Duplicated Genes on Homologous Chromosomes Decipher the Dominant Epistasis of the Fiberless Mutant in Cotton
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Agricultural Traits Evaluation of the fblSHZ
2.3. Genetic Mapping of the fblSHZ
2.4. Gene Cloning, Vector Construction, and Cotton Transformation
2.5. On-Target Analysis of Gene-Edited Plants
2.6. Scanning Electron Microscopy (SEM)
2.7. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
2.8. Subcellular Localization
2.9. Transcriptome Sequencing
3. Results
3.1. Phenotype and Inheritance of the Fiberless Mutant fblSHZ
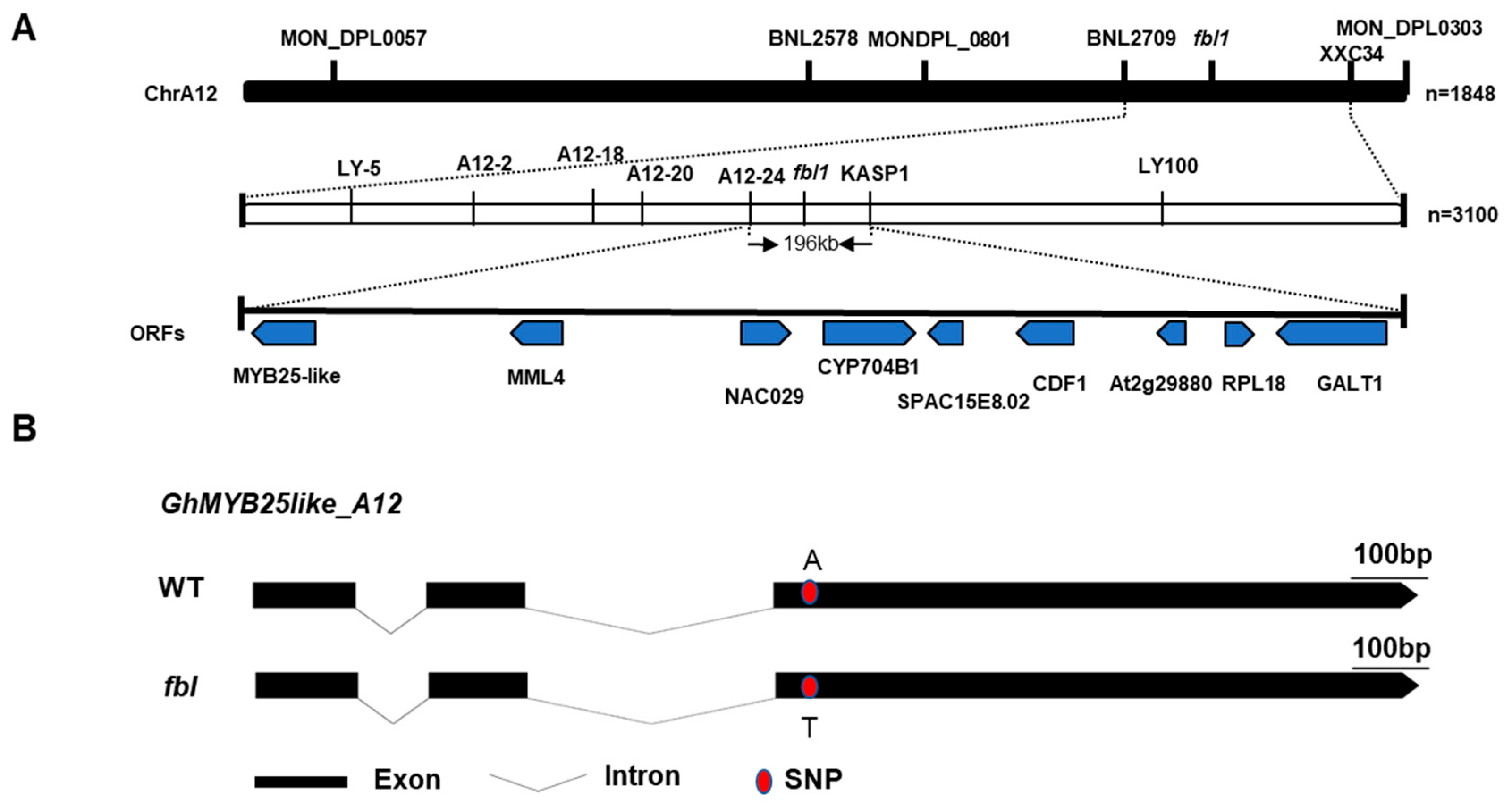
3.2. Genetic Mapping of the fblSHZ Gene
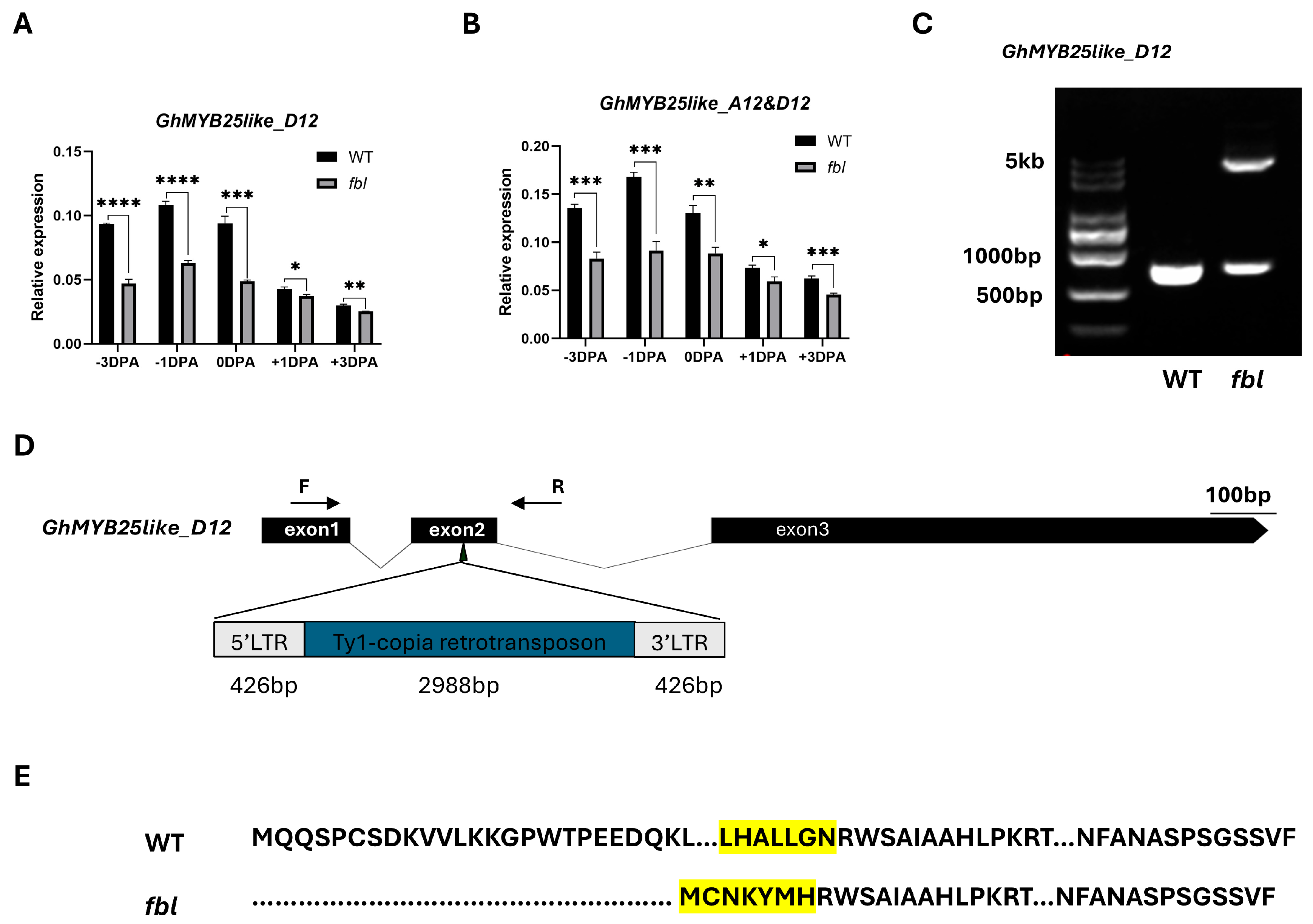
3.3. Cloning of the fblSHZ Gene
3.4. Functional Verification of GhMYB25like
3.5. The Fuzz Gene Was Dominant Epistatic to the Lint Gene
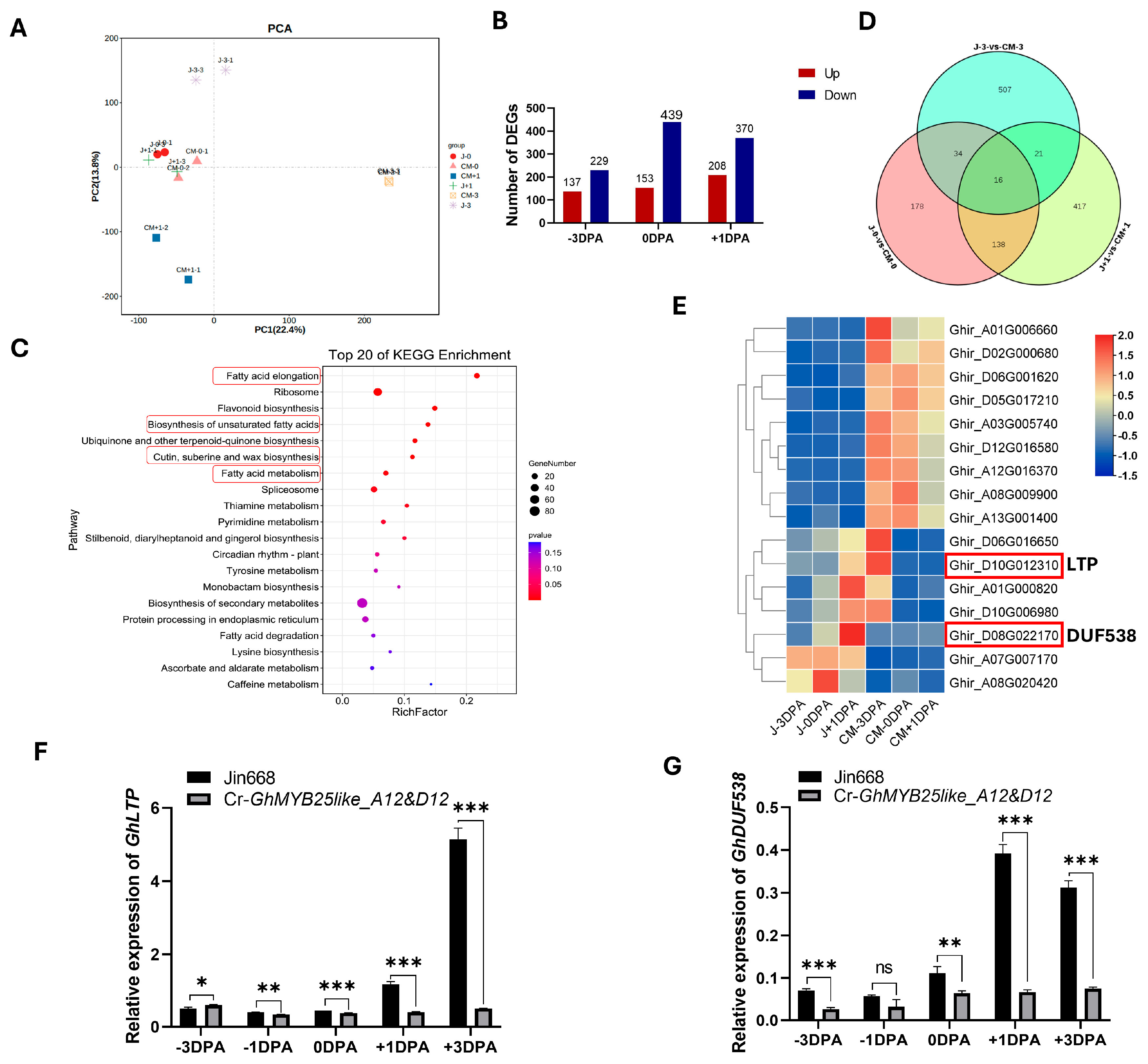
3.6. GhMYB25like Is Involved in Regulating Fiber Initiation in Multiple Pathways
4. Discussion
4.1. The Duplicated GhMYB25like Genes Regulate Cotton Lint and Fuzz Fiber Initiation
4.2. The Dominant Epistasis of the Fiberless Mutant in Cotton
4.3. GhMYB25like Regulates Cotton Fiber Initiation Through Multiple Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CDS | Coding Sequence |
| CRISPR-Cas9 | Clustered Regularly Interspaced Short Palindromic Repeptides-Cas9 |
| CTAB | Hexadecyltrimethylammonium bromide |
| DEG | Differentially Expressed Gene |
| DPA | Days Post-Anthesis |
| GATK | Genome Analysis Toolkit |
| GFP | Green Fluorescent Protein |
| Hi-Tom | High-Throughput Tracking of Mutations |
| InDel | Insertion/Deletion |
| KASP | Kompetitive Allele-Specific PCR |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MML | MYB-MIXTA-like |
| NGS | Next-Generation Sequencing |
| PCR | Polymerase Chain Reaction |
| RNA-seq | RNA sequencing |
| RT-qPCR | Real-Time quantitative PCR |
| SEM | Scanning Electron Microscopy |
| SNP | Single Nucleotide Polymorphism |
| SSR | Simple Sequence Repeat |
| TE | Transposable Element |
| TF | Transcription Factor |
| WT | Wild Type |
References
- Lang, A.G. The origin of lint and fuzz hairs of cotton. J. Agric. Res. 1938, 56, 507–521. [Google Scholar]
- Pu, L.; Li, Q.; Fan, X.; Yang, W.; Xue, Y. The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 2008, 180, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.; Wu, Y.; Yang, Y.; Llewellyn, D.J.; Dennis, E.S. The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 2009, 59, 52–62. [Google Scholar] [CrossRef]
- Walford, S.-A.; Wu, Y.; Llewellyn, D.J.; Dennis, E.S. GhMYB25-like: A key factor in early cotton fibre development. Plant J. 2011, 65, 785–797. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.W.; Yu, N.; Li, C.H.; Luo, B.; Gou, J.Y.; Wang, L.J.; Chen, X.Y. Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 2004, 16, 2323–2334. [Google Scholar] [CrossRef]
- Bedon, F.; Ziolkowski, L.; Walford, S.A.; Dennis, E.S.; Llewellyn, D.J. Members of the MYBMIXTA-like transcription factors may orchestrate the initiation of fiber development in cotton seeds. Front. Plant Sci. 2014, 5, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Wan, Q.; Guan, X.; Yang, N.; Wu, H.; Pan, M.; Liu, B.; Fang, L.; Yang, S.; Hu, Y.; Ye, W.; et al. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol. 2016, 210, 1298–1310. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Yuan, Y.M.; Stiller, W.; Jia, Y.H.; Wang, P.P.; Pan, Z.E.; Du, X.M.; Llewellyn, D.; Wilson, I. Genetic dissection of the fuzzless seed trait in Gossypium barbadense. J. Exp. Bot. 2018, 69, 997–1009. [Google Scholar] [CrossRef]
- Wu, H.; Tian, Y.; Wan, Q.; Fang, L.; Guan, X.; Chen, J.; Hu, Y.; Ye, W.; Zhang, H.; Guo, W.; et al. Genetics and evolution of MIXTA genes regulating cotton lint fiber development. New Phytol. 2018, 217, 883–895. [Google Scholar] [CrossRef]
- Chen, R.; Zhang, J.; Li, J.; Chen, J.; Dai, F.; Tian, Y.; Hu, Y.; Zhu, Q.H.; Zhang, T. Two duplicated GhMML3 genes coordinately control development of lint and fuzz fibers in cotton. Plant Commun. 2025, 6, 101281. [Google Scholar] [CrossRef]
- Chen, W.; Li, Y.; Zhu, S.; Fang, S.; Zhao, L.; Guo, Y.; Wang, J.; Yuan, L.; Lu, Y.; Liu, F.; et al. A retrotransposon insertion in GhMML3_D12 is likely responsible for the lintless locus li3 of tetraploid cotton. Front. Plant Sci. 2020, 11, 593679. [Google Scholar] [CrossRef]
- Qin, Y.; Sun, M.; Li, W.; Xu, M.; Shao, L.; Liu, Y.; Zhao, G.; Liu, Z.; Xu, Z.; You, J.; et al. Single-cell RNA-seq reveals fate determination control of an individual fibre cell initiation in cotton (Gossypium hirsutum). Plant Biotechnol. J. 2022, 20, 2372–2388. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.N.; Le, Y.; Sun, M.L.; Xu, J.W.; Qin, Y.; Men, S.; Ye, Z.X.; Tan, H.Z.; Hu, H.Y.; You, J.Q.; et al. A dominant negative mutation of GhMYB25-like alters cotton fiber initiation, reducing lint and fuzz. Plant Cell 2024, 36, 2759–2777. [Google Scholar] [CrossRef]
- Paterson, A.H.; Brubaker, C.L.; Wendel, J.F. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Rep. 1993, 11, 122–127. [Google Scholar] [CrossRef]
- Li, X.; Jin, X.; Wang, H.; Zhang, X.; Lin, Z. Structure, evolution, and comparative genomics of tetraploid cotton based on a high-density genetic linkage map. DNA Res. 2016, 23, 283–293. [Google Scholar] [CrossRef]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Zhang, R.; Shen, C.; Zhu, D.; Le, Y.; Wang, N.; Li, Y.; Zhang, X.; Lin, Z. Fine-mapping and candidate gene analysis of qFL-c10-1 controlling fiber length in upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 2022, 135, 4483–4494. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, J.; Sun, L.; Ma, Y.; Xu, J.; Liang, S.; Deng, J.; Tan, J.; Zhang, Q.; Tu, L.; et al. High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 2018, 16, 137–150. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, C.; Jiao, X.; Zhang, H.; Song, L.; Li, Y.; Gao, C.; Wang, K. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019, 62, 1–7. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Walford, S.A.; Wu, Y.; Llewellyn, D.J.; Dennis, E.S. Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. Plant J. 2012, 71, 464–478. [Google Scholar] [CrossRef]
- Salminen, T.A.; Blomqvist, K.; Edqvist, J. Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 2016, 244, 971–997. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.C.; Cheng, P.T.; Peng, P.; Lyu, P.C.; Sun, Y.J. Lipid binding in rice nonspecific lipid transfer protein-1 complexes from Oryza sativa. Protein Sci. 2009, 13, 2304–2315. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Yao, H.; Wang, J.; Wang, J.; Xue, H.; Zuo, K. GhLTPG1, a cotton GPI-anchored lipid transfer protein, regulates the transport of phosphatidylinositol monophosphates and cotton fiber elongation. Sci. Rep. 2016, 6, 26829. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Shang, X.; He, Q.; Zhu, L.; Li, W.; Song, X.; Guo, W. LIPID TRANSFER PROTEIN4 regulates cotton ceramide content and activates fiber cell elongation. Plant Physiol. 2023, 193, 1816–1833. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, A. DUF538 protein superfamily is predicted to be chlorophyll hydrolyzing enzymes in plants. Physiol. Mol. Biol. Plants 2016, 22, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Yoshikawa, M.; Kamada, A.; Ohtsuki, T.; Uchida, A.; Nakayama, K.; Satoh, H. The photoconvertible water-soluble chlorophyll-binding protein of Chenopodium album is a member of DUF538, a superfamily that distributes in embryophyta. J. Plant Physiol. 2013, 170, 1549–1552. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Sharma, O.; Nguyen, P.H.T.; Hartono, C.D.; Kanehara, K. A pair of DUF538 domain-containing proteins modulates plant growth and trichome development through the transcriptional regulation of GLABRA1 in Arabidopsis thaliana. Plant J. 2021, 108, 992–1004. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, Y.; Xiong, X.; Xu, Z.; Chen, M.; Li, Y.; Fu, C.; You, C.; Lin, Z. Duplicated Genes on Homologous Chromosomes Decipher the Dominant Epistasis of the Fiberless Mutant in Cotton. Biology 2025, 14, 983. https://doi.org/10.3390/biology14080983
Le Y, Xiong X, Xu Z, Chen M, Li Y, Fu C, You C, Lin Z. Duplicated Genes on Homologous Chromosomes Decipher the Dominant Epistasis of the Fiberless Mutant in Cotton. Biology. 2025; 14(8):983. https://doi.org/10.3390/biology14080983
Chicago/Turabian StyleLe, Yu, Xingchen Xiong, Zhiyong Xu, Meilin Chen, Yuanxue Li, Chao Fu, Chunyuan You, and Zhongxu Lin. 2025. "Duplicated Genes on Homologous Chromosomes Decipher the Dominant Epistasis of the Fiberless Mutant in Cotton" Biology 14, no. 8: 983. https://doi.org/10.3390/biology14080983
APA StyleLe, Y., Xiong, X., Xu, Z., Chen, M., Li, Y., Fu, C., You, C., & Lin, Z. (2025). Duplicated Genes on Homologous Chromosomes Decipher the Dominant Epistasis of the Fiberless Mutant in Cotton. Biology, 14(8), 983. https://doi.org/10.3390/biology14080983






