Prognostic Significance of WWOX/HIF1A Ratio in Cancer Subtypes: Insights into Metabolism, ECM, and EMT
Simple Summary
Abstract
1. Introduction
1.1. WWOX/HIF1A and Breast Cancer
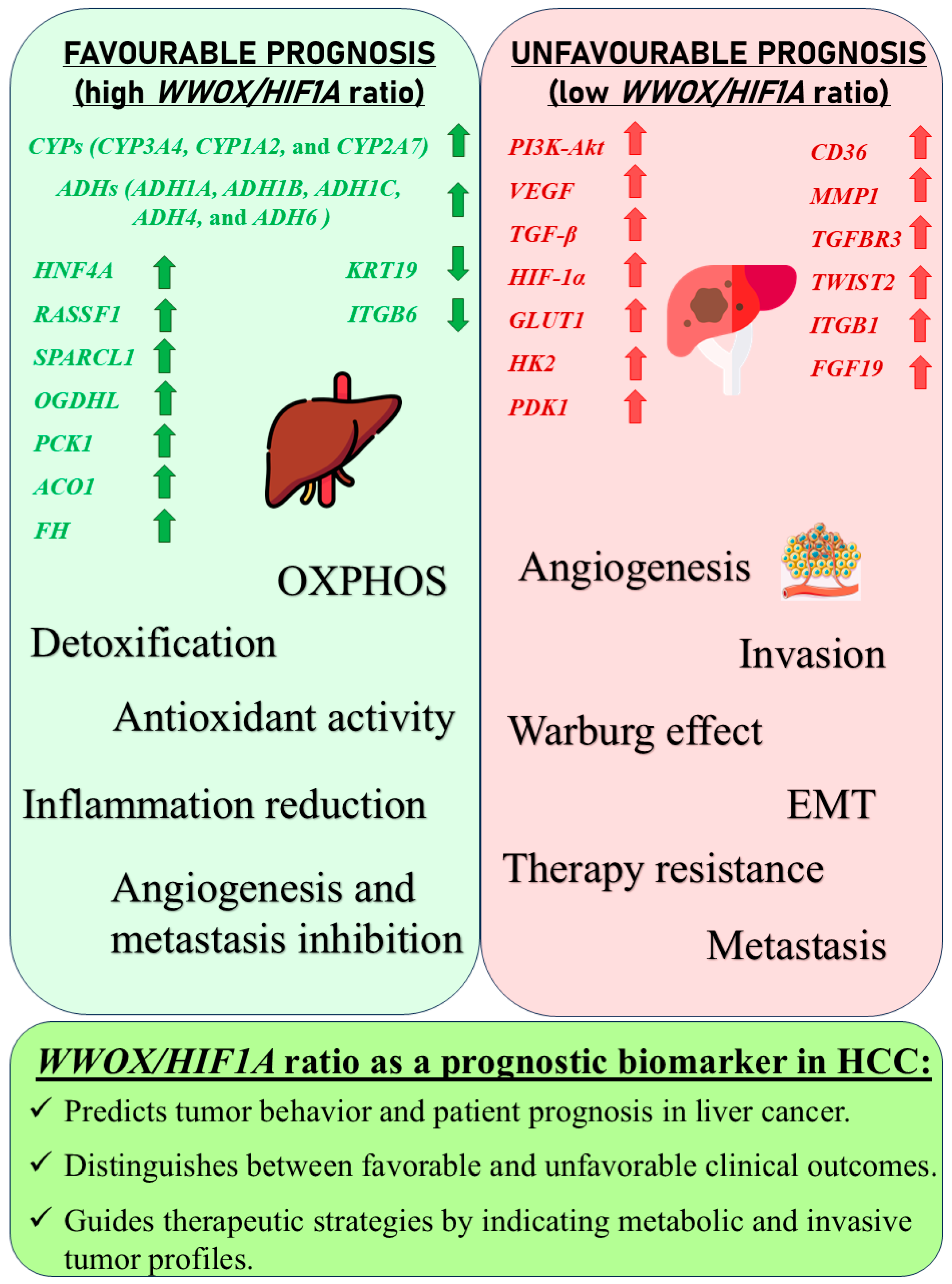
1.2. WWOX/HIF1A and Hepatocellular Carcinoma
1.3. WWOX/HIF1A and Brain Tumours
2. Materials and Methods
2.1. Data Extraction
2.2. Cutpoint Determination
2.3. Multivariate Analysis
2.4. Differential Gene Expression Analysis
2.5. Association Analysis Between DEGs and Patient Prognosis
2.6. Reproducibility and Data Availability
3. Results
3.1. Breast Cancer-Basal Subtype
3.2. Breast Cancer-HER2 Subtype
3.3. Breast Cancer-Luminal A Subtype
3.4. Breast Cancer-Luminal B Subtype
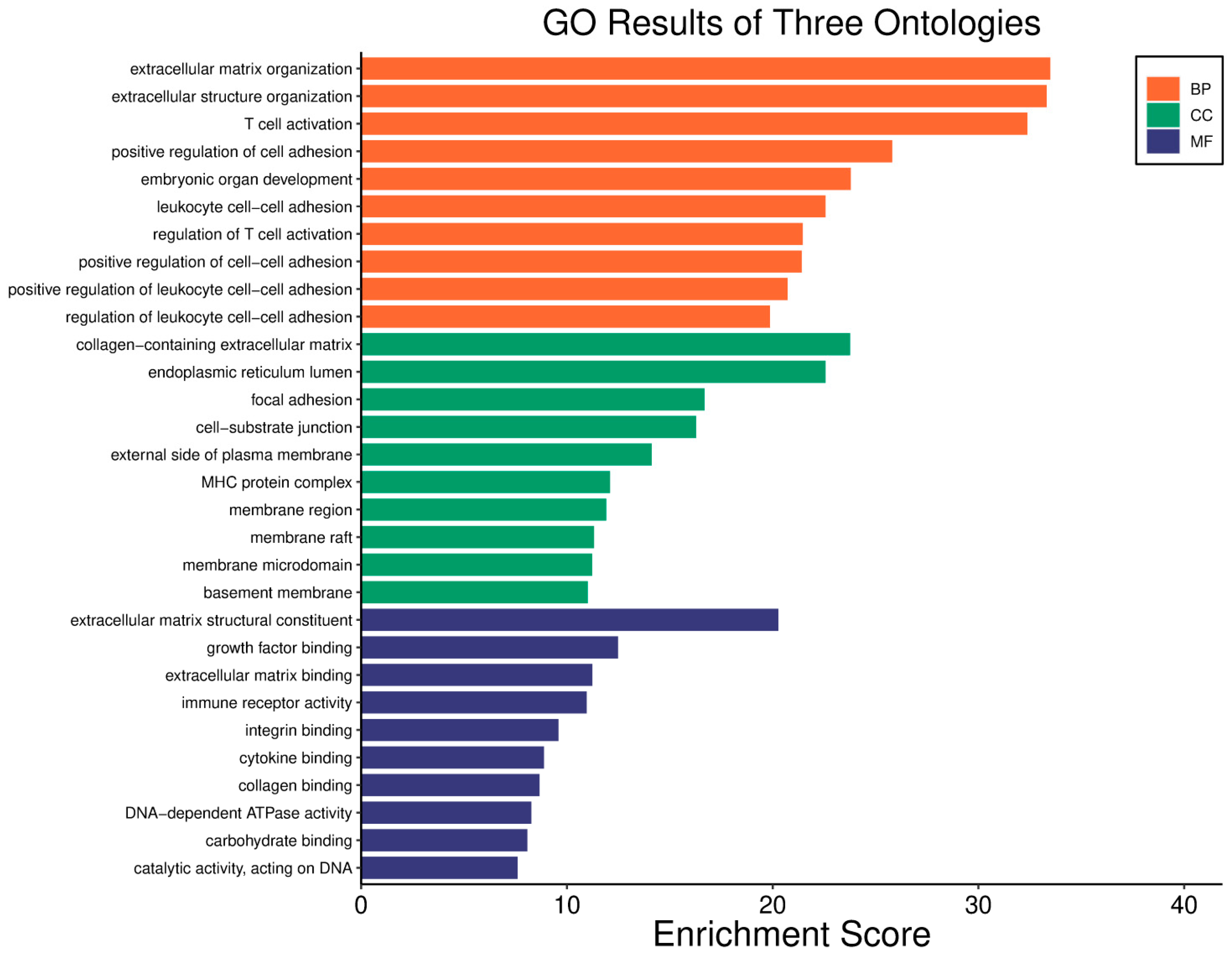
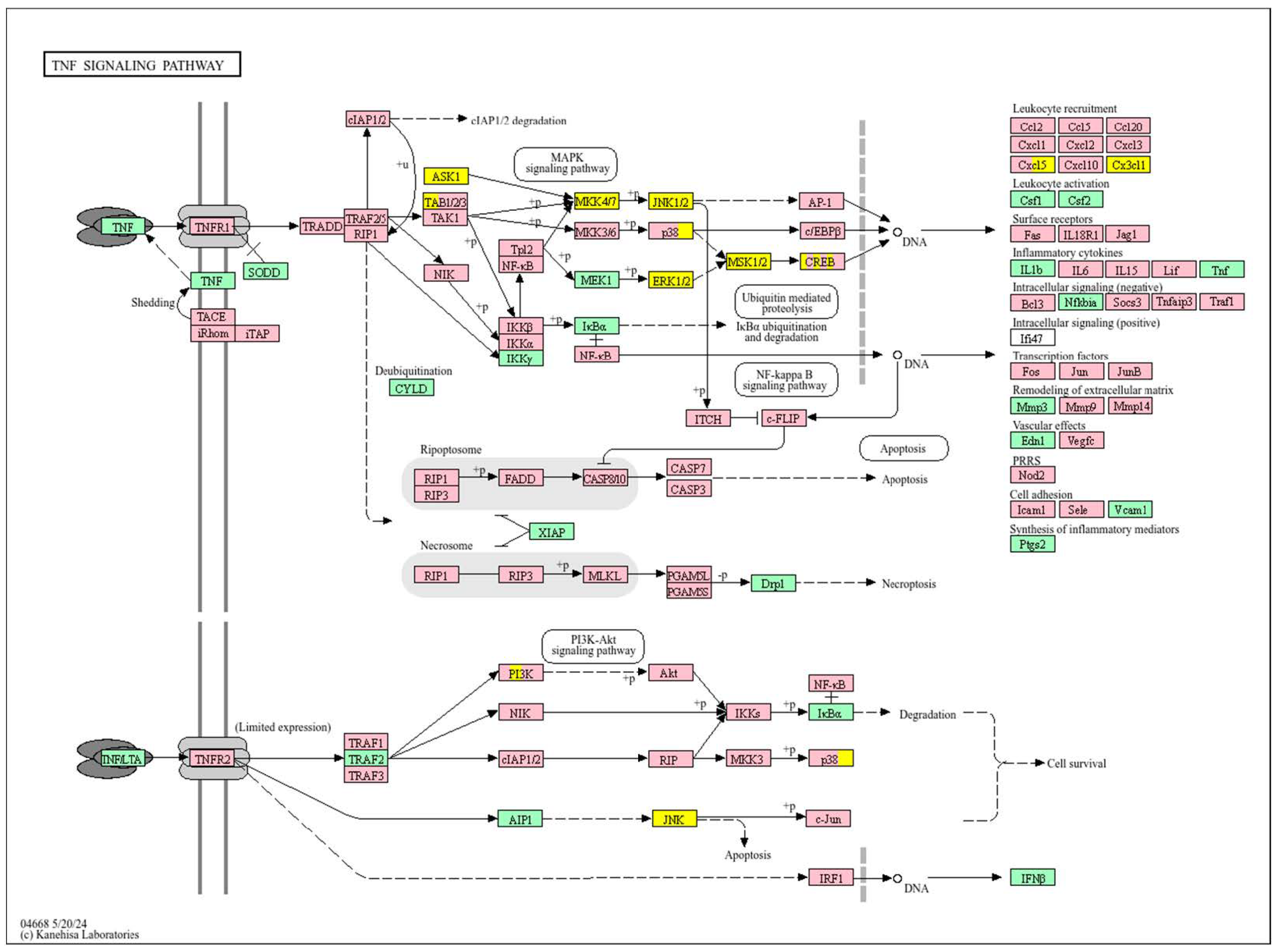
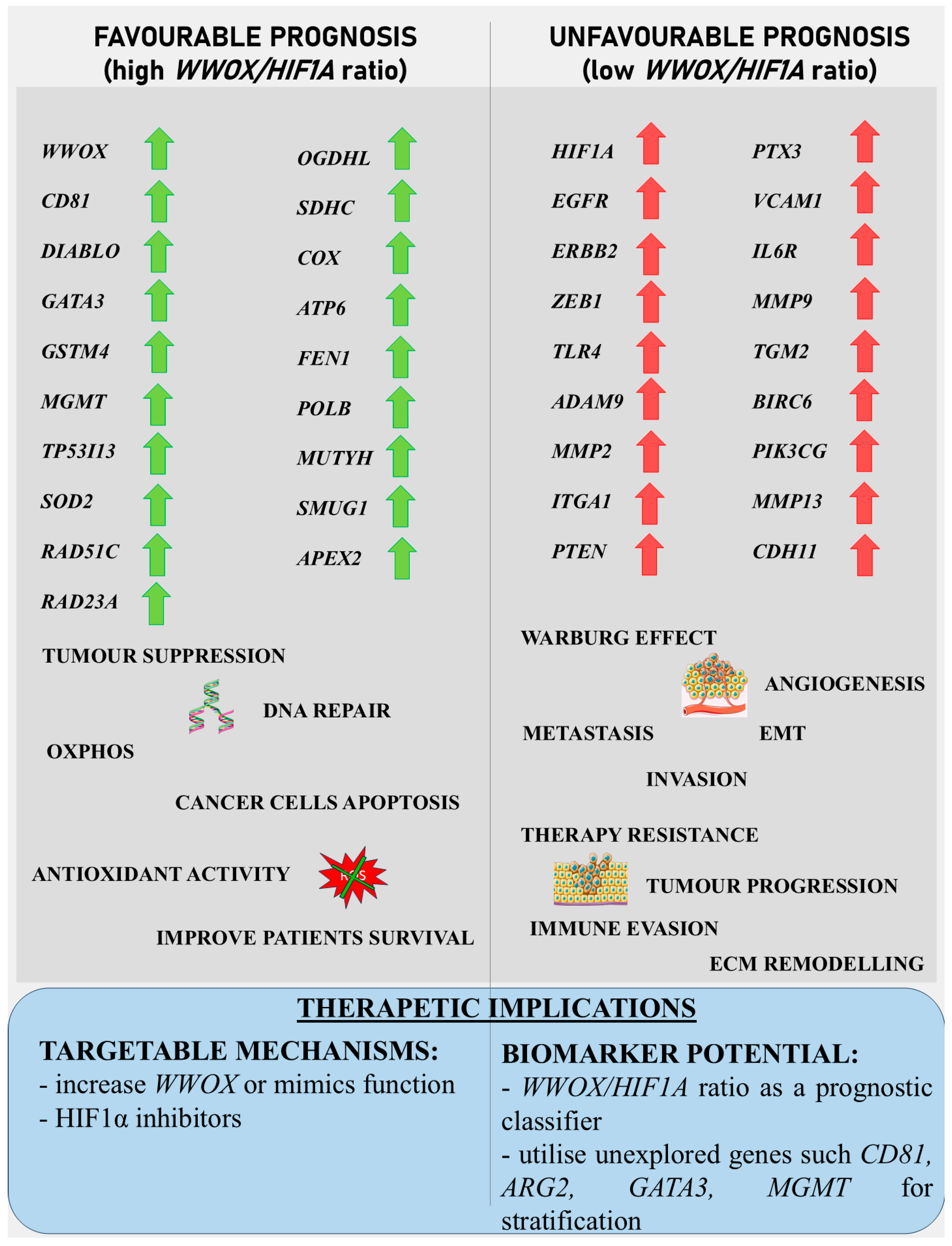
3.5. Hepatocellular Carcinoma
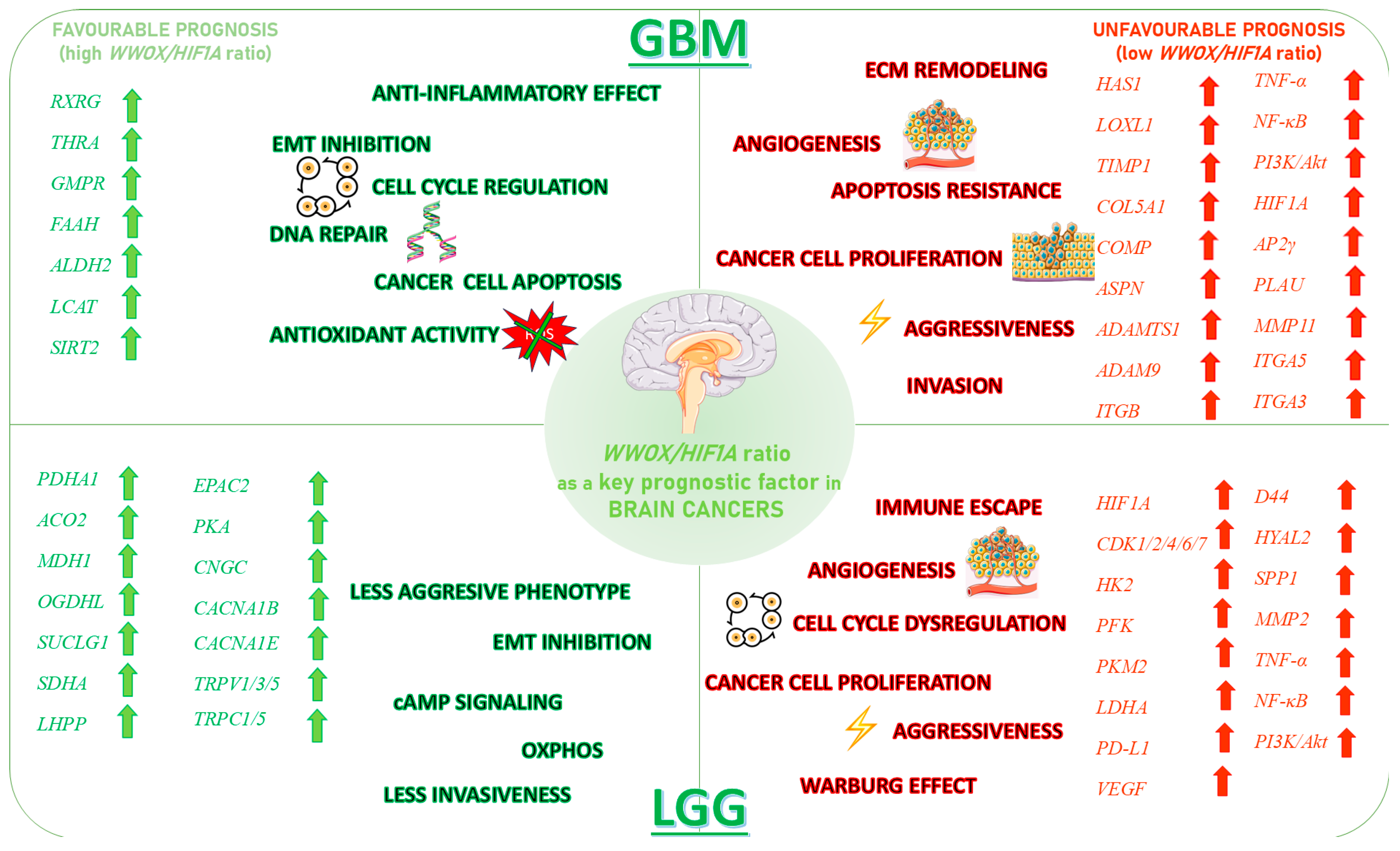
3.6. Glioblastoma
3.7. Low Grade Glioma
4. Discussion
4.1. Metabolic Reprogramming, EMT, Invasiveness, and Angiogenesis Across Tumour Types
4.2. Cell Proliferation and Signalling Pathways Across Tumour Types
4.3. Genomic Integrity and DNA Repair Across Tumour Types
4.4. Immune Regulation and Inflammation Across Tumour Types
4.5. Underexplored Genes and Clinical Potential Across Tumour Types
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GBM | Glioblastoma multiforme |
| LGG | Low-Grade Glioma |
| HCC | Hepatocellular Carcinoma |
| ECM | Extracellular matrix |
| VHL | Von Hippel–Lindau |
| OXPHOS | Oxidative Phosphorylation |
| TME | Tumour Microenvironment |
| NK | Natural Killer |
| TNBC | Triple Negative Breast Cancer |
| NAFLD | Non-alcoholic Fatty Liver Disease |
| TCGA | The Cancer Genome Atlas |
| GDC | Genomic Data Commons |
| FDR | False Discovery Rate |
| EMT | Epithelial–Mesenchymal Transition |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| TCA | Tricarboxylic Acid Cycle |
| ROS | Reactive Oxygen Species |
References
- Brown, J.S.; Amend, S.R.; Austin, R.H.; Gatenby, R.A.; Hammarlund, E.U.; Pienta, K.J. Updating the Definition of Cancer. Mol. Cancer Res. 2023, 21, 1142–1147. [Google Scholar] [CrossRef]
- Marbaniang, C.; Kma, L. Dysregulation of Glucose Metabolism by Oncogenes and Tumor Suppressors in Cancer Cells. Asian Pac. J. Cancer Prev. 2018, 19, 2377–2390. [Google Scholar] [CrossRef]
- Tufail, M.; Jiang, C.-H.; Li, N. Altered Metabolism in Cancer: Insights into Energy Pathways and Therapeutic Targets. Mol. Cancer 2024, 23, 203. [Google Scholar] [CrossRef]
- Tufail, M.; Hu, J.-J.; Liang, J.; He, C.-Y.; Wan, W.-D.; Huang, Y.-Q.; Jiang, C.-H.; Wu, H.; Li, N. Hallmarks of Cancer Resistance. iScience 2024, 27, 109979. [Google Scholar] [CrossRef] [PubMed]
- Bhat, G.R.; Sethi, I.; Sadida, H.Q.; Rah, B.; Mir, R.; Algehainy, N.; Albalawi, I.A.; Masoodi, T.; Subbaraj, G.K.; Jamal, F.; et al. Cancer Cell Plasticity: From Cellular, Molecular, and Genetic Mechanisms to Tumor Heterogeneity and Drug Resistance. Cancer Metastasis Rev. 2024, 43, 197–228. [Google Scholar] [CrossRef] [PubMed]
- Bose, S.; Zhang, C.; Le, A. Glucose Metabolism in Cancer: The Warburg Effect and Beyond. In The Heterogeneity of Cancer Metabolism; Le, A., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2021; Volume 1311, pp. 3–15. ISBN 978-3-030-65767-3. [Google Scholar]
- Jaworska, M.; Szczudło, J.; Pietrzyk, A.; Shah, J.; Trojan, S.E.; Ostrowska, B.; Kocemba-Pilarczyk, K.A. The Warburg Effect: A Score for Many Instruments in the Concert of Cancer and Cancer Niche Cells. Pharmacol. Rep. 2023, 75, 876–890. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Jose, C.; Bellance, N.; Rossignol, R. Choosing Between Glycolysis and Oxidative Phosphorylation: A Tumor’s Dilemma? Biochim. Biophys. Acta (BBA)-Bioenerg. 2011, 1807, 552–561. [Google Scholar] [CrossRef]
- Zhao, Y.; Xing, C.; Deng, Y.; Ye, C.; Peng, H. HIF-1α Signaling: Essential Roles in Tumorigenesis and Implications in Targeted Therapies. Genes Dis. 2024, 11, 234–251. [Google Scholar] [CrossRef]
- Abu-Remaileh, M.; Aqeilan, R.I. Tumor Suppressor WWOX Regulates Glucose Metabolism via HIF1α Modulation. Cell Death Differ. 2014, 21, 1805–1814. [Google Scholar] [CrossRef] [PubMed]
- Pospiech, K.; Płuciennik, E.; Bednarek, A.K. WWOX Tumor Suppressor Gene in Breast Cancer, a Historical Perspective and Future Directions. Front. Oncol. 2018, 8, 345. [Google Scholar] [CrossRef]
- Bednarek, A.K.; Keck-Waggoner, C.L.; Daniel, R.L.; Laflin, K.J.; Bergsagel, P.L.; Kiguchi, K.; Brenner, A.J.; Aldaz, C.M. WWOX, the FRA16D Gene, Behaves as a Suppressor of Tumor Growth. Cancer Res. 2001, 61, 8068–8073. [Google Scholar] [PubMed]
- Baryła, I.; Styczeń-Binkowska, E.; Bednarek, A.K. Alteration of WWOX in Human Cancer: A Clinical View. Exp. Biol. Med. 2015, 240, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Abu-Remaileh, M.; Abu-Remaileh, M.; Akkawi, R.; Knani, I.; Udi, S.; Pacold, M.E.; Tam, J.; Aqeilan, R.I. WWOX Somatic Ablation in Skeletal Muscles Alters Glucose Metabolism. Mol. Metab. 2019, 22, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Lee, J.; Abba, M.C.; Chen, J.; Aldaz, C.M. Delineating WWOX Protein Interactome by Tandem Affinity Purification-Mass Spectrometry: Identification of Top Interactors and Key Metabolic Pathways Involved. Front. Oncol. 2018, 8, 591. [Google Scholar] [CrossRef]
- Baryła, I.; Styczeń-Binkowska, E.; Płuciennik, E.; Kośla, K.; Bednarek, A.K. The WWOX/HIF1A Axis Downregulation Alters Glucose Metabolism and Predispose to Metabolic Disorders. Int. J. Mol. Sci. 2022, 23, 3326. [Google Scholar] [CrossRef]
- Gnocchi, D.; Sabbà, C.; Massimi, M.; Mazzocca, A. Metabolism as a New Avenue for Hepatocellular Carcinoma Therapy. Int. J. Mol. Sci. 2023, 24, 3710. [Google Scholar] [CrossRef]
- Cortes Ballen, A.I.; Amosu, M.; Ravinder, S.; Chan, J.; Derin, E.; Slika, H.; Tyler, B. Metabolic Reprogramming in Glioblastoma Multiforme: A Review of Pathways and Therapeutic Targets. Cells 2024, 13, 1574. [Google Scholar] [CrossRef]
- Barba, I.; Carrillo-Bosch, L.; Seoane, J. Targeting the Warburg Effect in Cancer: Where Do We Stand? Int. J. Mol. Sci. 2024, 25, 3142. [Google Scholar] [CrossRef]
- Duraj, T.; García-Romero, N.; Carrión-Navarro, J.; Madurga, R.; Ortiz De Mendivil, A.; Prat-Acin, R.; Garcia-Cañamaque, L.; Ayuso-Sacido, A. Beyond the Warburg Effect: Oxidative and Glycolytic Phenotypes Coexist within the Metabolic Heterogeneity of Glioblastoma. Cells 2021, 10, 202. [Google Scholar] [CrossRef]
- Chen, S.; Xu, Y.; Zhuo, W.; Zhang, L. The Emerging Role of Lactate in Tumor Microenvironment and Its Clinical Relevance. Cancer Lett. 2024, 590, 216837. [Google Scholar] [CrossRef]
- Lee, T.-Y. Lactate: A Multifunctional Signaling Molecule. Yeungnam Univ. J. Med. 2021, 38, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Certo, M.; Llibre, A.; Lee, W.; Mauro, C. Understanding Lactate Sensing and Signalling. Trends Endocrinol. Metab. 2022, 33, 722–735. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, W.; Tang, C.; Hu, Y.; Yi, K.; Xu, X.; Chen, Z. Silencing AREG Enhances Sensitivity to Irradiation by Suppressing the PI3K/AKT Signaling Pathway in Colorectal Cancer Cells. Biol. Targets Ther. 2024, 18, 273–284. [Google Scholar] [CrossRef]
- Végran, F.; Boidot, R.; Michiels, C.; Sonveaux, P.; Feron, O. Lactate Influx through the Endothelial Cell Monocarboxylate Transporter MCT1 Supports an NF-κB/IL-8 Pathway That Drives Tumor Angiogenesis. Cancer Res. 2011, 71, 2550–2560. [Google Scholar] [CrossRef] [PubMed]
- Conroy, S.; Kruyt, F.A.E.; Wagemakers, M.; Bhat, K.P.L.; Den Dunnen, W.F.A. IL-8 Associates with a pro-Angiogenic and Mesenchymal Subtype in Glioblastoma. Oncotarget 2018, 9, 15721–15731. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Santucci, C.; Mignozzi, S.; Malvezzi, M.; Boffetta, P.; Collatuzzo, G.; Levi, F.; La Vecchia, C.; Negri, E. European Cancer Mortality Predictions for the Year 2024 with Focus on Colorectal Cancer. Ann. Oncol. 2024, 35, 308–316. [Google Scholar] [CrossRef]
- Zhi, S.; Chen, C.; Huang, H.; Zhang, Z.; Zeng, F.; Zhang, S. Hypoxia-Inducible Factor in Breast Cancer: Role and Target for Breast Cancer Treatment. Front. Immunol. 2024, 15, 1370800. [Google Scholar] [CrossRef]
- Saheed, E.S.; Aromolaran, R.F.; Atoyebi, A.D.; Adeleke, F.C.; Otuyalo, A.I.; Edozie, P.K. Mechanism of the Warburg Effect and Its Role in Breast Cancer Immunotherapy. Discov. Med. 2024, 1, 110. [Google Scholar] [CrossRef]
- Liu, S.; Li, Y.; Yuan, M.; Song, Q.; Liu, M. Correlation Between the Warburg Effect and Progression of Triple-Negative Breast Cancer. Front. Oncol. 2023, 12, 1060495. [Google Scholar] [CrossRef]
- Piard, J.; Hawkes, L.; Milh, M.; Villard, L.; Borgatti, R.; Romaniello, R.; Fradin, M.; Capri, Y.; Héron, D.; Nougues, M.-C.; et al. The Phenotypic Spectrum of WWOX-Related Disorders: 20 Additional Cases of WOREE Syndrome and Review of the Literature. Genet. Med. 2019, 21, 1308–1318. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Fabbri, M.; Druck, T.; Qin, H.R.; Han, S.-Y.; Huebner, K. Inhibition of Breast Cancer Cell Growth In Vitro and In Vivo: Effect of Restoration of Wwox Expression. Clin. Cancer Res. 2007, 13, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ge, F.; Chen, W.; Yang, R.; Zhou, Z.; Chang, N.; Chen, C.; Zou, T.; Liu, R.; Tan, J.; Ren, G. WWOX Suppresses KLF5 Expression and Breast Cancer Cell Growth. Chin. J. Cancer Res. 2014, 26, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Abdeen, S.K.; Aqeilan, R.I. Decoding the Link Between WWOX and P53 in Aggressive Breast Cancer. Cell Cycle 2019, 18, 1177–1186. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Li, P.; Zhou, C.; Liu, P. The Downregulation of WWOX Induces Epithelial–Mesenchymal Transition and Enhances Stemness and Chemoresistance in Breast Cancer. Exp. Biol. Med. 2018, 243, 1066–1073. [Google Scholar] [CrossRef]
- Ludes-Meyers, J.H.; Bednarek, A.K.; Popescu, N.C.; Bedford, M.; Aldaz, C.M. WWOX, the Common Chromosomal Fragile Site, FRA16D, Cancer Gene. Cytogenet. Genome Res. 2003, 100, 101–110. [Google Scholar] [CrossRef]
- Darawadi, B. Role of Tumor Suppressor P53 Family in Glucose Metabolism in Association with Diabetes. Adv. Cancer Chemother. Pharmacol. 2023, 1, 1–7. [Google Scholar] [CrossRef]
- Gao, Y.; Zhou, H.; Liu, G.; Wu, J.; Yuan, Y.; Shang, A. Tumor Microenvironment: Lactic Acid Promotes Tumor Development. J. Immunol. Res. 2022, 2022, 3119375. [Google Scholar] [CrossRef] [PubMed]
- Suteau, V.; Bukasa-Kakamba, J.; Virjogh-Cenciu, B.; Adenis, A.; Sabbah, N.; Drak Alsibai, K. Pathological Significance of GLUT-1 Expression in Breast Cancer Cells in Diabetic and Obese Patients: The French Guiana Study. Cancers 2022, 14, 437. [Google Scholar] [CrossRef]
- Abu-Remaileh, M.; Khalaileh, A.; Pikarsky, E.; Aqeilan, R.I. WWOX Controls Hepatic HIF1α to Suppress Hepatocyte Proliferation and Neoplasia. Cell Death Dis. 2018, 9, 511. [Google Scholar] [CrossRef]
- Chen, H.; Wu, Q.; Peng, L.; Cao, T.; Deng, M.-L.; Liu, Y.-W.; Huang, J.; Hu, Y.; Fu, N.; Zhou, K.-B.; et al. Mechanism, Clinical Significance, and Treatment Strategy of Warburg Effect in Hepatocellular Carcinoma. J. Nanomater. 2021, 2021, 5164100. [Google Scholar] [CrossRef]
- Nekvindova, J.; Mrkvicova, A.; Zubanova, V.; Hyrslova Vaculova, A.; Anzenbacher, P.; Soucek, P.; Radova, L.; Slaby, O.; Kiss, I.; Vondracek, J.; et al. Hepatocellular Carcinoma: Gene Expression Profiling and Regulation of Xenobiotic-Metabolizing Cytochromes P450. Biochem. Pharmacol. 2020, 177, 113912. [Google Scholar] [CrossRef]
- Collino, A.; Termanini, A.; Nicoli, P.; Diaferia, G.; Polletti, S.; Recordati, C.; Castiglioni, V.; Caruso, D.; Mitro, N.; Natoli, G.; et al. Sustained Activation of Detoxification Pathways Promotes Liver Carcinogenesis in Response to Chronic Bile Acid-Mediated Damage. PLoS Genet. 2018, 14, e1007380. [Google Scholar] [CrossRef]
- Guo, Y.; Xiao, Z.; Yang, L.; Gao, Y.; Zhu, Q.; Hu, L.; Huang, D.; Xu, Q. Hypoxia-inducible Factors in Hepatocellular Carcinoma (Review). Oncol. Rep. 2019, 43, 3–15. [Google Scholar] [CrossRef]
- Cho, K.J.; Choi, N.K.; Shin, M.H.; Chong, A.R. Clinical Usefulness of FDG-PET in Patients with Hepatocellular Carcinoma Undergoing Surgical Resection. Ann. Hepato-Biliary-Pancreat. Surg. 2017, 21, 194. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, Y.; Xiong, Y.; Gong, Y.; Lu, J.; Zhang, Y.; Wang, D.; Liu, Z.; Shi, X. Research Progress of Warburg Effect in Hepatocellular Carcinoma. Front. Biosci.-Landmark 2024, 29, 178. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Chen, W.; Sun, J.; Atyah, M.; Yin, Y.; Zhang, W.; Guo, L.; Ye, Q.; Dong, Q.; Shi, Y.; et al. Low Expression of WW Domain-containing Oxidoreductase Associates with Hepatocellular Carcinoma Aggressiveness and Recurrence after Curative Resection. Cancer Med. 2018, 7, 3031–3043. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, C.; Zheng, X.; Li, S.; Zhang, W.; Kang, Z.; Yin, S.; Chen, J.; Chen, F.; Li, W. Warburg Effect-Related Risk Scoring Model to Assess Clinical Significance and Immunity Characteristics of Glioblastoma. Cancer Med. 2023, 12, 20639–20654. [Google Scholar] [CrossRef] [PubMed]
- Ming, C.; Zheng, L.; Zhang, M.; Wu, J.; Feng, Y.; Wu, Y.; Wang, X.; Wang, X. The Diagnostic and Prognositic Significances of HIF1A in Glioma Patients, a Meta-Analysis and Bioinformatics Approach. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Li, H.; Niu, X.; Cheng, R. Prevalence, Prognostic and Clinicopathological Value of HIF-1α in Glioblastoma Patients: A Systematic Review and Meta-Analysis. Neurosurg. Rev. 2024, 47, 860. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Tian, R.; Liu, W.; Fei, Z.; Long, Q.; Wang, X.; Zhang, X. The Expression and Significance of HIF-1α and GLUT-3 in Glioma. Brain Res. 2009, 1304, 149–154. [Google Scholar] [CrossRef]
- Li, J.; Zhu, S.; Tong, J.; Hao, H.; Yang, J.; Liu, Z.; Wang, Y. Suppression of Lactate Dehydrogenase A Compromises Tumor Progression by Downregulation of the Warburg Effect in Glioblastoma. NeuroReport 2016, 27, 110–115. [Google Scholar] [CrossRef]
- Zhang, S.; Lachance, B.B.; Mattson, M.P.; Jia, X. Glucose Metabolic Crosstalk and Regulation in Brain Function and Diseases. Prog. Neurobiol. 2021, 204, 102089. [Google Scholar] [CrossRef]
- Șovrea, A.S.; Boșca, B.; Melincovici, C.S.; Constantin, A.-M.; Crintea, A.; Mărginean, M.; Dronca, E.; Jianu, M.E.; Suflețel, R.; Gonciar, D.; et al. Multiple Faces of the Glioblastoma Microenvironment. Int. J. Mol. Sci. 2022, 23, 595. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Cao, R.; Wang, X.; Zhang, Y.; Wang, P.; Gao, H.; Li, C.; Yang, F.; Zeng, R.; Wei, P.; et al. Mitochondrial PKM2 Regulates Oxidative Stress-Induced Apoptosis by Stabilizing Bcl2. Cell Res. 2017, 27, 329–351. [Google Scholar] [CrossRef]
- Zhao, J.; Ma, X.; Gao, P.; Han, X.; Zhao, P.; Xie, F.; Liu, M. Advancing Glioblastoma Treatment by Targeting Metabolism. Neoplasia 2024, 51, 100985. [Google Scholar] [CrossRef]
- Kośla, K.; Nowakowska, M.; Pospiech, K.; Bednarek, A.K. WWOX Modulates the Gene Expression Profile in the T98G Glioblastoma Cell Line Rendering Its Phenotype Less Malignant. Oncol. Rep. 2014, 32, 1362–1368. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kosla, K.; Pluciennik, E.; Kurzyk, A.; Jesionek-Kupnicka, D.; Kordek, R.; Potemski, P.; Bednarek, A.K. Molecular Analysis of WWOX Expression Correlation with Proliferation and Apoptosis in Glioblastoma Multiforme. J. Neurooncol. 2011, 101, 207–213. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kałuzińska-Kołat, Ż.; Kołat, D.; Kośla, K.; Płuciennik, E.; Bednarek, A.K. Molecular Landscapes of Glioblastoma Cell Lines Revealed a Group of Patients That Do Not Benefit from WWOX Tumor Suppressor Expression. Front. Neurosci. 2023, 17, 1260409. [Google Scholar] [CrossRef]
- Ogłuszka, M.; Orzechowska, M.; Jędroszka, D.; Witas, P.; Bednarek, A.K. Evaluate Cutpoints: Adaptable Continuous Data Distribution System for Determining Survival in Kaplan-Meier Estimator. Comput. Methods Programs Biomed. 2019, 177, 133–139. [Google Scholar] [CrossRef]
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D.; Van Den Brand, T. Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics, version 3.5.2; Comprehensive R Archive Network (CRAN): Vienna, Austria, 2007. [Google Scholar] [CrossRef]
- Husson, F.; Josse, J.; Le, S.; Mazet, J. FactoMineR: Multivariate Exploratory Data Analysis and Data Mining, version 2.11; Comprehensive R Archive Network (CRAN): Vienna, Austria, 2006. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. Gplots: Various R Programming Tools for Plotting Data, version 3.2.0; Comprehensive R Archive Network (CRAN): Vienna, Austria, 2005. [Google Scholar] [CrossRef]
- Therneau, T.M.; Lumley, T.; Atkinson, E.; Crowson, C. Survival: Survival Analysis, version 3.8-3; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Wickham, H. ggplot2, Elegant Graphics for Data Analysis, 2nd ed.; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A Free Online Platform for Data Visualization and Graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG Mapping Tools for Uncovering Hidden Features in Biological Data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, G.P.; Nozell, S.E.; Benveniste, E.N. NF-κB and STAT3 Signaling in Glioma: Targets for Future Therapies. Expert Rev. Neurother. 2010, 10, 575–586. [Google Scholar] [CrossRef]
- Jalouli, M. Emerging Role of Hypoxia-Inducible Factors (HIFs) in Modulating Autophagy: Perspectives on Cancer Therapy. Int. J. Mol. Sci. 2025, 26, 1752. [Google Scholar] [CrossRef] [PubMed]
- Appelhoff, R.J.; Tian, Y.-M.; Raval, R.R.; Turley, H.; Harris, A.L.; Pugh, C.W.; Ratcliffe, P.J.; Gleadle, J.M. Differential Function of the Prolyl Hydroxylases PHD1, PHD2, and PHD3 in the Regulation of Hypoxia-Inducible Factor. J. Biol. Chem. 2004, 279, 38458–38465. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Yang, L.; Peng, X.; Wei, S.; Fan, Q.; Yang, S.; Li, X.; Li, B.; Jin, H.; Wu, B.; et al. Autonomous Glucose Metabolic Reprogramming of Tumour Cells under Hypoxia: Opportunities for Targeted Therapy. J. Exp. Clin. Cancer Res. 2020, 39, 185. [Google Scholar] [CrossRef]
- Baryła, I.; Kośla, K.; Bednarek, A.K. WWOX and Metabolic Regulation in Normal and Pathological Conditions. J. Mol. Med. 2022, 100, 1691–1702. [Google Scholar] [CrossRef]
- Xu, Y.; Yan, Y.-C.; Hu, Y.-K.; Fang, L.-S.; Li, Q.; Xu, J.; Yan, H.-C. WWOX Regulates the Elf5/Snail1 Pathway to Effect Epithelial-Mesenchymal Transition of Ovarian Carcinoma Cells in Vitro. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1041–1053. [Google Scholar] [CrossRef]
- Płuciennik, E.; Nowakowska, M.; Pospiech, K.; Stępień, A.; Wołkowicz, M.; Gałdyszyńska, M.; Popęda, M.; Wójcik-Krowiranda, K.; Bieńkiewicz, A.; Bednarek, A.K. The Role of WWOX Tumor Suppressor Gene in the Regulation of EMT Process via Regulation of CDH1-ZEB1-VIM Expression in Endometrial Cancer. Int. J. Oncol. 2015, 46, 2639–2648. [Google Scholar] [CrossRef]
- Zhang, P.; Sun, Y.; Ma, L. ZEB1: At the Crossroads of Epithelial-Mesenchymal Transition, Metastasis and Therapy Resistance. Cell Cycle 2015, 14, 481–487. [Google Scholar] [CrossRef]
- Muz, B.; De La Puente, P.; Azab, F.; Azab, A.K. The Role of Hypoxia in Cancer Progression, Angiogenesis, Metastasis, and Resistance to Therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular Principles of Metastasis: A Hallmark of Cancer Revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Hypoxia-Inducible Factors in Physiology and Medicine. Cell 2012, 148, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Broadfield, L.A.; Pane, A.A.; Talebi, A.; Swinnen, J.V.; Fendt, S.-M. Lipid Metabolism in Cancer: New Perspectives and Emerging Mechanisms. Dev. Cell 2021, 56, 1363–1393. [Google Scholar] [CrossRef]
- Jia, X.; He, X.; Huang, C.; Li, J.; Dong, Z.; Liu, K. Protein Translation: Biological Processes and Therapeutic Strategies for Human Diseases. Signal Transduct. Target. Ther. 2024, 9, 44. [Google Scholar] [CrossRef]
- Martinez-Ruiz, G.; Maldonado, V.; Ceballos-Cancino, G.; Grajeda, J.P.R.; Melendez-Zajgla, J. Role of Smac/DIABLO in Cancer Progression. J. Exp. Clin. Cancer Res. 2008, 27, 48. [Google Scholar] [CrossRef]
- Bacha, R.; Alwisi, N.; Ismail, R.; Pedersen, S.; Al-Mansoori, L. Unveiling GATA3 Signaling Pathways in Health and Disease: Mechanisms, Implications, and Therapeutic Potential. Cells 2024, 13, 2127. [Google Scholar] [CrossRef]
- Chatterjee, A.; Gupta, S. The Multifaceted Role of Glutathione S-Transferases in Cancer. Cancer Lett. 2018, 433, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zhang, L.; Wei, Q.; Shao, A. O6-Methylguanine-DNA Methyltransferase (MGMT): Challenges and New Opportunities in Glioma Chemotherapy. Front. Oncol. 2020, 9, 1547. [Google Scholar] [CrossRef]
- Choudhury, F.K. Mitochondrial Redox Metabolism: The Epicenter of Metabolism during Cancer Progression. Antioxidants 2021, 10, 1838. [Google Scholar] [CrossRef]
- Bobustuc, G.C.; Kassam, A.B.; Rovin, R.A.; Jeudy, S.; Smith, J.S.; Isley, B.; Singh, M.; Paranjpe, A.; Srivenugopal, K.S.; Konduri, S.D. MGMT Inhibition in ER Positive Breast Cancer Leads to CDC2, TOP2A, AURKB, CDC20, KIF20A, Cyclin A2, Cyclin B2, Cyclin D1, ERα and Survivin Inhibition and Enhances Response to Temozolomide. Oncotarget 2018, 9, 29727–29742. [Google Scholar] [CrossRef]
- Rutkovsky, A.C.; Yeh, E.S.; Guest, S.T.; Findlay, V.J.; Muise-Helmericks, R.C.; Armeson, K.; Ethier, S.P. Eukaryotic Initiation Factor 4E-Binding Protein as an Oncogene in Breast Cancer. BMC Cancer 2019, 19, 491. [Google Scholar] [CrossRef]
- Sung, H.-C.; Chang, K.-S.; Chen, S.-T.; Hsu, S.-Y.; Lin, Y.-H.; Hou, C.-P.; Feng, T.-H.; Tsui, K.-H.; Juang, H.-H. Metallothionein 2A with Antioxidant and Antitumor Activity Is Upregulated by Caffeic Acid Phenethyl Ester in Human Bladder Carcinoma Cells. Antioxidants 2022, 11, 1509. [Google Scholar] [CrossRef] [PubMed]
- Stefkovich, M.; Titchenell, P.M. Location Matters: Mitochondrial Arginase 2 as a Treatment for Metabolic Disease? Cell. Mol. Gastroenterol. Hepatol. 2024, 17, 883–884. [Google Scholar] [CrossRef]
- Wolosowicz, M.; Prokopiuk, S.; Kaminski, T.W. The Complex Role of Matrix Metalloproteinase-2 (MMP-2) in Health and Disease. Int. J. Mol. Sci. 2024, 25, 13691. [Google Scholar] [CrossRef]
- Jiang, J.; Ren, H.; Xu, Y.; Wudu, M.; Wang, Q.; Liu, Z.; Su, H.; Jiang, X.; Zhang, Y.; Zhang, B.; et al. TRIM67 Promotes the Proliferation, Migration, and Invasion of Non-Small-Cell Lung Cancer by Positively Regulating the Notch Pathway. J. Cancer 2020, 11, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Tekpli, X.; Lien, T.; Røssevold, A.H.; Nebdal, D.; Borgen, E.; Ohnstad, H.O.; Kyte, J.A.; Vallon-Christersson, J.; Fongaard, M.; Due, E.U.; et al. An Independent Poor-Prognosis Subtype of Breast Cancer Defined by a Distinct Tumor Immune Microenvironment. Nat. Commun. 2019, 10, 5499. [Google Scholar] [CrossRef]
- Storch, J.; Corsico, B. The Multifunctional Family of Mammalian Fatty Acid–Binding Proteins. Annu. Rev. Nutr. 2023, 43, 25–54. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Y.; Wang, Y.; Wang, C.; Chen, X.; Xiong, Y.; Liu, L.; Yuan, X.; Tang, H.; Shu, C.; et al. CYP4F2-Catalyzed Metabolism of Arachidonic Acid Promotes Stromal Cell-Mediated Immunosuppression in Non–Small Cell Lung Cancer. Cancer Res. 2022, 82, 4016–4030. [Google Scholar] [CrossRef] [PubMed]
- Bonin, S.; Pracella, D.; Barbazza, R.; Dotti, I.; Boffo, S.; Stanta, G. PI3K/AKT Signaling in Breast Cancer Molecular Subtyping and Lymph Node Involvement. Dis. Markers 2019, 2019, 7832376. [Google Scholar] [CrossRef]
- Vleugel, M.M.; Greijer, A.E.; Shvarts, A.; Van Der Groep, P.; Van Berkel, M.; Aarbodem, Y.; Van Tinteren, H.; Harris, A.L.; Van Diest, P.J.; Van Der Wall, E. Differential Prognostic Impact of Hypoxia Induced and Diffuse HIF-1α Expression in Invasive Breast Cancer. J. Clin. Pathol. 2005, 58, 172–177. [Google Scholar] [CrossRef]
- Liu, X.; Li, T.; Kong, D.; You, H.; Kong, F.; Tang, R. Prognostic Implications of Alcohol Dehydrogenases in Hepatocellular Carcinoma. BMC Cancer 2020, 20, 1204. [Google Scholar] [CrossRef]
- Favero, A.; Segatto, I.; Capuano, A.; Mattevi, M.C.; Rampioni Vinciguerra, G.L.; Musco, L.; D’Andrea, S.; Dall’Acqua, A.; Gava, C.; Perin, T.; et al. Loss of the Extracellular Matrix Glycoprotein EMILIN1 Accelerates Δ16HER2-Driven Breast Cancer Initiation in Mice. Npj Breast Cancer 2024, 10, 5. [Google Scholar] [CrossRef]
- Chen, T.-H.; Wang, H.-C.; Chang, C.-J.; Lee, S.-Y. Mitochondrial Glutathione in Cellular Redox Homeostasis and Disease Manifestation. Int. J. Mol. Sci. 2024, 25, 1314. [Google Scholar] [CrossRef]
- Rascio, F.; Spadaccino, F.; Rocchetti, M.T.; Castellano, G.; Stallone, G.; Netti, G.S.; Ranieri, E. The Pathogenic Role of PI3K/AKT Pathway in Cancer Onset and Drug Resistance: An Updated Review. Cancers 2021, 13, 3949. [Google Scholar] [CrossRef] [PubMed]
- Kośla, K.; Płuciennik, E.; Styczeń-Binkowska, E.; Nowakowska, M.; Orzechowska, M.; Bednarek, A.K. The WWOX Gene Influences Cellular Pathways in the Neuronal Differentiation of Human Neural Progenitor Cells. Front. Cell. Neurosci. 2019, 13, 391. [Google Scholar] [CrossRef] [PubMed]
- Ashida, R.; Okamura, Y.; Ohshima, K.; Kakuda, Y.; Uesaka, K.; Sugiura, T.; Ito, T.; Yamamoto, Y.; Sugino, T.; Urakami, K.; et al. CYP3A4 Gene Is a Novel Biomarker for Predicting a Poor Prognosis in Hepatocellular Carcinoma. Cancer Genom. Proteom. 2017, 14, 445–453. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, G.; Tan, X.; Ke, K.; Zhao, B.; Cheng, N.; Dang, Y.; Liao, N.; Wang, F.; Zheng, X.; et al. MT1G Serves as a Tumor Suppressor in Hepatocellular Carcinoma by Interacting with P53. Oncogenesis 2019, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiang, L.; Hu, Y.; Xiao, C.; Xu, N.; Zhou, J.; Zhou, X. Metallothionein 1H (MT1H) Functions as a Tumor Suppressor in Hepatocellular Carcinoma through Regulating Wnt/β-Catenin Signaling Pathway. BMC Cancer 2017, 17, 161. [Google Scholar] [CrossRef]
- Yang, R.; Roshani, D.; Gao, B.; Li, P.; Shang, N. Metallothionein: A Comprehensive Review of Its Classification, Structure, Biological Functions, and Applications. Antioxidants 2024, 13, 825. [Google Scholar] [CrossRef]
- Ozensoy Guler, O.; Ercan, E.; Uysal, T.K. GSH-Related Enzyme Activity and Tumor Relation: Glutathione Peroxidase and Glutathione Reductase Status under Hypoxia in HepG2 Cells. Turk. J. Biochem. 2024, 49, 252–258. [Google Scholar] [CrossRef]
- Hsiao, Y.-F.; Cheng, S.-B.; Lai, C.-Y.; Liu, H.-T.; Huang, S.-C.; Huang, Y.-C. The Prognostic Role of Glutathione and Its Related Antioxidant Enzymes in the Recurrence of Hepatocellular Carcinoma. Nutrients 2021, 13, 4071. [Google Scholar] [CrossRef]
- Zhao, Z.; Cui, T.; Wei, F.; Zhou, Z.; Sun, Y.; Gao, C.; Xu, X.; Zhang, H. Wnt/β-Catenin Signaling Pathway in Hepatocellular Carcinoma: Pathogenic Role and Therapeutic Target. Front. Oncol. 2024, 14, 1367364. [Google Scholar] [CrossRef]
- Tian, L.-Y.; Smit, D.J.; Jücker, M. The Role of PI3K/AKT/mTOR Signaling in Hepatocellular Carcinoma Metabolism. Int. J. Mol. Sci. 2023, 24, 2652. [Google Scholar] [CrossRef] [PubMed]
- Paskeh, M.D.A.; Ghadyani, F.; Hashemi, M.; Abbaspour, A.; Zabolian, A.; Javanshir, S.; Razzazan, M.; Mirzaei, S.; Entezari, M.; Goharrizi, M.A.S.B.; et al. Biological Impact and Therapeutic Perspective of Targeting PI3K/Akt Signaling in Hepatocellular Carcinoma: Promises and Challenges. Pharmacol. Res. 2023, 187, 106553. [Google Scholar] [CrossRef]
- Fontana, F.; Giannitti, G.; Marchesi, S.; Limonta, P. The PI3K/Akt Pathway and Glucose Metabolism: A Dangerous Liaison in Cancer. Int. J. Biol. Sci. 2024, 20, 3113–3125. [Google Scholar] [CrossRef]
- Xie, B.; Zen, Q.; Wang, X.; He, X.; Xie, Y.; Zhang, Z.; Li, H. ACK1 Promotes Hepatocellular Carcinoma Progression via Downregulating WWOX and Activating AKT Signaling. Int. J. Oncol. 2015, 46, 2057–2066. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Luo, X.; Li, W.; Zhong, J.; Cao, J.; Zhu, S.; Chen, X.; Zhou, R.; Shang, C.; Chen, Y. TNF-α Is a Potential Therapeutic Target to Overcome Sorafenib Resistance in Hepatocellular Carcinoma. eBioMedicine 2019, 40, 446–456. [Google Scholar] [CrossRef]
- Sinha, R.A.; Yen, P.M. Metabolic Messengers: Thyroid Hormones. Nat. Metab. 2024, 6, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Leal, A.S.; Hung, P.-Y.; Chowdhury, A.S.; Liby, K.T. Retinoid X Receptor Agonists as Selective Modulators of the Immune System for the Treatment of Cancer. Pharmacol. Ther. 2023, 252, 108561. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ni, F.; Sun, D.; Peng, Y.; Zhao, Y.; Wu, X.; Li, S.; Qi, X.; He, X.; Li, M.; et al. Glucagon Enhances Chemotherapy Efficacy By Inhibition of Tumor Vessels in Colorectal Cancer. Adv. Sci. 2024, 11, 2307271. [Google Scholar] [CrossRef]
- Hao, J.; Chen, Q.; Feng, Y.; Jiang, Q.; Sun, H.; Deng, B.; Huang, X.; Guan, J.; Chen, Q.; Liu, X.; et al. Combination Treatment with FAAH Inhibitors/URB597 and Ferroptosis Inducers Significantly Decreases the Growth and Metastasis of Renal Cell Carcinoma Cells via the PI3K-AKT Signaling Pathway. Cell Death Dis. 2023, 14, 247. [Google Scholar] [CrossRef]
- Liu, M.; Xu, Z.; Du, Z.; Wu, B.; Jin, T.; Xu, K.; Xu, L.; Li, E.; Xu, H. The Identification of Key Genes and Pathways in Glioma by Bioinformatics Analysis. J. Immunol. Res. 2017, 2017, 1278081. [Google Scholar] [CrossRef] [PubMed]
- Groblewska, M.; Litman-Zawadzka, A.; Mroczko, B. The Role of Selected Chemokines and Their Receptors in the Development of Gliomas. Int. J. Mol. Sci. 2020, 21, 3704. [Google Scholar] [CrossRef]
- Lin, C.; Chen, J.; Su, Z.; Liu, P.; Liu, Z.; Zhu, C.; Xu, D.; Lin, Z.; Xu, P.; Liu, G.; et al. A Calcium-Related Immune Signature in Prognosis Prediction of Patients With Glioma. Front. Cell Dev. Biol. 2021, 9, 723103. [Google Scholar] [CrossRef]
- Chou, P.-Y.; Lin, S.-R.; Lee, M.-H.; Schultz, L.; Sze, C.-I.; Chang, N.-S. A P53/TIAF1/WWOX Triad Exerts Cancer Suppression but May Cause Brain Protein Aggregation Due to P53/WWOX Functional Antagonism. Cell Commun. Signal. 2019, 17, 76. [Google Scholar] [CrossRef]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt Signal Transduction for Cancer Therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef]
- Lee, S.; Rauch, J.; Kolch, W. Targeting MAPK Signaling in Cancer: Mechanisms of Drug Resistance and Sensitivity. Int. J. Mol. Sci. 2020, 21, 1102. [Google Scholar] [CrossRef]
- Cuesta, C.; Arévalo-Alameda, C.; Castellano, E. The Importance of Being PI3K in the RAS Signaling Network. Genes 2021, 12, 1094. [Google Scholar] [CrossRef]
- Tomuleasa, C.; Tigu, A.-B.; Munteanu, R.; Moldovan, C.-S.; Kegyes, D.; Onaciu, A.; Gulei, D.; Ghiaur, G.; Einsele, H.; Croce, C.M. Therapeutic Advances of Targeting Receptor Tyrosine Kinases in Cancer. Signal Transduct. Target. Ther. 2024, 9, 201. [Google Scholar] [CrossRef]
- Cooper, O.; Bonert, V.S.; Rudnick, J.; Pressman, B.D.; Lo, J.; Salvatori, R.; Yuen, K.C.J.; Fleseriu, M.; Melmed, S. EGFR/ErbB2-Targeting Lapatinib Therapy for Aggressive Prolactinomas. J. Clin. Endocrinol. Metab. 2021, 106, e917–e925. [Google Scholar] [CrossRef]
- Sonnenblick, A.; Venet, D.; Brohée, S.; Pondé, N.; Sotiriou, C. pAKT Pathway Activation Is Associated with PIK3CA Mutations and Good Prognosis in Luminal Breast Cancer in Contrast to P-mTOR Pathway Activation. npj Breast Cancer 2019, 5, 7. [Google Scholar] [CrossRef]
- Yang, S.X.; Polley, E.; Lipkowitz, S. New Insights on PI3K/AKT Pathway Alterations and Clinical Outcomes in Breast Cancer. Cancer Treat. Rev. 2016, 45, 87–96. [Google Scholar] [CrossRef]
- Marei, H.E.; Althani, A.; Afifi, N.; Hasan, A.; Caceci, T.; Pozzoli, G.; Morrione, A.; Giordano, A.; Cenciarelli, C. P53 Signaling in Cancer Progression and Therapy. Cancer Cell Int. 2021, 21, 703. [Google Scholar] [CrossRef]
- Bai, W.; Ren, J.-S.; Li, K.; Jiang, Q. An Integrated Analysis Revealing the Angiogenic Function of TP53I11 in Tumor Microenvironment. Heliyon 2024, 10, e29504. [Google Scholar] [CrossRef] [PubMed]
- Orhan, E.; Velazquez, C.; Tabet, I.; Sardet, C.; Theillet, C. Regulation of RAD51 at the Transcriptional and Functional Levels: What Prospects for Cancer Therapy? Cancers 2021, 13, 2930. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Xu, J.; Cui, H. The Recent Research Progress of NF-κB Signaling on the Proliferation, Migration, Invasion, Immune Escape and Drug Resistance of Glioblastoma. Int. J. Mol. Sci. 2023, 24, 10337. [Google Scholar] [CrossRef] [PubMed]
- Fassl, A.; Geng, Y.; Sicinski, P. CDK4 and CDK6 Kinases: From Basic Science to Cancer Therapy. Science 2022, 375, eabc1495. [Google Scholar] [CrossRef]
- Ezzati, S.; Salib, S.; Balasubramaniam, M.; Aboud, O. Epidermal Growth Factor Receptor Inhibitors in Glioblastoma: Current Status and Future Possibilities. Int. J. Mol. Sci. 2024, 25, 2316. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, S.; Xin, Q.; Zhang, Y.; Wang, K.; Li, M. Recent Progress of CDK4/6 Inhibitors’ Current Practice in Breast Cancer. Cancer Gene Ther. 2024, 31, 1283–1291. [Google Scholar] [CrossRef]
- Pellarin, I.; Dall’Acqua, A.; Favero, A.; Segatto, I.; Rossi, V.; Crestan, N.; Karimbayli, J.; Belletti, B.; Baldassarre, G. Cyclin-Dependent Protein Kinases and Cell Cycle Regulation in Biology and Disease. Signal Transduct. Target. Ther. 2025, 10, 11. [Google Scholar] [CrossRef]
- Richard, S.A. EPAC2: A New and Promising Protein for Glioma Pathogenesis and Therapy. Oncol. Rev. 2020, 14, 446. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ge, H.; Lemon, B.; Vonderfecht, S.; Weiszmann, J.; Hecht, R.; Gupte, J.; Hager, T.; Wang, Z.; Lindberg, R.; et al. FGF19-Induced Hepatocyte Proliferation Is Mediated through FGFR4 Activation. J. Biol. Chem. 2010, 285, 5165–5170. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Chen, Y.; Zhu, J. LHPP Suppresses Proliferation, Migration, and Invasion and Promotes Apoptosis in Pancreatic Cancer. Biosci. Rep. 2020, 40, BSR20194142. [Google Scholar] [CrossRef]
- Farhan, M.; Silva, M.; Xingan, X.; Huang, Y.; Zheng, W. Role of FOXO Transcription Factors in Cancer Metabolism and Angiogenesis. Cells 2020, 9, 1586. [Google Scholar] [CrossRef]
- Wang, M.; Hu, T.; Xie, K.-Y. Dihydrofolate Reductase as a Predictor for Poor Response to Platinum-Based Chemotherapy in Epithelial Ovarian Cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 1723–1730. [Google Scholar]
- Gulliver, C.; Hoffmann, R.; Baillie, G.S. The Enigmatic Helicase DHX9 and Its Association With the Hallmarks of Cancer. Future Sci. OA 2021, 7, FSO650. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Xue, C.; Zeng, Y.; Yuan, X.; Chu, Q.; Jiang, S.; Wang, J.; Zhang, Y.; Zhu, D.; Li, L. Notch Signaling Pathway in Cancer: From Mechanistic Insights to Targeted Therapies. Signal Transduct. Target. Ther. 2024, 9, 128. [Google Scholar] [CrossRef]
- Soler, M.F.; Abaurrea, A.; Azcoaga, P.; Araujo, A.M.; Caffarel, M.M. New Perspectives in Cancer Immunotherapy: Targeting IL-6 Cytokine Family. J. Immunother. Cancer 2023, 11, e007530. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Y.; Fu, Y. The Critical Role of Toll-like Receptor-Mediated Signaling in Cancer Immunotherapy. Med. Drug Discov. 2022, 14, 100122. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, H.; Zheng, J.; Zeng, X.; Yu, W.; Liu, W.; Huang, G.; Zhang, Y.; Fu, W. Identification of Immune-Related Prognostic Biomarkers Based on the Tumor Microenvironment in 20 Malignant Tumor Types With Poor Prognosis. Front. Oncol. 2020, 10, 1008. [Google Scholar] [CrossRef]
- Lin, Y.; Song, Y.; Zhang, Y.; Li, X.; Kan, L.; Han, S. New Insights on Anti-Tumor Immunity of CD8+ T Cells: Cancer Stem Cells, Tumor Immune Microenvironment and Immunotherapy. J. Transl. Med. 2025, 23, 341. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Ke, S.; Chen, J.; Qin, Z.; Zhang, W.; Yuan, Y.; Meng, D.; Zhao, G.; Wu, K.; Li, B.; et al. FOXP3+ Regulatory T Cells and the Immune Escape in Solid Tumours. Front. Immunol. 2022, 13, 982986. [Google Scholar] [CrossRef]
- Mei, J.; Cai, Y.; Xu, R.; Yang, X.; Zhou, W.; Wang, H.; Liu, C. Characterization of the Clinical Significance of PD-1/PD-Ls Expression and Methylation in Patients With Low-Grade Glioma. Technol. Cancer Res. Treat. 2021, 20, 15330338211011970. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Song, G.; Yu, J. The Prognostic Significance of PD-L1 Expression in Patients with Glioma: A Meta-Analysis. Sci. Rep. 2017, 7, 4231. [Google Scholar] [CrossRef]
- Ding, X.; Wang, L.; Zhang, X.; Xu, J.; Li, P.; Liang, H.; Zhang, X.; Xie, L.; Zhou, Z.; Yang, J.; et al. The Relationship Between Expression of PD-L1 and HIF-1α in Glioma Cells Under Hypoxia. J. Hematol. Oncol. 2021, 14, 92. [Google Scholar] [CrossRef]
- Zhu, X.; Shi, G.; Lu, J.; Qian, X.; Wang, D. Potential Regulatory Mechanism of TNF-α/TNFR1/ANXA1 in Glioma Cells and Its Role in Glioma Cell Proliferation. Open Life Sci. 2022, 17, 208–220. [Google Scholar] [CrossRef]
- Puliyappadamba, V.T.; Hatanpaa, K.J.; Chakraborty, S.; Habib, A.A. The Role of NF-κB in the Pathogenesis of Glioma. Mol. Cell. Oncol. 2014, 1, e963478. [Google Scholar] [CrossRef]
- Xiao, Y.; Cui, G.; Ren, X.; Hao, J.; Zhang, Y.; Yang, X.; Wang, Z.; Zhu, X.; Wang, H.; Hao, C.; et al. A Novel Four-Gene Signature Associated With Immune Checkpoint for Predicting Prognosis in Lower-Grade Glioma. Front. Oncol. 2020, 10, 605737. [Google Scholar] [CrossRef]
- Morris, N.L.; Yeligar, S.M. Role of HIF-1α in Alcohol-Mediated Multiple Organ Dysfunction. Biomolecules 2018, 8, 170. [Google Scholar] [CrossRef]
- Vences-Catalán, F.; Rajapaksa, R.; Kuo, C.-C.; Miller, C.L.; Lee, A.; Ramani, V.C.; Jeffrey, S.S.; Levy, R.; Levy, S. Targeting the Tetraspanin CD81 Reduces Cancer Invasion and Metastasis. Proc. Natl. Acad. Sci. USA 2021, 118, e2018961118. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, C.; Pruneri, G.; Possanzini, P.; Manzotti, M.; Barile, M.; Feroce, I.; Colleoni, M.; Bonanni, B.; Maisonneuve, P.; Radice, P.; et al. Methylation of O 6-Methylguanine-DNA Methyltransferase (MGMT) Promoter Gene in Triple-Negative Breast Cancer Patients. Breast Cancer Res. Treat. 2012, 134, 131–137. [Google Scholar] [CrossRef]
- Fandy, T.E.; Shankar, S.; Srivastava, R.K. Smac/DIABLO Enhances the Therapeutic Potential of Chemotherapeutic Drugs and Irradiation, and Sensitizes TRAIL-Resistant Breast Cancer Cells. Mol. Cancer 2008, 7, 60. [Google Scholar] [CrossRef]
- Das, S.C.; Tasnim, W.; Rana, H.K.; Acharjee, U.K.; Islam, M.M.; Khatun, R. Comprehensive Bioinformatics and Machine Learning Analyses for Breast Cancer Staging Using TCGA Dataset. Brief. Bioinform. 2024, 26, bbae628. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Reprogramming of Fatty Acid Metabolism in Cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Bensaad, K.; Favaro, E.; Lewis, C.A.; Peck, B.; Lord, S.; Collins, J.M.; Pinnick, K.E.; Wigfield, S.; Buffa, F.M.; Li, J.-L.; et al. Fatty Acid Uptake and Lipid Storage Induced by HIF-1α Contribute to Cell Growth and Survival after Hypoxia-Reoxygenation. Cell Rep. 2014, 9, 349–365. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-T.; Ye, W.-T.; Wang, Y.-C.; Chen, P.-M.; Liu, J.-Y.; Tai, C.-K.; Tang, F.-Y.; Li, J.-R.; Liu, C.-C.; Chiang, E.-P.I. MTHFR Knockdown Assists Cell Defense against Folate Depletion Induced Chromosome Segregation and Uracil Misincorporation in DNA. Int. J. Mol. Sci. 2021, 22, 9392. [Google Scholar] [CrossRef]
- Xiong, X.; Zheng, L.-W.; Ding, Y.; Chen, Y.-F.; Cai, Y.-W.; Wang, L.-P.; Huang, L.; Liu, C.-C.; Shao, Z.-M.; Yu, K.-D. Breast Cancer: Pathogenesis and Treatments. Signal Transduct. Target. Ther. 2025, 10, 49. [Google Scholar] [CrossRef]
- Cornen, S.; Guille, A.; Adélaïde, J.; Addou-Klouche, L.; Finetti, P.; Saade, M.-R.; Manai, M.; Carbuccia, N.; Bekhouche, I.; Letessier, A.; et al. Candidate Luminal B Breast Cancer Genes Identified by Genome, Gene Expression and DNA Methylation Profiling. PLoS ONE 2014, 9, e81843. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Ren, J.; Zheng, J.; Zhang, F.; Han, M. Prognosis of Tumor Microenvironment in Luminal B-Type Breast Cancer. Dis. Markers 2022, 2022, 5621441. [Google Scholar] [CrossRef]
- Dalal, H.; Dahlgren, M.; Gladchuk, S.; Brueffer, C.; Gruvberger-Saal, S.K.; Saal, L.H. Clinical Associations of ESR2 (Estrogen Receptor Beta) Expression across Thousands of Primary Breast Tumors. Sci. Rep. 2022, 12, 4696. [Google Scholar] [CrossRef] [PubMed]
- Höller, A.; Nguyen-Sträuli, B.D.; Frauchiger-Heuer, H.; Ring, A. Diagnostic and Prognostic Biomarkers of Luminal Breast Cancer: Where Are We Now? Breast Cancer Targets Ther. 2023, 15, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Mamoor, S. Differential Expression of P53 Inducible Protein 11 (TP53I11) in Cancers of the Breast. OSF Preprints 2021. [Google Scholar] [CrossRef]
- Nieto-Jiménez, C.; Alcaraz-Sanabria, A.; Páez, R.; Pérez-Peña, J.; Corrales-Sánchez, V.; Pandiella, A.; Ocaña, A. DNA-Damage Related Genes and Clinical Outcome in Hormone Receptor Positive Breast Cancer. Oncotarget 2017, 8, 62834–62841. [Google Scholar] [CrossRef]
- Ito, K.; Kitajima, Y.; Kai, K.; Matsufuji, S.; Yamada, K.; Egawa, N.; Kitagawa, H.; Okuyama, K.; Tanaka, T.; Noshiro, H. Matrix Metalloproteinase-1 Expression Is Regulated by HIF-1-dependent and Epigenetic Mechanisms and Serves a Tumor-suppressive Role in Gastric Cancer Progression. Int. J. Oncol. 2021, 59, 102. [Google Scholar] [CrossRef]
- Cohen, A.L.; Holmen, S.L.; Colman, H. IDH1 and IDH2 Mutations in Gliomas. Curr. Neurol. Neurosci. Rep. 2013, 13, 345. [Google Scholar] [CrossRef]
- Liu, Y.; Tong, X.; Hu, W.; Chen, D. HDAC11: A Novel Target for Improved Cancer Therapy. Biomed. Pharmacother. 2023, 166, 115418. [Google Scholar] [CrossRef]
- Chen, S.; Cao, J.; Li, K.; Wan, S.; Yang, L. BIN1 in Cancer: Biomarker and Therapeutic Target. J. Cancer Res. Clin. Oncol. 2023, 149, 7933–7944. [Google Scholar] [CrossRef]
- Lin, D.-C.; Xu, L.; Chen, Y.; Yan, H.; Hazawa, M.; Doan, N.; Said, J.W.; Ding, L.-W.; Liu, L.-Z.; Yang, H.; et al. Genomic and Functional Analysis of the E3 Ligase PARK2 in Glioma. Cancer Res. 2015, 75, 1815–1827. [Google Scholar] [CrossRef]
- Kouri, F.M.; Jensen, S.A.; Stegh, A.H. The Role of Bcl-2 Family Proteins in Therapy Responses of Malignant Astrocytic Gliomas: Bcl2L12 and Beyond. Sci. World J. 2012, 2012, 838916. [Google Scholar] [CrossRef] [PubMed]
- Saccon, R.A.; Bunton-Stasyshyn, R.K.A.; Fisher, E.M.C.; Fratta, P. Is SOD1 Loss of Function Involved in Amyotrophic Lateral Sclerosis? Brain 2013, 136, 2342–2358. [Google Scholar] [CrossRef]
- Fernández-Calle, R.; Konings, S.C.; Frontiñán-Rubio, J.; García-Revilla, J.; Camprubí-Ferrer, L.; Svensson, M.; Martinson, I.; Boza-Serrano, A.; Venero, J.L.; Nielsen, H.M.; et al. APOE in the Bullseye of Neurodegenerative Diseases: Impact of the APOE Genotype in Alzheimer’s Disease Pathology and Brain Diseases. Mol. Neurodegener. 2022, 17, 62. [Google Scholar] [CrossRef]
- Guerrero-Rodríguez, S.L.; Mata-Cruz, C.; Pérez-Tapia, S.M.; Velasco-Velázquez, M.A. Role of CD36 in Cancer Progression, Stemness, and Targeting. Front. Cell Dev. Biol. 2022, 10, 1079076. [Google Scholar] [CrossRef]
- Lian, G.-Y.; Wang, Q.-M.; Mak, T.S.-K.; Huang, X.-R.; Yu, X.-Q.; Lan, H.-Y. Inhibition of Tumor Invasion and Metastasis by Targeting TGF-β-Smad-MMP2 Pathway with Asiatic Acid and Naringenin. Mol. Ther.-Oncolytics 2021, 20, 277–289. [Google Scholar] [CrossRef]
- Moustakas, A.; Heldin, C.-H. Mechanisms of TGFβ-Induced Epithelial–Mesenchymal Transition. J. Clin. Med. 2016, 5, 63. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Mo, J.; Dong, S.; Liao, Z.; Zhang, B.; Zhu, P. Integrinβ-1 in Disorders and Cancers: Molecular Mechanisms and Therapeutic Targets. Cell Commun. Signal. 2024, 22, 71. [Google Scholar] [CrossRef]
- DeLaForest, A.; Di Furio, F.; Jing, R.; Ludwig-Kubinski, A.; Twaroski, K.; Urick, A.; Pulakanti, K.; Rao, S.; Duncan, S.A. HNF4A Regulates the Formation of Hepatic Progenitor Cells from Human iPSC-Derived Endoderm by Facilitating Efficient Recruitment of RNA Pol II. Genes 2018, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Dubois, F.; Bergot, E.; Zalcman, G.; Levallet, G. RASSF1A, Puppeteer of Cellular Homeostasis, Fights Tumorigenesis, and Metastasis—An Updated Review. Cell Death Dis. 2019, 10, 928. [Google Scholar] [CrossRef] [PubMed]
- Naschberger, E.; Liebl, A.; Schellerer, V.S.; Schütz, M.; Britzen-Laurent, N.; Kölbel, P.; Schaal, U.; Haep, L.; Regensburger, D.; Wittmann, T.; et al. Matricellular Protein SPARCL1 Regulates Tumor Microenvironment–Dependent Endothelial Cell Heterogeneity in Colorectal Carcinoma. J. Clin. Investig. 2016, 126, 4187–4204. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.D.; Sarker, D.; Meyer, T.; Yau, T.; Macarulla, T.; Park, J.-W.; Choo, S.P.; Hollebecque, A.; Sung, M.W.; Lim, H.-Y.; et al. First-in-Human Phase I Study of Fisogatinib (BLU-554) Validates Aberrant FGF19 Signaling as a Driver Event in Hepatocellular Carcinoma. Cancer Discov. 2019, 9, 1696–1707. [Google Scholar] [CrossRef] [PubMed]
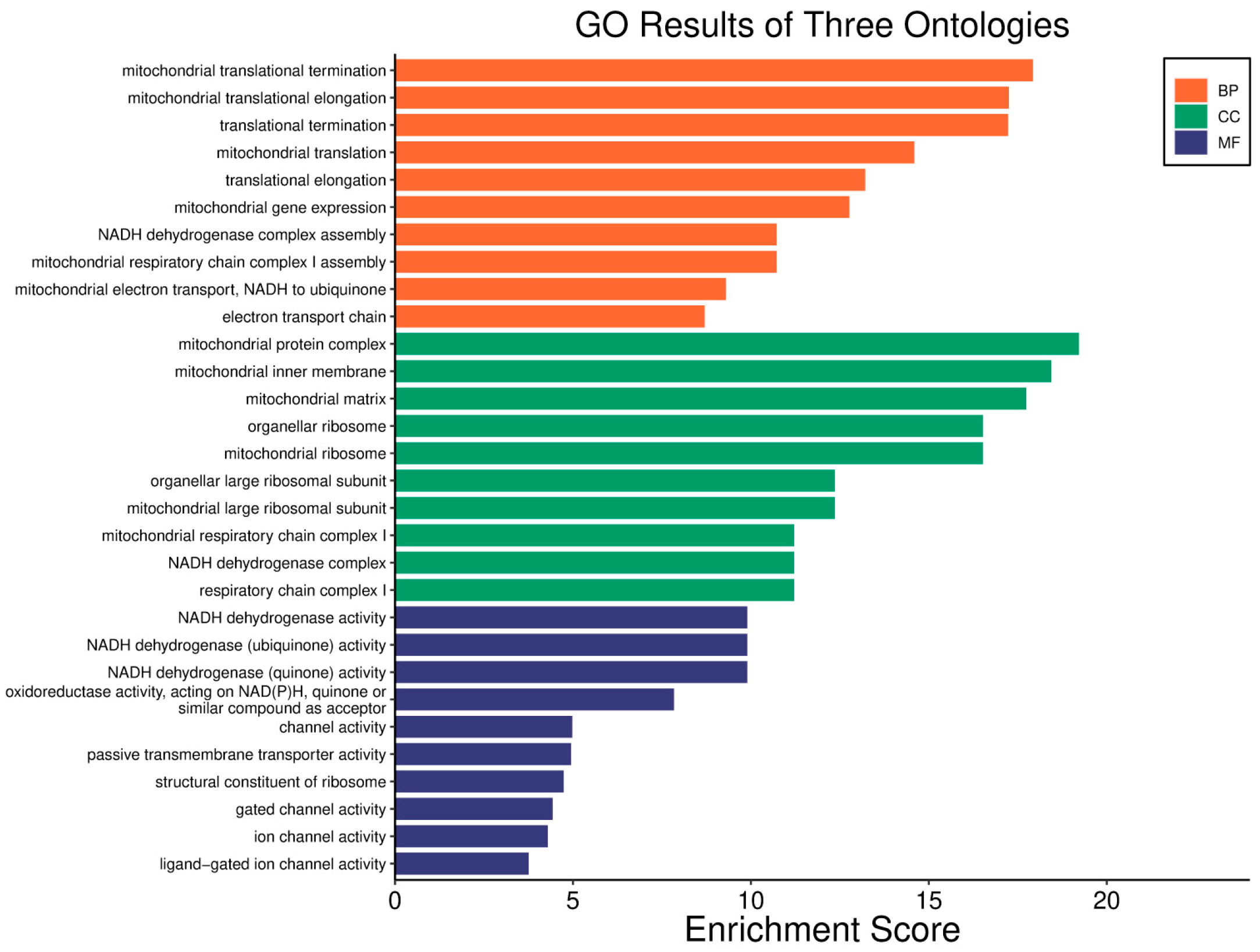
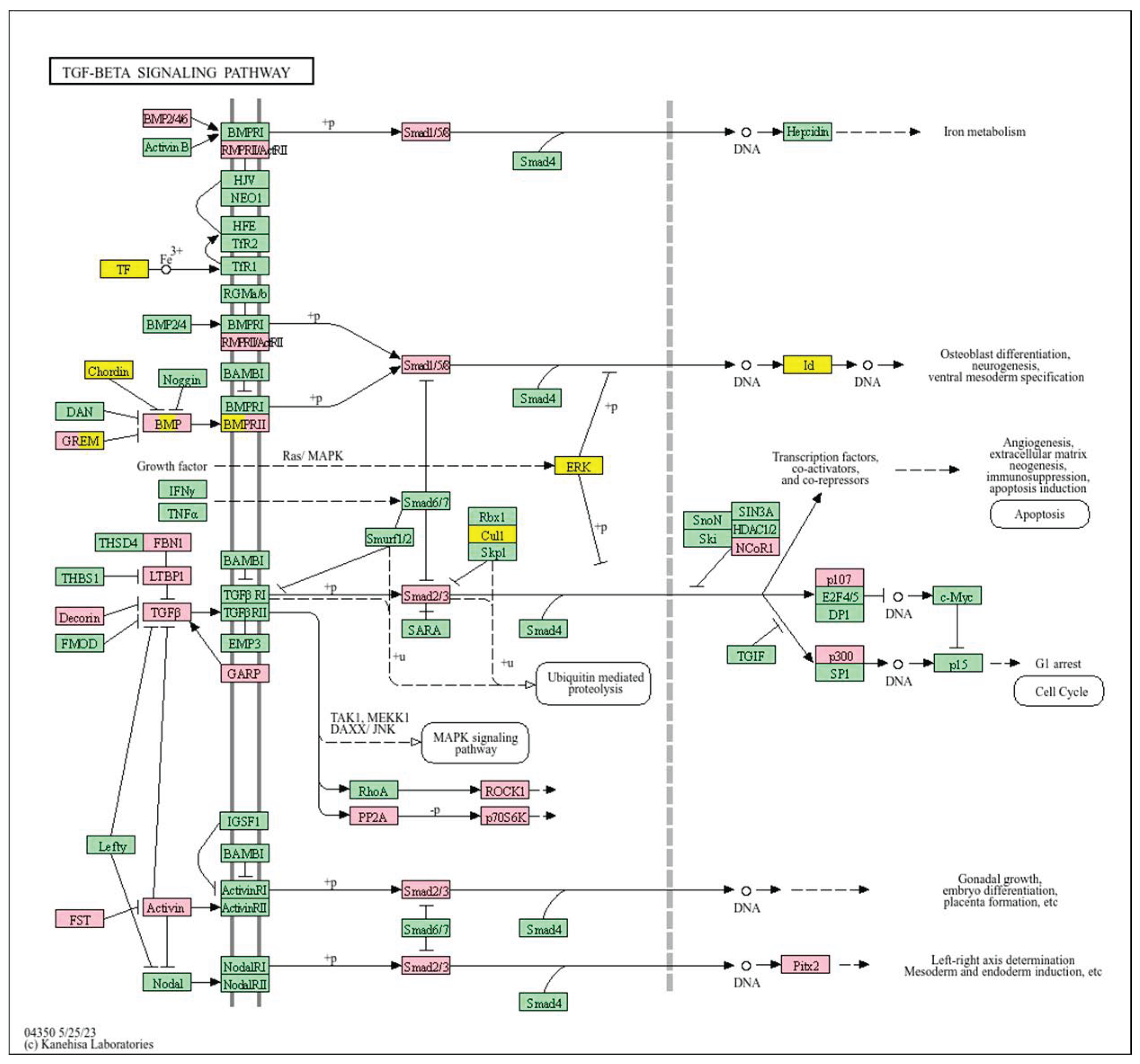
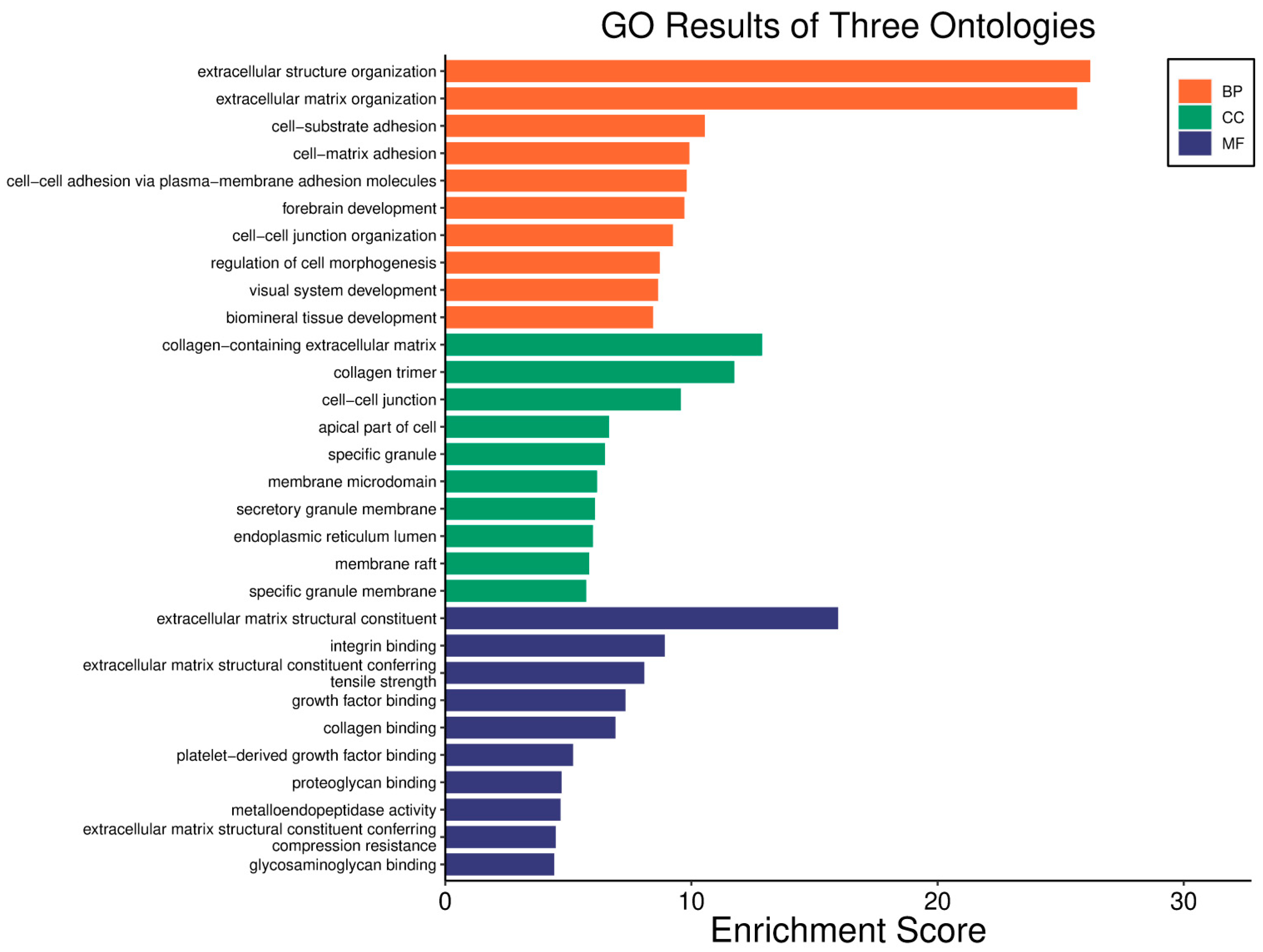
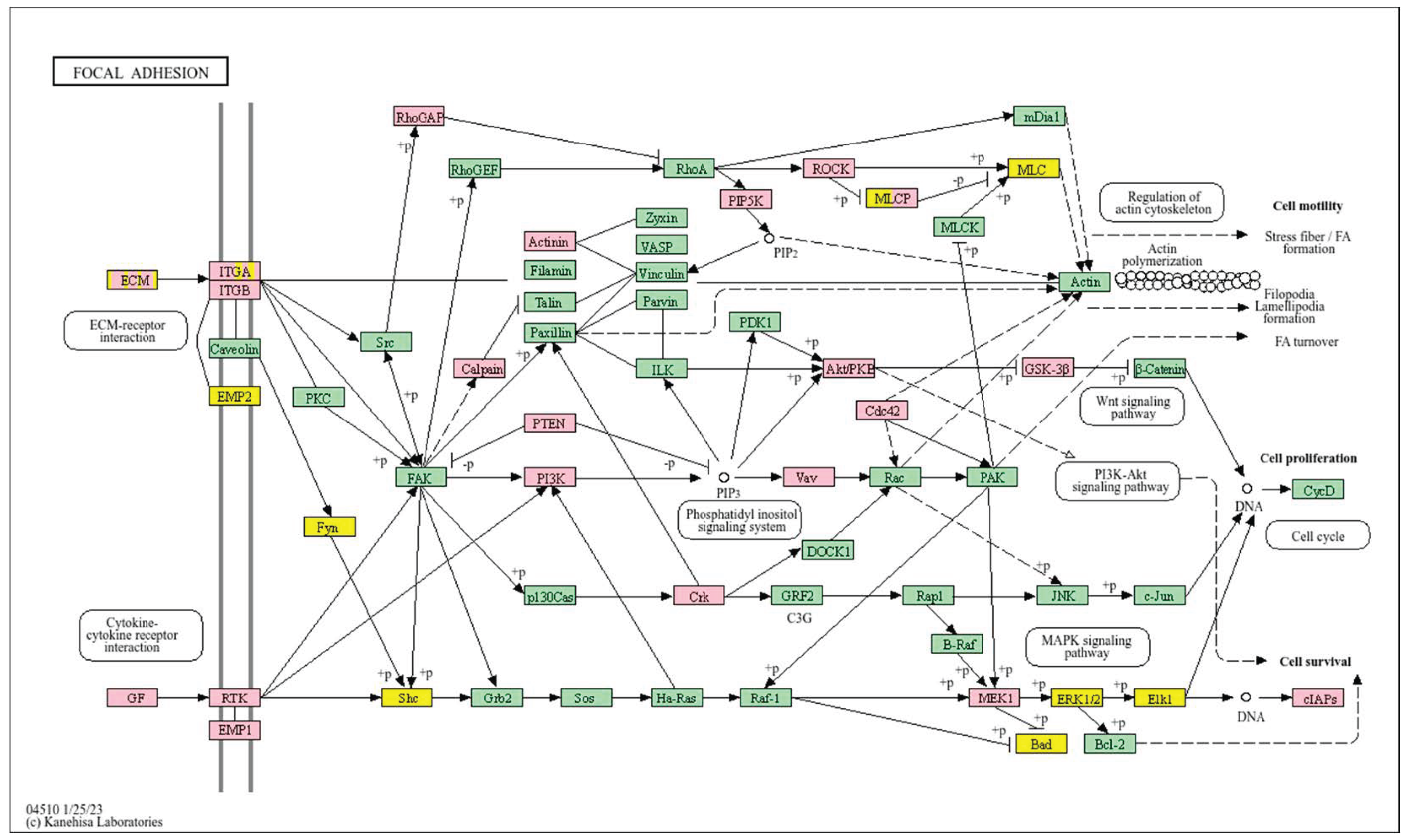
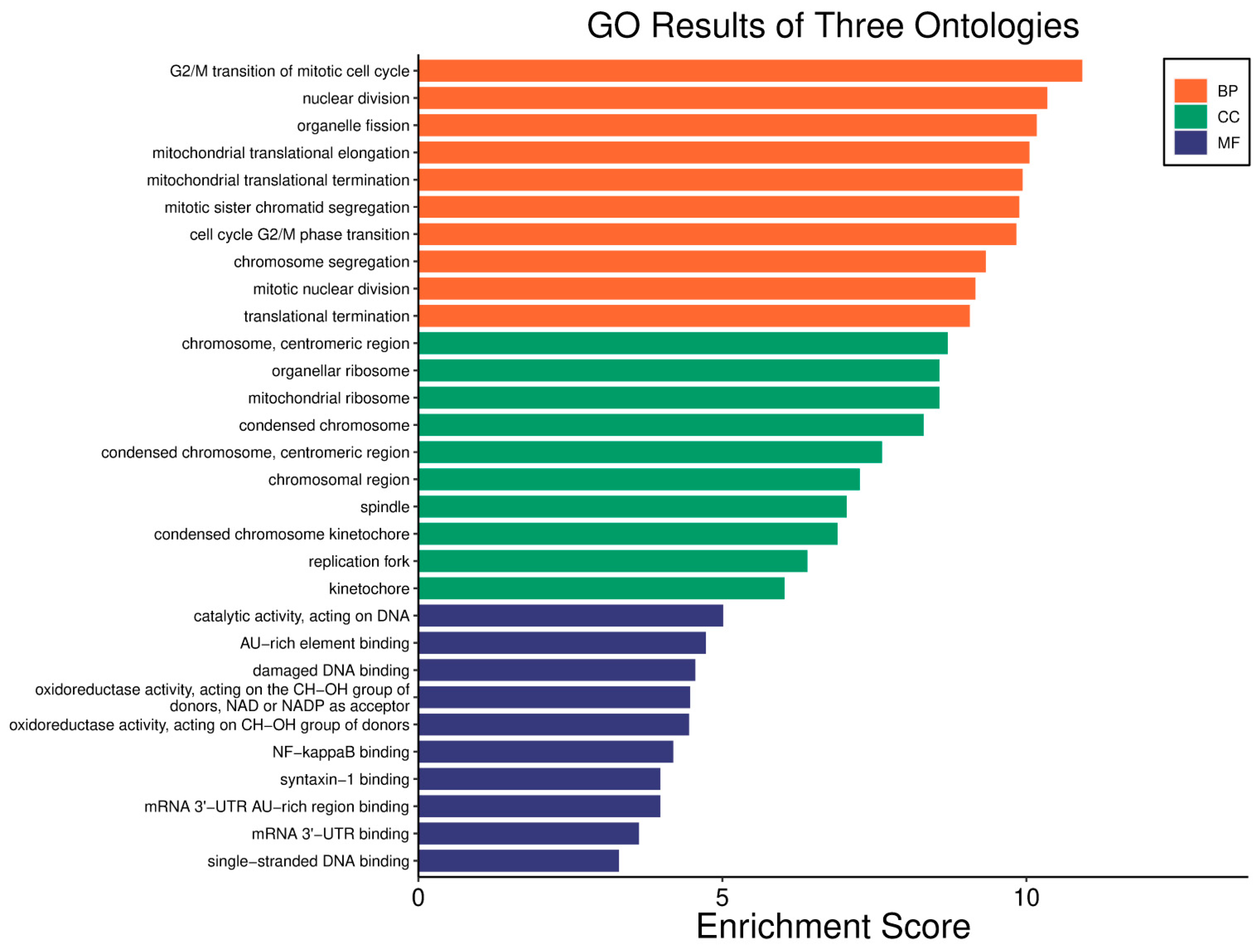
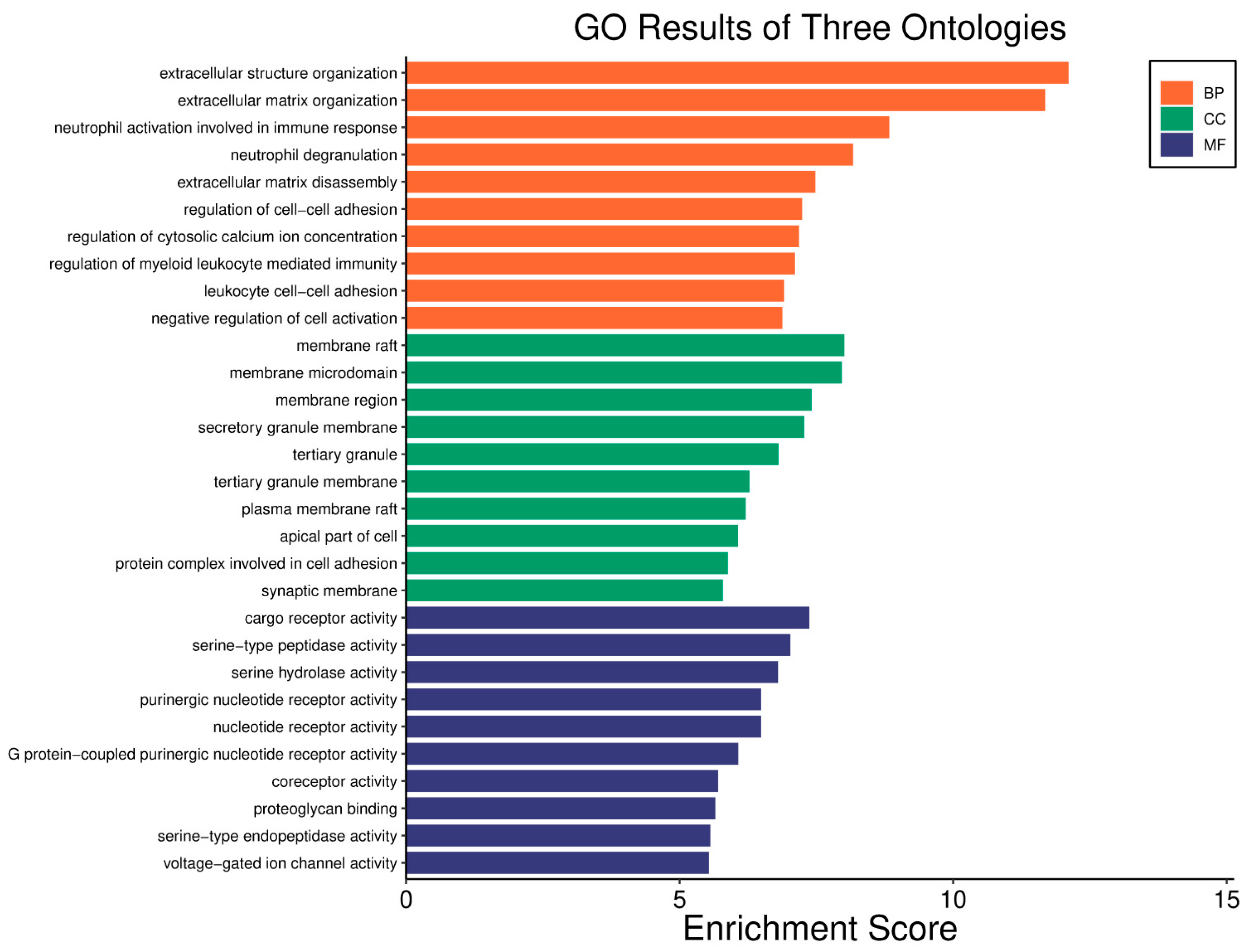
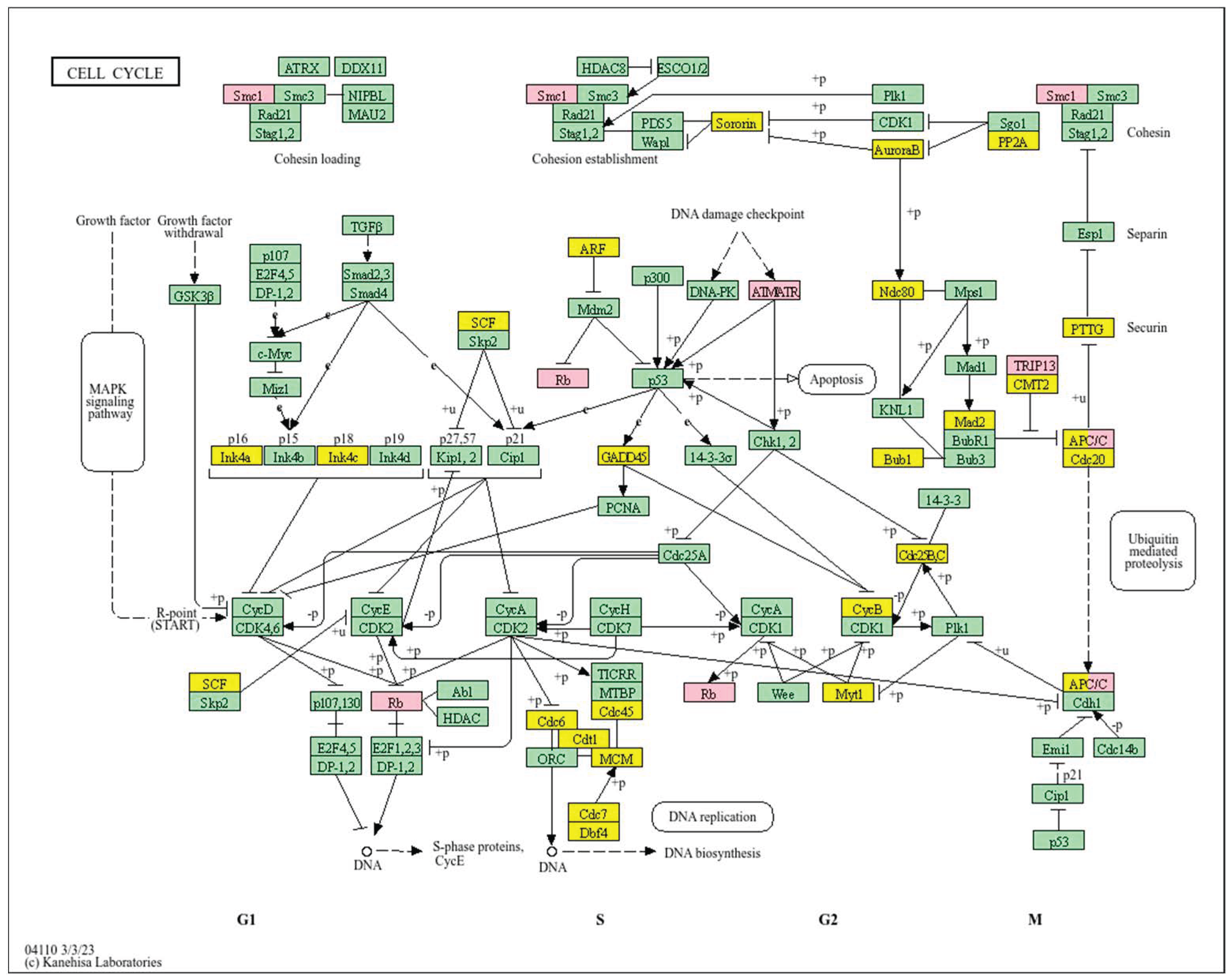
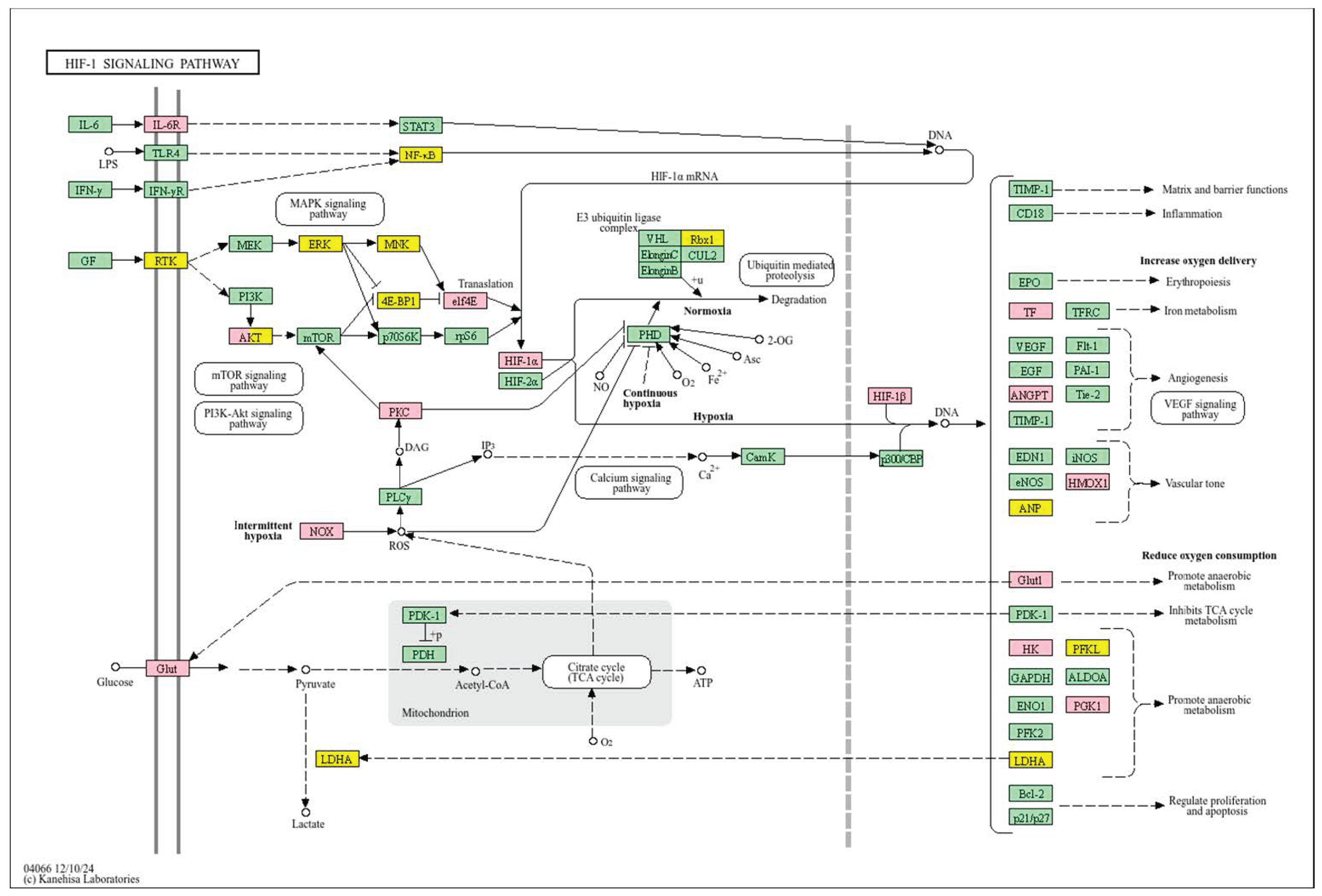
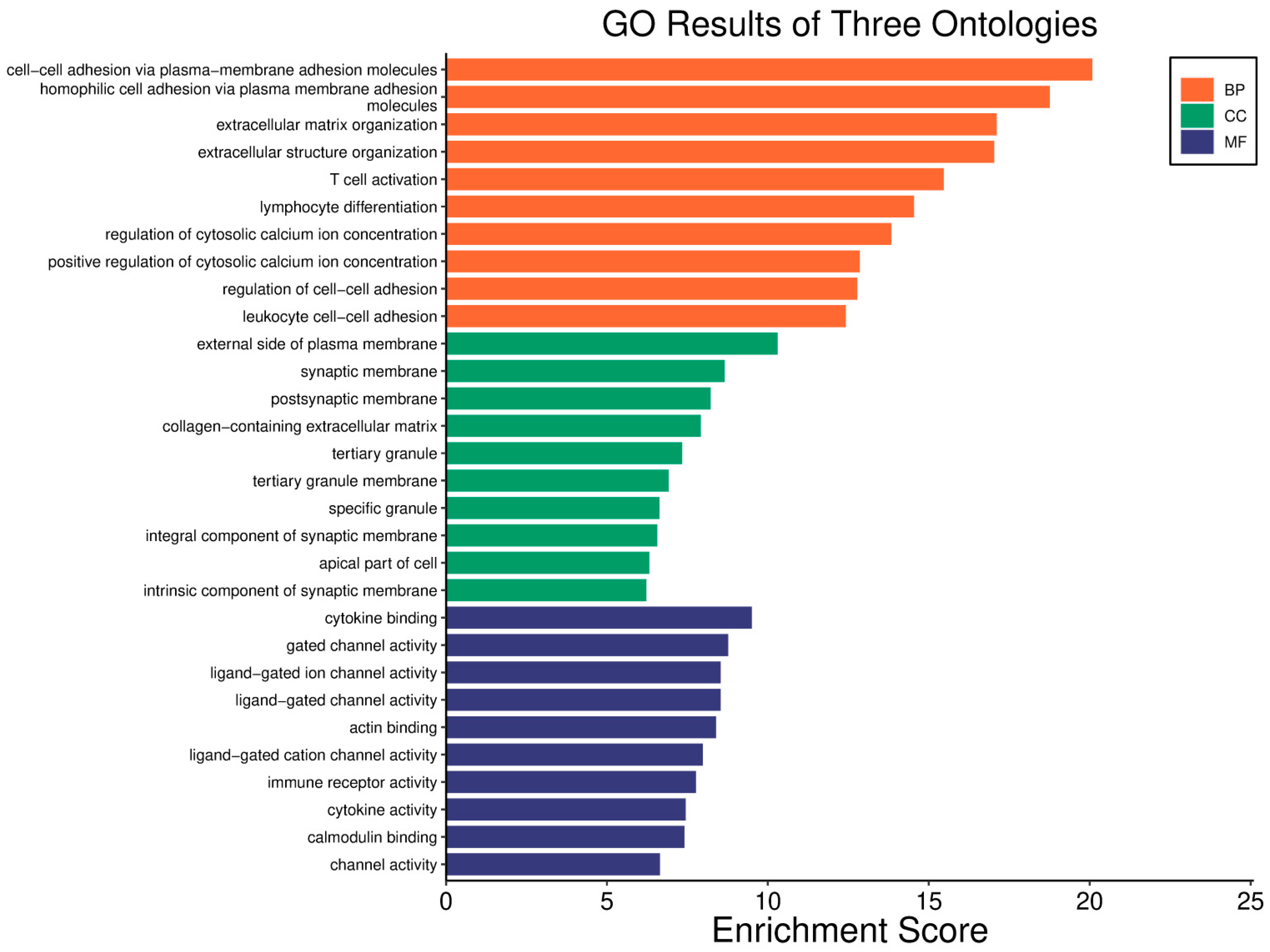
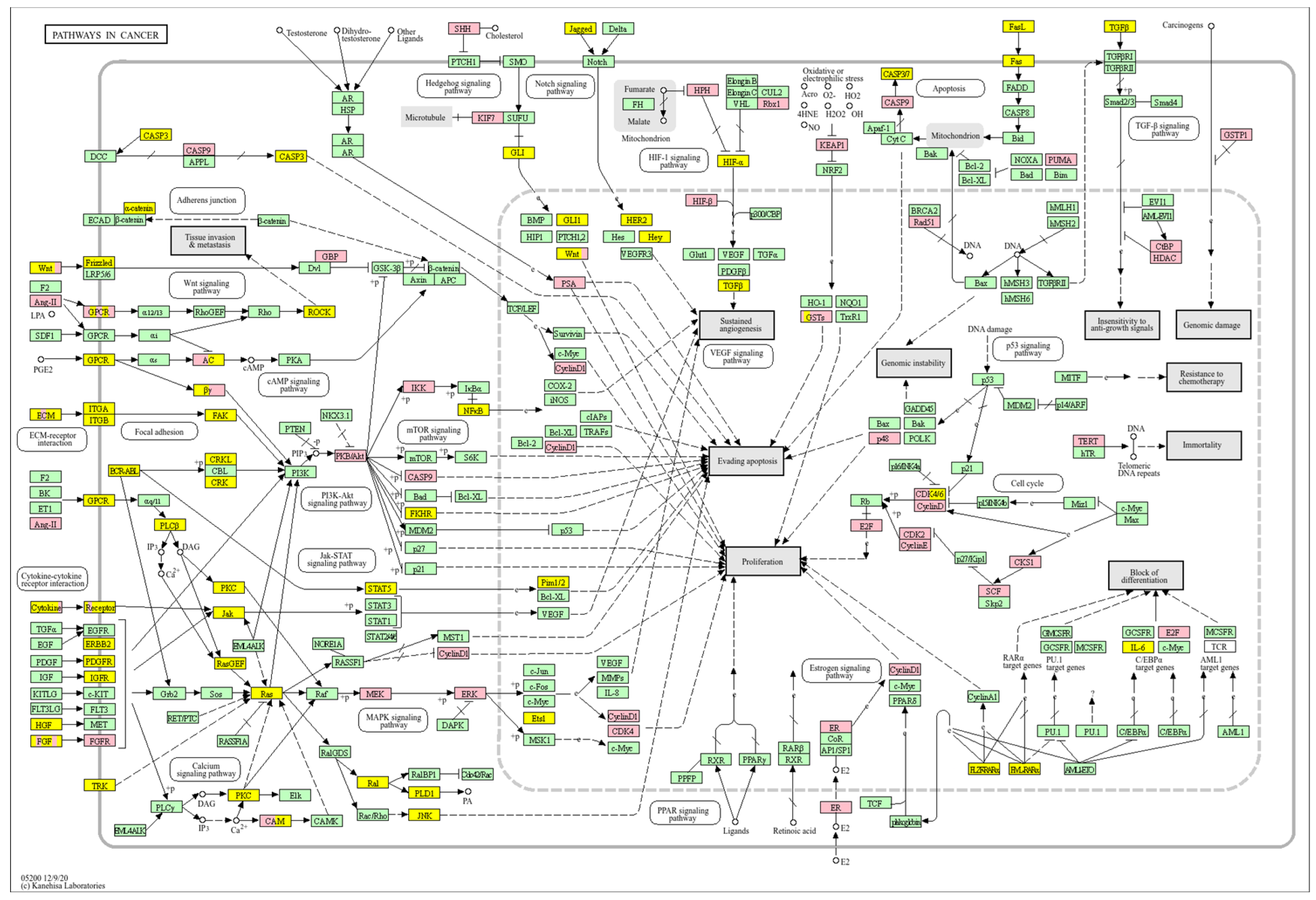
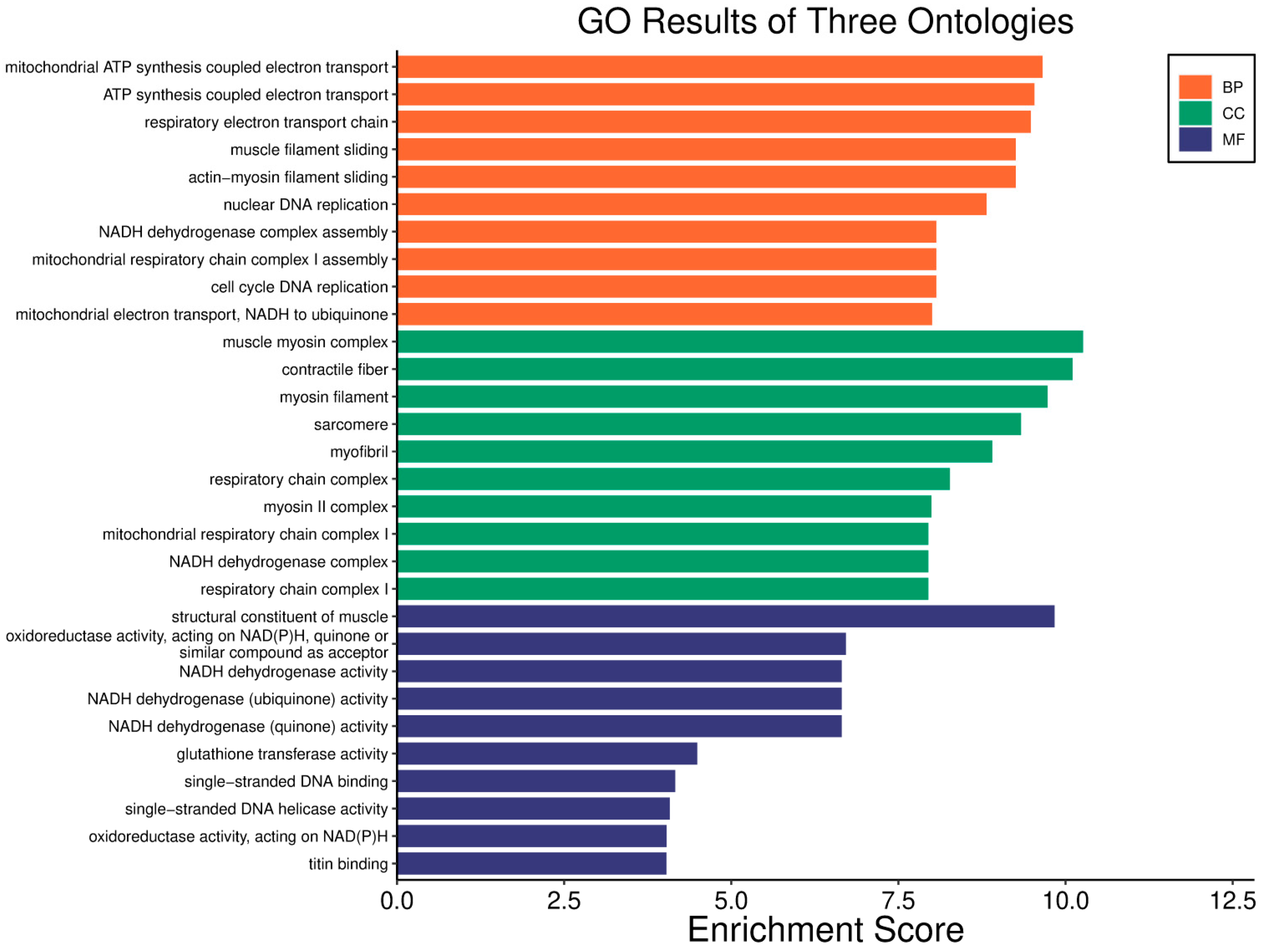
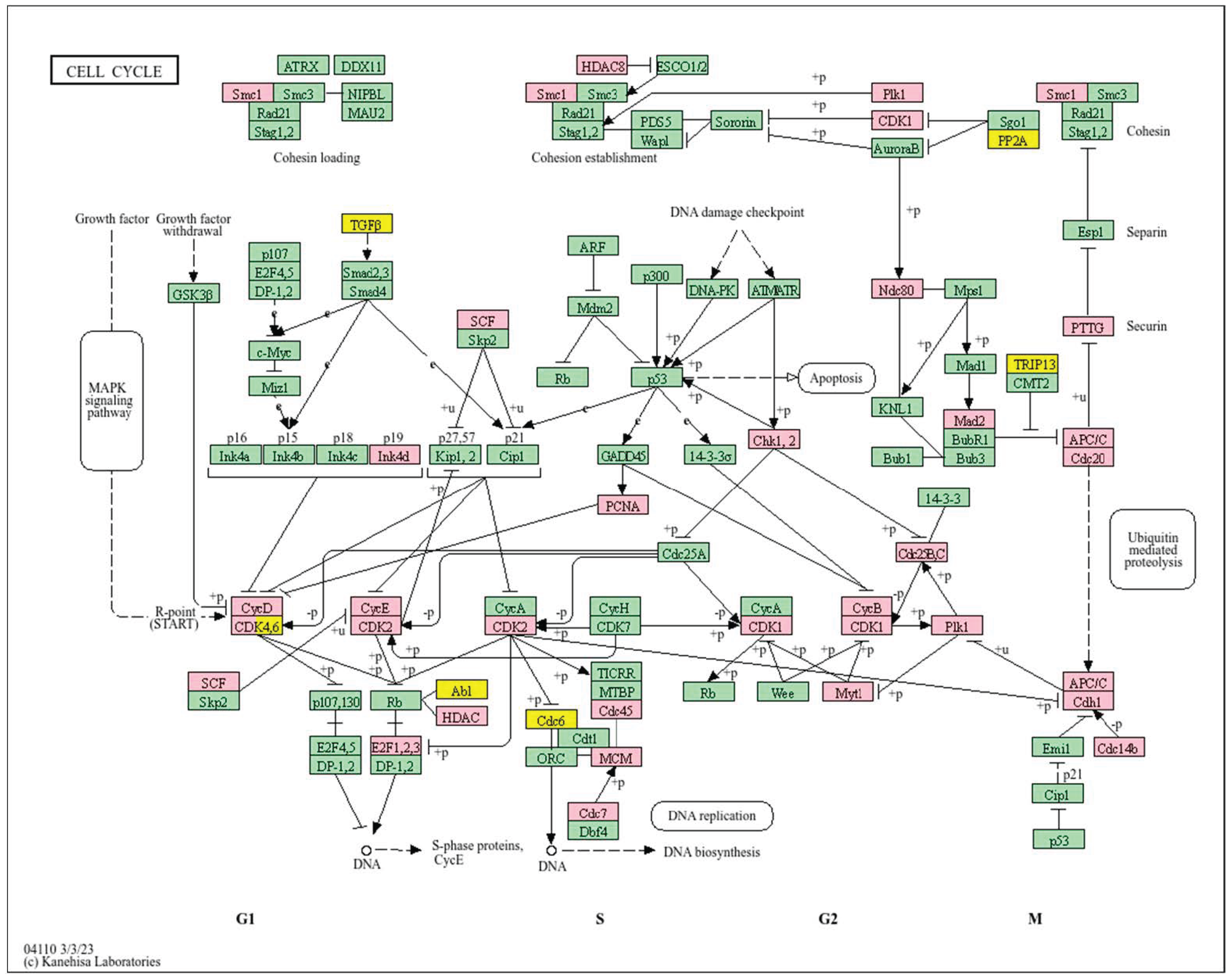
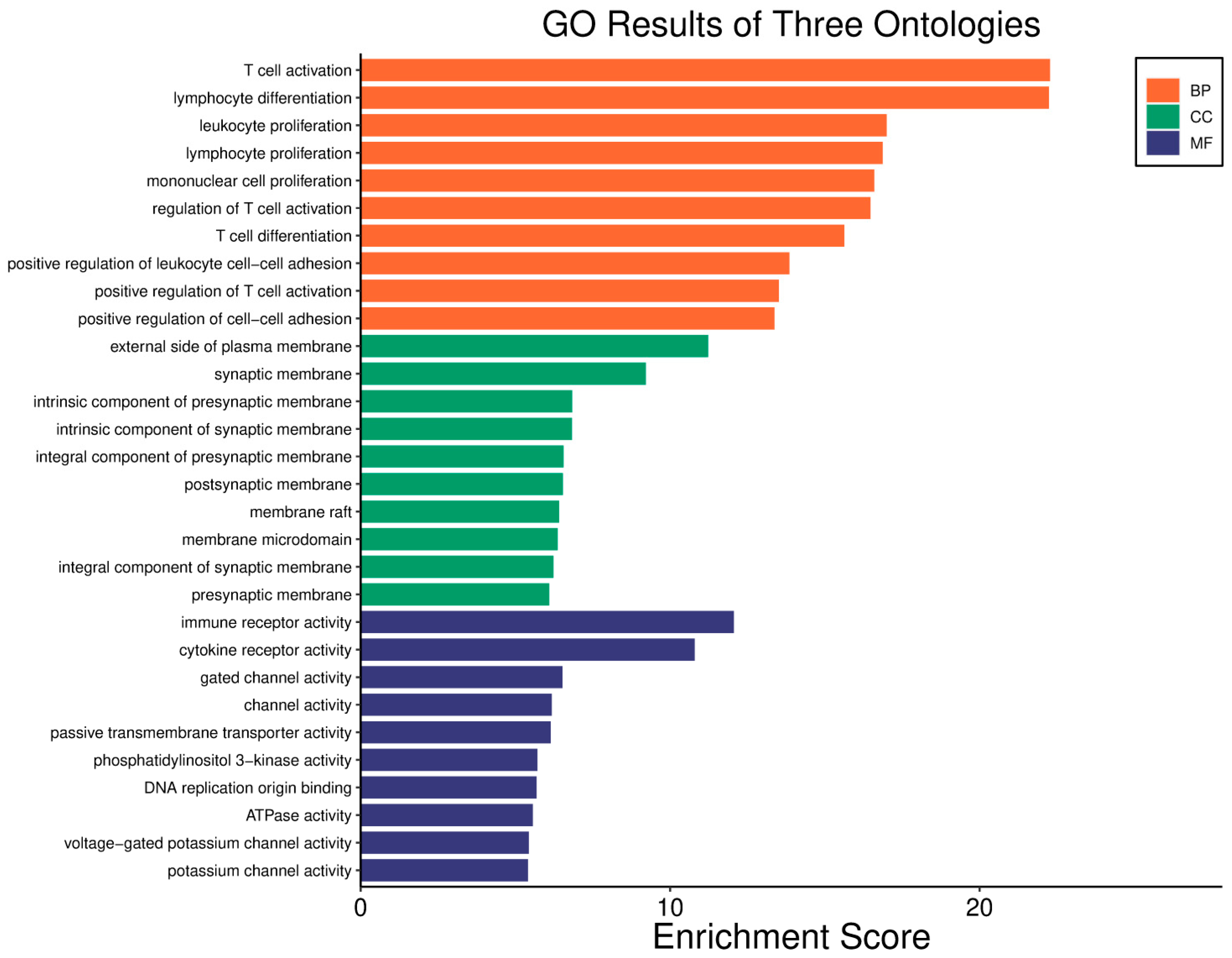
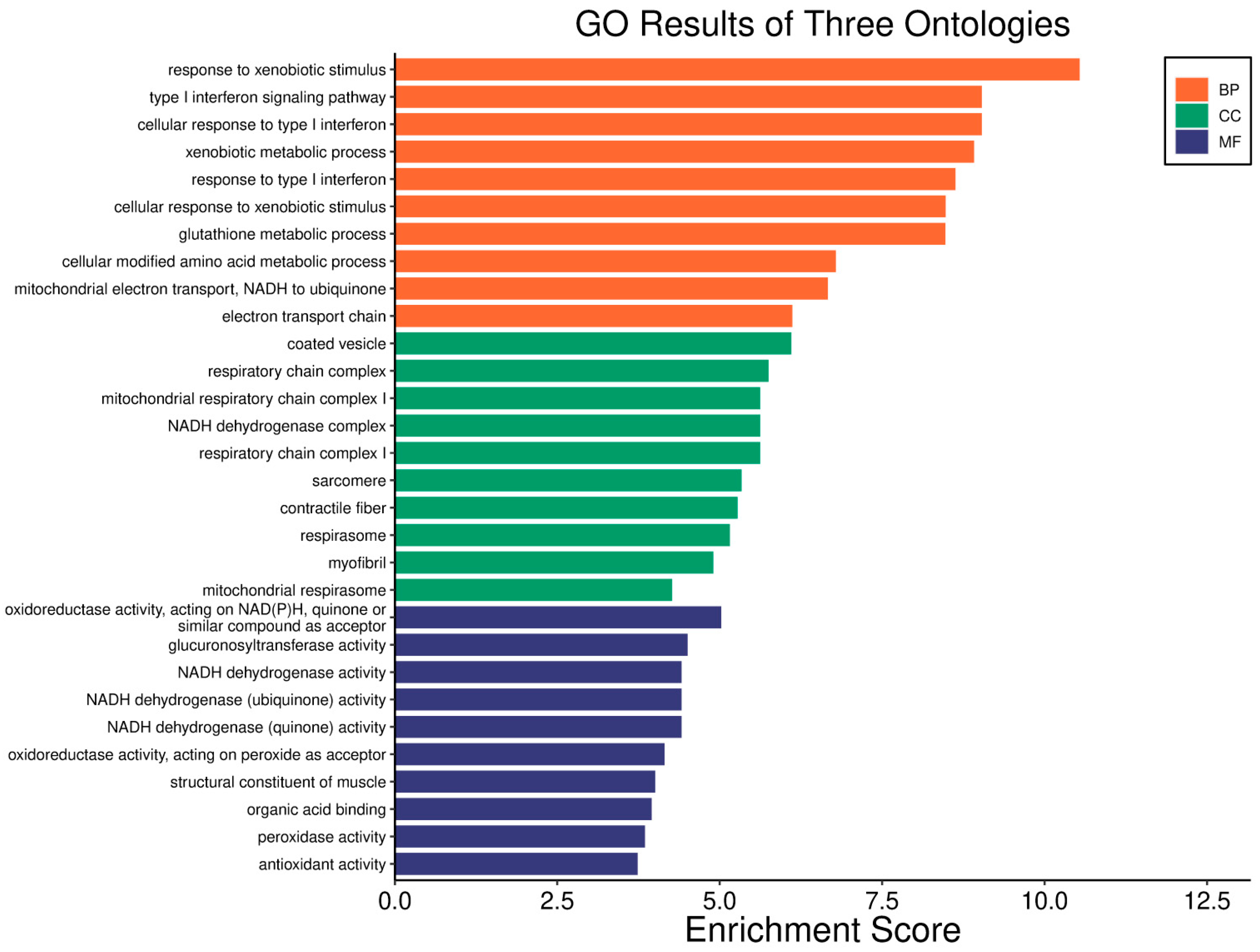
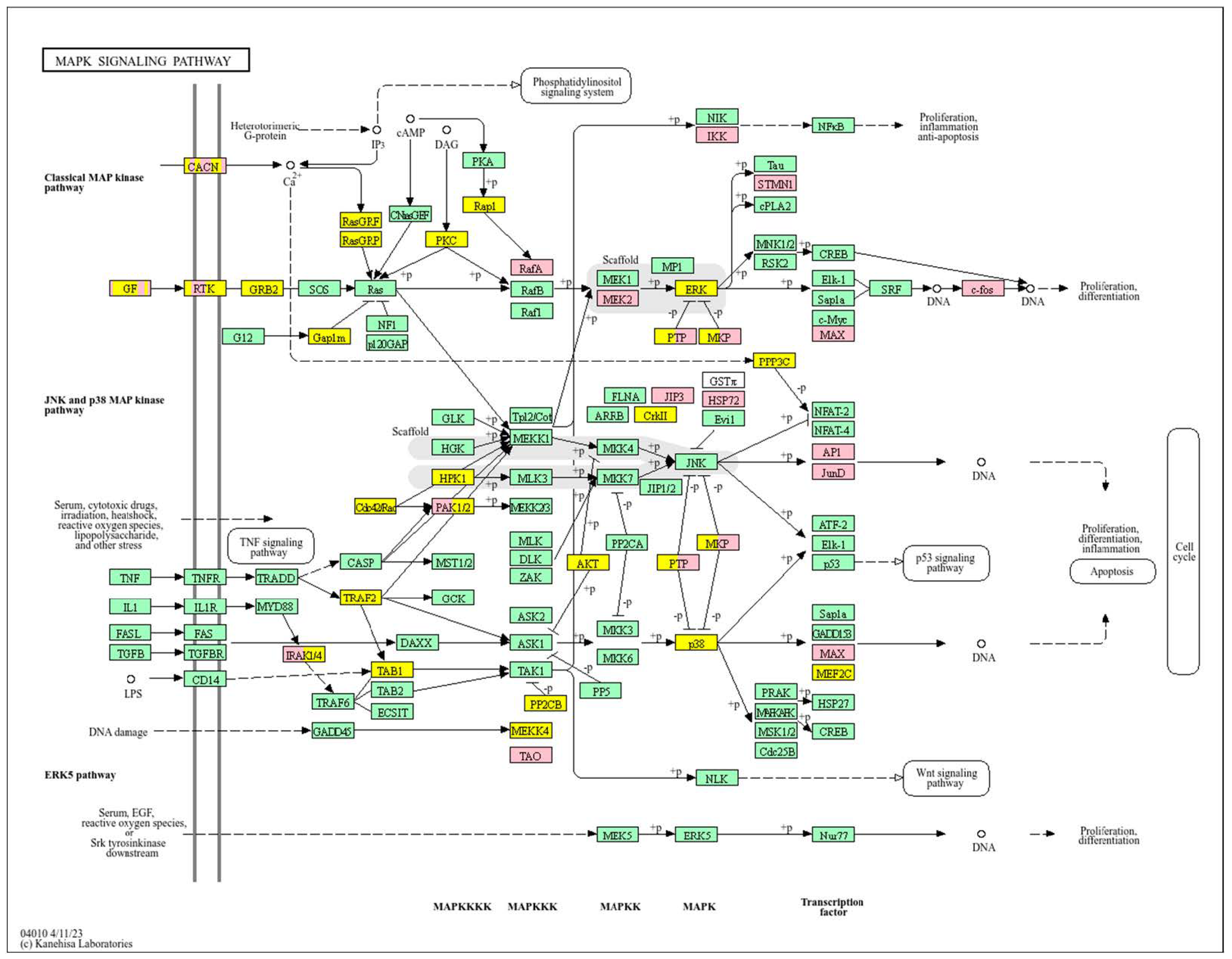
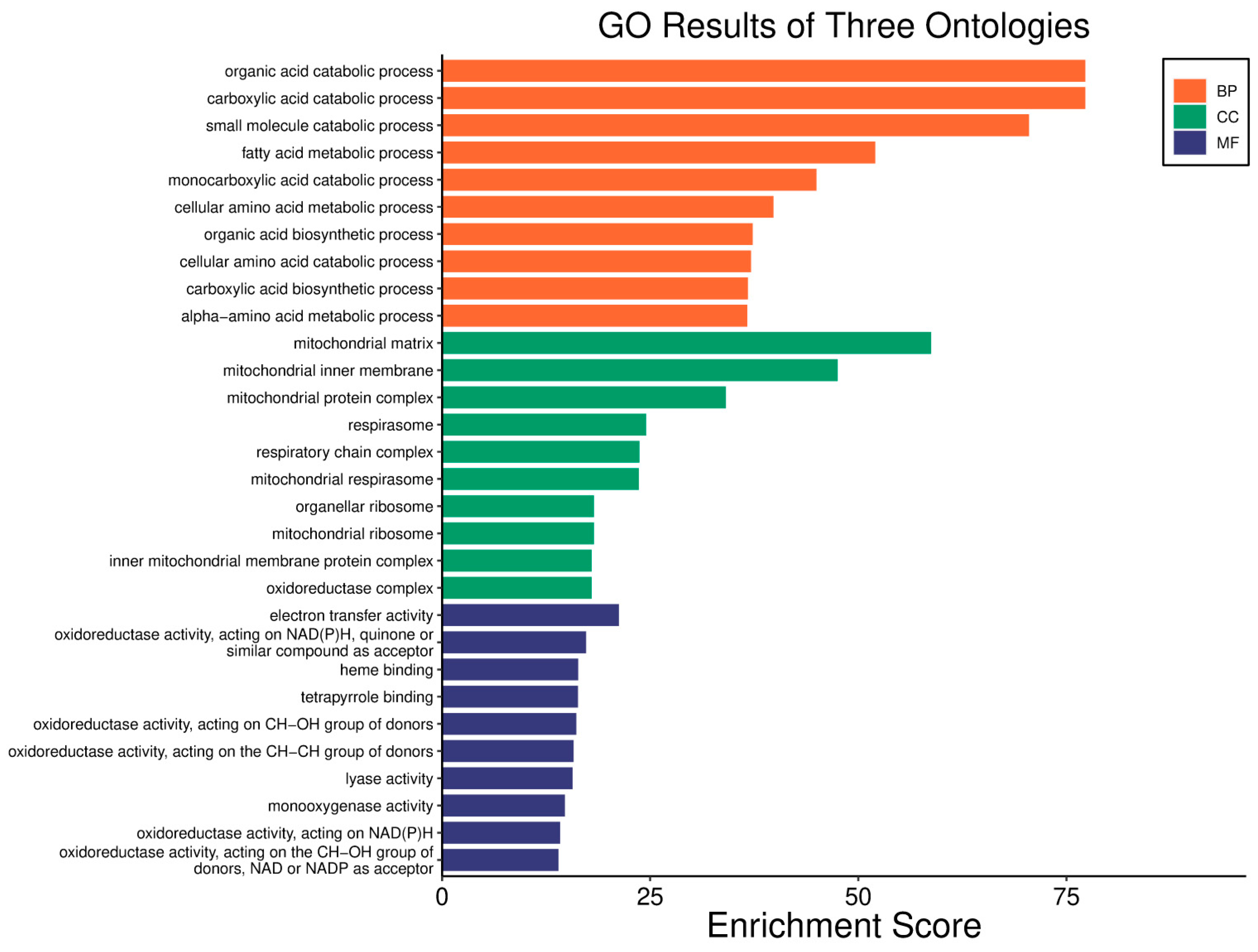
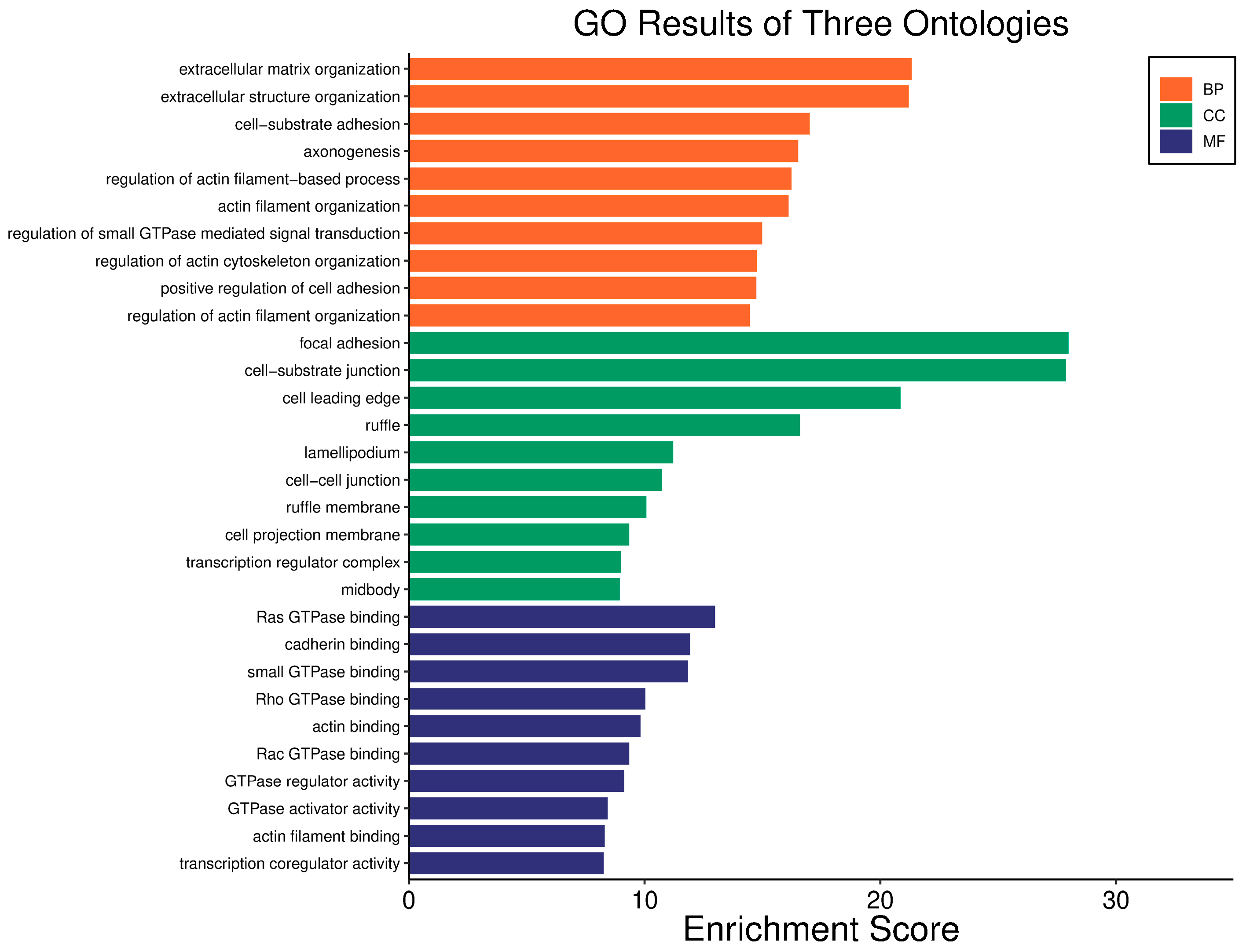
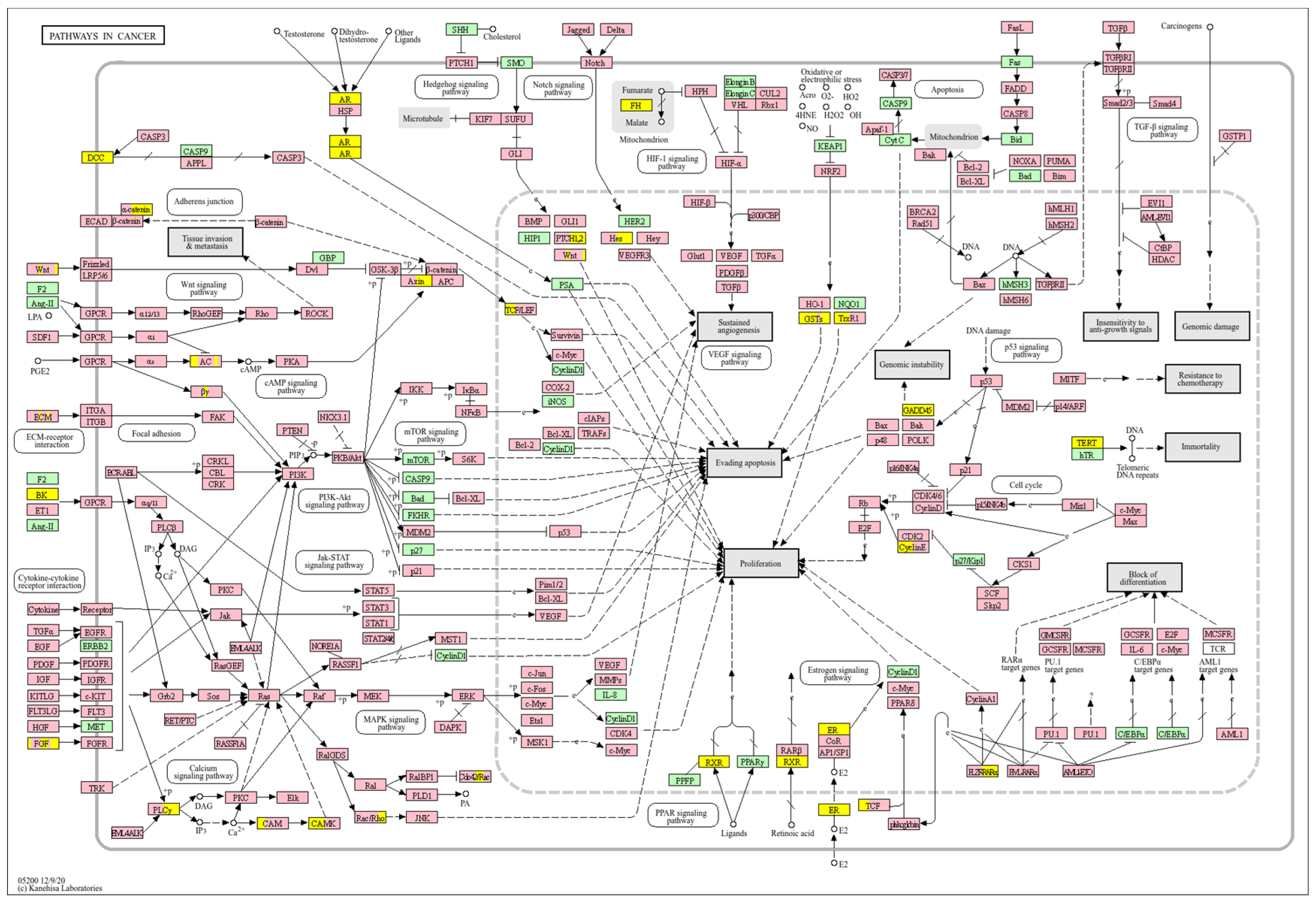
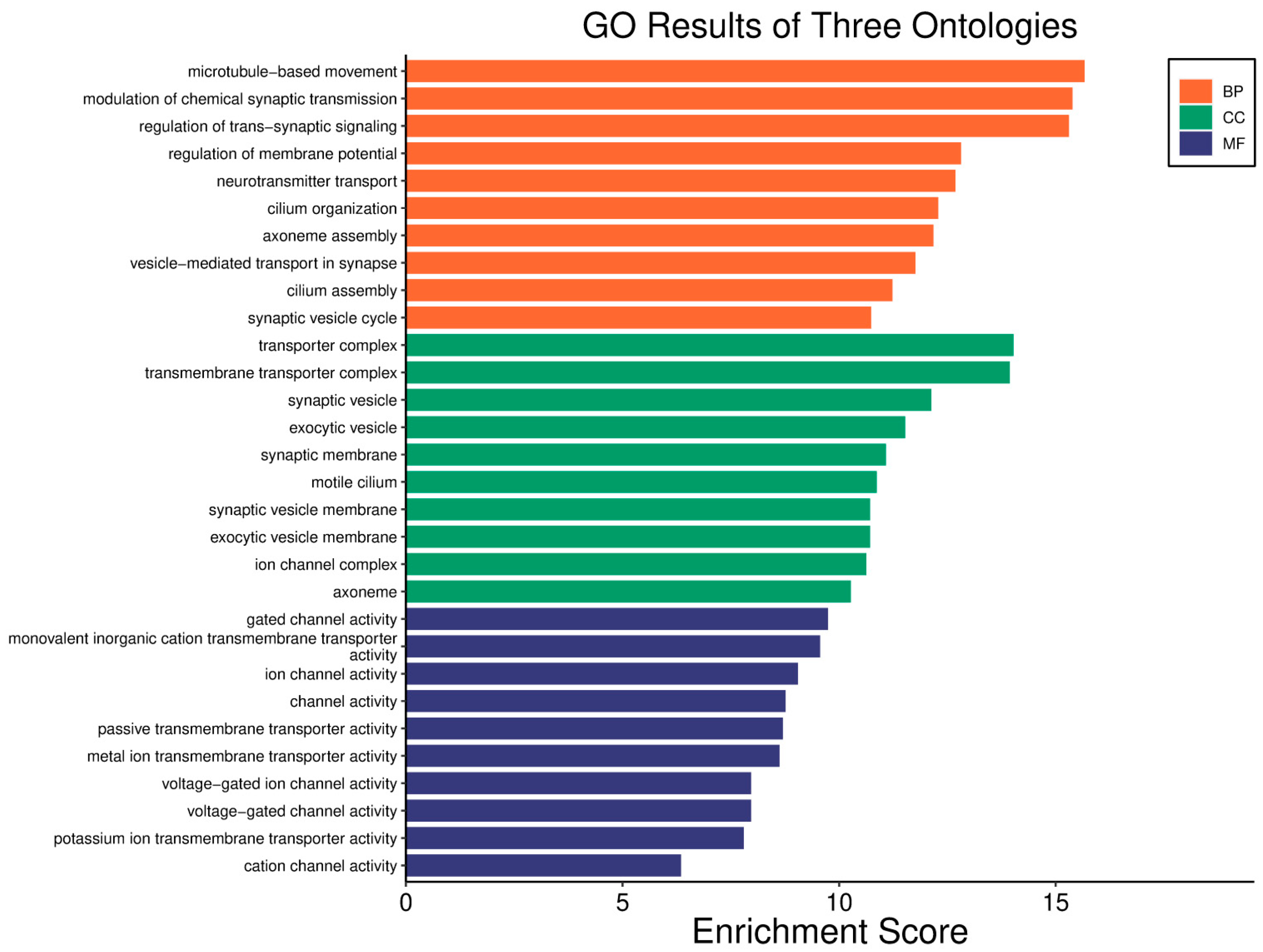
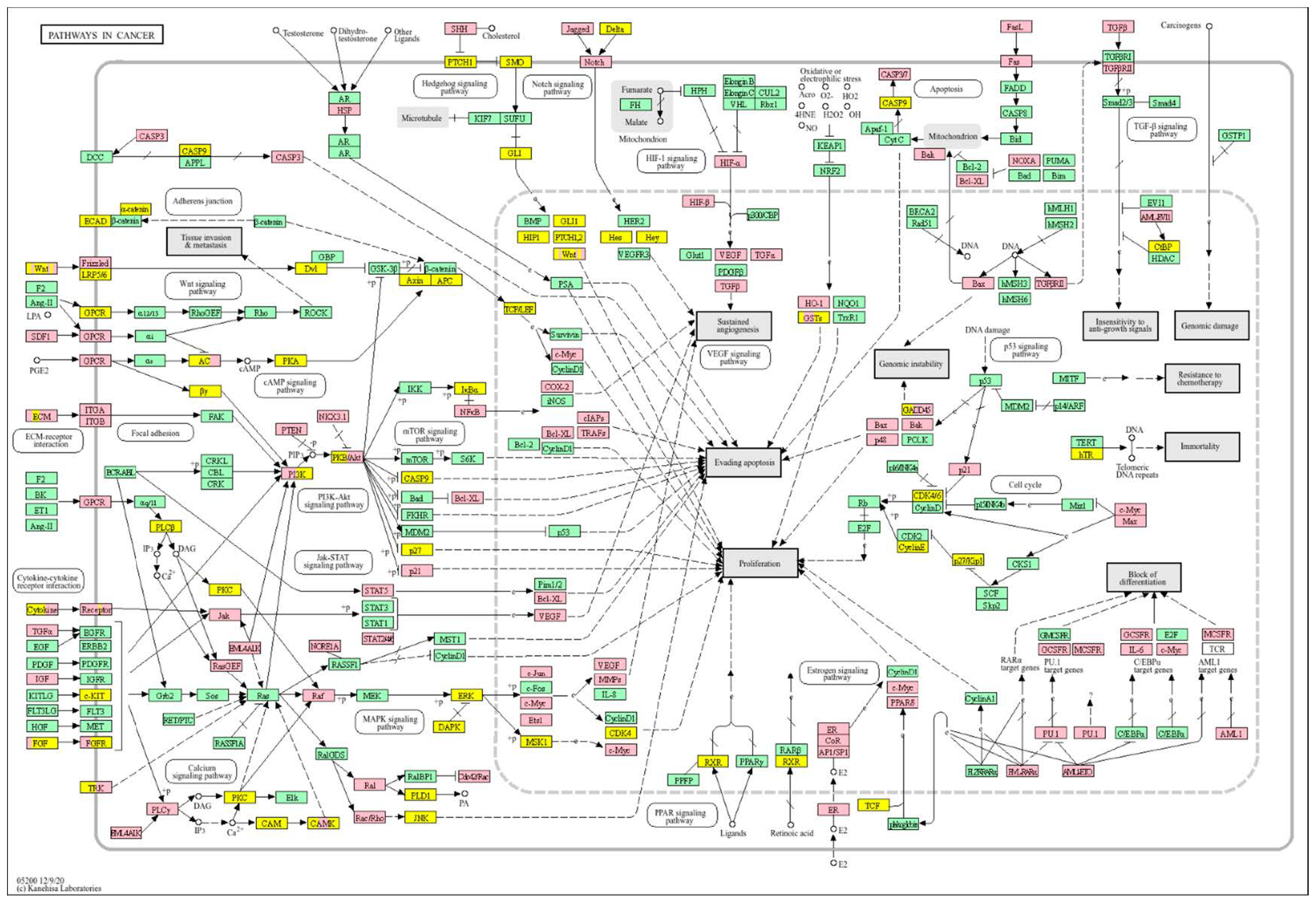
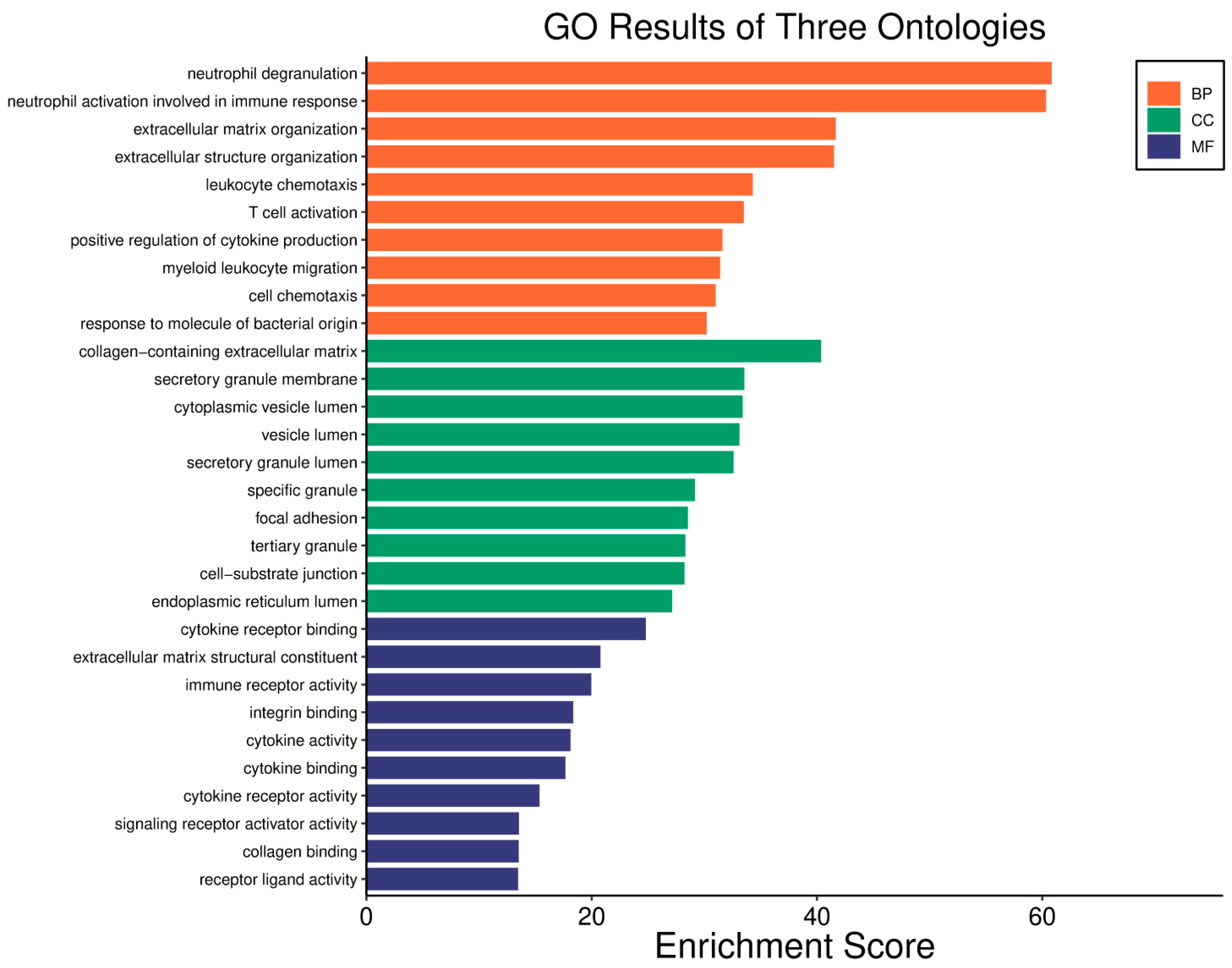
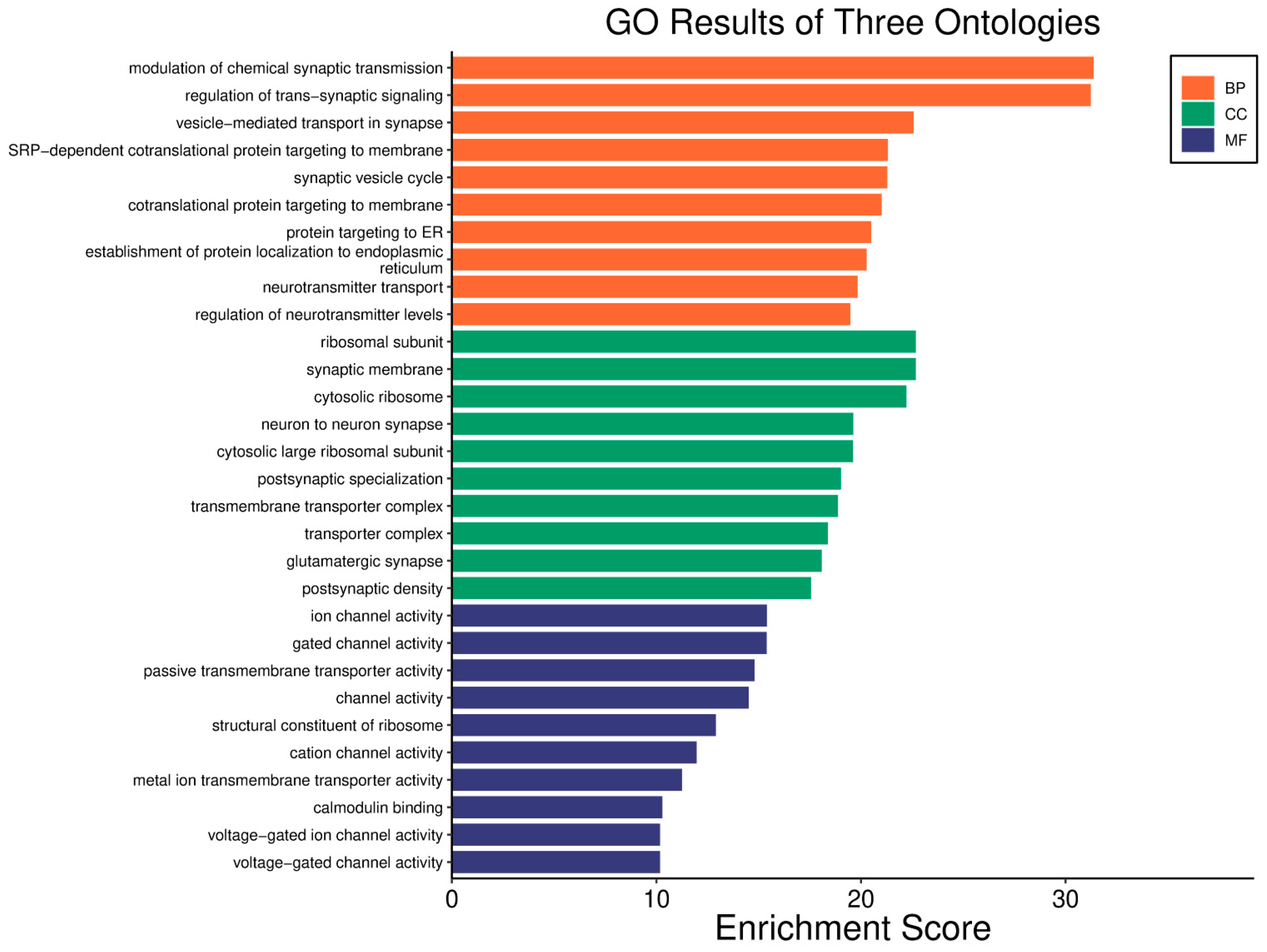
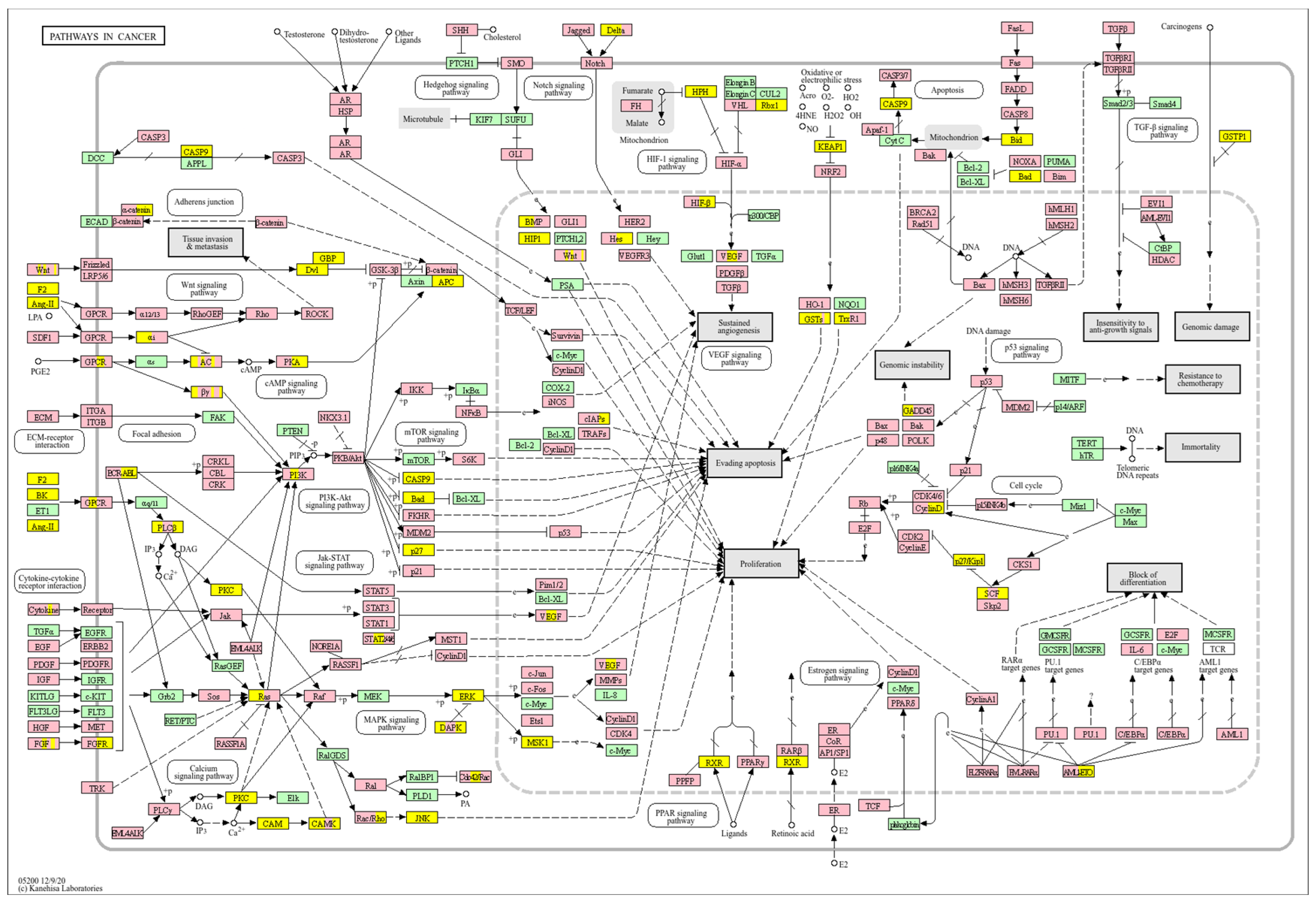
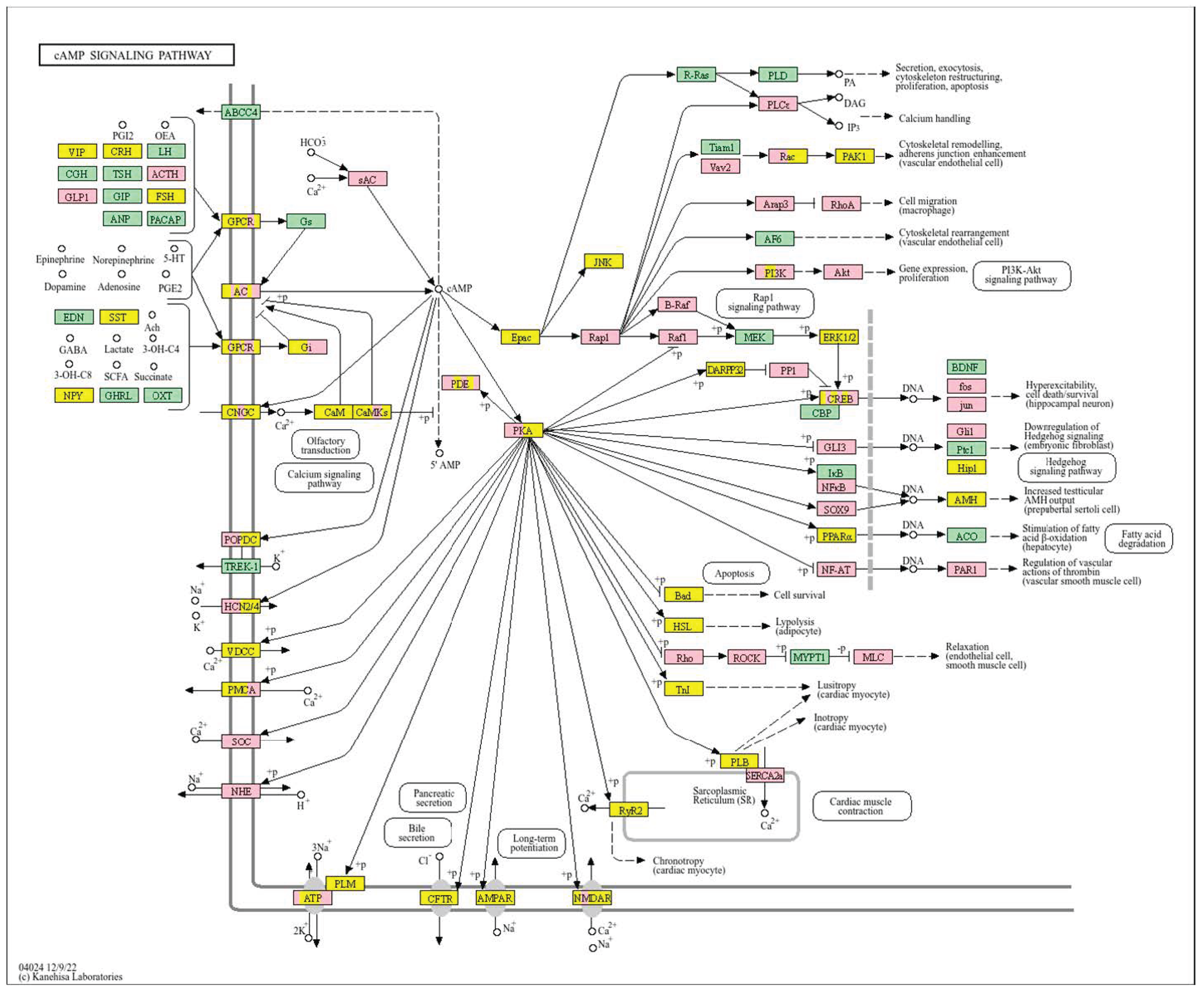
| Clinical Relevance | Basal BC | HER2 BC | Luminal A BC | Luminal B BC | GBM | LGG | HCC | |
|---|---|---|---|---|---|---|---|---|
| WWOX/HIF1A prognostic Ratio | higher associated with good prognosis | higher associated with good prognosis | lower associated with good prognosis | lower associated with good prognosis | higher associated with good prognosis | higher associated with good prognosis | higher associated with good prognosis | |
| Pathway Specificity (Good Prognosis) | Lipid and Inflammatory Pathways, DNA Repair and Stability, Central Metabolism, Reactive Oxygen Species (ROS) and Damage, Energy Production | DNA Replication and Repair, Cell Cycle Regulation, Metabolism, Protein Synthesis, Reactive Oxygen Species, Lipid and Sugar Metabolism | Growth and Proliferation, Second Messenger Systems, Immune and Inflammatory Response, Cell Adhesion and Cytoskeleton Dynamics, Cancer-Specific Pathways, Infection-Related Pathways | Immune System and Inflammation, Cell Cycle and DNA Repair, Signal Transduction, Cancer-Specific Pathways, Neuroactive and Hormonal Signalling, Cell Adhesion and Cytoskeleton Dynamics, Metabolism and Biosynthesis, Gap Junctions and Vascular Function | Signal Transduction Metabolism, Cellular Processes, Stem Cell Regulation, Carbohydrate Metabolism | Energy Production and Metabolism, Amino Acid and Nitrogen Metabolism, Signal Transduction, Hormonal Regulation, Lipid and Arachidonic Acid Metabolism, Cellular Dynamics and Motor Proteins, Biosynthesis and Cofactors, Neuroactive Processes | Energy Production and Metabolism, Lipid and Fatty Acid Metabolism, Amino Acid Metabolism, Nitrogen and Sulphur Metabolism, Carbohydrate Metabolism, Vitamin and Cofactor Metabolism, Drug and Xenobiotic Metabolism, Cellular functions | |
| Pathway Specificity (Poor Prognosis) | Growth and Proliferation, ECM and Cell Adhesion, Hypoxia and Metabolism, Immune and Inflammatory, Apoptosis and Senescence | Hormonal and Reproductive Signalling, Cytoskeleton and Cell Adhesion, Immune and Inflammatory Signalling, Cancer and Cellular Processes, Stem Cell and Longevity Regulation, Metabolism and Biosynthesis, Vascular and Muscle Function, Genetic Information Processing, Diabetes and Endocrine Disorders | DNA Replication and Repair, Cell Cycle Regulation, Reactive Oxygen Species (ROS) and Damage, Drug Metabolism, Nitrogen and Carbon Metabolism, Nucleotide and Cofactor Biosynthesis, Protein and Motor Functions, Hormonal Regulation, Histidine Metabolism, Endocannabinoid Signalling | DNA Replication and Repair, Cell Cycle and Senescence, Reactive Oxygen Species (ROS) and Damage, Drug Metabolism, Metabolism and Biosynthesis, Hormonal Regulation, Immune and Cellular Processes, Neuroactive and Cellular Signalling, Energy Production and Intermediates, Motor Proteins and Cellular Dynamics | Immune and Inflammatory Response, Cancer-Specific Pathways, Signal Transduction, Cell Adhesion and Cytoskeleton Dynamics, Cell Cycle and Apoptosis, Metabolism and Biosynthesis, Endocytosis and Intracellular Transport, Hormonal and Reproductive Signalling, Diabetes and Complications, Neuroactive Processes | Cell Adhesion and Extracellular Matrix, Immune and Inflammatory Response, DNA Replication and Repair, Cell Cycle and Apoptosis, Signal Transduction, Cancer-Specific Pathways, Metabolism and Biosynthesis, Diabetes and Complications | Signal Transduction, Immune and Inflammatory Response, Cancer-Specific Pathways, Cell Adhesion and Cytoskeleton Dynamics, Cell Cycle and Apoptosis, Metabolism and Biosynthesis, Neuroactive Processes, Pathogen Interaction and Resistance, Diabetes and Complications, Chromatin and Gene Regulation | |
| Therapeutic/Clinical Relevance | Markers with Direct Therapeutic Application | ERBB2, EGFR, PTEN, HIF1A, IL1RAPL1, NOTCH1 | EGFR, PTEN, HER2 | ESR1,PGR, FGFR2 | PIK3CA | PTEN | NA | VEGF-D, FGF19 |
| Markers with Potential Therapeutic Application | CD44, BCAR4, PIK3CA, MMP2, PARP, MMP9, VEGF | MMP15, ADCYAP1, HIF1A, LYN, PRKG1, STK39, PLXNC1 | CDK6, MMP10, PIK3CG, GPER, LYN, WWOX, ID01 | KCNC1 | ADAMTS1, CD44, IL6, NRP1, HIF1A, CD70, LYN | LYN, BRAF | BCL2L15, HNF4A, MMP1, TGFBR3 | |
| Functional Role in Tumourigenesis | Markers Promoting Tumour Growth | HIF1A, EGFR, ERBB2, PIK3CA, PTEN, MYCBP2, ITGA1, MMP2, ZEB1, ERBB2, TLR4, CCNE2 | EGFR, PTTG1, ERBB2, MMP15, ADCYAP1, HIF1A, CDC6 | ERBB2, CCND1, ADCYAP1 | AURKA, CCNE2, HIF1A, MYCBP2, NRC1 | HIF1A, EGFR, PIK3CD, AKT1, PTK7, MKNK2, RELB, S100A8, PTGS2, COL1A1, CD44, AXL, PLAU | IDH1, EGFR, CDK4/6, FGFR | FGF19, MYCL1, CD36 |
| Markers Promoting Tumour Suppression | CD81, GATA3, MGMT, DIABLO, SOD2, TP53I13, BARD1, PTEN | PTEN, RB1 | CDH1, WWOX, CILP, TP53 | FAS, CDH1 | PTEN | TP53 | HNF4A, RASSF1 | |
| Markers Involved in Metastasis | ITGA1, MMP2, ZEB1, CD44 | CD44, PLXNC1, ITGA4 | CD44, MMP10, FAT1, ITGA4 | KLK13, ITGA1 | SPP1, MMP13, LOXL1, MMP14, ITGA4 | MMP2, VEGF | TWIST2, MMP1, CDH6, TGFBR3 | |
| Identified genes from this study contributing to prognosis according to WWOX/HIF1A ratio | ↑ CD81, ↑ GATA3, ↑ MGMT | ↑ ARG2, ↑ S100A1, ↑ MT2A, ↑ EIF4EBP1, ↑ KLF4, ↑ BTG3, ↑ BAG1 | ↑ HK3, ↑ LDHAL6A, ↑ ADH6, ↑ PRKCB, ↓ MYH7, ↓ FABP3, ↓ CYP4F2, ↓ MTHFR | ↓ WWOX, ↓ TRIM67, ↑ FOXO3, ↑ DHFR, ↑ CD8A, ↑ ESR2, ↑ TP53I11, ↑ DHX9, ↑ RAD51, ↑ FOXP3 | ↓ MMP1, ↑ PTCH1, ↑ CDK4, ↑ CDKN1B | ↑ WWOX, ↑ HDAC11, ↑ BIN1, ↑ BCL2L2, ↑ PARK2, ↑ SOD1, ↑ APOE | ↓ MMP1, ↑ CD36, ↑ TWIST2, ↑ FGF19, ↑ TGFBR3, ↓ ITGB1, ↑ HNF4A, ↓ RASSF1, ↑ SPARCL1, ↑ LHPP | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baryła, I.; Hammouz, R.Y.; Maciejek, K.; Bednarek, A.K. Prognostic Significance of WWOX/HIF1A Ratio in Cancer Subtypes: Insights into Metabolism, ECM, and EMT. Biology 2025, 14, 1151. https://doi.org/10.3390/biology14091151
Baryła I, Hammouz RY, Maciejek K, Bednarek AK. Prognostic Significance of WWOX/HIF1A Ratio in Cancer Subtypes: Insights into Metabolism, ECM, and EMT. Biology. 2025; 14(9):1151. https://doi.org/10.3390/biology14091151
Chicago/Turabian StyleBaryła, Izabela, Raneem Y. Hammouz, Kinga Maciejek, and Andrzej K. Bednarek. 2025. "Prognostic Significance of WWOX/HIF1A Ratio in Cancer Subtypes: Insights into Metabolism, ECM, and EMT" Biology 14, no. 9: 1151. https://doi.org/10.3390/biology14091151
APA StyleBaryła, I., Hammouz, R. Y., Maciejek, K., & Bednarek, A. K. (2025). Prognostic Significance of WWOX/HIF1A Ratio in Cancer Subtypes: Insights into Metabolism, ECM, and EMT. Biology, 14(9), 1151. https://doi.org/10.3390/biology14091151





