li, H.; Luo, H.; Ni, Q.; Yao, Y.; Xu, H.; Zeng, B.; Li, Y.; Wei, Z.; et al. The revised complete mitogenome sequence of the tree frog Polypedates megacephalus (Anura, Rhacophoridae) by next-generation sequencing and phylogenetic analysis. PeerJ 2019, 7, e7415. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.M.; Prendini, E.; Wu, Y.H.; Zhang, B.L.; Suwannapoom, C.; Chen, H.M.; Jin, J.Q.; Lemmon, E.M.; Lemmon, A.R.; Stuart, B.L.; et al. An integrative phylogenomic approach illuminates the evolutionary history of Old World tree frogs (Anura: Rhacophoridae). Mol. Phylogenet. Evol. 2020, 145, 106724. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Che, J.; Bain, R.H.; Zhao, E.M.; Zhang, Y.P. Molecular phylogeny of Rhacophoridae (Anura): A framework of taxonomic reassignment of species within the genera Aquixalus, Chiromantis, Rhacophorus, and Philautus. Mol. Phylogenet. Evol. 2008, 48, 302–312. [Google Scholar] [CrossRef]
- Mahony, S.; Foley, N.M.; Biju, S.D.; Teeling, E.C. Teeling, evolutionary history of the Asian horned frogs (Megophryinae):Integrative approaches to timetree dating in the absence of a fossil record. Mol. Biol. Evol. 2017, 34, 744–771. [Google Scholar]
- Jiang, D.; Jiang, K.; Ren, J.; Wu, J.; Li, J. Resurrection of the genus Leptomantis, with description of a new genus to the family Rhacophoridae (Amphibia: Anura). Asian Herpetol. Res. 2019, 10, 1–12. [Google Scholar]
- An, H.; Haijun, L.; Hongdi, L.; Qingyong, N.; Yongfang, Y.; Huailiang, X.; Bo, Z.; Ying, L.; Zhimin, W.; Mingwang, Z. The complete mitochondrial genome of the tree frog, Polypedates braueri (Anura, Rhacophoridae). Mitochondrial DNA B 2019, 4, 1739–1740. [Google Scholar]
- Liu, Y.; Yang, M.; Jiang, Y.; Han, F.; Li, Y.; Ni, Q.; Yao, Y.; Xu, H.; Zhang, M. The near complete mitochondrial genome of white-lipped Treefrog, Polypedates braueri (Anura, Rhacophoridae). Mitochondrial DNA Part A 2017, 28, 271–272. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Kuraishi, N.; Matsui, M.; Hamidy, A.; Belabut, D.M.; Ahmad, N.; Panha, S.; Sudin, A.; Yong, H.S.; Jiang, J.; Ota, H.; et al. Phylogenetic and taxonomic relationships of the Polypedates leucomystax complex (Amphibia). Zool. Scr. 2012, 42, 54–70. [Google Scholar] [CrossRef]
- Yuan, L.M.; Deng, X.L.; Jiang, D.C.; Klaus, S.; Orlov, N.L.; Yang, K.; Li, J.T. Geographical range evolution of the genus Polypedates (Anura: Rhacophoridae) from the Oligocene to present. Zool. Res. 2021, 42, 116–123. [Google Scholar] [CrossRef]
- Fedorin, N.D.; Chuykova, O.V.; Eprintsev, T.A. Modification of the Method for Isolating MicroRNA from Plants by Phenol–Chloroform Extraction Using Polyethylene Glycol 1500. Biochem. Moscow. Suppl. Ser. B. 2023, 17, 26–30. [Google Scholar] [CrossRef]
- Kurabayashi, A.; Sumida, M. PCR primers for the neobatrachian mitochondrial genome. Curr. Herpetol. 2009, 28, 1–11. [Google Scholar] [CrossRef]
- Xia, X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Tamura, K.; Glen, S.; Sudhir, K. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 302–307. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, B.A.; Hoksza, D.; Nawrocki, E.P.; Ribas, C.E.; Madeira, F.; Cannone, J.J.; Gutell, R.; Maddala, A.; Meade, C.D.; Williams, L.D.; et al. R2DT is a framework for predicting and visualising RNA secondary structure using templates. Nat. Commun. 2021, 12, 3494. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Li, W.X.; Jakovli’c, I.; Zou, H.; Zhang, J.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012, 9, 772. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhou, H.; Chen, Y.; Liu, Y.; Qu, L. Mitogenomic perspectives on the origin and phylogeny of living amphibians. Syst. Biol. 2005, 54, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating Selective Pressure on Coding and Non-Coding Sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhou, H.; Liang, D.; Liu, Y.; Chen, Y.; Qu, L. The complete mitochondrial genome of a tree frog, Polypedates megacephalus (Amphibia: Anura: Rhacophoridae), and a novel gene organization in living amphibians. Gene 2005, 346, 133–143. [Google Scholar] [CrossRef]
- Varani, G.; McClain, W.H. The G x UWobble Base Pair. A Fundamental Building Block of RNA Structure Crucial to RNA Function in Diverse Biological Systems. EMBO Rep. 2000, 1, 18–23. [Google Scholar] [CrossRef]
- Zheng, C.; Nie, L.; Wang, J.; Zhou, H.; Hou, H.; Wang, H.; Liu, J. Recombination and evolution of duplicate control regions in the mitochondrial genome of the Asian big-headed turtle, Platysternon megacephalum. PLoS ONE 2013, 8, e82854. [Google Scholar] [CrossRef][Green Version]
- Akiyama, T.; Nishida, C.; Momose, K.; Onuma, M.; Takami, K.; Masuda, R. Gene duplication and concerted evolution of mitochondrial DNA in crane species. Mol. Phylogenet. Evol. 2017, 106, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Cheng, C.H. ND6 gene lost and found: Evolution of mitochondrial gene rearrangement in antarctic notothenioids. Mol. Biol. Evol. 2010, 27, 1391–1403. [Google Scholar] [CrossRef] [PubMed]
- Pyron, R.A.; Wiens, J.J. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 2011, 61, 543–583. [Google Scholar] [CrossRef]
- Meiklejohn, C.D.; Montooth, K.L.; Rand, D.M. Positive and negative selection on the mitochondrial genome. Trends Genet. 2007, 23, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Lu, G.; Bork, P.; Hu, S.; Lercher, M.J. Energy efficiency trade-offs drive nucleotide usage in transcribed regions. Nat. Commun. 2016, 7, 11334. [Google Scholar] [CrossRef]
- Li, X.; Yan, L.; Pape, T.; Gao, Y.; Zhang, D. Evolutionary insights into bot flies (Insecta: Diptera: Oestridae) from comparative analysis of the mitochondrial genomes. Int. J. Biol. Macromol. 2020, 149, 371–380. [Google Scholar] [CrossRef]
- Yokobori, S.; Fukuda, N.; Nakamura, M.; Aoyama, T.; Oshima, T. Long-term conservation of six duplicated structural genes in cephalopod mitochondrial genomes. Mol. Biol. Evol. 2004, 21, 2034–2046. [Google Scholar] [CrossRef]
- Zhou, M.; Yu, J.; Li, B.; Ouyang, B.; Yang, J. The complete mitochondrial genome of Budorcas taxicolor tibetana (Artiodactyla: Bovidae) and comparison with other Caprinae species: Insight into the phylogeny of the genus Budorcas. Int. J. Biol. Macromol. 2019, 121, 223–232. [Google Scholar] [CrossRef]
- Sun, C.; Liu, H.; Min, X.; Lu, C. Mitogenome of the little owl Athene noctua and phylogenetic analysis of Strigidae. Int. J. Biol. Macromol. 2020, 151, 924–931. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Miya, M.; Kawaguchi, A.; Nishida, M. Mitogenomic exploration of higher teleostean phylogenies: A case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol. Biol. Evol. 2001, 18, 1993–2009. [Google Scholar] [CrossRef]
- Steinberg, S.; Cedergren, R. Structural compensation in atypical mitochondrial tRNAs. Nat. Struct. Biol. 1994, 1, 507–510. [Google Scholar] [CrossRef]
- Kurabayashi, A.; Usuki, C.; Mikami, N.; Fujii, T.; Yonekawa, H.; Sumida, M.; Hasegawa, M. Complete nucleotide sequence of the mitochondrial genome of a Malagasy poison frog Mantella madagascariensis: Evolutionary implications on mitochondrial genomes of higher anuran groups. Mol. Phylogenet. Evol. 2006, 39, 223–236. [Google Scholar] [CrossRef]
- Igawa, T.; Kurabayashi, A.; Usuki, C.; Fujii, T.; Sumida, M. Complete mitochondrial genomes of three neobatrachian anurans: A case study of divergence time estimation using different data and calibration settings. Gene 2008, 407, 116–129. [Google Scholar] [CrossRef]
- Kurabayashi, A.; Yoshikawa, N.; Sato, N.; Hayashi, Y.; Oumi, S.; Fujii, T.; Sumida, M. Complete mitochondrial DNA sequence of the endangered frog Odorrana ishikawae (family Ranidae) and unexpected diversity of mt gene arrangements in ranids. Mol. Phylogenet. Evol. 2010, 56, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Kurabayashi, A.; Sumida, M. Afrobatrachian mitochondrial genomes: Genome reorganization, gene rearrangement mechanisms, and evolutionary trends of duplicated and rearranged genes. BMC Genom. 2013, 14, 633. [Google Scholar] [CrossRef] [PubMed]
- Ladoukakis, E.D.; Zouros, E. Recombination in animal mitochondrial DNA: Evidence from published sequences. Mol. Biol. Evol. 2001, 18, 2127–2131. [Google Scholar] [CrossRef][Green Version]
- Meegaskumbura, M.; Senevirathne, G.; Biju, S.; Garg, S.; Meegaskumbura, S.; Pethiyagoda, R.; Hanken, J.; Schneider, C.J. Patterns of reproductive-mode evolution in Old World tree frogs (Anura, Rhacophoridae). Zool. Scr. 2015, 44, 509–522. [Google Scholar] [CrossRef]
- Feng, Y.; Blackburn, D.C.; Liang, D.; Hillis, D.M.; Wake, D.B.; Cannatella, D.C.; Zhang, P. Phylogenomics reveals rapid, simultaneous diversification of three major clades of Gondwanan frogs at the Cretaceous–Paleogene boundary. Proc. Natl. Acad. Sci. USA 2017, 114, E5864–E5870. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Klaus, S.; Rao, D.; Hillis, D.M.; Zhang, Y. Diversification of rhacophorid frogs provides evidence for accelerated faunal exchange between India and Eurasia during the Oligocene. Proc. Natl. Acad. Sci. USA 2013, 110, 3441–3446. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhang, B.; Raxworthy, C.J.; Weisrock, D.W.; Hime, P.M.; Jin, J.; Lemmon, E.M.; Lemmon, A.R.; Holland, S.D.; Kortyna, M.L. Natatanuran frogs used the Indian Plate to step-stone disperse and radiate across the Indian Ocean. Natl. Sci. Rev. 2018, 6, 10–14. [Google Scholar] [CrossRef]
- Chen, G.; Wang, B.; Liu, J.; Xie, F.; Jiang, J. Complete mitochondrial genome of Nanorana pleskei (Amphibia: Anura: Dicroglossidae) and evolutionary characteristics. Curr. Zool. 2011, 57, 785–805. [Google Scholar] [CrossRef]
- Ren, Z.; Zhu, B.; Ma, E.; Wen, J.; Tu, T.; Cao, Y.; Hasegawa, M.; Zhong, Y. Complete nucleotide sequence and gene arrangement of the mitochondrial genome of the crab-eating frog Fejervarya cancrivora and evolutionary implications. Gene 2009, 441, 148–155. [Google Scholar] [CrossRef]
- Li, J.; Liang, D.; Wang, Y.; Guo, P.; Huang, S.; Zhang, P. A large-scale systematic framework of Chinese snakes based on a unified multilocus marker system. Mol. Phylogenet. Evol. 2020, 148, 106807. [Google Scholar] [CrossRef]
- Ballard, J.W.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef]
- Pan, T.; Zhang, Y.; Wang, H.; Wu, J.; Kang, X.; Qian, L.; Chen, J.; Rao, D.; Jiang, J.; Zhang, B. The reanalysis of biogeography of the Asian tree frog, Rhacophorus (Anura: Rhacophoridae): Geographic shifts and climatic change influenced the dispersal process and diversification. PeerJ 2017, 5, e3995. [Google Scholar] [CrossRef]
- Scouras, A.; Beckenbach, K.; Arndt, A.; Smith, M.J. Complete mitochondrial genome DNA sequence for two ophiuroids and a holothuroid: The utility of protein gene sequence and gene maps in the analyses of deep deuterostome phylogeny. Mol. Phylogenet. Evol. 2004, 31, 50–65. [Google Scholar] [CrossRef]
- Bain, R.H.; Hurley, M.M. A biogeographic synthesis of the amphibians and reptiles of Indochina. Bull. Am. Mus. Nat. Hist. 2011, 360, 1–138. [Google Scholar] [CrossRef]
- Zhang, D.X.; Hewitt, G.M. Highly conserved nuclear copies of the mitochondrial control region in the desert locust Schistocerca gregaria: Some implications for population studies. Mol Ecol. 1996, 5, 295–300. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Brown, W.M. Trichinella spiralis mtDNA: A nematode mitochondrial genome that encodes a putative ATP8 and normally structured tRNAs and has a gene arrangement relatable to those of coelomate metazoans. Genetics 2001, 157, 621–637. [Google Scholar] [CrossRef]
- Uliano-Silva, M.; Americo, J.A.; Costa, I.; Schomaker-Bastos, A.; de Freitas Rebelo, M.; Prosdocimi, F. The complete mitochondrial genome of the golden mussel Limnoperna fortunei and comparative mitogenomics of Mytilidae. Gene 2016, 577, 202–208. [Google Scholar] [CrossRef]
- Suga, K.; Mark Welch, D.B.; Tanaka, Y.; Sakakura, Y.; Hagiwara, A. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol. Biol. Evol. 2008, 25, 1129–1137. [Google Scholar] [CrossRef]
- Steinauer, M.L.; Nickol, B.B.; Broughton, R.; Ortí, G. First sequenced mitochondrial genome from the phylum Acanthocephala (Leptorhynchoides thecatus)and its phylogenetic position within Metazoa. J. Mol. Evol. 2005, 60, 706–715. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Rand, D.M. Population genetics of the cytoplasm and the units of selection on mitochondrial DNA in Drosophila melanogaster. Genetica 2011, 139, 685–697. [Google Scholar] [CrossRef][Green Version]
- Castresana, J. Cytochrome b phylogeny and the taxonomy of great apes and mammals. Mol. Biol. Evol. 2001, 18, 465–471. [Google Scholar] [CrossRef]
- Gissi, C.; Iannelli, F.; Pesole, G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 2008, 101, 301–320. [Google Scholar] [CrossRef]
- Nee, S.; May, R.M.; Harvey, P.H. The reconstructed evolutionary process. Philos. Trans. R Soc. Lond. B Biol. Sci. 1994, 344, 305–311. [Google Scholar]
- Bromham, L. Why do species vary in their rate of molecular evolution? Biol. Lett. 2009, 5, 401–404. [Google Scholar] [CrossRef]
- da Fonseca, R.R.; Johnson, W.E.; O’Brien, S.J.; Ramos, M.J.; Antunes, A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genom. 2008, 9, 119. [Google Scholar] [CrossRef]
| Gene | Position | Size | Position | Size | Position | Size | Strand | |||
|---|---|---|---|---|---|---|---|---|---|---|
| From | To | From | To | From | To | |||||
| P.leucomystax | P. megacephalus | P.braueri | ||||||||
| tRNAThr | 1 | 70 | 70 | 1 | 71 | 71 | 1 | 72 | 72 | H |
| tRNALeu | 71 | 141 | 71 | 72 | 143 | 72 | 73 | 144 | 72 | H |
| tRNAPro | 147 | 215 | 69 | 149 | 217 | 69 | 148 | 216 | 69 | L |
| tRNAPhe | 217 | 286 | 70 | 219 | 288 | 70 | 218 | 287 | 70 | H |
| 12SrRNA | 287 | 1214 | 928 | 287 | 1216 | 930 | 286 | 1216 | 931 | H |
| tRNAVal | 1215 | 1283 | 69 | 1217 | 1285 | 69 | 1217 | 1285 | 69 | H |
| 16SrRNA | 1284 | 2857 | 1574 | 1286 | 2856 | 1571 | 1288 | 2861 | 1574 | H |
| tRNALeu | 2858 | 2931 | 74 | 2859 | 2932 | 74 | 2863 | 2936 | 74 | H |
| ND1 | 2935 | 3895 | 961 | 2936 | 3896 | 96 | 2940 | 3900 | 961 | H |
| tRNAIle | 3896 | 3966 | 71 | 3897 | 3967 | 71 | 3901 | 3971 | 71 | H |
| tRNAGln | 3966 | 4036 | 71 | 3967 | 4037 | 71 | 3971 | 4041 | 71 | L |
| tRNAMet | 4036 | 4104 | 69 | 4037 | 4105 | 69 | 4041 | 4109 | 69 | H |
| ND2 | 4078 | 5134 | 1057 | 4106 | 5141 | 1036 | 4110 | 5145 | 1036 | H |
| tRNATrp | 5141 | 5211 | 71 | 5142 | 5212 | 71 | 5146 | 5215 | 70 | H |
| tRNAAla | 5213 | 5282 | 70 | 5213 | 5282 | 70 | 5216 | 5285 | 70 | L |
| tRNAAsn | 5284 | 5356 | 73 | 5284 | 5356 | 73 | 5287 | 5359 | 73 | L |
| OL | 5359 | 5384 | 26 | 5359 | 5384 | 26 | 5362 | 5387 | 26 | H |
| tRNACys | 5384 | 5448 | 65 | 5384 | 5448 | 65 | 5387 | 5450 | 64 | L |
| tRNATyr | 5449 | 5515 | 67 | 5449 | 5515 | 67 | 5452 | 5518 | 67 | L |
| COX1 | 5520 | 7073 | 1554 | 5520 | 7073 | 1554 | 5523 | 7076 | 1554 | H |
| tRNASer | 7061 | 7131 | 71 | 7061 | 7131 | 71 | 7064 | 7134 | 71 | L |
| tRNAAsp | 7133 | 7201 | 69 | 7133 | 7201 | 69 | 7136 | 7204 | 69 | H |
| COX2 | 7202 | 7891 | 690 | 7202 | 7891 | 690 | 7205 | 7894 | 690 | H |
| tRNALys | 7897 | 7966 | 70 | 7897 | 7966 | 70 | 7900 | 7970 | 71 | H |
| ATP8 | 8676 | 8828 | 153 | 8680 | 8832 | 153 | — | — | — | H |
| ATP6 | 8816 | 9503 | 688 | 8829 | 9507 | 679 | 8138 | 8816 | 679 | H |
| COX3 | 9504 | 10,287 | 784 | 9508 | 10,291 | 784 | 8817 | 9600 | 784 | H |
| tRNAGly | 10,312 | 10,379 | 68 | 10,292 | 10,359 | 68 | 9601 | 9669 | 69 | H |
| ND3 | 10,380 | 10,719 | 340 | 10,360 | 10,699 | 340 | 9670 | 10,009 | 340 | H |
| tRNAArg | 10,720 | 10,789 | 70 | 10,700 | 10,768 | 69 | 10,010 | 10,078 | 69 | H |
| ND4L | 10,790 | 11,074 | 285 | 10,769 | 11,053 | 285 | 10,080 | 10,364 | 285 | H |
| ND4 | 11,068 | 12,427 | 1360 | 11,047 | 12,409 | 1363 | 10,358 | 11,720 | 1363 | H |
| tRNAHis | 12,431 | 12,499 | 69 | 12,410 | 12,478 | 69 | 11,721 | 11,789 | 69 | H |
| tRNASer | 12,500 | 12,567 | 68 | 12,479 | 12,546 | 68 | 11,801 | 11,856 | 56 | H |
| ND6 | 12,570 | 13,061 | 492 | 12,549 | 13,040 | 492 | 11,859 | 12,350 | 492 | L |
| tRNAGlu | 13,062 | 13,130 | 69 | 13,041 | 13,109 | 69 | 12,418 | 12,351 | 68 | L |
| CYTB | 13,135 | 14,304 | 1170 | 13,114 | 14,269 | 1156 | 12,423 | 13,572 | 1150 | H |
| CR1 | 14,305 | 15,828 | 1524 | 14,270 | 15,920 | 1651 | 13,573 | 15,325 | 1753 | H |
| ND5 | 15,829 | 17,607 | 1779 | 15,921 | 17,699 | 1779 | 15,326 | 17,107 | 1782 | H |
| CR2 | 17,608 | 20,794 | 3187 | 17,700 | 19,384 | 1685 | 17,108 | 20,254 | 3147 | H |
| ND5 | — | — | — | 19,385 | 21,163 | 1779 | — | — | — | H |
| CR3 | — | — | — | 21,164 | 24,103 | 2940 | — | — | — | H |
| Gene | Codon | Codon | Codon | |||
|---|---|---|---|---|---|---|
| Start | Stop | Start | Stop | Start | Stop | |
| P. leucomystax | P. megacephalus | P. braueri | ||||
| ND1 | ATG | T-- | ATG | T-- | ATG | T-- |
| ND2 | ATG | T-- | ATT | T-- | ATT | T-- |
| COX1 | ATA | AGG | ATA | AGG | ATA | AGG |
| COX2 | ATA | TAA | ATA | TAA | ATG | TAA |
| ATP8 | ATG | TAG | ATG | TAG | --- | --- |
| ATP6 | ATG | T-- | ATA | T-- | ATA | T-- |
| COX3 | ATG | T-- | ATG | T-- | ATG | T-- |
| ND3 | ATG | T-- | ATG | T-- | ATG | T-- |
| ND4L | ATG | TAA | ATG | TAA | ATG | TAA |
| ND4 | GTG | T-- | GTG | T-- | ATG | T-- |
| ND6 | ATG | AGG | ATG | AGG | ATG | AGG |
| CYTP | ATG | T-- | ATG | T-- | ATG | T-- |
| ND5 | ATG | TAA | ATG | TAA | ATG | TAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Huang, H.; Shan, S.; Li, C.; Huang, K.; Xu, X.; Jiang, L. Characterization of the Complete Mitogenome of Polypedates braueri (Anura, Rhacophoridae, Polypedates) and Insights into the Phylogenetic Relationships of Rhacophoridae. Biology 2025, 14, 1299. https://doi.org/10.3390/biology14091299
Chen S, Huang H, Shan S, Li C, Huang K, Xu X, Jiang L. Characterization of the Complete Mitogenome of Polypedates braueri (Anura, Rhacophoridae, Polypedates) and Insights into the Phylogenetic Relationships of Rhacophoridae. Biology. 2025; 14(9):1299. https://doi.org/10.3390/biology14091299
Chicago/Turabian StyleChen, Simin, Huiling Huang, Siqi Shan, Chengmin Li, Kaiyuan Huang, Xinyi Xu, and Lichun Jiang. 2025. "Characterization of the Complete Mitogenome of Polypedates braueri (Anura, Rhacophoridae, Polypedates) and Insights into the Phylogenetic Relationships of Rhacophoridae" Biology 14, no. 9: 1299. https://doi.org/10.3390/biology14091299
APA StyleChen, S., Huang, H., Shan, S., Li, C., Huang, K., Xu, X., & Jiang, L. (2025). Characterization of the Complete Mitogenome of Polypedates braueri (Anura, Rhacophoridae, Polypedates) and Insights into the Phylogenetic Relationships of Rhacophoridae. Biology, 14(9), 1299. https://doi.org/10.3390/biology14091299






