Whole Genome Sequencing and Extracellular Metabolite Profiling of Lactiplantibacillus plantarum FRT4: Insights into Probiotic Functionality
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Lp. plantarum FRT4 Strain and Its Supernatant
2.2. Genomic DNA Extraction
2.3. Whole Genome Sequencing and Library Preparation
2.4. Sequencing and Assembly
2.5. Genome Annotation and Bioinformatic Analysis
2.6. Untargeted Metabolomics Analysis
3. Results
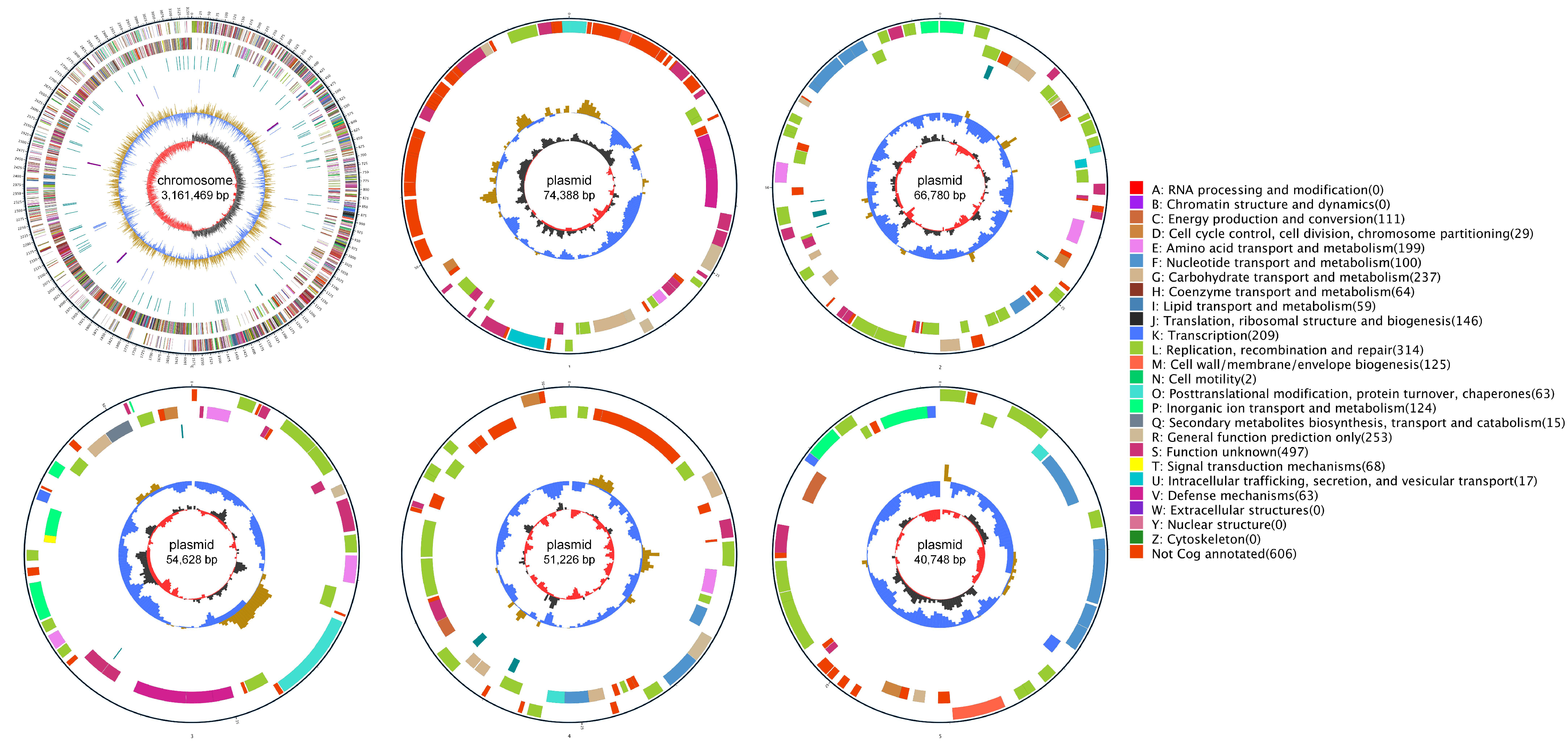
3.1. Genomic Features of Lp. plantarum FRT4
3.2. Functional Annotation Analysis of Lp. plantarum FRT4
3.2.1. Functional Annotation Based on KEGG Database
3.2.2. Functional Annotation Based on GO Database
3.2.3. Functional Annotation Based on eggNOG Database
3.2.4. Functional Annotation of CAZy Database
3.2.5. Antibiotic Resistance Gene-Annotation Based on CARD
3.2.6. Virulence Factor Annotation Based on the VFDB
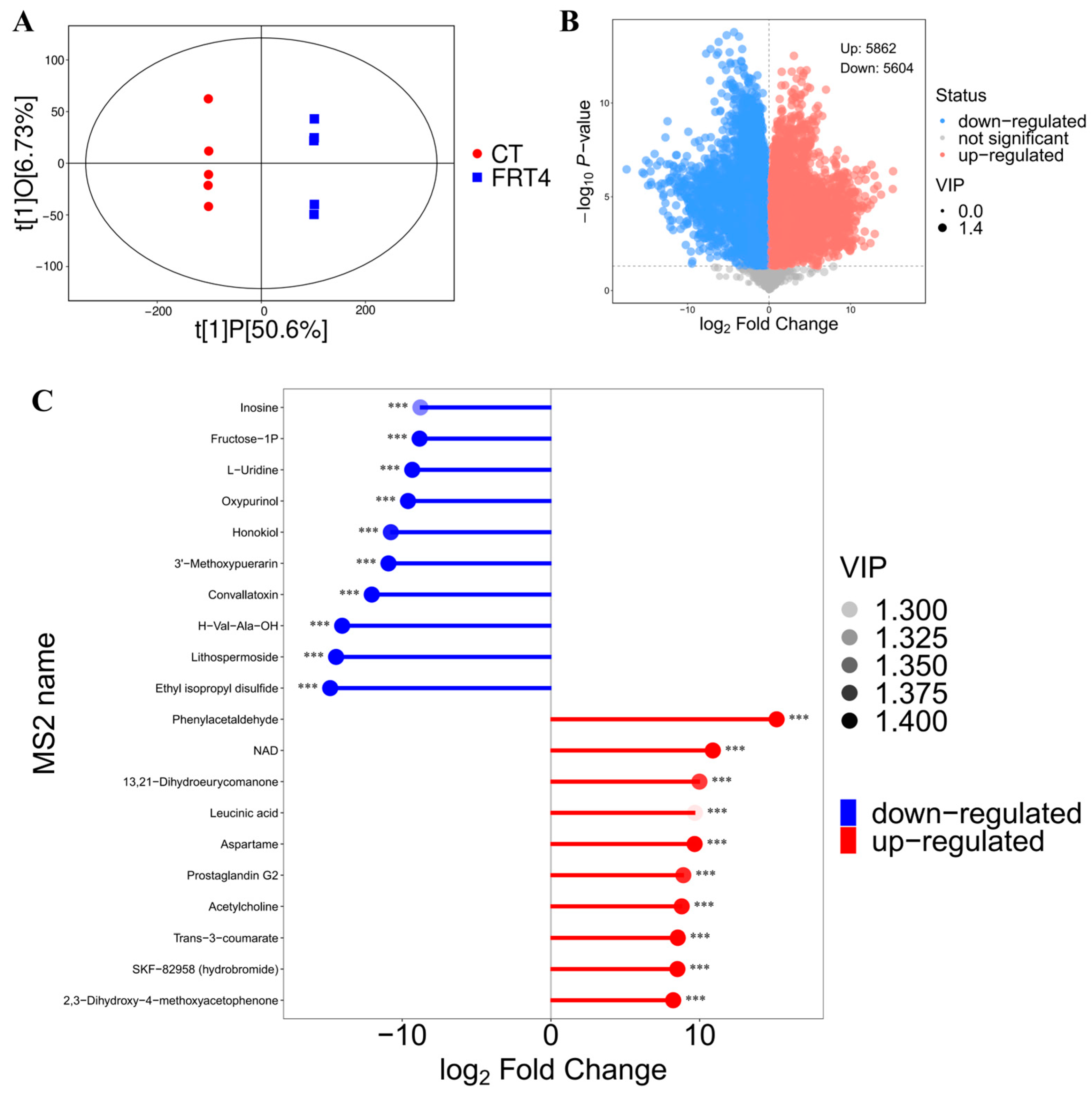
3.3. Untargeted Metabolomics Reveals Significant Extracellular Metabolic Changes in FRT4 Supernatant
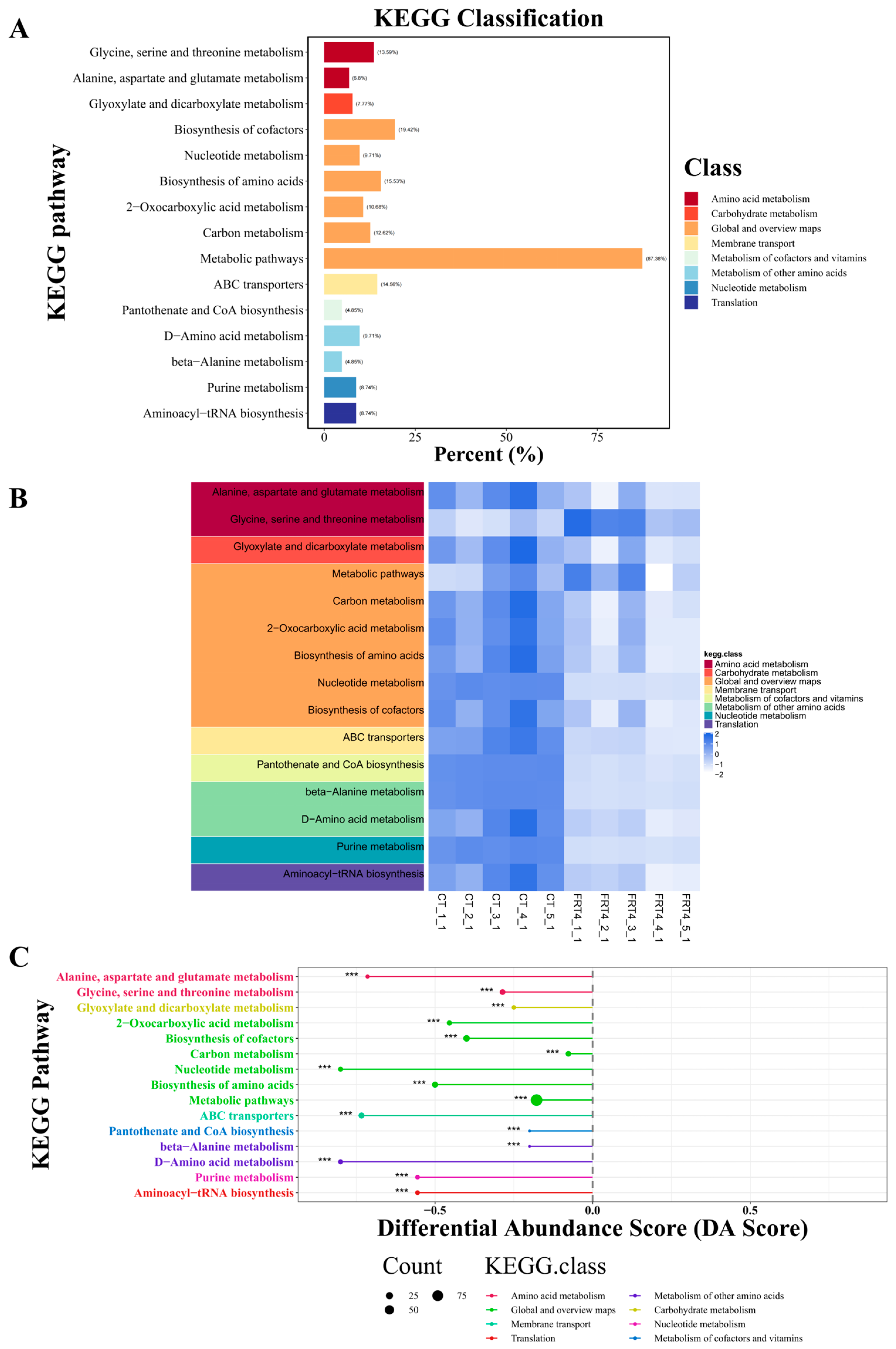
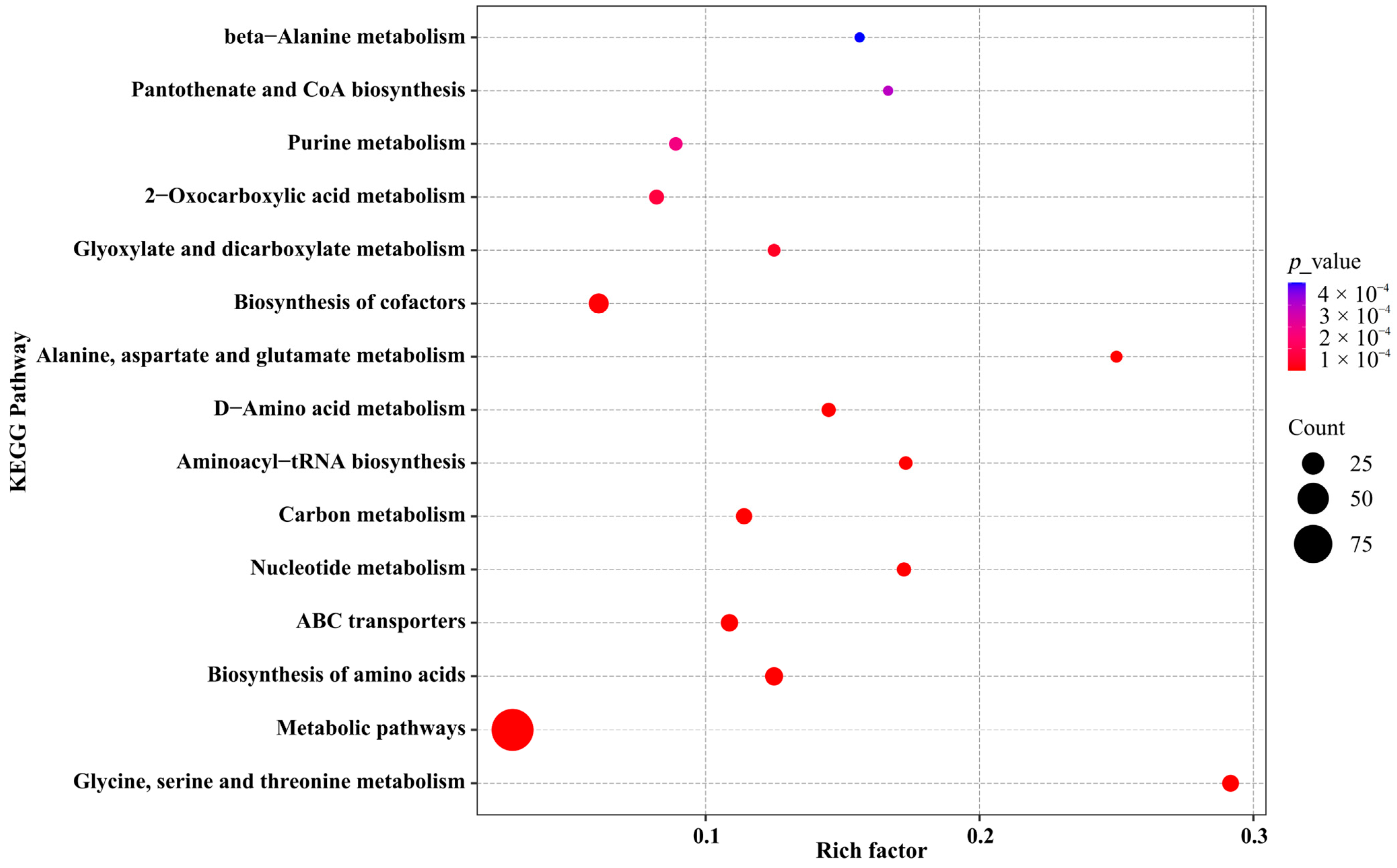
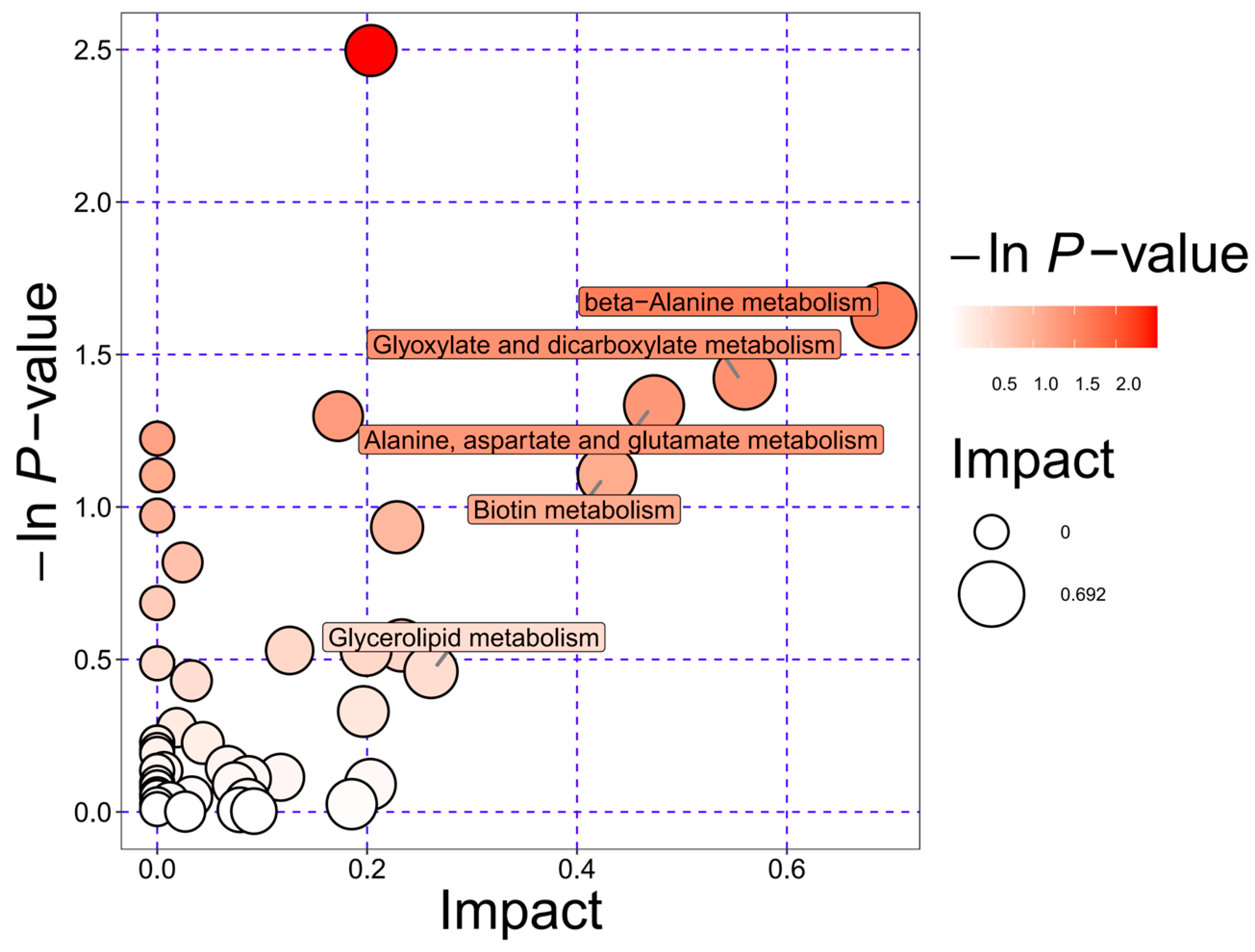
3.4. KEGG Pathway Enrichment Analysis and Functional Implications of the Metabolome
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gruber, T.; Lechner, F.; Krieger, J.-P.; García-Cáceres, C. Neuroendocrine Gut-Brain Signaling in Obesity. Trends Endocrinol. Metab. 2025, 36, 42–54. [Google Scholar] [CrossRef]
- Shen, Z.; Cui, T.; Liu, Y.; Wu, S.; Han, C.; Li, J. Astragalus membranaceus and Salvia miltiorrhiza Ameliorate Diabetic Kidney Disease via the “Gut-Kidney Axis”. Phytomedicine 2023, 121, 155129. [Google Scholar] [CrossRef]
- Kronsten, V.T.; Tranah, T.H.; Pariante, C.; Shawcross, D.L. Gut-Derived Systemic Inflammation as a Driver of Depression in Chronic Liver Disease. J. Hepatol. 2022, 76, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.A. Modulating Brain Function with Microbiota. Science 2022, 376, 936–937. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Bae, M.; Cassilly, C.D.; Jabba, S.V.; Thorpe, D.W.; Martin, A.M.; Lu, H.-Y.; Wang, J.; Thompson, J.D.; Lickwar, C.R.; et al. Enteroendocrine Cells Sense Bacterial Tryptophan Catabolites to Activate Enteric and Vagal Neuronal Pathways. Cell Host Microbe 2021, 29, 179–196.e9. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, T.L.; Eckstrom, K.; Lile, K.H.; Caldwell, S.; Heney, E.R.; Lahue, K.G.; D’Alessandro, A.; Wargo, M.J.; Krementsov, D.N. Lactobacillus reuteri Tryptophan Metabolism Promotes Host Susceptibility to CNS Autoimmunity. Microbiome 2022, 10, 198. [Google Scholar] [CrossRef]
- Cho, M.-Y.; Eom, J.-H.; Choi, E.-M.; Yang, S.-J.; Lee, D.; Kim, Y.Y.; Kim, H.-S.; Hwang, I. Recent Advances in Therapeutic Probiotics: Insights from Human Trials. Clin. Microbiol. Rev. 2025, 38, e0024024. [Google Scholar] [CrossRef]
- Fentie, E.G.; Lim, K.; Jeong, M.; Shin, J.-H. A Comprehensive Review of the Characterization, Host Interactions, and Stabilization Advancements on Probiotics: Addressing the Challenges in Functional Food Diversification. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13424. [Google Scholar] [CrossRef]
- Kim, M.-J.; Shin, S.-K.; Han, J.-W.; Kim, J.E.; Lee, M.J.; Bae, H.R.; Kwon, E.-Y. Lactobacillus paragasseri SBT2055 Attenuates Obesity via the Adipose Tissue-Muscle-Gut Axis in Obese Mice. Microbiol. Res. 2025, 290, 127972. [Google Scholar] [CrossRef]
- De Filippis, F.; Pasolli, E.; Ercolini, D. The Food-Gut Axis: Lactic Acid Bacteria and Their Link to Food, the Gut Microbiome and Human Health. FEMS Microbiol. Rev. 2020, 44, 454–489. [Google Scholar] [CrossRef]
- Choi, Y.; Park, E.; Kim, S.; Ha, J.; Oh, H.; Kim, Y.; Lee, Y.; Seo, Y.; Kang, J.; Lee, S.; et al. Fermented Milk with Lactobacillus curvatus SMFM2016-NK Alleviates Periodontal and Gut Inflammation, and Alters Oral and Gut Microbiota. J. Dairy Sci. 2021, 104, 5197–5207. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Chen, Z.; Liu, N.; Xia, C.; Zhou, Q.; Li, P. Lactiplantibacillus plantarum ZJ316-Fermented Milk Ameliorates Dextran Sulfate Sodium-Induced Chronic Colitis by Improving the Inflammatory Response and Regulating Intestinal Microbiota. J. Dairy Sci. 2023, 106, 7352–7366. [Google Scholar] [CrossRef] [PubMed]
- Echegaray, N.; Yilmaz, B.; Sharma, H.; Kumar, M.; Pateiro, M.; Ozogul, F.; Lorenzo, J.M. A Novel Approach to Lactiplantibacillus plantarum: From Probiotic Properties to the Omics Insights. Microbiol. Res. 2023, 268, 127289. [Google Scholar] [CrossRef] [PubMed]
- Arellano-García, L.; Trepiana, J.; Martínez, J.A.; Portillo, M.P.; Milton-Laskibar, I. Beneficial Effects of Viable and Heat-Inactivated Lactobacillus rhamnosus GG Administration on Oxidative Stress and Inflammation in Diet-Induced NAFLD in Rats. Antioxidants 2023, 12, 717. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, V.; Pramanik, J.; Rustagi, S.; Prajapati, B.; Jebreen, A.; Pande, R. Lactiplantibacillus plantarum as a Complementary Approach for Diabetes Treatment and Management. Curr. Nutr. Rep. 2025, 14, 72. [Google Scholar] [CrossRef]
- Duarte Luiz, J.; Manassi, C.; Magnani, M.; da Cruz, A.G.; Pimentel, T.C.; Verruck, S. Lactiplantibacillus plantarum as a Promising Adjuvant for Neurological Disorders Therapy through the Brain-Gut Axis and Related Action Pathways. Crit. Rev. Food Sci. Nutr. 2025, 65, 715–727. [Google Scholar] [CrossRef]
- Chen, C.-M.; Wu, C.-C.; Kim, Y.; Hsu, W.-Y.; Tsai, Y.-C.; Chiu, S.-L. Enhancing Social Behavior in an Autism Spectrum Disorder Mouse Model: Investigating the Underlying Mechanisms of Lactiplantibacillus plantarum Intervention. Gut Microbes 2024, 16, 2359501. [Google Scholar] [CrossRef]
- Liang, S.; Sun, J.; Gu, X.; Zhao, Y.; Wang, X.; Tao, H.; Wang, Z.; Zhong, Y.; Wang, J.; Han, B. Lactobacillus plantarum L11 and Lactobacillus reuteri LR: Ameliorate Obesity via AMPK Pathway. Nutrients 2024, 17, 4. [Google Scholar] [CrossRef]
- Stojanov, S.; Plavec, T.V.; Zupančič, Š.; Berlec, A. Modified Vaginal Lactobacilli Expressing Fluorescent and Luminescent Proteins for More Effective Monitoring of Their Release from Nanofibers, Safety and Cell Adhesion. Microb. Cell Factories 2024, 23, 333. [Google Scholar] [CrossRef]
- Kaushik, J.K.; Kumar, A.; Duary, R.K.; Mohanty, A.K.; Grover, S.; Batish, V.K. Functional and Probiotic Attributes of an Indigenous Isolate of Lactobacillus plantarum. PLoS ONE 2009, 4, e8099. [Google Scholar] [CrossRef]
- Yadav, R.; Singh, P.K.; Puniya, A.K.; Shukla, P. Catalytic Interactions and Molecular Docking of Bile Salt Hydrolase (BSH) from L. Plantarum RYPR1 and Its Prebiotic Utilization. Front. Microbiol. 2016, 7, 2116. [Google Scholar] [CrossRef]
- Rocchetti, M.T.; Russo, P.; Capozzi, V.; Drider, D.; Spano, G.; Fiocco, D. Bioprospecting Antimicrobials from Lactiplantibacillus plantarum: Key Factors Underlying Its Probiotic Action. Int. J. Mol. Sci. 2021, 22, 12076. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.O.; Goldman, E.A.; Brooks, J.T.; Golomb, B.L.; Yim, I.S.; Gotcheva, V.; Angelov, A.; Kim, E.B.; Marco, M.L. Strain Diversity of Plant-Associated Lactiplantibacillus plantarum. Microb. Biotechnol. 2021, 14, 1990–2008. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Q.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Strain-Specific Regulative Effects of Lactobacillus plantarum on Intestinal Barrier Dysfunction Are Associated with Their Capsular Polysaccharides. Int. J. Biol. Macromol. 2022, 222, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.L.; Thorsen, L.; Schwan, R.F.; Jespersen, L. Strain-Specific Probiotics Properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis Isolates from Brazilian Food Products. Food Microbiol. 2013, 36, 22–29. [Google Scholar] [CrossRef]
- Yang, J.; Shang, P.; Zhang, B.; Wang, J.; Du, Z.; Wang, S.; Xing, J.; Zhang, H. Genomic and Metabonomic Methods Reveal the Probiotic Functions of Swine-Derived Ligilactobacillus salivarius. BMC Microbiol. 2023, 23, 242. [Google Scholar] [CrossRef]
- Li, D.; Meng, K.; Liu, G.; Wen, Z.; Han, Y.; Liu, W.; Xu, X.; Song, L.; Cai, H.; Yang, P. Lactiplantibacillus plantarum FRT4 Protects against Fatty Liver Hemorrhage Syndrome: Regulating Gut Microbiota and FoxO/TLR-4/NF-κB Signaling Pathway in Laying Hens. Microbiome 2025, 13, 88. [Google Scholar] [CrossRef]
- Li, D.; Cai, H.; Liu, G.; Han, Y.; Qiu, K.; Liu, W.; Meng, K.; Yang, P. Lactiplantibacillus plantarum FRT4 Attenuates High-Energy Low-Protein Diet-Induced Fatty Liver Hemorrhage Syndrome in Laying Hens through Regulating Gut-Liver Axis. J. Anim. Sci. Biotechnol. 2024, 15, 31. [Google Scholar] [CrossRef]
- Cai, H.; Wen, Z.; Xu, X.; Wang, J.; Li, X.; Meng, K.; Yang, P. Serum Metabolomics Analysis for Biomarkers of Lactobacillus plantarum FRT4 in High-Fat Diet-Induced Obese Mice. Foods 2022, 11, 184. [Google Scholar] [CrossRef]
- Cai, H.; Wen, Z.; Zhao, L.; Yu, D.; Meng, K.; Yang, P. Lactobacillus plantarum FRT4 Alleviated Obesity by Modulating Gut Microbiota and Liver Metabolome in High-Fat Diet-Induced Obese Mice. Food Nutr. Res. 2022, 66, 10-29219. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k-Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to Identify Repetitive Elements in Genomic Sequences. Curr. Protoc. Bioinform. 2009, 25, 4.10.1–4.10.14. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-Fold Faster RNA Homology Searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Bland, C.; Ramsey, T.L.; Sabree, F.; Lowe, M.; Brown, K.; Kyrpides, N.C.; Hugenholtz, P. CRISPR Recognition Tool (CRT): A Tool for Automatic Detection of Clustered Regularly Interspaced Palindromic Repeats. BMC Bioinform. 2007, 8, 209. [Google Scholar] [CrossRef]
- Bertelli, C.; Brinkman, F.S.L. Improved Genomic Island Predictions with IslandPath-DIMOB. Bioinformatics 2018, 34, 2161–2167. [Google Scholar] [CrossRef]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A Novel Algorithm for Finding Prophages in Bacterial Genomes That Combines Similarity- and Composition-Based Strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Zhou, Z.; Luo, M.; Zhang, H.; Yin, Y.; Cai, Y.; Zhu, Z.-J. Metabolite Annotation from Knowns to Unknowns through Knowledge-Guided Multi-Layer Metabolic Networking. Nat. Commun. 2022, 13, 6656. [Google Scholar] [CrossRef] [PubMed]
- Davray, D.; Bawane, H.; Kulkarni, R. Non-Redundant Nature of Lactiplantibacillus plantarum Plasmidome Revealed by Comparative Genomic Analysis of 105 Strains. Food Microbiol. 2023, 109, 104153. [Google Scholar] [CrossRef] [PubMed]
- Wardman, J.F.; Bains, R.K.; Rahfeld, P.; Withers, S.G. Carbohydrate-Active Enzymes (CAZymes) in the Gut Microbiome. Nat. Rev. Microbiol. 2022, 20, 542–556. [Google Scholar] [CrossRef]
- Carpi, F.M.; Coman, M.M.; Silvi, S.; Picciolini, M.; Verdenelli, M.C.; Napolioni, V. Comprehensive Pan-Genome Analysis of Lactiplantibacillus plantarum Complete Genomes. J. Appl. Microbiol. 2022, 132, 592–604. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Niu, H.; Qu, Q.; Guo, D.; Wan, X.; Yang, Q.; Mo, Z.; Tan, S.; Xiang, Q.; Tian, X.; et al. Advancements in Lactiplantibacillus plantarum: Probiotic Characteristics, Gene Editing Technologies and Applications. Crit. Rev. Food Sci. Nutr. 2025, 1–22. [Google Scholar] [CrossRef]
- Kok, C.R.; Rose, D.; Hutkins, R. Predicting Personalized Responses to Dietary Fiber Interventions: Opportunities for Modulation of the Gut Microbiome to Improve Health. Annu. Rev. Food Sci. Technol. 2023, 14, 157–182. [Google Scholar] [CrossRef]
- White, B.A.; Lamed, R.; Bayer, E.A.; Flint, H.J. Biomass Utilization by Gut Microbiomes. Annu. Rev. Microbiol. 2014, 68, 279–296. [Google Scholar] [CrossRef]
- Raba, G.; Luis, A.S. Mucin Utilization by Gut Microbiota: Recent Advances on Characterization of Key Enzymes. Essays Biochem. 2023, 67, 345–353. [Google Scholar] [CrossRef]
- Peng, Z.; Ehrmann, M.A.; Waldhuber, A.; Niemeyer, C.; Miethke, T.; Frick, J.-S.; Xiong, T.; Vogel, R.F. Phosphotransferase Systems in Enterococcus faecalis OG1RF Enhance Anti-Stress Capacity in Vitro and in Vivo. Res. Microbiol. 2017, 168, 558–566. [Google Scholar] [CrossRef]
- Quazi, F.; Molday, R.S. Lipid Transport by Mammalian ABC Proteins. Essays Biochem. 2011, 50, 265–290. [Google Scholar] [CrossRef]
- van Hoek, M.L.; Hoang, K.V.; Gunn, J.S. Two-Component Systems in Francisella Species. Front. Cell. Infect. Microbiol. 2019, 9, 198. [Google Scholar] [CrossRef] [PubMed]
- Groisman, E.A. Feedback Control of Two-Component Regulatory Systems. Annu. Rev. Microbiol. 2016, 70, 103–124. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Song, H.; Luo, Z.; Wu, H.; Chen, L.; Wang, Y.; Cui, H.; Zhang, Y.; Wang, B.; Li, W.; et al. Acetylcholine Ameliorates Colitis by Promoting IL-10 Secretion of Monocytic Myeloid-Derived Suppressor Cells through the nAChR/ERK Pathway. Proc. Natl. Acad. Sci. USA 2021, 118, e2017762118. [Google Scholar] [CrossRef] [PubMed]
- Ananth, M.R.; Rajebhosale, P.; Kim, R.; Talmage, D.A.; Role, L.W. Basal Forebrain Cholinergic Signalling: Development, Connectivity and Roles in Cognition. Nat. Rev. Neurosci. 2023, 24, 233–251. [Google Scholar] [CrossRef]
- Zapata-Pérez, R.; Wanders, R.J.A.; van Karnebeek, C.D.M.; Houtkooper, R.H. NAD+ Homeostasis in Human Health and Disease. EMBO Mol. Med. 2021, 13, e13943. [Google Scholar] [CrossRef]
- Wohl, J.; Petersen, M. Phenolic Metabolism in the Hornwort Anthoceros Agrestis: 4-Coumarate CoA Ligase and 4-Hydroxybenzoate CoA Ligase. Plant Cell Rep. 2020, 39, 1129–1141. [Google Scholar] [CrossRef]
- Zhang, G.; Lian, Y.; Li, Q.; Zhou, S.; Zhang, L.; Chen, L.; Tang, J.; Liu, H.; Li, N.; Pan, Q.; et al. Vagal Pathway Activation Links Chronic Stress to Decline in Intestinal Stem Cell Function. Cell Stem Cell 2025, 32, 778–794.e10. [Google Scholar] [CrossRef]
- Hosomi, K.; Saito, M.; Park, J.; Murakami, H.; Shibata, N.; Ando, M.; Nagatake, T.; Konishi, K.; Ohno, H.; Tanisawa, K.; et al. Oral Administration of Blautia wexlerae Ameliorates Obesity and Type 2 Diabetes via Metabolic Remodeling of the Gut Microbiota. Nat. Commun. 2022, 13, 4477. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, R.-S.; Handy, D.E.; Loscalzo, J. NAD(H) and NADP(H) Redox Couples and Cellular Energy Metabolism. Antioxid. Redox Signal 2018, 28, 251–272. [Google Scholar] [CrossRef]
- Myakala, K.; Wang, X.X.; Shults, N.V.; Krawczyk, E.; Jones, B.A.; Yang, X.; Rosenberg, A.Z.; Ginley, B.; Sarder, P.; Brodsky, L.; et al. NAD Metabolism Modulates Inflammation and Mitochondria Function in Diabetic Kidney Disease. J. Biol. Chem. 2023, 299, 104975. [Google Scholar] [CrossRef]
- Abdellatif, M.; Sedej, S.; Kroemer, G. NAD+ Metabolism in Cardiac Health, Aging, and Disease. Circulation 2021, 144, 1795–1817. [Google Scholar] [CrossRef] [PubMed]
- Yaku, K.; Nakagawa, T. NAD+ Precursors in Human Health and Disease: Current Status and Future Prospects. Antioxid. Redox Signal 2023, 39, 1133–1149. [Google Scholar] [CrossRef] [PubMed]
- Chini, C.C.S.; Cordeiro, H.S.; Tran, N.L.K.; Chini, E.N. NAD Metabolism: Role in Senescence Regulation and Aging. Aging Cell 2024, 23, e13920. [Google Scholar] [CrossRef]
- Strefeler, A.; Blanco-Fernandez, J.; Jourdain, A.A. Nucleosides Are Overlooked Fuels in Central Carbon Metabolism. Trends Endocrinol. Metab. 2024, 35, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Idzko, M.; Ferrari, D.; Riegel, A.-K.; Eltzschig, H.K. Extracellular Nucleotide and Nucleoside Signaling in Vascular and Blood Disease. Blood 2014, 124, 1029–1037. [Google Scholar] [CrossRef]
- Kaur, T.; Weadick, B.; Mace, T.A.; Desai, K.; Odom, H.; Govindarajan, R. Nucleoside Transporters and Immunosuppressive Adenosine Signaling in the Tumor Microenvironment: Potential Therapeutic Opportunities. Pharmacol. Ther. 2022, 240, 108300. [Google Scholar] [CrossRef]
- Proffitt, C.; Bidkhori, G.; Lee, S.; Tebani, A.; Mardinoglu, A.; Uhlen, M.; Moyes, D.L.; Shoaie, S. Genome-Scale Metabolic Modelling of the Human Gut Microbiome Reveals Changes in the Glyoxylate and Dicarboxylate Metabolism in Metabolic Disorders. iScience 2022, 25, 104513. [Google Scholar] [CrossRef]
- Chen, G.; Ye, G.; Zhang, X.; Liu, X.; Tu, Y.; Ye, Z.; Liu, J.; Guo, Q.; Wang, Z.; Wang, L.; et al. Metabolomics Reveals Protection of Resveratrol in Diet-Induced Metabolic Risk Factors in Abdominal Muscle. Cell. Physiol. Biochem. 2018, 45, 1136–1148. [Google Scholar] [CrossRef]
- Prochownik, E.V.; Wang, H. The Metabolic Fates of Pyruvate in Normal and Neoplastic Cells. Cells 2021, 10, 762. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, H. Reprogramming of Glucose, Fatty Acid and Amino Acid Metabolism for Cancer Progression. Cell. Mol. Life Sci. 2016, 73, 377–392. [Google Scholar] [CrossRef]
- Liao, Y.; Chen, Q.; Liu, L.; Huang, H.; Sun, J.; Bai, X.; Jin, C.; Li, H.; Sun, F.; Xiao, X.; et al. Amino Acid Is a Major Carbon Source for Hepatic Lipogenesis. Cell Metab. 2024, 36, 2437–2448.e8. [Google Scholar] [CrossRef]
- Asakura, J.; Nagao, M.; Shinohara, M.; Hosooka, T.; Kuwahara, N.; Nishimori, M.; Tanaka, H.; Satomi-Kobayashi, S.; Matsui, S.; Sasaki, T.; et al. Impaired Cardiac Branched-Chain Amino Acid Metabolism in a Novel Model of Diabetic Cardiomyopathy. Cardiovasc. Diabetol. 2025, 24, 167. [Google Scholar] [CrossRef]
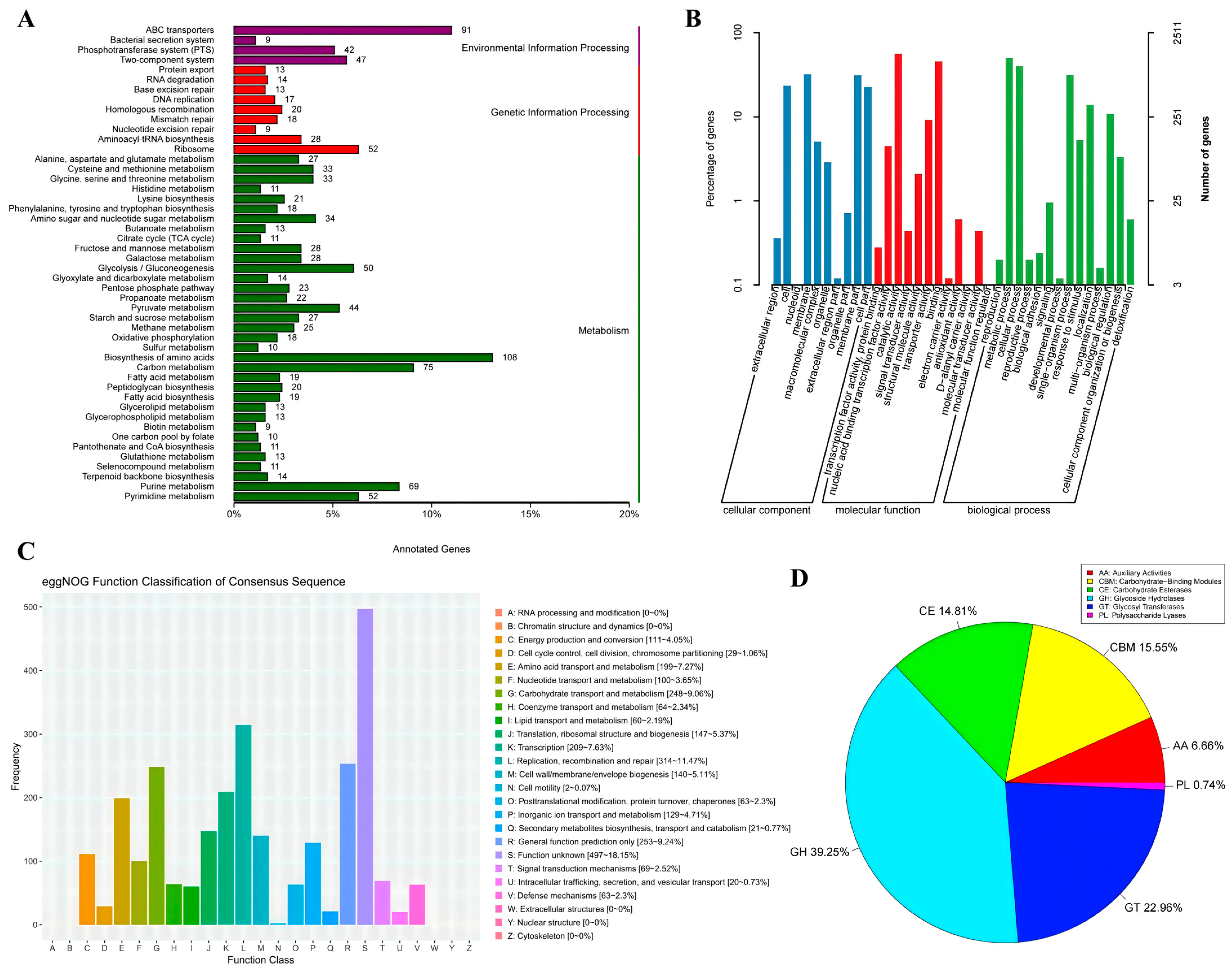
| Type | Number | Percentage (%) |
|---|---|---|
| AA | 9 | 6.66 |
| CBM | 21 | 15.55 |
| CE | 20 | 14.81 |
| GH | 53 | 39.25 |
| GT | 31 | 22.96 |
| PL | 1 | 0.74 |
| VFDB Gene Name | VFDB Gene Function | Virulence Factor Name | Virulence Factor ID | VFDB Target ID | E-Value | Percent Identity (%) |
|---|---|---|---|---|---|---|
| hasC | UDP-glucose pyrophosphorylase | Hyaluronic acid capsule | VF0244 | VFG000964 | 3.5572 × 10−161 | 75.34 |
| clpP | ATP-dependent Clp protease proteolytic subunit | ClpP | VF0074 | VFG000077 | 1.13222 × 10−98 | 69.79 |
| cpsA | undecaprenyl diphosphate synthase | Capsule | VF0361 | VFG002190 | 8.90991 × 10−116 | 67.51 |
| bsh | bile salt hydrolase | BSH | VF0350 | VFG002162 | 2.25858 × 10−154 | 67.11 |
| cpsI | UDP-galactopyranose mutase | Capsule | VF0361 | VFG002182 | 1.31649 × 10−180 | 65.3 |
| msrA/B(pilB) | trifunctional thioredoxin/methionine sulfoxide reductase A/B protein | MsrAB | VF0456 | VFG037100 | 5.17036 × 10−60 | 64.03 |
| gnd | 6-phosphogluconate dehydrogenase | Capsule | VF0560 | VFG048830 | 0 | 62.87 |
| clpE | ATP-dependent protease | ClpE | VF0073 | VFG000080 | 0 | 60.76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Meng, K.; Liu, G.; Chen, Z.; Han, Y.; Yang, P.; Zhang, R.; Cai, H. Whole Genome Sequencing and Extracellular Metabolite Profiling of Lactiplantibacillus plantarum FRT4: Insights into Probiotic Functionality. Biology 2025, 14, 1167. https://doi.org/10.3390/biology14091167
Huang Y, Meng K, Liu G, Chen Z, Han Y, Yang P, Zhang R, Cai H. Whole Genome Sequencing and Extracellular Metabolite Profiling of Lactiplantibacillus plantarum FRT4: Insights into Probiotic Functionality. Biology. 2025; 14(9):1167. https://doi.org/10.3390/biology14091167
Chicago/Turabian StyleHuang, Yuyin, Kun Meng, Guohua Liu, Zhimin Chen, Yunsheng Han, Peilong Yang, Rui Zhang, and Hongying Cai. 2025. "Whole Genome Sequencing and Extracellular Metabolite Profiling of Lactiplantibacillus plantarum FRT4: Insights into Probiotic Functionality" Biology 14, no. 9: 1167. https://doi.org/10.3390/biology14091167
APA StyleHuang, Y., Meng, K., Liu, G., Chen, Z., Han, Y., Yang, P., Zhang, R., & Cai, H. (2025). Whole Genome Sequencing and Extracellular Metabolite Profiling of Lactiplantibacillus plantarum FRT4: Insights into Probiotic Functionality. Biology, 14(9), 1167. https://doi.org/10.3390/biology14091167








