Simple Summary
Plants can be colonized by numerous and diverse bacteria. Studies of microorganisms isolated from the roots of Chamaecytisus albus (Spanish broom) plants identified two bacterial strains with characteristics different from those previously identified. Detailed genetic and morphological studies as well as analysis of selected organic factors indicate that these strains are related to known Phyllobacterium species, but differ sufficiently from them to be considered distinct species. For these two new species, we propose the names Phyllobacterium chamaecytisi (from Chamaecytisus, the host plant from which strains were isolated) and Phyllobacterium lublinensis (referring to Lublin, the region in eastern Poland where the host plant was collected). Interestingly, although these strains do not belong to the group of rhizobia, i.e., bacteria establishing symbiosis with legumes, which also includes Chamaecytisus, they were found in root nodules - the part of the plant colonized by symbiotic bacteria. The research conducted has expanded our knowledge of the species richness of bacteria from the genus Phyllobacterium and the biodiversity of microbial communities inhabiting plant tissues.
Abstract
The taxonomic status of two bacterial strains, KW56T and 2063T, isolated from root nodules of Chamaecytisus albus (Spanish broom), was investigated using a polyphasic approach. Both isolates belong to the genus Phyllobacterium, yet exhibit significant genotypic and phenotypic differences from all currently described species. Whole-genome comparisons revealed that strain KW56T is most closely related to Phyllobacterium trifolii PETP 02T, while strain 2063T is related to Phyllobacterium brassicacearum strains STM 196T and 29-15. However, digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI) values between the new isolates and their closest relatives were below established species delineation thresholds, supporting their recognition as novel species. Phenotypic analyses confirmed morphological and growth characteristics typical for Phyllobacterium, while also revealing several discriminatory physiological traits. Fatty acid profiling showed that cyclopropyl 19:0 was the major fatty acid in both strains, though each exhibited a unique fatty acid composition. Chemotaxonomic markers included 3-OH 14:0, a19:1, and 21:0 for strain KW56T, and 3-OH 14:0, 3-OH 17:0, and 3-OH 18:0 for strain 2063T. Based on the genomic, phenotypic, and chemotaxonomic data, we propose that strains KW56T and 2063T represent two novel species, for which the names Phyllobacterium chamaecytisi sp. nov. (DSM 113831) and Phyllobacterium lublinensis sp. nov. (DSM 113830) are proposed.
1. Introduction
Bacteria belonging to the genus Phyllobacterium can be commonly found on the rhizoplane and phylloplane of plants, as well as inside plant tissues. The genus Phyllobacterium was originally described by Knösel [1,2,3], and the first well-characterized species was P. myrsinacearum originating from Ardisia leaf nodules [4].
Until now, fourteen recognized Phyllobacterium species have been described. Numerous type strains were isolated from root nodules of different legumes like Trifolium pratense (P. trifolii) [5], Lathyrus numidicus (P. ifriquiense) [6], Astragalus algerianus (P. leguminum) [6], Phaseolus vulgaris (P. endophyticus) [7], Lotus corniculatus (P. loti) [8], Sophora florescens (P. sophorae) [9], Lotus lancerottensis (P. salinisoli) [10], or Oxytropis triphylla (P. zundukense) [11]. However, the capacity of these isolates to induce nodulation was demonstrated only for P. trifolii and P. sophorae [5,9] suggesting that most of mentioned bacteria are non-symbiotic plant endophytes. Some Phyllobacterium species like P. bourgognense or P. brassicacearum were found inside tissues of non-leguminous plants; other were isolated from soil (P. phragmitis, P. pellucidum) [12,13] or from volcanic rock (P. catacumbae) [14]. Moreover, different Phyllobacterium strains have also been found in water [15] or associated with unicellular Eukaryotes [16,17], which shows the great variety of habitats which can be colonized by these microorganisms, and the ability of phyllobacteria to adaptation to free-living, associative, and even endosymbiotic lifestyles.
Chamaecytisus albus (white Spanish broom) is a legume shrub that can be found in only one natural habitat, near Hrubieszow in south-eastern Poland. The Polish natural habitat of C. albus is still shrinking, and so is the number of living plants in this environment; therefore, this species was described as “critically endangered” in 2016 in the “Polish Red List of Fern and Flowering Plants” [18]. Bacterial endophytes of these plants were recently isolated and described [19].
The aim of the current study was to demonstrate the distinct taxonomic position of the Phyllobacterium-related isolates KW56 and 2063, based on 16S rRNA gene sequencing, whole-genome sequence analysis, the comparison of morphological and physiological features, and comparative whole-cell fatty acid analysis.
2. Materials and Methods
2.1. Strains Isolation
Strains were isolated from root nodules of Chamaecytisus albus (a plant growing near Hrubieszow in the South-Eastern region of Poland, 50°48′09″ N, 23°53′31″ E). Nodules were harvested and surface-sterilized; they were rinsed several times with sterile water, incubated with 0.1% HgCl2 for 5 min, rinsed several times with sterile water again, incubated with 70% ethanol for 5 min, and finally rinsed several times with sterile water. The suspension was made by crushing single nodules in sterile water. It was transfered on yeast–mannitol medium (YEM) and incubated at 28 °C for 7 days [20]. Bacteria isolated from Chamaecytisus albus root nodules were purified by streaking several times on YEM agar. The purified strains were kept in YEM medium at 4 °C as well as at −70 °C in YEM medium with 15% (v/v) glycerol.
2.2. Whole-Genome Sequencing
DNA was extracted from overnight cultures using a Genomic Mini AX Bacteria Spin Kit (A&A Biotechnology, Gdansk, Poland). Extractions were performed as described in the manufacturer’s protocol for Gram-negative bacteria. Genomic DNA was sequenced via StarSEQ GmbH, Mainz, Germany.
Illumina raw pair-end 150 bp reads were quality controlled and trimmed via FastQC v0.11.9 (https://github.com/s-andrews/FastQC, accessed on 12 October 2023), FastQ Screen v0.14.1 (https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen, accessed on 12 October 2023), and Trimmomatic [21], using standard parameters. Filtered reads were assembled into contigs using Spades (3.13.0) [22]. The draft genomes assemblies were deposited at GenBank under accession numbers GCA_020164435.1 and GCA_020164455.1.
2.3. Genome Annotation
The genome was annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) [23]. For comparison, the genome was also submitted to the Comprehensive Genome Analysis Service at PATRIC (www.patricbrc.org) [24] and annotated using RAST tool kit v1.073 [25]. The genome was checked for completeness and contamination using EvalG and EvalCon [26]. The genome maps were prepared using the circos v0.69-9 tool as part of the Comprehensive Genome Analysis service on PATRIC [24].
2.4. Phylogenomic Analyses
Single-gene phylogenies based on 16S rRNA gene sequences were inferred by the GGDC web server [27] available at http://ggdc.dsmz.de/ using the DSMZ phylogenomics pipeline [28] adapted to single genes. A multiple sequence alignment was created with MUSCLE v3.8.31 [29]. Maximum likelihood (ML) and maximum parsimony (MP) trees were inferred from the alignment with RAxML v8 [30] and TNT [31], respectively. For ML, rapid bootstrapping in conjunction with the autoMRE bootstrapping criterion [32] and subsequent search for the best tree was used; for MP, 1000 bootstrapping replicates were used in conjunction with tree-bisection-and-reconnection branch swapping and ten random sequence addition replicates. The sequences were checked for a compositional bias using the Χ2 test as implemented in PAUP* v4.0 Beta [33].
Whole-genome phylogenomic analysis utilizing digital DNA-DNA hybridization (dDDH) was calculated using the Type Strain Genome Server (TYGS) available at https://tygs.dsmz.de/ [34] based on the available genomes of closely related neighbors. For comparison and confirmation, the ANI analysis was also performed using the JSpeciesWS webserver [35] using the same set of genomes. Default parameters were used for all software unless otherwise specified.
2.5. Phenotypic Analyses
The pure cultures of Chamaecytisus albus isolates KW56 and 2063 were phenotypically characterized by the Biolog GEN III system (Biolog Inc., Hayward, CA, USA), following the manufacturer’s instructions. The GEN III MicroPlates™ (Biolog Inc., Hayward, CA, USA) enables micro-testing of bacteria, assessing the ability to metabolize 71 carbon sources and containing 23 chemical sensitivity assays. The GEN III plates contained tetrazolium redox dye, which was used to colorimetrically indicate positive reactions. Bacterial colonies were transferred to inoculating fluid A (IFA) to generate bacterial cell suspensions, the transmittance of which was adjusted to achieve a 95% using a turbidimeter (Laxco Inc., Mill Creek, WA, USA). 100 µL of the cell suspension was dispensed into each well. The color development was monitored every 24 h as absorbance with Synergy™ H1 microplate reader (Agilent Technologies, Santa Clara, CA, USA) at a wavelength of 590 and 750 nm over three days. The most consistent readings came from three-day-old Biolog plates, and these data were used in the analyses. Enzymatic characterization of bacterial isolates was performed using API ZYM strips (BioMérieux, Marcy-l’Étoile, France) according to the manufacturer’s protocol.
2.6. Preparation of Fatty Acid Methyl Esters (FAME)
The strains were cultivated for 48 h in a liquid TY medium (tryptone 5.0 g, yeast extract 3.0 g, CaCl2 × 2 H2O 0.9 g, distilled water 1000.0 mL, pH 7) at 28 °C with aeration by vigorous shaking. Then, 5 mg of bacterial mass was suspended in 1 mL of 2 M methanolic HCl [(prepared from acetyl chloride, Sigma-Aldrich, St. Louis, MO, USA)]. The samples were heated at 85 °C for 18 h. After removing the excess reagent on the evaporator (Büchi Labortechnik AG, Flawil, Switzerland), FAMEs were extracted with a mixture of chloroform water (1:2 v/v). Extraction of FAMEs was repeated twice, and the pooled chloroform phases dehydrated on columns with anhydrous sodium sulfate were dried in a nitrogen stream. FAMEs were converted into trimethylsilyl (TMS) ether derivatives by adding 30 µL TMS reagent [HMDS/TMCS/pyridine (3:1:9, v/v/v), Sigma-Aldrich, St. Louis, MO, USA]. The FAME samples were analyzed by GC/MS using an Agilent 7890A-5975C instrument equipped with an HP-5MS capillary column (30 m × 0.25 mm; Supelco, St. Louis, MO, USA). Helium was used as carrier gas and the temperature program was initially 150 °C for 5 min, then raised to 310 °C at a ramp rate of 3 °C/min, final time of 20 min.
FAMEs were identified mainly based on their chromatographic and mass spectral characteristics, as well as comparison of their retention times with those of authentic standards. The relative content (%) of each fatty acid was calculated from the ratio of the area of its peak to the total area of all peaks. FAMEs of the studied strains were prepared in three independent experiments.
3. Results
3.1. The Origination of Strains
Strains KW56 and 2063 were obtained from root nodules of Chamaecytisus albus growing in Hrubieszów, Poland, during the study of rhizobia and other nodule endophytes of this shrub, in May 2019. Phenotypic traits of these strains were different; however, none of them was able to nodulate C. albus plants [19].
3.2. Genetic Analyses
Genetic analyses included traditional 16S rRNA gene sequences comparison as well as an overall genome-related index (OGRI) using parameters like dDDH (digital DNA-DNA hybridization) and ANI (average nucleotide identity) which are considered appropriate tools for delineating new species [36].
The comparison of 16S rDNA sequences of studied strains and Phyllobacterium type strains revealed that KW56T and 2063T have different closest relatives (Figure 1); Phyllobacterium strain KW 56T was located in one clade together with P. loti S 658T and P. trifolii CECT 7015T, whereas Phyllobacterium strain 2063T was located in a clade containing also P. sophorae CCBAU 03422T, P. brassicacearum STM 196T, and P. brassicacearum 29-15.
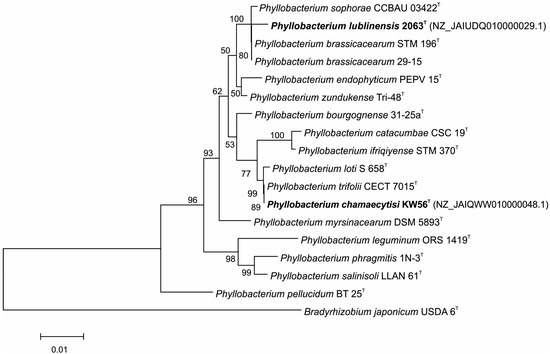
Figure 1.
Phylogenetic tree based on 16S rRNA gene sequences of studied and reference Phyllobacterium strains.
For the 2063T strain, 16S rRNA gene sequence similarity varied from 96.7% (P. catacumbae CSC 19T) to 99.6% (P. brassicacearum 29-15 and P. brassicacearum STM 196T). These values calculated for the KW56 strain ranged from 96.1% (P. leguminum ORS 1419T) to 100% (P. trifolii CECT 7015T and P. loti S 658T) (Table 1).

Table 1.
16S rDNA sequence similarity calculated for Phyllobacterium reference strains and KW56 as well as 2063 strains.
Features of the KW56T and 2063T genomes are summarized in Table 2. In total, 93 contigs for KW56T were obtained, with 100-fold coverage. The average contig length was 58,081 bp, with the largest contig being 733,223 bp and the shortest being 6055 bp. The KW56 has a genome of 6.4 Mb (226.4 kb scaffold N50) with 5857 protein-coding genes and 54 RNA genes (3 rRNAs, 47 tRNAs, 4 ncRNAs) (Table 2, Figure S1).

Table 2.
Genome organization.
In the case of 2063T strain in total, 45 contigs were obtained, with 100-fold coverage. The average contig length was 187.708 bp, with the largest contig being 598,383 bp and the shortest being 202 bp. The 2063T strain has a genome of 4.5 Mb (188.4 kb scaffold N50) with 4231 protein-coding genes and 52 RNA genes (3 rRNA, 45 tRNAs, 4 ncRNAs) (Table 2, Figure S1).
The DNA G+C content of strains KW56T and 2063T were 56 and 57.5 mol%, respectively. These values are within the range of DNA G+C content reported for members of the genus Phyllobacterium (51–58 mol%) [6].
The predicted genes were functionally categorized using the SEED subsystems [34] at the RAST server. The genome of strains KW56T and 2063T were additionally mapped to the seed subsystem to attain high-quality genome annotation via BlastKoala. The distributions of genes linked to subsystems in 11 different categories are shown in Figure 2A,B.
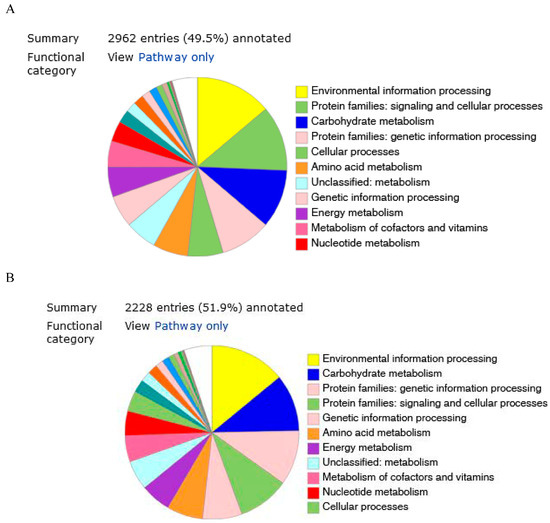
Figure 2.
Genome annotation of 2063T (A) and KW56T (B) by BlastKoala.
Most of Phyllobacterium genomes available in databases are draft genomes; moreover, they differ from each other in size, therefore “formula d4” (i.e., sum of all identities found in high-scoring pairs divided by total genome length), which is independent of genome length, was used in dDDH comparisons. These analyses revealed that dDDH similarity level for KW56T compared with other strains varied from 19.7 (P. phragmitis 1N-3T) to 58.6% (P. trifolii CECT 7015T), and this value for 2063T varied from 19.5% (P. salinisoli LLAN 61T) to 28.3% (P. brassicacearum 29-15 and P. brassicacearum STM 196T) (Table 3).

Table 3.
The comparison of genetic traits of KW56T and 2063T strains with other sequenced Phyllobacterium genomes.
Studying ANIb values showed that KW56T strain revealed the highest similarity to P. trifolii CECT 7015T (93.52%) and the lowest to P. leguminum ORS 1419T (71.72%). The same analyses performed for the 2063 strain revealed the highest similarity to P. brassicacearum 29-15 (84.08%) and the lowest to P. salinisoli LLAN 61T (72.31%) (Table 3).
3.3. Phenotypic Analyses
Strain KW56T has the general characteristics of the genus Phyllobacterium. It is a Gram-negative, aerobic rod. The optimal growth temperature is 28 °C. Growth on YMA occurs at 37 °C but not at 40 °C. Growth occurs in YM broth with 1, 2, or 3% NaCl and YM broth at pH 6–8, but not at pH 5. Catalase and oxidase activities are positive. Phosphate solubilization, siderophores, and IAA production is positive [19].
Strain KW56T utilizes the following sugars: D-maltose, D-cellobiose, gentiobiose, sucrose, D-turanose, N-acetyl-d-glucosamine, N-acetyl-β-d-mannosamine, N-acetyl-neuraminic acid, α-d-glucose, D-mannose, D-fructose, D-galactose, D-fucose, L-fucose, L-rhamnose. It uses such polyvalent alcohols as D-sorbitol, D-mannitol, D-arbitol, myo-inositol, glycerol. Assimilation of hexose acids such as D-galacturonic acid, L-galactonic acid lactone, D-glucuronic acid, glucuronamide, quinic acid, and D-saccharic acid was positive. Among the carboxylic acids, esters, and fatty acids tested, substrates such as L-lactic acid, D-malic acid, L-malic acid, bromo-succinic acid, tween 40, α-hydroxy-butyric acid, β-hydroxy-D, L-butyric acid, α-keto-butyric acid, acetoacetic acid, propionic acid, acetic acid, and formic acid were used. The strain KW56 also used different amino acids: glycyl-L-proline, L-alanine, L-apartic acid, L-serine.
This strain was sensitive to pH 5, 8% NaCl, and niaproof 4. No positive reaction was observed for aztreonam, sodium butyrate, lithium chloride, potassium tellurite, fusidic acid, troleandomycin, rifamycin SV, minocycline, lincomycin, or guanidine HCl. An API ZYM kit showed positive enzyme activities for alkaline phosphatase, esterase (C4), leucine arylamidase, acid phosphatase, and naphthol-AS-BI-phosphohydrolase, but not for lipase (C14), esterase lipase (C8), valine arylamidase, cystine arylamidase, trypsin, α-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, or α fucosidase.
Strain 2063T has the general characteristics of the genus Phyllobacterium. It is a Gram-negative, aerobic rod. The optimal growth temperature is 28 °C. Growth on YMA occurs at 37 °C but not at 40 °C. Growth occurs in YM broth with 1, 2, or 4% NaCl and YM broth at pH 6–8, but not at pH 5. Catalase and oxidase activities are positive. Siderophores and IAA production is positive [19].
This strain utilizes the following sugars: D-maltose, D-trehalose, D-cellobiose, gentiobiose, D-turanose, D-glucose, D-mannose, D-fructose, D-galactose, D-fucose, L-fucose, N-acetyl-d-glucosamine. It uses such polyvalent alcohols as D-sorbitol, D-mannitol, D-arabitol, myo-inositol, and glycerol. Assimilation of L-glutamic acid, L-histidine, quinic acid and glucuronamide is positive. Among the carboxylic acids, esters and fatty acids tested, L-lactic acid, keto-glutaric acid, acetoacetic acid, propionic acid, acetic acid, and formic acid were used. No positive reaction was observed for fusidic acid, minocycline, or guanidine HCl. The 2063T strain was sensitive to pH 5, 8% NaCl, minocycline, and niaproof 4.
API ZYM kit, showed positive enzyme activities for alkaline phosphatase, esterase (C4), leucine arylamidase, trypsin, acid phosphatase, α-glucosidase, and naphthol-AS-BI-phosphohydrolase, but not for lipase (C14), esterase lipase (C8), valine arylamidase, cystine arylamidase, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase or α fucosidase.
The discriminatory characters for KW56 and 2063 strains, as well as closely related Phyllobacterium species P. brassicacearum LMG 22836T and P. trifolii PETP02T are listed in Table 4.

Table 4.
Physiological traits of KW56T, 2063T, P. brassicacearum LMG 22836T and P. trifolii PETP 02T [5,6,8].
3.4. Fatty Acid Porofiles of KW56T and 2063T Strains
Strain KW56T synthesized 21 different fatty acids with 14 to 21 carbon atoms, including saturated, unsaturated, branched, hydroxy acids as well as FAs with a cyclopropane ring (Table 5). The 16:0 FA was dominant among the saturated acids. Monounsaturated FA comprised 16:1, 17:1, 18:1, 19:1. This strain contained two cyclopropyl fatty acids, of which cyclopropyl 19:0 was the predominant acid. The second most abundant acid was 3-OH 16:0. This strain produced also 2-OH 19:1 and 11-OH 19:0 fatty acids.

Table 5.
Fatty acid composition and their relative content (%) in cells of KW56T and 2063T strain.
Analysis of FAMEs performed for 2063T strain showed the presence of a varied composition including straight-chain saturated and unsaturated, branched-chain, cyclopropyl-ringed, and hydroxy FAs. The dominant components in this strain were cyclopropyl 19:0 (42%) and 18:1 (16%) (Table 3). This strain was characterized by a high content of hydroxy fatty acids, which together accounted for approx. 20%. Among hydroxy FAs, 3-OH was the predominant acid. The fatty acid 16:0 was the most abundant of the saturated FAs of the 2063T strain.
4. Discussion
As presented above, the 16S rDNA sequences of the studied strains did not differ from those obtained from some reference strains. However, the whole-genome analysis showed large differences between compared genomes. The threshold proposed for delineation of new species is <70% for dDDH and <95–96% for ANI [36,37], and values obtained for KW56T and 2063T were below this threshold; therefore, these strains can be claimed as new Phyllobacterium species according to the obtained OGRI values.
Based on the RAST annotation, the individual predicted genes were assigned to general functional categories. The assignment of individual CDSs to functional categories was illustrated in the genomic maps for both analyzed strains (Online resource 1). For both genomes, the number of genes in each category was similar; however, differences were found. The genes involved in basic metabolism were the largest group (880 for KW56T and 764 for 2063T), followed by energy production (337 and 248 respectively) and protein processing (226 and 220).
Differences found between the genomes of KW56T, 2063T, and their closest relatives (P. trifolii and P. brassicacearum, respectively) were supported in the physiological and metabolic properties of the strains. the studied strains differed from the reference strains in general physiological traits like growth temperature or pH and salt tolerance, as well as in metabolic potential (e.g., utilization of numerous specific organic acids, aminoacids and other carbon and energy sources).
Comparative analysis of cellular fatty acids showed a similarity of KW56T strain to its closest relative P. trifolii CECT 7015T characterized by major acids 16:0, 18:1ω7c, and 19:0 cyclopropyl ω8c (Table 3). The KW56T strain differed from the P. trifolii strain in the amount of 19:0 cyclopropyl, which was three times higher in the novel strain, in the amount of 18:1, which was lower [5]. The presence of significant amounts of 3-OH 14:0, branched 19:0, and 21:0 fatty acids clearly distinguished the KW56T strain from P. trifolii CECT 7015T. These fatty acids may be chemotaxonomic markers of this novel strain.
The predominance of fatty acids such as 16:0 and cyclopropyl 19:0 was similar to that of the closest related P. brassicacearum 29-15 strain [8]. The 2063T strain contained a significant amount of 18:1 fatty acid, which was not present in the P. brassicacearum 29-15 strain. A unique signature fatty acid, a19:1, was found only in the 2063T strain and represented 3% of the total. This novel strain could be differentiated from P. brassicacearum 29-15 also due to the presence of 3-OH 14:0, 3-OH 17:0, 3-OH 18:0.
In conclusion, the phenotypic and genotypic data presented in this study demonstrate that the KW56T and 2063T strains belong to a separate species in the genus Phyllobacterium. Therefore, we conclude that the KW56T and 2063T strains are representatives of a new species, for which we propose the name P. chamaecytisi KW56T, with strain KW56T as the type strain. The 2063T strain is representative of a second new species, for which we propose the name P. lublinensis 2063T, with strain 2063T as the type strain.
5. Conclusions
The bacteria Phyllobacterium chamaecytisi KW56T and Phyllobacterium lublinensis 2063T, isolated from root nodules of Chamaecytisus albus, represent novel species within the genus Phyllobacterium. Both strains are capable of forming endophytic associations with their leguminous host plant. Genomic analyses, including ANI and dDDH, confirmed their distinction from previously described Phyllobacterium species.
P. chamaecytisi KW56T and P. lublinensis 2063T differ in genome size, GC content, and fatty acid profiles, particularly in the proportions of hydroxy and cyclopropyl fatty acids. Phenotypic characterization revealed distinct metabolic and enzymatic traits, including specific carbon source utilization and enzymatic activities.
Both strains exhibit plant-associated traits and may contribute to plant health and nitrogen metabolism through endophytic colonization. This research expands current knowledge of the genus Phyllobacterium, particularly in relation to symbiotic bacteria associated with rare legume species.
The discovery of P. chamaecytisi KW56T and P. lublinensis 2063T provides new insights into bacterial diversity within root nodule microbiomes and lays the foundation for further studies on plant–microbe interactions and potential agricultural applications.
Protologue description of Phyllobacterium chamaecytisi sp. nov. and Phyllobacterium lublinensis sp. nov.
- Phyllobacterium chamaecytisi sp. nov.
Phyllobacterium chamaecytisi (cha.mae.cy.ti’si. N.L. gen. n. chamaecytisi, from Chamaecytisus, the host plant from which the type strain was isolated).
Phyllobacterium chamaecytisi sp. nov. is represented by strain KW56ᵀ (=DSM 113831ᵀ = GCA_020164455.1), which was isolated in May 2019 from root nodules of Chamaecytisus albus collected in Hrubieszów, Poland (50°48′09″ N, 23°53′31″ E). Cells are Gram-negative, non-spore-forming rods. The strain grows aerobically on yeast extract mannitol agar (YMA) at 28 °C and 37 °C but not at 40 °C. It tolerates up to 3% (w/v) NaCl and grows at pH values between 6.0 and 8.0. The strain is catalase- and oxidase-positive. It does not fix nitrogen, and no nodABC genes are present in the genome.
The major fatty acid is cyclopropyl 19:0 (36%), with additional components including 3-OH 14:0, anteiso 19:1, and 21:0, which collectively distinguish the strain from Phyllobacterium trifolii. The 16S rRNA gene sequence (NZ_JAIQWW010000048.1) of strain KW56ᵀ is identical (100%) to that of P. trifolii CECT7015ᵀ. However, genome-based metrics support its designation as a novel species, with an average nucleotide identity (ANI) of 93.5% and digital DNA–DNA hybridization (dDDH) value of 58.6% compared to P. trifolii.
Strain KW56ᵀ differs from P. trifolii in several phenotypic traits. It can grow at 4% NaCl and pH 5.0, is urease-positive, and is capable of utilizing L-serine, L-alanine, propionate, acetic acid, and Tween 40 as carbon sources. Biolog GENIII and API ZYM enzymatic profiles clearly differentiate this strain from its closest phylogenetic relative.
- Phyllobacterium lublinensis sp. nov.
Phyllobacterium lublinensis (lu.bli.nen’sis. N.L. fem. adj. lublinensis, referring to Lublin, the region in eastern Poland where the host plant was collected).
Phyllobacterium lublinensis sp. nov. is represented by strain 2063ᵀ (=DSM 113830ᵀ = GCA_020164435.1), which was isolated in May 2019 from root nodules of Chamaecytisus albus collected in Hrubieszów, Poland. Cells are Gram-negative, rod-shaped, and non-spore-forming. The strain grows aerobically on YMA at 28 °C and 37 °C, but not at 40 °C. It tolerates up to 4% (w/v) NaCl and grows in a pH range of 6.0–8.0. The strain is catalase and oxidase-positive. It does not fix nitrogen, and no nodABC genes are present in the genome.
The major fatty acids are cyclopropyl 19:0 (42%) and 18:1. Additional key components, including 3-OH 14:0, 3-OH 17:0, and 3-OH 18:0, are present and allow differentiation from Phyllobacterium brassicacearum, its closest phylogenetic neighbor. The 16S rRNA gene sequence (NZ_JAIUDQ010000029.1) of strain 2063ᵀ shares 99.6% identity with that of P. brassicacearum STM 196T. However, ANI (84.1%) and dDDH (28.3%) values are below the species-level thresholds, confirming its status as a novel species.
Strain 2063ᵀ is distinguishable from P. brassicacearum by its ability to grow in the presence of 4% NaCl and to metabolize α-ketoglutarate, D-glucuronate, acetic acid, and propionate. It also exhibits trypsin activity. Biolog and API ZYM profiles further support its differentiation from P. brassicacearum.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology14081024/s1, Figure S1. A circular maps of Phyllobacterium lublinensis 2063T (A) and Phyllobacterium chamaecytisi KW56T (B) genomes. Graphical display of the distribution of the genome annotations is provided. This includes, from outer to inner rings, the contigs, CDS on the forward strand, CDS on the reverse strand, RNA genes, CDS with homology to known antimicrobial resistance genes, CDS with homology to know virulence factors, GC content and GC skew. The colors of the CDS on the forward and reverse strand indicate the RAST subsystem that these genes belong to. The percentage share of individual subsystems in the total number of annotated genes with the color legend is shown in the form of a pie chart under the genomic map.
Author Contributions
Conceptualization, J.W., S.W.-W. and M.M.-K.; methodology, S.W.-W., M.P.-S. and P.K.; software, P.K.; formal analysis, K.W.-C., S.W.-W., M.P.-S. and P.K.; investigation, K.W.-C., M.P.-S., M.M.-K. and S.W.-W.; resources, J.W., S.W.-W. and M.M.-K.; data curation, J.W., S.W.-W., M.P.-S. and P.K.; writing—original draft preparation, J.W., S.W.-W., M.P.-S. and P.K.; writing—review and editing, J.W. and S.W.-W.; visualization, S.W.-W., M.P.-S. and P.K.; supervision, J.W. and S.W.-W.; project administration, J.W. and S.W.-W.; funding acquisition, J.W., K.W.-C., M.M.-K. and S.W.-W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Provincial Fund for Environmental Protection and Water Management in Lublin, title of the project: “The use of rhizobia in biological protection of Chamaecytisus albus” (11/2020/D/IN).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article; further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Knösel, D. Prüfung von bakterien auf fähigkeit zur sternbildung. Zentralblatt Bakteriol. Parasitenkd. Infekt. Hyg. II Abt. 1962, 116, 79–100. [Google Scholar]
- Knösel, D. Genus IV Phyllobacterium (Ex Knösel 1962) nom. rev. (Phyllobacterium Knösel 1962, 96). Bergey’s Man. Syst. Bacteriol. 1984, 1, 254–256. [Google Scholar]
- Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Int. J. Syst. Evol. Microbiol. 2001, 51, 1619–1620. [CrossRef]
- Mergaert, J. Phyllobacterium myrsinacearum (subjective synonym Phyllobacterium rubiacearum) emend. Int. J. Syst. Evol. Microbriol. 2002, 52, 1821–1823. [Google Scholar] [CrossRef] [PubMed]
- Valverde, A.; Velázquez, E.; Fernández-Santos, F.; Vizcaíno, N.; Rivas, R.; Mateos, P.F.; Martínez-Molina, E.; Igual, J.M.; Willems, A. Phyllobacterium trifolii sp. nov., nodulating Trifolium and Lupinus in Spanish soils. Int. J. Syst. Evol. Microbiol. 2005, 55, 1985–1989. [Google Scholar] [CrossRef] [PubMed]
- Mantelin, S.; Fischer-Le Saux, M.; Zakhia, F.; Béna, G.; Bonneau, S.; Jeder, H.; de Lajudie, P.; Cleyet-Marel, J.-C. Emended description of the genus Phyllobacterium and description of four novel species associated with plant roots: Phyllobacterium bourgognense sp. nov., Phyllobacterium ifriqiyense sp. nov., Phyllobacterium leguminum sp. nov. and Phyllobacterium brassicacearum sp. nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Flores-Félix, J.D.; Carro, L.; Velázquez, E.; Valverde, Á.; Cerda-Castillo, E.; García-Fraile, P.; Rivas, R. Phyllobacterium endophyticum sp. nov., isolated from nodules of Phaseolus vulgaris. Int. J. Syst. Evol. Microbiol. 2013, 63, 821–826. [Google Scholar] [CrossRef]
- Sánchez, M.; Ramírez-Bahena, M.H.; Peix, A.; Lorite, M.J.; Sanjuán, J.; Velázquez, E.; Monza, J. Phyllobacterium loti sp. nov. isolated from nodules of Lotus corniculatus. Int. J. Syst. Evol. Microbiol. 2014, 64, 781–786. [Google Scholar] [CrossRef]
- Jiao, Y.S.; Yan, H.; Ji, Z.J.; Liu, Y.H.; Sui, X.H.; Zhang, X.X.; Wang, E.T.; Chen, W.F. Phyllobacterium sophorae sp. nov., a symbiotic bacterium isolated from root nodules of Sophora flavescens. Int. J. Syst. Evol. Microbiol. 2015, 65, 399–406. [Google Scholar] [PubMed]
- León-Barrios, M.; Ramírez-Bahena, M.H.; Igual, J.M.; Peix, Á.; Velázquez, E. Phyllobacterium salinisoli sp. nov., isolated from a Lotus lancerottensis root nodule in saline soil from Lanzarote. Int. J. Syst. Evol. Microbiol. 2018, 68, 1085–1089. [Google Scholar] [CrossRef]
- Safronova, V.I.; Sazanova, A.L.; Kuznetsova, I.G.; Belimov, A.A.; Andronov, E.E.; Chirak, E.R.; Popova, J.P.; Verkhozina, A.V.; Willems, A.; Tikhonovich, L.A. Phyllobacterium zundukense sp. nov., a novel species of rhizobia isolated from root nodules of the legume species Oxytropis triphylla (Pall.) Pers. Int. J. Syst. Evol. Microbiol. 2018, 68, 1644–1651. [Google Scholar] [CrossRef]
- Liang, L.X.; Sun, Q.W.; Hui, N.; Zhang, X.X.; Li, L.B.; Liu, L. Phyllobacterium phragmitis sp. nov., an endophytic bacterium isolated from Phragmites australis rhizome in Kumtag desert. Antonie Leeuwenhoek 2019, 112, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Ten, L.N.; Maeng, S.; Chang, Y.; Jung, H.Y.; Kim, M.K. Phyllobacterium pellucidum sp. nov., isolated from soil. Arch. Microbiol. 2021, 203, 2647–2652. [Google Scholar] [CrossRef] [PubMed]
- Jurado, V.; Laiz, L.; Gonzalez, J.M.; Hernandez-Marine, M.; Valens, M.; Saiz-Jimenez, C. Phyllobacterium catacumbae sp. nov., a member of the order “Rhizobiales” isolated from Roman catacombs. Int. J. Syst. Evol. Microbiol. 2005, 55, 1487–1490. [Google Scholar] [CrossRef]
- Mergaert, J.; Boley, A.; Cnockaert, M.C.; Müller, W.R.; Swings, J. Identity and potential functions of heterotrophic bacterial isolates from a continuous-upflow fixed-bed reactor for denitrification of drinking water with bacterial polyester as source of carbon and electron donor. Syst. Appl. Microbiol. 2001, 24, 303–310. [Google Scholar] [CrossRef]
- Gonzalez-Bashan, L.E.; Lebsky, V.K.; Hernandez, J.P.; Bustillos, J.J.; Bashan, Y. Changes in the metabolism of the microalga Chlorella vulgaris when coimmobilized in alginate with the nitrogen-fixing Phyllobacterium myrsinacearum. Can. J. Microbiol. 2000, 46, 653–659. [Google Scholar] [CrossRef]
- Alavi, M.; Miller, T.; Erlandson, K.; Schneider, R.; Belas, R. Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 2001, 3, 380–396. [Google Scholar] [CrossRef]
- Kaźmierczakowa, R.; Bloch-Orłowska, J.; Celka, Z.; Cwener, A.; Dajdok, Z.; Michalska-Hejduk, D.; Pawlikowski, P.; Szczęśniak, E.; Ziarnek, K. Polish Red List of Pteridophytes and Flowering Plants; Instytut Ochrony Przyrody Polskiej Akademii Nauk: Kraków, Poland, 2016. [Google Scholar]
- Włodarczyk, K.; Wdowiak-Wróbel, S.; Marek-Kozaczuk, M.; Wielbo, J. Genetic and physiological diversity of white Spanish broom (Chamaecytisus albus) endophytes. Acta Biochim. Pol. 2021, 68, 419–426. [Google Scholar] [CrossRef]
- Vincent, J. International Biological Programme. Burgess and Son, Berkshire. In A manual for the Practical Study of Root Nodule Bacteria; Blackwell Scientific Publications: Oxford, UK, 1970. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Parrello, B.; Butler, R.; Chlenski, P.; Olson, R.; Overbeek, J.; Pusch, G.D.; Vonstein, V.; Overbeek, R. A machine learning-based service for estimating quality of genomes using PATRIC. BMC Bioinform. 2019, 20, 486. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Pattengale, N.D.; Alipour, M.; Bininda-Emonds, O.R.P.; Moret, B.M.E.; Stamatakis, A. How many bootstrap replicates are necessary? J. Comput. Biol. 2010, 17, 337–354. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods); Version 4.0 Beta; Sinauer Associates: Sunderland, UK, 2001. [Google Scholar]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Hayashi Sant’Anna, F.; Bach, E.; Porto, R.Z.; Guella, F.; Hayashi Sant’Anna, E.; Passaglia, L.M.P. Genomic metrics made easy: What to do and where to go in the new era of bacterial taxonomy. Crit. Rev. Microbiol. 2019, 45, 182–200. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).