Transcriptomic Analysis of Peripheral Blood Mononuclear Cells During Ostertagia ostertagi Infection in Cattle Highlights a Generalized Host Immune Reaction
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Design
2.2. Sample Collection and Preparation
2.3. Sequencing and RNA Expression Analysis
2.4. Gene Ontology and Pathway Analysis
3. Results
3.1. Gene Expression Analysis Results for 0 Dpi vs. 14 Dpi Timepoint Comparison
3.2. Gene Expression Analysis Results for 0 Dpi vs. 21 Dpi Timepoint Comparison
3.3. Gene Expression Analysis Results for 0 Dpi vs. 26 Dpi Timepoint Comparison
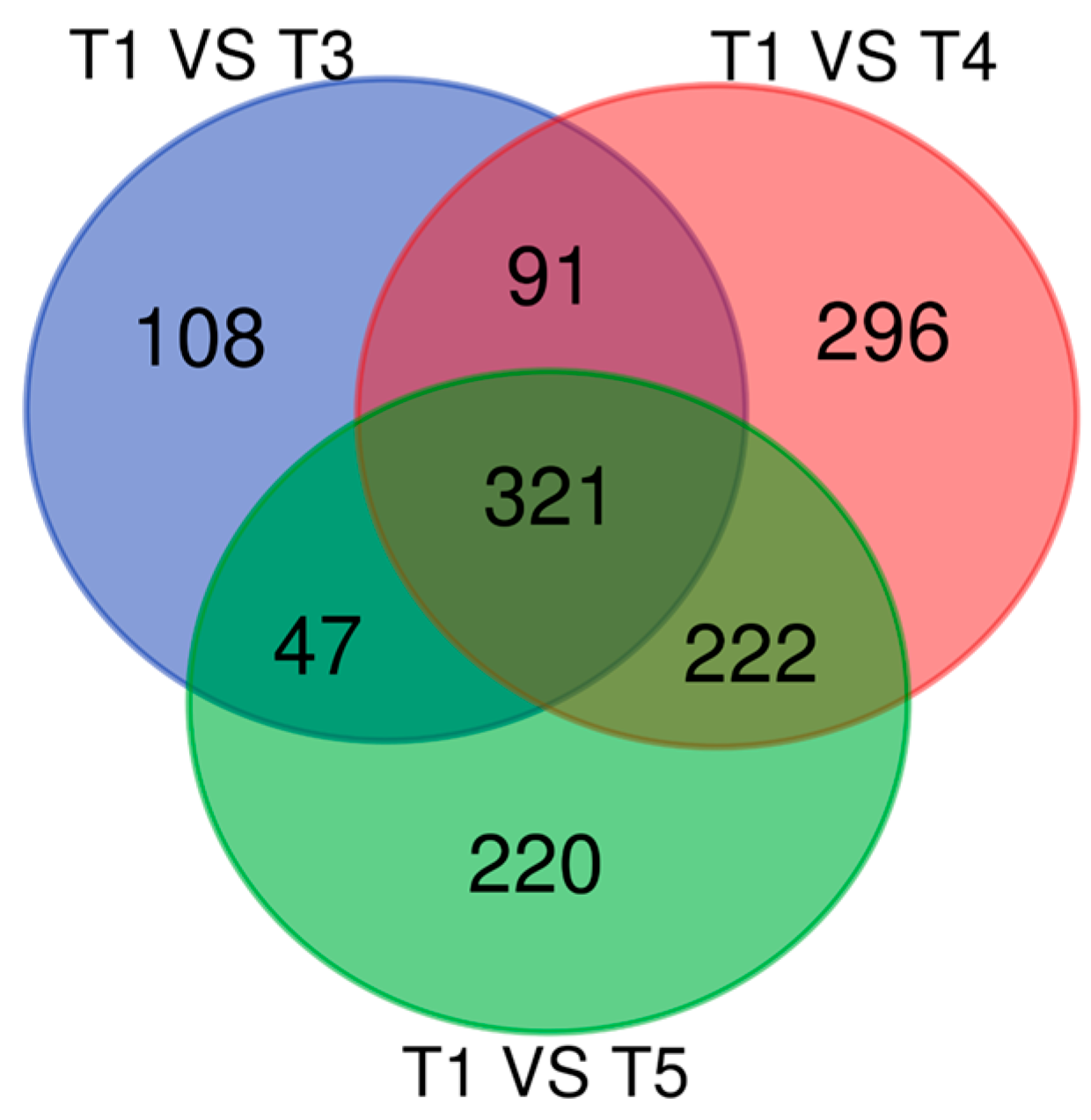
3.4. Gene Expression Analysis Results for Genes That Overlapped Across All Timepoints
| T1 vs. T3 | G.O./PATHWAY ANALYSIS | ID | Genes |
|---|---|---|---|
| Killing by host of symbiont cells | GO:0051873 | ARG1, ELANE, ROMO1 | |
| Apoptotic signaling pathway | GO:0097190 | NUPR1, SNAI1, PHLDA3, BBC3, SHISA5, FADD, HRAS, GPX1, ZNF385A, LMNA, BAK1, CEBPB, CDKN1A, SIVA1, RBCK1, GRINA, CD74, PPM1F, PRELID1, ENO1, TGFB1, BAD, YJEFN3, PELI3, PHIP, ATM, BRCA2 | |
| Abnormality of the gastrointestinal tract | HP:0011024 | ELANE, AGRN, ELN, TUBB4A, ENSBTAG00000050645, HSPG2, HRAS, SREBF1, ALDH4A1, CYBA, SLC25A1, PLXND1, NECTIN1, VWF, IFT27, LMNA, PLOD1, PSAP, ECM1, MRPS34, TUBA1A, MIF, NOTCH3, ENSBTAG00000011704, CDKN1A, CORO1A, RBCK1, GRN, AP2S1, FLNA, TREX1, GLYCTK, PAX8, CHCHD10, ARPC1B, B9D2, TGFB1, GNB2, PHGDH, ENSBTAG00000027075, HSD3B7, REV3L, SCLT1, DYNC2H1, PHIP, ATM, ARL6, ENSBTAG00000068505, LIG4, CEP290, ATRX, BRCA2, VPS13A, TMTC3 | |
| Epithelium migration | GO:0090132 | SNAI1, HB-EGF, GPX1, PLXND1, ZNF580, RRAS, NR4A1, CORO1A, CORO1B, PPM1F, TGFB1, VASH1 | |
| Efferocytosis | KEGG:04148 | ARG1, DUSP2, C1QA, NR1H3, AXL, CEBPB, LRP1, DUSP4, MFGE8, TGFB1 | |
| Biological process involved in interspecies interaction between organisms | GO:0044419 | ISG15, SIGLEC1, RSAD2, ARG1, CMPK2, ENSBTAG00000008021, ELANE, ISG20, IRF7, ZDHHC1, DHX58, ENSBTAG00000053806, CFD, ENSBTAG00000046944, FADD, HRAS, ZFP36, CFP, EMILIN1, GPX1, SLC15A3, CYBA, C1QA, NR1H3, NR4A1, ENSBTAG00000032450, AXL, ADAM15, MIF, BAK1, GSN, CEBPB, CORO1A, RBCK1, GRN, TREX1, BATF3, ENSBTAG00000017645, CD74, ZBED1, MAPKAPK3, NECTIN2, ENO1, TGFB1, TOLLIP, FCN1, PC, ROMO1, VAMP8, EEA1, ENSBTAG00000068505, GZMA, STXBP4, NEDD4, ENSBTAG00000047632 | |
| Epithelial cell migration | GO:0010631 | SNAI1, HB-EGF, GPX1, PLXND1, ZNF580, RRAS, NR4A1, CORO1A, CORO1B, PPM1F, TGFB1, VASH1 | |
| Defense response to symbiont | GO:0140546 | ISG15, RSAD2, ARG1, ENSBTAG00000008021, ELANE, ISG20IRF7, ZDHHC1, DHX58, ENSBTAG00000053806, CFD, ENSBTAG00000046944, FADD, CFP, SLC15A3, CYBA, C1QA, NR1H3, ENSBTAG00000032450, ADAM15, MIF, GSN, CORO1A, GRN, TREX1, ENSBTAG00000017645, CD74, MAPKAPK3, NECTIN2, TOLLIP, FCN1, ROMO1, STXBP4, NEDD4, ENSBTAG00000047632 | |
| T1 vs. T4 | Defense response to symbiont | GO:0140546 | CYBA, CFD, MUL1, GRN, TOLLIP, CD74, TFEB, FADD, CORO1A, TREX1, CFP, MAPKAPK3, ADAM15, GSN, SLC15A3, ZYX, ROMO1, NLRX1, NR1H3, IFI35, CDC42EP4, FGR, MARCHF2, MIF, LAMP1, ENSBTAG00000046944, SENP7, PYCARD, NEDD4, MASP2, ENSBTAG00000017645, ISG15, IRF7, MOV10, NECTIN2, DHX58, ENSBTAG00000011961, UBA7, CXCL3, RAB20, FCN1, SRC, GRO1, TRIM59, RSAD2, C1QA, ISG20, MX2, ENSBTAG00000047632, S100A8, ZBP1, RAB7B, CXCL2, IFI6, ENSBTAG00000053806, CSF1, IFIT2, ARG1, OAS1Y, IFITM3 |
| Epithelial cell migration | GO:0010631 | GPX1, CORO1B, RRAS, CORO1A, AKT3, TGFB1, PPM1F, HSPB1, HMOX1, ZNF580, VASH1, ACVRL1, PLXND1, SRC, JUP, S100A2, SNAI1 | |
| Abnormality of the gastrointestinal tract | HP:0011024 | HRAS, GNB2, CYBA, AP2S1, ARPC1B, ALDH4A1, SLC35C1, GRN, SREBF1, CRKLSLC25A1, RBCK1, CORO1A, TREX1, MRPS34, PSAP, IFT74, SPINT2, VWF, EDEM3, ECE1, FLNA, TTBK2, COMT, GLYCTK, PHIP, ABCA3, TGFB1, NCF1, TNFRSF1A, AHCY, SELENON, MYL9, B9D2, MIF, TYROBP, TMTC3, CSF1R, HMOX1, BRCA2PLOD1, NECTIN1, NOTCH3, REV3L, ATRX, SGO1, DYNC2H1, SPIB, SLC13A5, ENSBTAG00000007816, IL1RN, ECM1, GP1BB, ACVRL1, ELN, CENPF, SERPINE1, MASP2, CEP290, DYSF, PYGM, VPS13A, AGRN, ENSBTAG00000027075, ARL6, CHCHD10, PLXND1, LAMB2, LMNA, BOLA-DQB, HSPG2, ENPP1, SRC, ENSBTAG00000050645, EPCAM, IFT56, CDKN2A, TUBB4A | |
| Antioxidant activity | GO:0016209 | GPX1, GPX4, HBB, HBA, GPX3, ALOX5AP, LTC4S, S100A8 | |
| Biological process involved in interspecies interaction between organisms | GO:0044419 | HRAS, CYBA, GPX1, CFD, MUL1, ZBED1, ENO1, VAMP8, GRN, TOLLIP, CD74, TFEB, FADD, VPS18, RBCK1CORO1A, TREX1, CFP, MAPKAPK3, ADAM15, BAK1, GSN, SLC15A3, ZYX, ROMO1, NLRX1, TGFB1, NR1H3, IFI35, CDC42EP4, IL17RC, FGR, TNFRSF1A, MARCHF2, HSPB1, MIF, LAMP1, CSF1R, ENSBTAG00000046944, EEA1, SENP7, PYCARD, CEBPB, NEDD4, CXCL8, CCDC186, MASP2, ENSBTAG00000017645, ISG15, IRF7, BATF3, SIGLEC1, MOV10, NECTIN2, PC, DHX58, ENSBTAG00000011961, UBA7, SMAD6, CXCL3, RAB20, FCN1, SRC, GRO1, EMILIN1, TRIM59, RSAD2, C1QA, ISG20, MX2, ENSBTAG00000047632, S100A8, ZBP1, RAB7B, CXCL2, IFI6, C5AR1, ENSBTAG00000053806, CSF1, CMPK2, IFIT2, ARG1, OAS1Y, IFITM3 | |
| Metabolism of vitamins and cofactors | REAC:R-BTA-196854 | LRP10, SLC19A1, TCN2, MOCS1, PARP10, PDXK, LRP1, AMN, AGRN, PC, ENPP1, SDC3, SDC4 | |
| T1 vs. T5 | Wound healing | GO:0042060 | HRAS, GPX1, VWF, HPS6, GP9, FLNA, CORO1B, MPIG6B, HMOX1, GP1BB, ENSBTAG00000047175, TUBB1, DYSF, SELP, SDC4, FGF2 |
| Functional abnormality of the gastrointestinal tract | HP:0012719 | GNB2, HRAS, SLC35C1, ARPC1B, MRPS34, CORO1A, VWF, SLC25A1, ACTB, MIF, ALDH4A1, IFT74, RBCK1, TREX1, MYL9, GP9, FLNA, NFKBIA, TGFB1, TTBK2, PSAP, TNFAIP3, TMTC3, IL7R, ECE1, TUBB4A, HMOX1, SPIB, NOTCH3, ECM1, LMNA, CEP290, ATRXGP1BB, MASP2, NCF1, ELN, MVK, DTYMK, VPS13A, HSPG2, PHGDH, BRCA2, AGRN, CDKN2A, DYSF, NRCAM, PYGM | |
| Malaria | KEGG:05144 | CXCL8, TGFB1, GYPC, HBA, LRP1, HBB, SELP | |
| Defense response to symbiont | GO:0140546 | MUL1, FADD, TOLLIP, CORO1A, CYBA, EXOSC4, CXCL3, ROMO1, CFD, MIF, NFKBIZ, NR1H3, CD74, TREX1, CFP, GRO1, NFKBIA, MAPKAPK3, MSRB1, CDC42EP4, GSN, SLC15A3, CCL4 | |
| Biological process involved in interspecies interaction between organisms | GO:0044419 | HRAS, ENO1, BAK1, ZBED1, MUL1, CXCL8, FADD, TOLLIP, CORO1A, GPX1, CYBA, EXOSC4, CXCL3, ROMO1, CFD, MIF, NFKBIZ, RBCK1, NR1H3, THOC6, CD74, TREX1, CFP, GRO1, NFKBIA, TGFB1, MAPKAPK3, MSRB1, CDC42EP4, GSN, SLC15A3, CCL4, ENSBTAG00000032450, ENSBTAG00000011961, IFI35, MARCHF2, TNFAIP3, IL7R, HYAL2, ENSBTAG00000017645, IER3, IL17RC, CXCL2, MOV10, MASP2, IRF7, ISG15, ISG20, PYCARD, RAB7B, CDC42EP2, ABCA1, SIGLEC1, BATF, ENSBTAG00000008021, RSAD2, C1QA, NECTIN2, ENSBTAG00000053806, FCN1, RAB20, ENSBTAG00000046944, CMPK2, S100A8, UBA7, IFIT2, IFIT3, ARG1, IFITM3, MX2, ZBP1, IFI6, ZDHHC1, DHX58, LRRC19, LAG3, IL27 |
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DPI | Days post infection |
| PBMC | Peripheral Blood Mononuclear Cell |
References
- Klesius, P.H. Immunity to Ostertagia ostertagi. Vet. Parasitol. 1988, 27, 159–167. [Google Scholar] [CrossRef]
- Fox, M.T. Pathophysiology of infection with Ostertagia ostertagi in cattle. Vet. Parasitol. 1993, 46, 143–158. [Google Scholar] [CrossRef]
- Abubucker, S.; Zarlenga, D.S.; Martin, J.; Yin, Y.; Wang, Z.; McCarter, J.P.; Gasbarree, L.; Wilson, R.K.; Mitreva, M. The transcriptomes of the cattle parasitic nematode Ostertagia ostartagi. Vet. Parasitol. 2009, 162, 89–99. [Google Scholar] [CrossRef]
- Rose, J.H. The development of the parasitic stages of Ostertagia ostertagi. J. Helminthol. 1969, 43, 173–184. [Google Scholar] [CrossRef]
- Bakshi, M.; Hebert, D.; Gulbronson, C.; Bauchan, G.; Tuo, W.; Zarlenga, D. Ostertagia ostertagi Mediates Early Host Immune Responses via Macrophage and Toll-Like Receptor Pathways. Infect. Immun. 2021, 89, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, R.W.; Hou, Y.; Li, C.; Gasbarre, L.C. Localized complement activation in the development of protective immunity against Ostertagia ostertagi infections in cattle. Vet. Parasitol. 2010, 174, 247–256. [Google Scholar] [CrossRef]
- Maizels, R.M.; Smits, H.H.; McSorley, H.J. Modulation of Host Immunity by Helminths: The Expanding Repertoire of Parasite Effector Molecules. Immunity 2018, 49, 801–818. [Google Scholar] [CrossRef]
- Zarlenga, D.S.; Nisbet, A.J.; Gasbarre, L.C.; Garrett, W.M. A calcium-activated nucleotidase secreted from Ostertagia ostertagi 4th-stage larvae is a member of the novel salivary apyrases present in blood-feeding arthropods. Parasitology 2011, 138, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Campos, A.; Correia, C.N.; Naranjo-Lucena, A.; Garza-Cuartero, L.; Farries, G.; Browne, J.A.; MacHugh, D.E.; Mulcahy, G. Fasciola hepatica Infection in Cattle: Analyzing Responses of Peripheral Blood Mononuclear Cells (PBMC) Using a Transcriptomics Approach. Front. Immunol. 2019, 10, 2081. [Google Scholar] [CrossRef] [PubMed]
- Niedziela, D.A.; Naranjo-Lucena, A.; Molina-Hernandez, V.; Browne, J.A.; Martinez-Moreno, A.; Perez, J.; MacHugh, D.E.; Mulcahy, G. Timing of Transcriptomic Peripheral Blood Mononuclear Cell Responses of Sheep to Fasciola hepatica Infection Differs From Those of Cattle, Reflecting Different Disease Phenotypes. Front. Immunol. 2021, 12, 729217. [Google Scholar] [CrossRef]
- Qu, G.; Fetterer, R.; Leng, L.; Du, X.; Zarlenga, D.; Shen, Z.; Han, W.; Bucala, R.; Tuo, W. Ostertagia ostertagi macrophage migration inhibitory factor is present in all developmental stages and may cross-regulate host functions through interaction with the host receptor. Int. J. Parasitol. 2014, 44, 355–367. [Google Scholar] [CrossRef]
- Waghorn, T.S.; Miller, C.M.; Leathwick, D.M. Confirmation of ivermectin resistance in Ostertagia ostertagi in cattle in New Zealand. Vet. Parasitol. 2016, 229, 139–143. [Google Scholar] [CrossRef]
- Rose Vineer, H.; Morgan, E.R.; Hertzberg, H.; Bartley, D.J.; Bosco, A.; Charlier, J.; Chartier, C.; Claerebout, E.; de Waal, T.; Hendrickx, G.; et al. Increasing importance of anthelmintic resistance in European livestock: Creation and meta-analysis of an open database. Parasite 2020, 27, 69. [Google Scholar] [CrossRef]
- Kaplan, R.M. Biology, Epidemiology, Diagnosis, and Management of Anthelmintic Resistance in Gastrointestinal Nematodes of Livestock. Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 17–30. [Google Scholar] [CrossRef]
- Tuo, W.; Zarlenga, D.; Bakshi, M.; Vinyard, B. Repeated, drug-truncated infections with Ostertagia ostertagi elicit strong humoral and cell-mediated immune responses and confer partial protection in cattle. Vet. Parasitol. 2021, 296, 109510. [Google Scholar] [CrossRef] [PubMed]
- Avramenko, R.W.; Redman, E.M.; Lewis, R.; Yazwinski, T.A.; Wasmuth, J.D.; Gilleard, J.S. Exploring the Gastrointestinal “Nemabiome”: Deep Amplicon Sequencing to Quantify the Species Composition of Parasitic Nematode Communities. PLoS ONE 2015, 10, e0143559. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Galaxy, C. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351, Correction in Nucleic Acids Res. 2022, 50, 8999. [Google Scholar]
- Newman, V.; Moore, B.; Sparrow, H.; Perry, E. The Ensembl Genome Browser: Strategies for Accessing Eukaryotic Genome Data. Methods Mol. Biol. 2018, 1757, 115–139. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Maglott, D.; Ostell, J.; Pruitt, K.D.; Tatusova, T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2011, 39, D52–D57. [Google Scholar] [CrossRef]
- Hendawy, S.H.M. Immunity to gastrointestinal nematodes in ruminants: Effector cell mechanisms and cytokines. J. Parasit. Dis. 2018, 42, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Pawlowska, M.; Mila-Kierzenkowska, C.; Szczegielniak, J.; Wozniak, A. Oxidative Stress in Parasitic Diseases-Reactive Oxygen Species as Mediators of Interactions between the Host and the Parasites. Antioxidants 2023, 13, 38. [Google Scholar] [CrossRef]
- Mihi, B.; van Meulder, F.; Vancoppernolle, S.; Rinaldi, M.; Chiers, K.; van den Broeck, W.; Goddeeris, B.M.; Vercruysse, J.; Claerebout, E.; Geldhof, P. Analysis of the mucosal immune responses induced by single and trickle infections with the bovine abomasal nematode Ostertagia ostertagi. Parasite Immunol. 2014, 36, 150–156. [Google Scholar] [CrossRef]
- Taylor, S.R.; Markesbery, M.G.; Harding, P.A. Heparin-binding epidermal growth factor-like growth factor (HB-EGF) and proteolytic processing by a disintegrin and metalloproteinases (ADAM): A regulator of several pathways. Semin. Cell Dev. Biol. 2014, 28, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, M.; Dreesen, L.; Hoorens, P.R.; Li, R.W.; Claerebout, E.; Goddeeris, B.; Vercruysse, J.; Van Den Broek, W.; Geldhof, P. Infection with the gastrointestinal nematode Ostertagia ostertagi in cattle affects mucus biosynthesis in the abomasum. Vet. Res. 2011, 42, 61. [Google Scholar] [CrossRef]
- Ge, Y.; Huang, M.; Yao, Y.M. Efferocytosis and Its Role in Inflammatory Disorders. Front. Cell Dev. Biol. 2022, 10, 839248. [Google Scholar] [CrossRef] [PubMed]
- Villalba, M.; Fredericksen, F.; Otth, C.; Olavarria, V.H. Molecular characterization of the bovine IER3 gene: Down-regulation of IL-8 by blocking NF-kappaB activity mediated by IER3 overexpression in MDBK cells infected with bovine viral diarrhea virus-1. Mol. Immunol. 2017, 92, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Roh, J.S.; Sohn, D.H. Damage-Associated Molecular Patterns in Inflammatory Diseases. Immune Netw. 2018, 18, e27. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Good, B.; Hanrahan, J.P.; McGettigan, P.; Browne, J.; Keane, O.M.; Bahar, B.; Mehta, J.; Markey, B.; Lohan, A.; et al. Variation in the Ovine Abomasal Lymph Node Transcriptome between Breeds Known to Differ in Resistance to the Gastrointestinal Nematode. PLoS ONE 2015, 10, e0124823. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, A.; van der Veen, A.G.; Bunyan, M.; Borg, A.; Frith, D.; Howell, S.; Kjaer, S.; Beling, A.; Snijders, A.P.; Knobeloch, K.P.; et al. Cysteine-Reactive Free ISG15 Generates IL-1β-Producing CD8α+ Dendritic Cells at the Site of Infection. J. Immunol. 2018, 201, 604–614. [Google Scholar] [CrossRef]
- Doolan, R.; Putananickal, N.; Tritten, L.; Bouchery, T. How to train your myeloid cells: A way forward for helminth vaccines? Front. Immunol. 2023, 14, 1163364. [Google Scholar] [CrossRef]
- Hayes, K.S.; Bancroft, A.J.; Grencis, R.K. Immune-mediated regulation of chronic intestinal nematode infection. Immunol. Rev. 2004, 201, 75–88. [Google Scholar] [CrossRef]
- Abd Ellah, M.R. Involvement of free radicals in parasitic infestations. J. Appl. Anim. Res. 2013, 41, 69–76. [Google Scholar] [CrossRef]
- Gause, W.C.; Wynn, T.A.; Allen, J.E. Type 2 immunity and wound healing: Evolutionary refinement of adaptive immunity by helminths. Nat. Rev. Immunol. 2013, 13, 607–614. [Google Scholar] [CrossRef]
| Biological process involved in interspecies interaction between organisms (GO:0044419) | ||||
| GENE ID | GENE NAME | T3 | T4 | T5 |
| ENSBTAG00000014707 | ISG15 | 3.69 | 4.71 | 4.33 |
| ENSBTAG00000013167 | SIGLEC1 | 3.12 | 3.36 | 3.01 |
| ENSBTAG00000016061 | RSAD2 | 2.52 | 3.04 | 3.16 |
| ENSBTAG00000019979 | CMPK2 | 2.36 | 2.64 | 2.76 |
| ENSBTAG00000014762 | ISG20 | 2.08 | 2.44 | 2.75 |
| ENSBTAG00000012403 | ARG1 | 2.37 | 2.42 | 2.34 |
| ENSBTAG00000047680 | IRF7 | 1.98 | 2.35 | 2.23 |
| ENSBTAG00000046944 | ENSBTAG00000046944 | 1.31 | 2.10 | 1.33 |
| ENSBTAG00000046580 | DHX58 | 1.55 | 2.06 | 1.62 |
| ENSBTAG00000054195 | GPX1 | 1.23 | 1.44 | 1.24 |
| ENSBTAG00000048122 | CFD | 1.40 | 1.43 | 1.15 |
| ENSBTAG00000015815 | CFP | 1.25 | 1.41 | 1.29 |
| ENSBTAG00000046644 | HRAS | 1.29 | 1.41 | 1.37 |
| ENSBTAG00000053806 | ENSBTAG00000053806 | 1.44 | 1.40 | 1.54 |
| ENSBTAG00000061619 | CYBA | 1.23 | 1.38 | 1.14 |
| ENSBTAG00000018274 | FADD | 1.29 | 1.35 | 1.40 |
| ENSBTAG00000015318 | NECTIN2 | 1.04 | 1.34 | 1.18 |
| ENSBTAG00000008406 | TREX1 | 1.06 | 1.32 | 1.19 |
| ENSBTAG00000018366 | SLC15A3 | 1.23 | 1.30 | 1.17 |
| ENSBTAG00000017002 | RBCK1 | 1.09 | 1.27 | 1.15 |
| ENSBTAG00000016532 | MAPKAPK3 | 1.04 | 1.22 | 1.07 |
| ENSBTAG00000015228 | CD74 | 1.05 | 1.21 | 1.00 |
| ENSBTAG00000061194 | ZBED1 | 1.05 | 1.20 | 1.17 |
| ENSBTAG00000019915 | GSN | 1.10 | 1.20 | 1.08 |
| ENSBTAG00000010681 | NR1H3 | 1.16 | 1.16 | 1.24 |
| ENSBTAG00000013411 | ENO1 | 1.03 | 1.12 | 1.19 |
| ENSBTAG00000008631 | CORO1A | 1.09 | 1.10 | 1.16 |
| ENSBTAG00000000428 | BAK1 | 1.11 | 1.10 | 1.37 |
| ENSBTAG00000007153 | C1QA | 1.22 | 1.07 | 1.10 |
| ENSBTAG00000008237 | TOLLIP | 1.02 | 1.06 | 1.02 |
| ENSBTAG00000017645 | ENSBTAG00000017645 | 1.06 | 1.04 | 1.14 |
| ENSBTAG00000048155 | FCN1 | 1.02 | 1.03 | 1.01 |
| ENSBTAG00000020457 | TGFB1 | 1.03 | 1.03 | 1.04 |
| ENSBTAG00000027361 | ROMO1 | 1.01 | 1.02 | 1.14 |
| ENSBTAG00000007375 | MIF | 1.12 | 1.00 | 1.20 |
| Defense response to symbiont (GO:0140546) | ||||
| GENE ID | GENE NAME | T3 | T4 | T5 |
| ENSBTAG00000014707 | ISG15 | 3.69 | 4.71 | 4.33 |
| ENSBTAG00000016061 | RSAD2 | 2.52 | 3.04 | 3.16 |
| ENSBTAG00000014762 | ISG20 | 2.08 | 2.44 | 2.75 |
| ENSBTAG00000012403 | ARG1 | 2.37 | 2.42 | 2.34 |
| ENSBTAG00000047680 | IRF7 | 1.98 | 2.35 | 2.23 |
| ENSBTAG00000046944 | ENSBTAG00000046944 | 1.31 | 2.10 | 1.33 |
| ENSBTAG00000046580 | DHX58 | 1.55 | 2.06 | 1.62 |
| ENSBTAG00000048122 | CFD | 1.40 | 1.43 | 1.15 |
| ENSBTAG00000015815 | CFP | 1.25 | 1.41 | 1.29 |
| ENSBTAG00000053806 | ENSBTAG00000053806 | 1.44 | 1.40 | 1.54 |
| ENSBTAG00000061619 | CYBA | 1.23 | 1.38 | 1.14 |
| ENSBTAG00000018274 | FADD | 1.29 | 1.35 | 1.40 |
| ENSBTAG00000015318 | NECTIN2 | 1.04 | 1.34 | 1.18 |
| ENSBTAG00000008406 | TREX1 | 1.06 | 1.32 | 1.19 |
| ENSBTAG00000018366 | SLC15A3 | 1.23 | 1.30 | 1.17 |
| ENSBTAG00000016532 | MAPKAPK3 | 1.04 | 1.22 | 1.07 |
| ENSBTAG00000015228 | CD74 | 1.05 | 1.21 | 1.00 |
| ENSBTAG00000019915 | GSN | 1.10 | 1.20 | 1.08 |
| ENSBTAG00000010681 | NR1H3 | 1.16 | 1.16 | 1.24 |
| ENSBTAG00000008631 | CORO1A | 1.09 | 1.10 | 1.16 |
| ENSBTAG00000007153 | C1QA | 1.22 | 1.07 | 1.10 |
| ENSBTAG00000008237 | TOLLIP | 1.02 | 1.06 | 1.02 |
| ENSBTAG00000017645 | ENSBTAG00000017645 | 1.06 | 1.04 | 1.14 |
| ENSBTAG00000048155 | FCN1 | 1.02 | 1.03 | 1.01 |
| ENSBTAG00000027361 | ROMO1 | 1.01 | 1.02 | 1.14 |
| ENSBTAG00000007375 | MIF | 1.12 | 1.00 | 1.20 |
| Abnormality of the gastrointestinal tract (HP:0011024) | ||||
| GENE ID | GENE NAME | T3 | T4 | T5 |
| ENSBTAG00000013191 | AGRN | 1.94 | 2.52 | 2.17 |
| ENSBTAG00000012265 | VWF | 1.18 | 2.32 | 2.55 |
| ENSBTAG00000019517 | ELN | 1.44 | 1.57 | 1.45 |
| ENSBTAG00000017574 | LMNA | 1.16 | 1.49 | 1.73 |
| ENSBTAG00000030335 | ALDH4A1 | 1.26 | 1.46 | 1.15 |
| ENSBTAG00000046644 | HRAS | 1.29 | 1.41 | 1.37 |
| ENSBTAG00000001814 | PLXND1 | 1.20 | 1.40 | 1.00 |
| ENSBTAG00000043971 | NOTCH3 | 1.12 | 1.38 | 1.31 |
| ENSBTAG00000017122 | HSPG2 | 1.31 | 1.38 | 1.45 |
| ENSBTAG00000061619 | CYBA | 1.23 | 1.38 | 1.14 |
| ENSBTAG00000008406 | TREX1 | 1.06 | 1.32 | 1.19 |
| ENSBTAG00000017002 | RBCK1 | 1.09 | 1.27 | 1.15 |
| ENSBTAG00000021499 | PSAP | 1.13 | 1.23 | 1.10 |
| ENSBTAG00000011190 | FLNA | 1.07 | 1.19 | 1.15 |
| ENSBTAG00000010584 | AP2S1 | 1.09 | 1.17 | 1.10 |
| ENSBTAG00000008528 | SLC25A1 | 1.21 | 1.17 | 1.11 |
| ENSBTAG00000003806 | ECM1 | 1.13 | 1.13 | 1.15 |
| ENSBTAG00000026415 | MRPS34 | 1.12 | 1.10 | 1.27 |
| ENSBTAG00000008631 | CORO1A | 1.09 | 1.10 | 1.16 |
| ENSBTAG00000023452 | B9D2 | 1.04 | 1.08 | 1.14 |
| ENSBTAG00000046248 | ARPC1B | 1.04 | 1.07 | 1.01 |
| ENSBTAG00000021013 | TUBB4A | 1.40 | 1.07 | 1.73 |
| ENSBTAG00000020457 | TGFB1 | 1.03 | 1.03 | 1.04 |
| ENSBTAG00000006495 | GNB2 | 1.03 | 1.02 | 1.07 |
| ENSBTAG00000007375 | MIF | 1.12 | 1.00 | 1.20 |
| ENSBTAG00000018745 | CEP290 | −1.06 | −1.12 | −1.17 |
| ENSBTAG00000017734 | VPS13A | −1.24 | −1.14 | −1.05 |
| ENSBTAG00000038434 | ATRX | −1.13 | −1.15 | −1.03 |
| ENSBTAG00000009023 | TMTC3 | −1.28 | −1.39 | −1.38 |
| ENSBTAG00000000988 | BRCA2 | −1.13 | −1.41 | −1.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fleming, D.S.; Bakshi, M.; Thompson, P.; Beshah, E.; Tuo, W. Transcriptomic Analysis of Peripheral Blood Mononuclear Cells During Ostertagia ostertagi Infection in Cattle Highlights a Generalized Host Immune Reaction. Biology 2025, 14, 1034. https://doi.org/10.3390/biology14081034
Fleming DS, Bakshi M, Thompson P, Beshah E, Tuo W. Transcriptomic Analysis of Peripheral Blood Mononuclear Cells During Ostertagia ostertagi Infection in Cattle Highlights a Generalized Host Immune Reaction. Biology. 2025; 14(8):1034. https://doi.org/10.3390/biology14081034
Chicago/Turabian StyleFleming, Damarius S., Mariam Bakshi, Peter Thompson, Ethiopia Beshah, and Wenbin Tuo. 2025. "Transcriptomic Analysis of Peripheral Blood Mononuclear Cells During Ostertagia ostertagi Infection in Cattle Highlights a Generalized Host Immune Reaction" Biology 14, no. 8: 1034. https://doi.org/10.3390/biology14081034
APA StyleFleming, D. S., Bakshi, M., Thompson, P., Beshah, E., & Tuo, W. (2025). Transcriptomic Analysis of Peripheral Blood Mononuclear Cells During Ostertagia ostertagi Infection in Cattle Highlights a Generalized Host Immune Reaction. Biology, 14(8), 1034. https://doi.org/10.3390/biology14081034







