Mechanistic Study of NT5E in Reg3β-Induced Macrophage Polarization and Cooperation with Plasma Proteins in Myocarditis Injury and Repair
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Cell Culture and RNA-Seq Technical Analysis
2.3. Cell Transfection and Grouping
2.4. Single-Cell RNA-Seq Analysis
2.5. Western Blot Detection of Relevant Protein Expression Levels
2.6. qRT–PCR Assay (Q-PCR)
2.7. MR Analysis
2.8. Mediation Analysis
2.9. GO and KEGG Enrichment Analysis
2.10. Protein–Protein Interaction (PPI) Network Construction and Core Gene Screening
2.11. Drug Screening
2.12. Molecular Docking Analysis
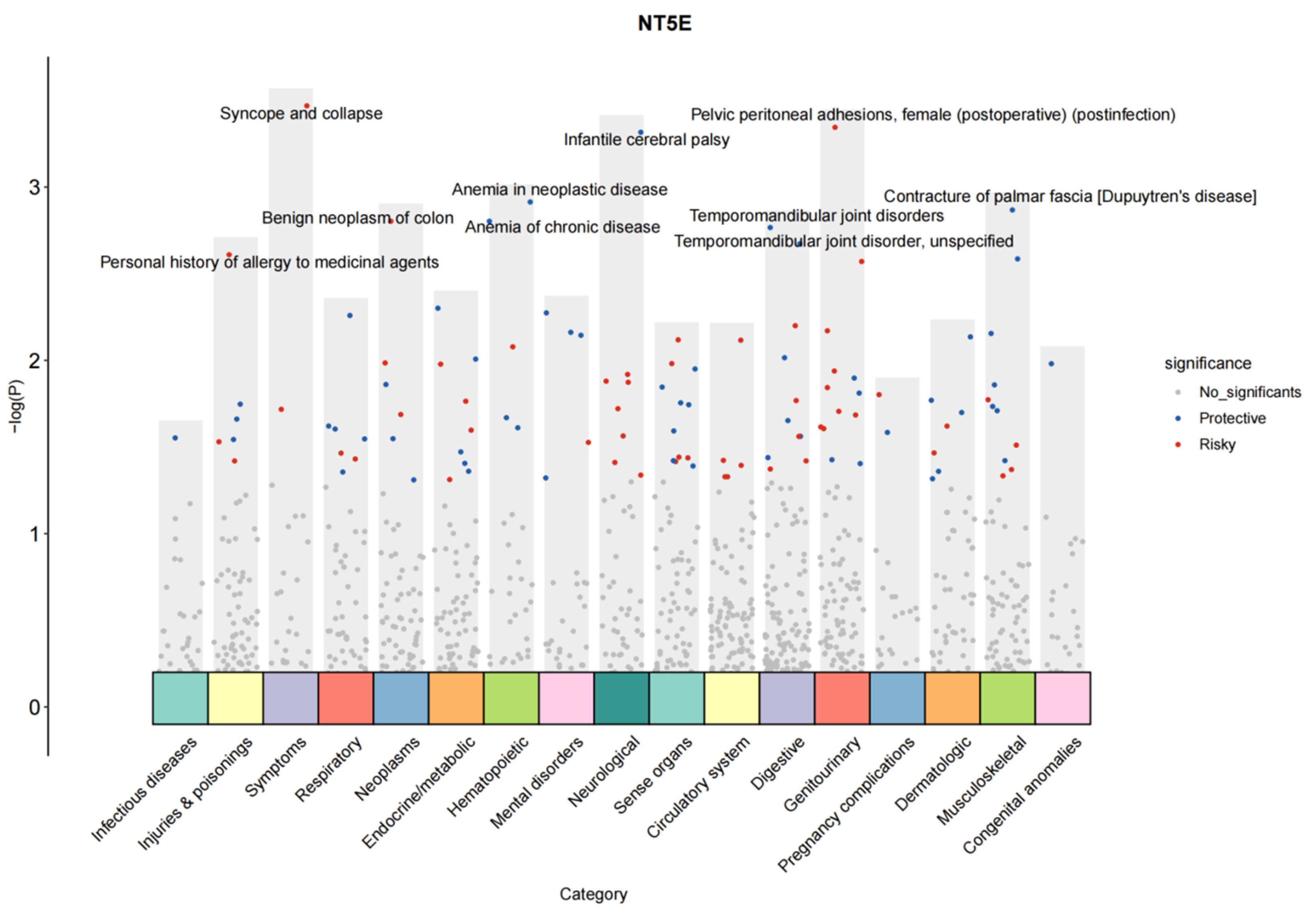
2.13. PheWAS Analysis
3. Results
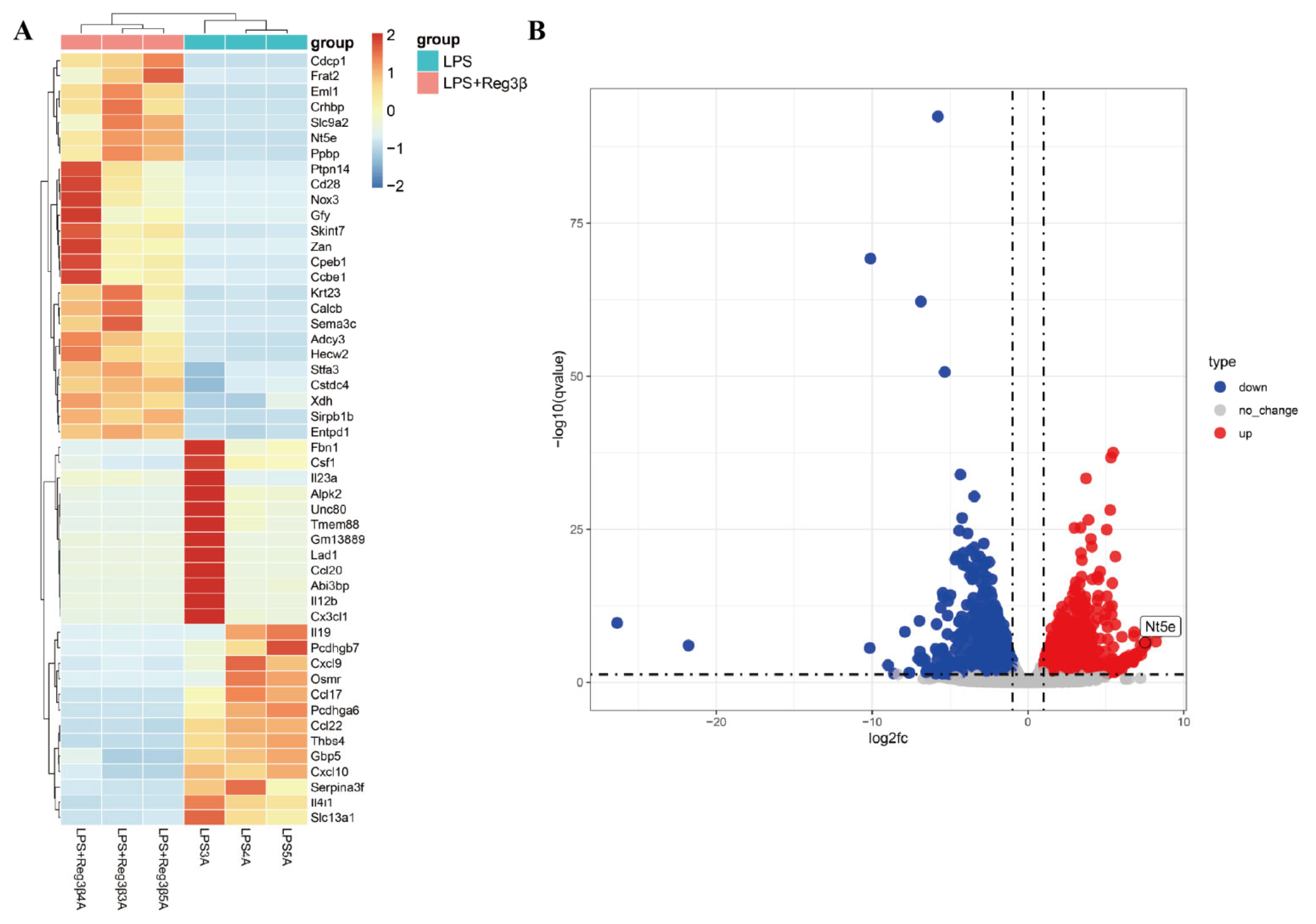
3.1. RNA-Seq Analysis
3.2. Results of Single-Cell RNA-Seq Analysis
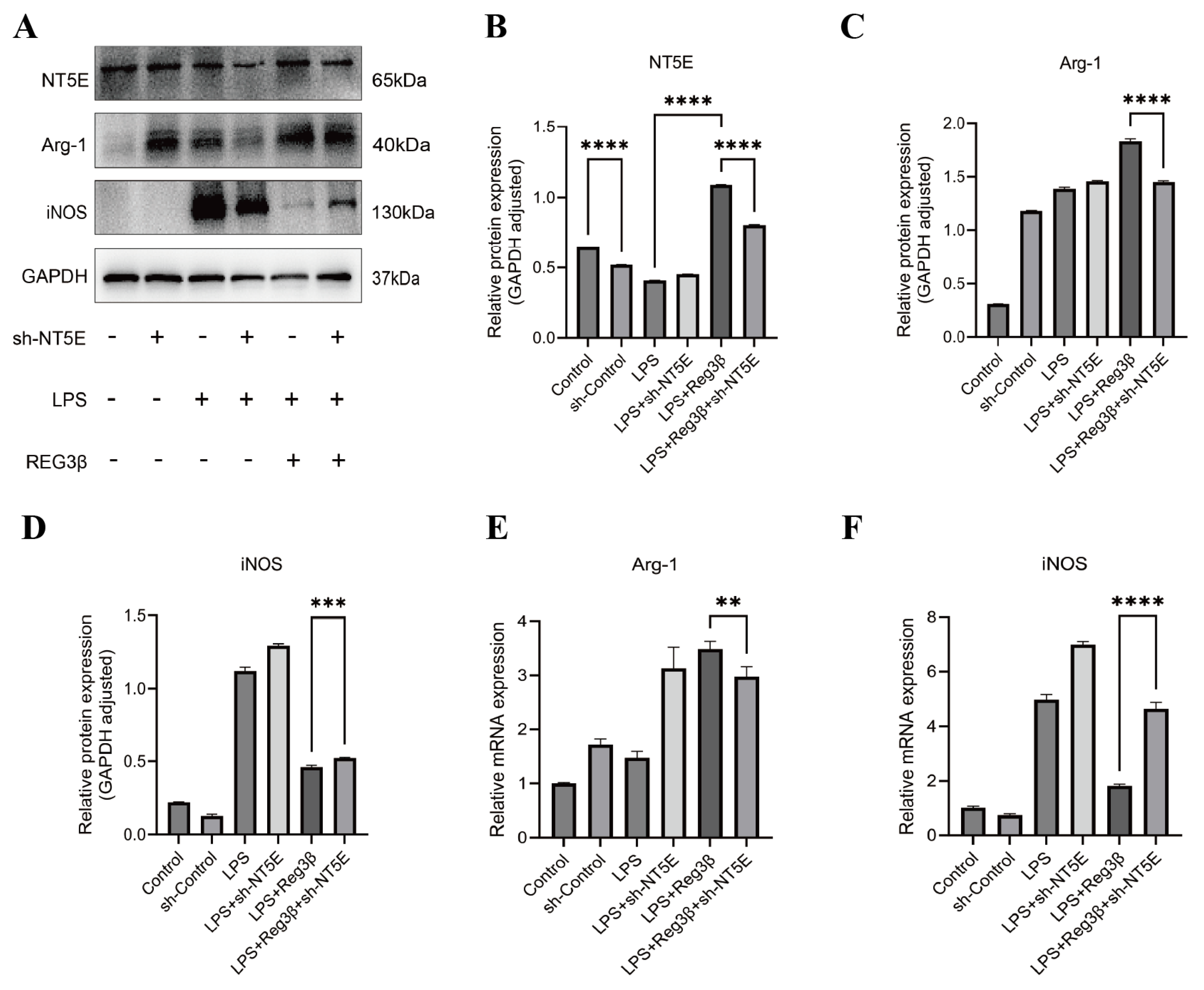
3.3. Validation of Biomarkers by Western Blot (WB) and Quantitative Polymerase Chain Reaction (Q-PCR)
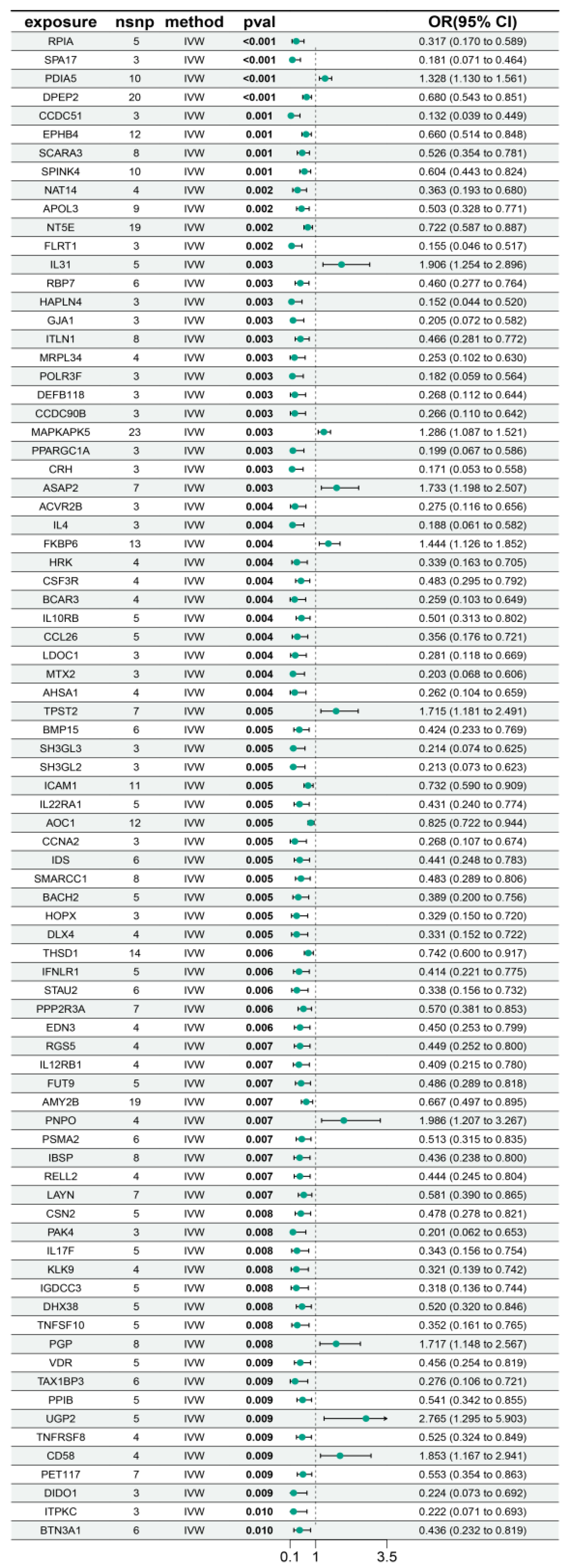
3.4. Identification of Candidate Proteins Associated with Myocarditis
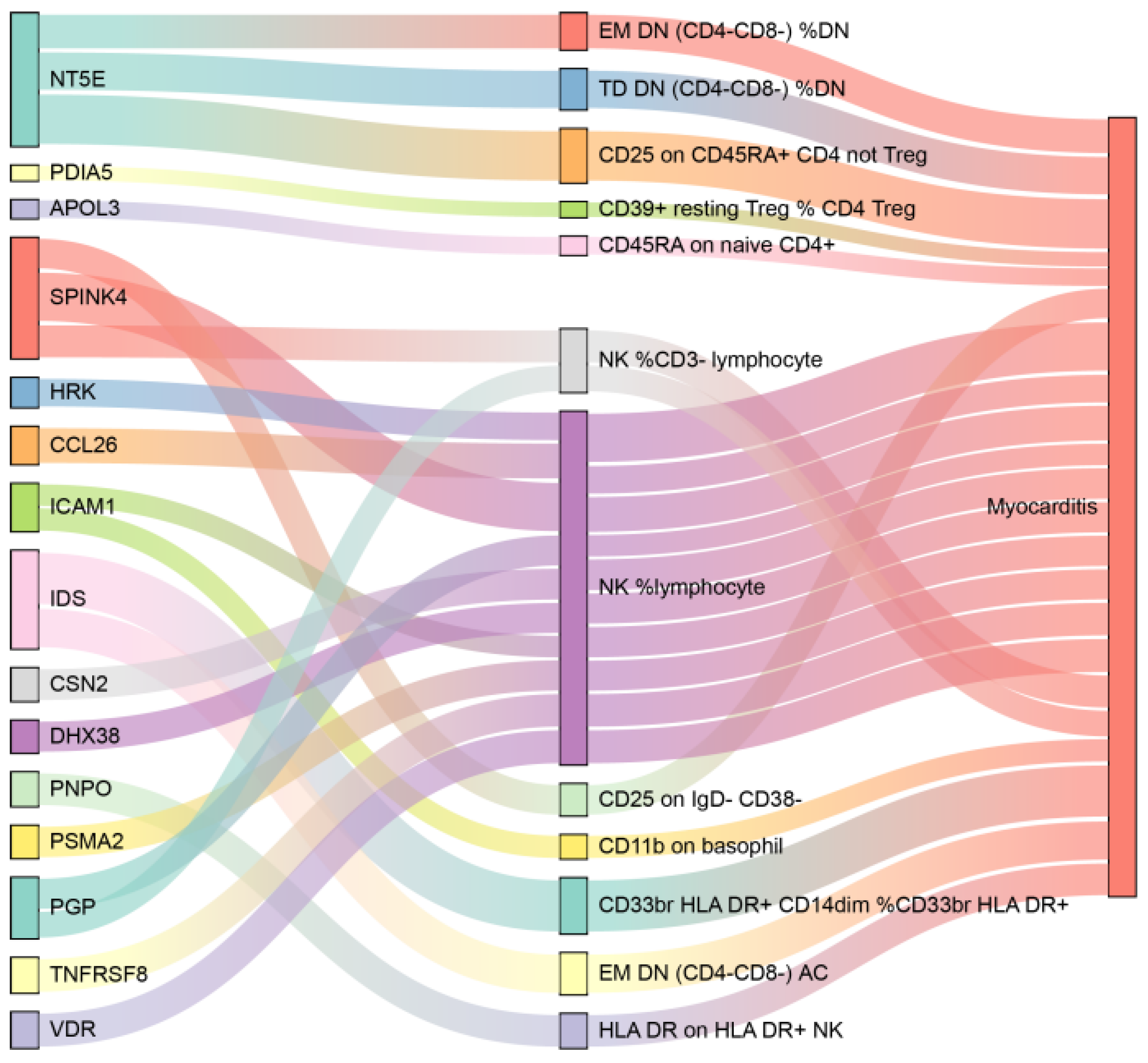
3.5. Mediation Analysis Results
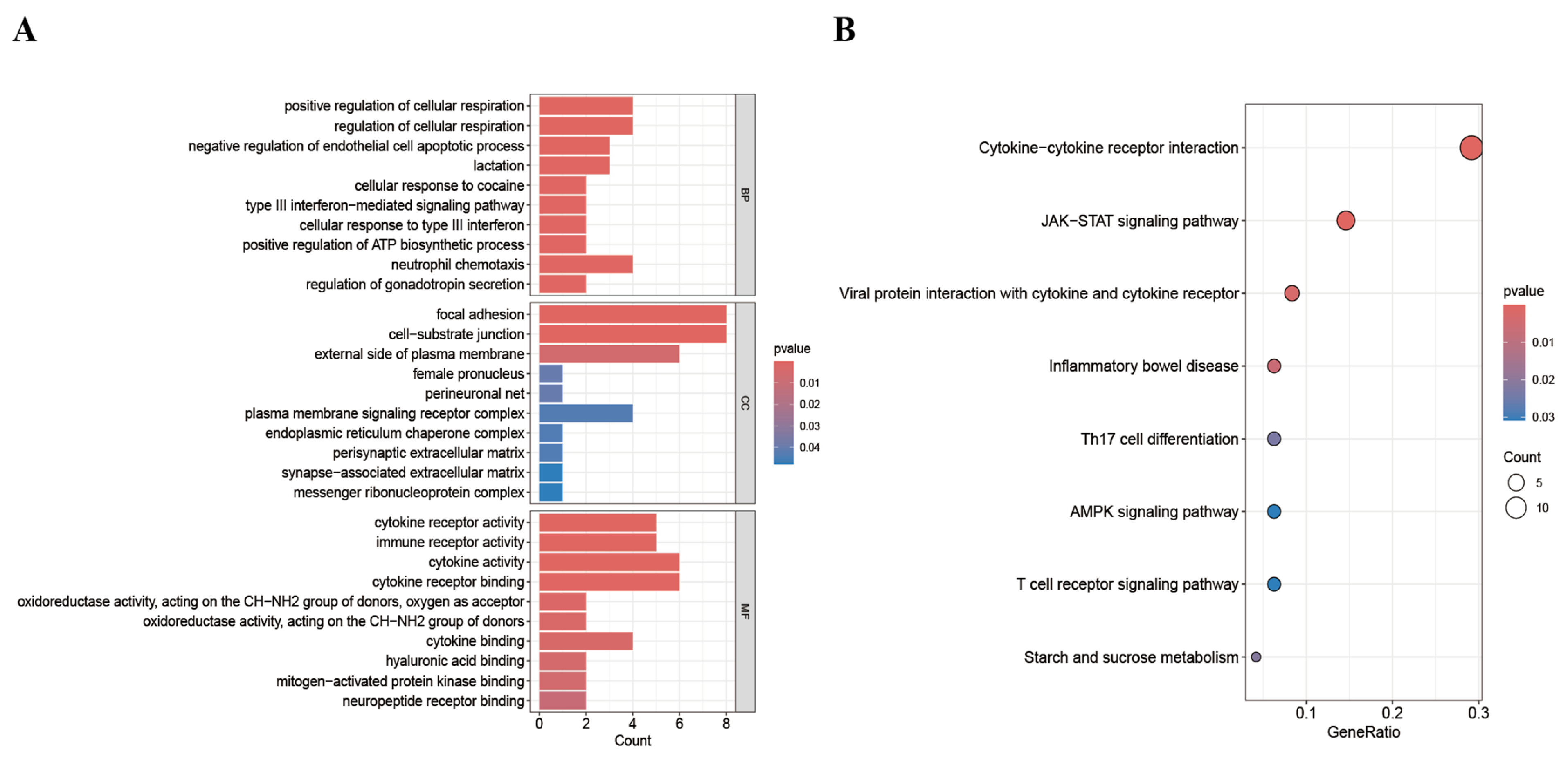
3.6. Enrichment Analysis
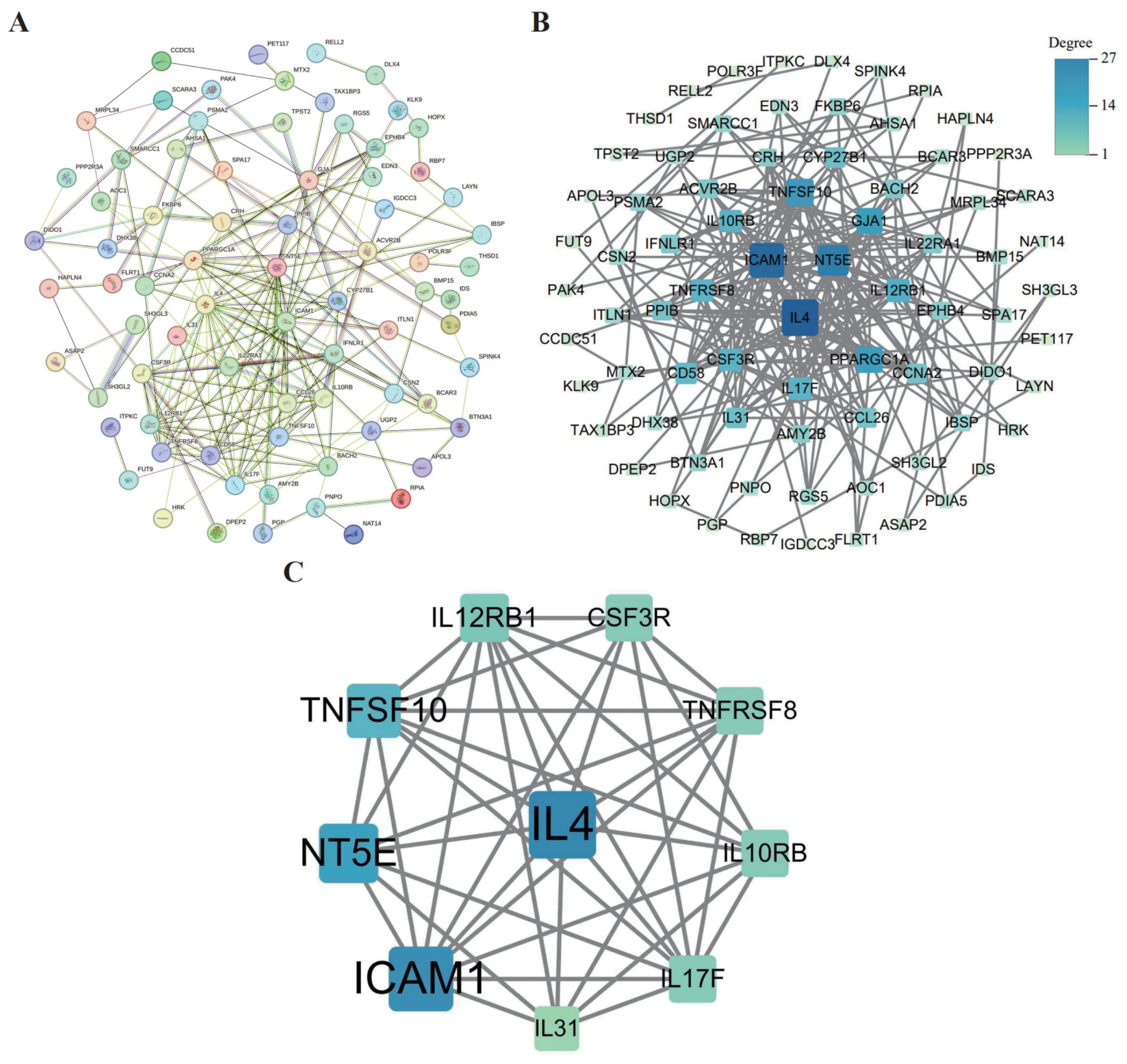
3.7. PPI Network Construction and Core Gene Screening
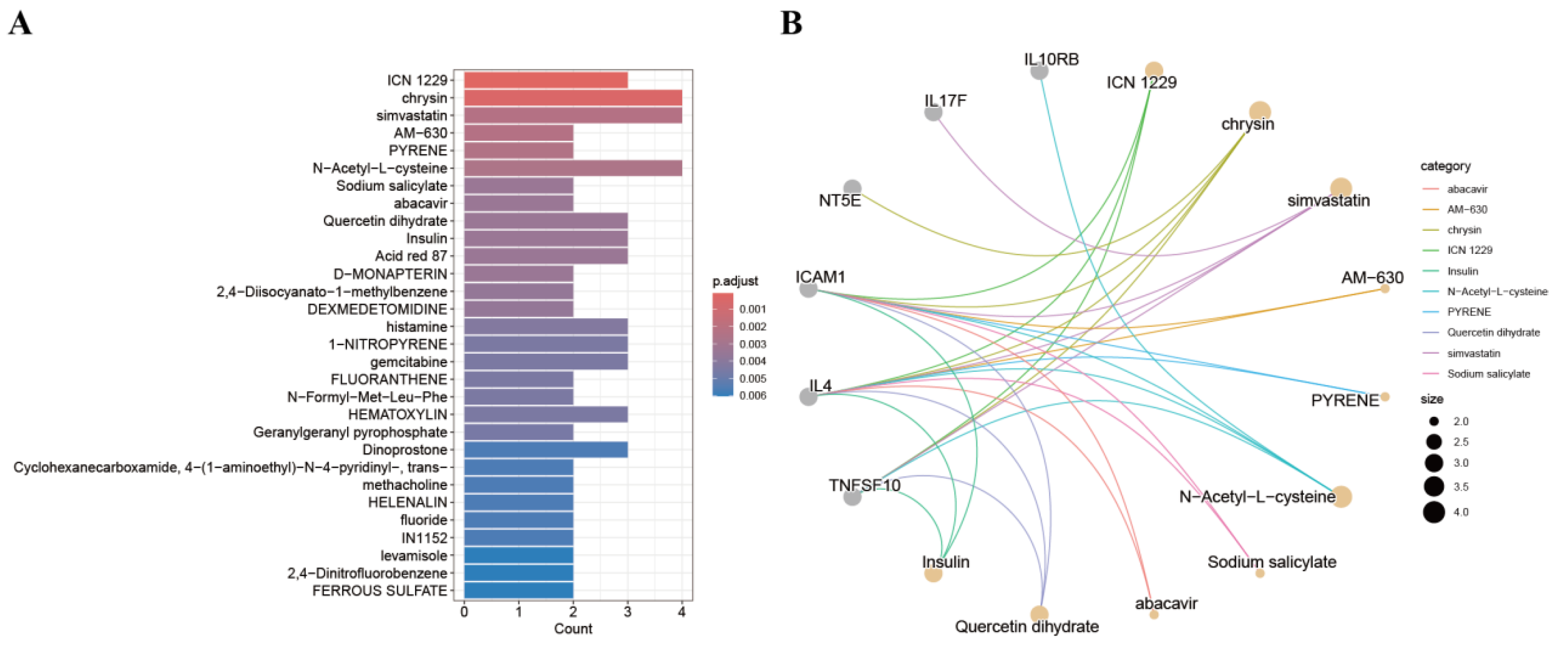
3.8. Screening for Candidate Drugs
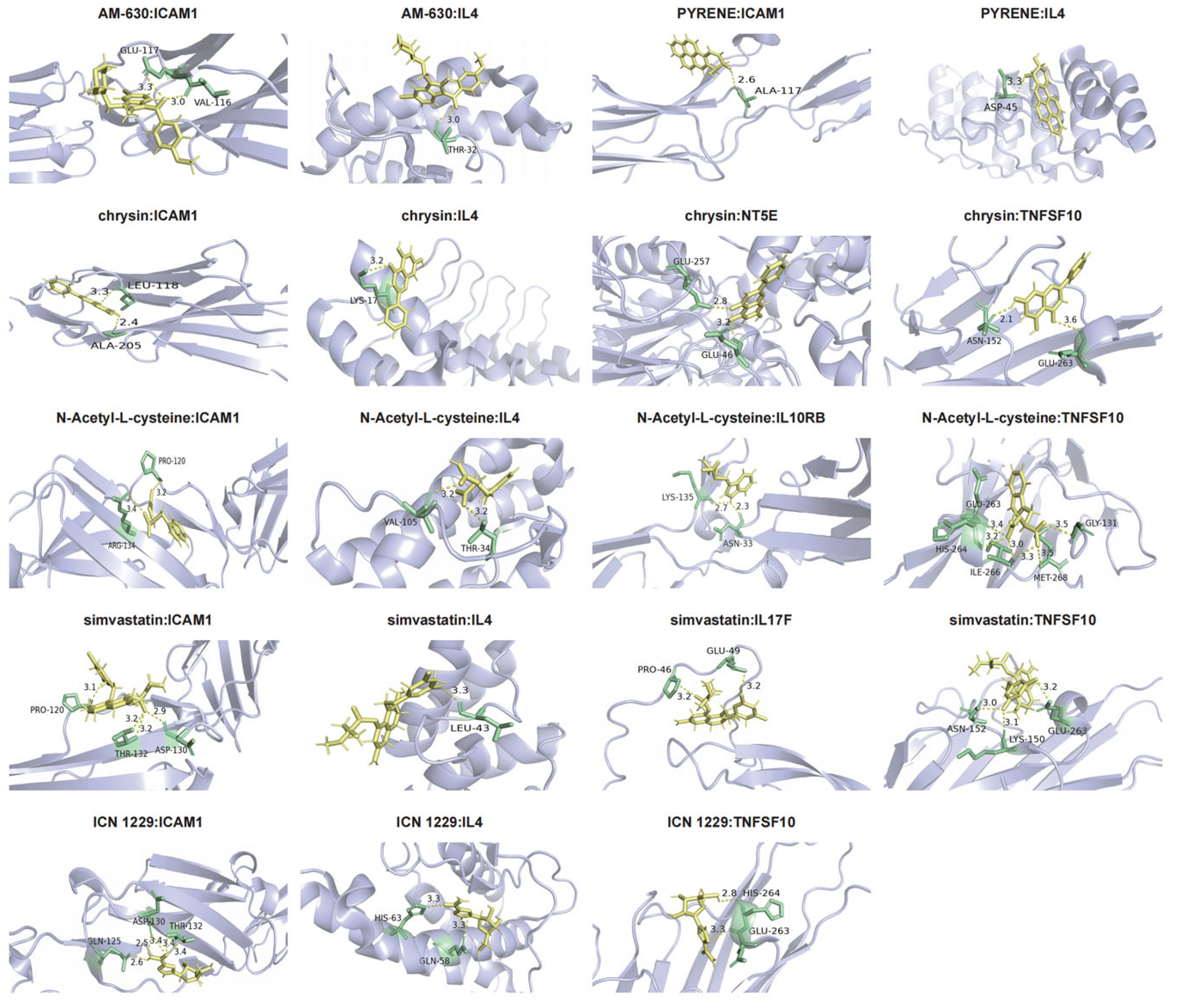
3.9. Molecular Docking
3.10. PheWAS Reveals the Potential Side Effects of Drugs Targeting Myocarditis-Related Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MR | Mendelian randomization |
| PheWAS | phenome-wide association studies |
| NT5E | extracellular-5′-nucleotidase |
| Reg3β | Regenerating islet-derived protein 3 beta |
| Arg-1 | Arginase-1 |
| Reg3α | Regenerating islet-derived 3 alpha |
| Reg3γ | Regenerating islet-derived 3 gamma |
| MyHC-α | α-myosin heavy chain |
| CK-MB | Creatine Kinase-MB |
| GWAS | Genome-wide association studies |
| pQTLs | protein quantitative trait loci |
| RNA-seq | RNA sequencing |
| GEO | Gene Expression Omnibus |
| SNPs | single nucleotide polymorphisms |
| IVs | instrumental variables |
| AC | absolute cell counts |
| RC | relative cell counts |
| MFI | median fluorescence intensities |
| MP | morphological parameters |
| FBS | fetal bovine serum |
| LPS | lipopolysaccharide |
| sh-NT5E | knockdown plasmid targeting NT5E |
| scRNA-seq | single-cell RNA sequencing |
| SDS–PAGE | sodium dodecyl sulfate-polyacrylamide gel |
| IVW | inverse variance weighted |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PPI | Protein-protein interaction |
| DSigDB | Drug Signatures Database |
| WB | Western blotting |
| Q-PCR | quantitative polymerase chain reaction |
| iNOS | Inducible Nitric Oxide Synthase |
| SPINK4 | Serine Peptidase Inhibitor Kazal Type 4 |
| HRK | Harakiri |
| CCL26 | C-C Motif Chemokine Ligand 26 |
| BP | biological processes |
| CC | cellular components |
| MF | molecular functions |
| JAK-STAT | Janus Kinase-Signal Transducer and Activator of Transcription |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| CCR2 | C-C Chemokine Receptor Type 2 |
| RT–qPCR | Reverse-Transcription Quantitative Polymerase Chain Reaction |
| ICAM-1 | Intercellular adhesion molecule-1 |
| TRAIL | Tumor Necrosis Factor Superfamily Member 10 |
| IL10RA | Interleukin-10 Receptor Alpha |
| IL10RB | Interleukin-10 Receptor Beta |
| JAK1 | Janus Kinase 1 |
| TYK2 | Tyrosine Kinase 2 |
| STAT3 | Signal Transducer and Activator of Transcription 3 |
| TNFRSF8 | Tumor Necrosis Factor Receptor Superfamily Member 8 |
| CSF3R | Colony-Stimulating Factor 3 Receptor |
| CVB3 | Coxsackievirus B3 |
| UKB | UK Biobank |
| ICN 1229 | ribavirin |
References
- He, W.; Zhou, L.; Xu, K.; Li, H.; Wang, J.J.; Chen, C.; Wang, D. Immunopathogenesis and immunomodulatory therapy for myocarditis. Sci. China Life Sci. 2023, 66, 2112–2137. [Google Scholar] [CrossRef]
- Zhang, B.; Thacker, D.; Zhou, T.; Zhang, D.; Lei, Y.; Chen, J.; Chrischilles, E.A.; Christakis, D.A.; Fernandez, S.; Garg, V.; et al. Cardiovascular post-acute sequelae of SARS-CoV-2 in children and adolescents: Cohort study using electronic health records. Nat. Commun. 2025, 16, 3445. [Google Scholar] [CrossRef] [PubMed]
- Kangel, D.; Ozyılmaz, İ.; Ozkok, S.; Özcanoğlu, H.D.; Güzelbağ, A.N.; Çevlik, B.; Tanıdır, İ.C.; Hatemi, A.C.; Öztürk, E. New Systemic Inflammatory Indices as Predictors of Fulminant Myocarditis in Children. Diagnostics 2025, 15, 961. [Google Scholar] [CrossRef] [PubMed]
- Peretto, G.; Sala, S.; Rizzo, S.; De Luca, G.; Campochiaro, C.; Sartorelli, S.; Benedetti, G.; Palmisano, A.; Esposito, A.; Tresoldi, M.; et al. Arrhythmias in myocarditis: State of the art. Heart Rhythm. 2019, 16, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Cooper, L.T. Myocarditis. N. Engl. J. Med. 2009, 360, 1526–1538. [Google Scholar] [CrossRef]
- Zadok, O.I.B.; O’Hare, M.J.; Nohria, A. Immune Checkpoint Inhibitor-Related Myocarditis With or Without Concomitant Myopathy: Clinical Findings and Cardiovascular Outcomes. JACC CardioOncol 2025, 7, 252–264. [Google Scholar] [CrossRef]
- Pinkert, S.; Dieringer, B.; Klopfleisch, R.; Savvatis, K.; Van Linthout, S.; Pryshliak, M.; Tschöpe, C.; Klingel, K.; Kurreck, J.; Beling, A.; et al. Early Treatment of Coxsackievirus B3-Infected Animals With Soluble Coxsackievirus-Adenovirus Receptor Inhibits Development of Chronic Coxsackievirus B3 Cardiomyopathy. Circ. Heart Fail. 2019, 12, e005250. [Google Scholar] [CrossRef]
- Epelman, S.; Liu, P.P.; Mann, D.L. Role of innate and adaptive immune mechanisms in cardiac injury and repair. Nat. Rev. Immunol. 2015, 15, 117–129. [Google Scholar] [CrossRef]
- Ghigo, A.; Franco, I.; Morello, F.; Hirsch, E. Myocyte signalling in leucocyte recruitment to the heart. Cardiovasc. Res. 2014, 102, 270–280. [Google Scholar] [CrossRef]
- Leor, J.; Palevski, D.; Amit, U.; Konfino, T. Macrophages and regeneration: Lessons from the heart. Semin. Cell Dev. Biol. 2016, 58, 26–33. [Google Scholar] [CrossRef]
- Wynn, T.A.; Vannella, K.M. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity 2016, 44, 450–462. [Google Scholar] [CrossRef]
- Chen, Z.; Downing, S.; Tzanakakis, E.S. Four Decades After the Discovery of Regenerating Islet-Derived (Reg) Proteins: Current Understanding and Challenges. Front. Cell Dev. Biol. 2019, 7, 235. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Tian, Y.; Liu, Y.; Su, Z. Reg3β: A Potential Therapeutic Target for Tissue Injury and Inflammation-Associated Disorders. Int. Rev. Immunol. 2022, 41, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Lörchner, H.; Hou, Y.; Adrian-Segarra, J.M.; Kulhei, J.; Detzer, J.; Günther, S.; Gajawada, P.; Warnecke, H.; Niessen, H.W.; Pöling, J.; et al. Reg proteins direct accumulation of functionally distinct macrophage subsets after myocardial infarction. Cardiovasc. Res. 2018, 114, 1667–1679. [Google Scholar] [CrossRef]
- Lörchner, H.; Pöling, J.; Gajawada, P.; Hou, Y.; Polyakova, V.; Kostin, S.; Adrian-Segarra, J.M.; Boettger, T.; Wietelmann, A.; Warnecke, H.; et al. Myocardial healing requires Reg3β-dependent accumulation of macrophages in the ischemic heart. Nat. Med. 2015, 21, 353–362. [Google Scholar] [CrossRef]
- Zhou, S.; Jiang, H.; Wang, H.; Lu, H.; Chen, R.; Xu, H.; Su, Z.; Shao, X. Reg3β from cardiomyocytes regulated macrophage migration, proliferation and functional skewing in experimental autoimmune myocarditis. Am. J. Clin. Exp. Immunol. 2018, 7, 8–15. [Google Scholar]
- Smith, J.G.; Gerszten, R.E. Emerging affinity-based proteomic technologies for large scale plasma profiling in cardiovascular disease. Circulation 2017, 135, 1651–1664. [Google Scholar] [CrossRef]
- Jacob, J.; Ngo, D.; Finkel, N.; Pitts, R.; Gleim, S.; Benson, M.D.; Keyes, M.J.; Farrell, L.A.; Morgan, T.; Jennings, L.L.; et al. Application of large scale aptamer-based proteomic profiling to “planned” myocardial infarctions. Circulation 2018, 137, 1270–1277. [Google Scholar] [CrossRef]
- Emilsson, V.; Ilkov, M.; Lamb, J.R.; Finkel, N.; Gudmundsson, E.F.; Pitts, R.; Hoover, H.; Gudmundsdottir, V.; Horman, S.R.; Aspelund, T.; et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018, 361, 769–773. [Google Scholar] [CrossRef]
- Sun, B.B.; Maranville, J.C.; Peters, J.E.; Stacey, D.; Staley, J.R.; Blackshaw, J.; Burgess, S.; Jiang, T.; Paige, E.; Surendran, P.; et al. Genomic atlas of the human plasma proteome. Nature 2018, 558, 73–79. [Google Scholar] [CrossRef]
- Suhre, K.; McCarthy, M.I.; Schwenk, J.M. Genetics meets proteomics: Perspectives for large population-based studies. Nat. Rev. Genet. 2021, 22, 19–37. [Google Scholar] [CrossRef]
- Gudjonsson, A.; Gudmundsdottir, V.; Axelsson, G.T.; Gudmundsson, E.F.; Jonsson, B.G.; Launer, L.J.; Lamb, J.R.; Jennings, L.L.; Aspelund, T.; Emilsson, V.; et al. A genome-wide association study of serum proteins reveals shared loci with common diseases. Nat. Commun. 2022, 13, 480. [Google Scholar] [CrossRef]
- Bourgault, J.; Abner, E.; Manikpurage, H.D.; Pujol-Gualdo, N.; Laisk, T.; Estonian Biobank Research Team; Gobeil, É.; Gagnon, E.; Girard, A.; Mitchell, P.L.; et al. Arsenault, Proteome-Wide Mendelian Randomization Identifies Causal Links Between Blood Proteins and Acute Pancreatitis. Gastroenterology 2023, 164, 953–965.e3. [Google Scholar] [CrossRef] [PubMed]
- Clough, E.; Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; et al. NCBI GEO: Archive for gene expression and epigenomics data sets: 23-year update. Nucleic Acids Res. 2024, 52, D138–D144. [Google Scholar] [CrossRef] [PubMed]
- Orrù, V.; Steri, M.; Sidore, C.; Marongiu, M.; Serra, V.; Olla, S.; Sole, G.; Lai, S.; Dei, M.; Mulas, A.; et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat. Genet. 2020, 52, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Stolarek, I.; Samelak-Czajka, A.; Figlerowicz, M.; Jackowiak, P. Dimensionality reduction by UMAP for visualizing and aiding in classification of imaging flow cytometry data. iScience 2022, 25, 105142. [Google Scholar] [CrossRef]
- Ference, B.A.; Holmes, M.V.; Smith, G.D. Using Mendelian Randomization to Improve the Design of Randomized Trials. Cold Spring Harb. Perspect. Med. 2021, 11, a040980. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Zhao, Q.; Lawlor, D.A.; Sheehan, N.A.; Thompson, J.; Smith, G.D. Improving the accuracy of two-sample summary-data Mendelian randomization: Moving beyond the NOME assumption. Int. J. Epidemiol. 2019, 48, 728–742. [Google Scholar] [CrossRef]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef]
- Chen, L.; Peters, J.E.; Prins, B.; Persyn, E.; Traylor, M.; Surendran, P.; Karthikeyan, S.; Yonova-Doing, E.; Di Angelantonio, E.; Roberts, D.J.; et al. Systematic Mendelian randomization using the human plasma proteome to discover potential therapeutic targets for stroke. Nat. Commun. 2022, 13, 6143. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.R.; Sanderson, E.; Hammerton, G.; Richmond, R.C.; Smith, G.D.; Heron, J.; Taylor, A.E.; Davies, N.M.; Howe, L.D. Mendelian randomisation for mediation analysis: Current methods and challenges for implementation. Eur. J. Epidemiol. 2021, 36, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Mosharaf, M.P.; Alam, K.; Gow, J.; Mahumud, R.A. Exploration of key drug target proteins highlighting their related regulatory molecules, functional pathways and drug candidates associated with delirium: Evidence from meta-data analyses. BMC Geriatr. 2023, 23, 767. [Google Scholar] [CrossRef]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef]
- Yoo, M.; Shin, J.; Kim, J.; Ryall, K.A.; Lee, K.; Lee, S.; Jeon, M.; Kang, J.; Tan, A.C. DSigDB: Drug signatures database for gene set analysis. Bioinformatics 2015, 31, 3069–3071. [Google Scholar] [CrossRef]
- Wu, C.H.; Apweiler, R.; Bairoch, A.; Natale, D.A.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; et al. The Universal Protein Resource (UniProt): An expanding universe of protein information. Nucleic Acids Res. 2006, 34, D187–D191. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15–Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Rosignoli, S.; Paiardini, A. Boosting the Full Potential of PyMOL with Structural Biology Plugins. Biomolecules 2022, 12, 1764. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Haberland, V.; Baird, D.; Walker, V.; Haycock, P.C.; Hurle, M.R.; Gutteridge, A.; Erola, P.; Liu, Y.; Luo, S.; et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. 2020, 52, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Allen, N.E.; Lacey, B.; Lawlor, D.A.; Pell, J.P.; Gallacher, J.; Smeeth, L.; Elliott, P.; Matthews, P.M.; Lyons, R.A.; Whetton, A.D.; et al. Prospective study design and data analysis in UK Biobank. Sci. Transl. Med. 2024, 16, eadf4428. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Liu, J.; Qin, J.; Lai, L.; Heo, G.S.; Luehmann, H.; Sultan, D.; Bredemeyer, A.; Bajapa, G.; Feng, G.; et al. Expansion of Pathogenic Cardiac Macrophages in Immune Checkpoint Inhibitor Myocarditis. Circulation 2024, 149, 48–66. [Google Scholar] [CrossRef]
- Bajpai, G.; Bredemeyer, A.; Li, W.; Zaitsev, K.; Koenig, A.L.; Lokshina, I.; Mohan, J.; Ivey, B.; Hsiao, H.-M.; Weinheimer, C.; et al. Tissue Resident CCR2- and CCR2+ Cardiac Macrophages Differentially Orchestrate Monocyte Recruitment and Fate Specification Following Myocardial Injury. Circ. Res. 2019, 124, 263–278. [Google Scholar] [CrossRef]
- Xia, C.; Yin, S.; To, K.K.W.; Fu, L. CD39/CD73/A2AR pathway and cancer immunotherapy. Mol. Cancer 2023, 22, 44. [Google Scholar] [CrossRef]
- Sun, Z.; Kang, J.; Yang, S.; Zhang, Y.; Huang, N.; Zhang, X.; Du, G.; Jiang, J.; Ning, B. CD73 inhibits titanium particle-associated aseptic loosening by alternating activation of macrophages. Int. Immunopharmacol. 2023, 122, 110561. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Li, S.; Zhang, T.; Chen, C.; Hu, J.; Sun, D.; Lu, H. Mechanical stimulation improves rotator cuff tendon-bone healing via activating IL-4/JAK/STAT signaling pathway mediated macrophage M2 polarization. J. Orthop. Transl. 2022, 37, 78–88. [Google Scholar] [CrossRef]
- Yang, P.; Li, F.; Tang, J.; Tian, Q.; Zheng, Z. ET-1 receptor type B (ETBR) overexpression associated with ICAM-1 downregulation leads to inflammatory attenuation in experimental autoimmune myocarditis. PeerJ 2023, 11, e16320. [Google Scholar] [CrossRef]
- Yoon, C.-H.; Hur, J.; Oh, I.-Y.; Park, K.-W.; Kim, T.-Y.; Shin, J.-H.; Kim, J.-H.; Lee, C.-S.; Chung, J.-K.; Park, Y.-B.; et al. Intercellular adhesion molecule-1 is upregulated in ischemic muscle, which mediates trafficking of endothelial progenitor cells. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 1066–1072. [Google Scholar] [CrossRef]
- Wu, Y.; Ip, J.E.; Huang, J.; Zhang, L.; Matsushita, K.; Liew, C.-C.; Pratt, R.E.; Dzau, V.J. Essential Role of ICAM-1/CD18 in Mediating EPC Recruitment, Angiogenesis, and Repair to the Infarcted Myocardium. Circ. Res. 2006, 99, 315–322. [Google Scholar] [CrossRef]
- Angerfors, A.; Brännmark, C.; Lagging, C.; Tai, K.; Svedberg, R.M.; Andersson, B.; Jern, C.; Stanne, T.M. Proteomic profiling identifies novel inflammation-related plasma proteins associated with ischemic stroke outcome. J. Neuroinflammation 2023, 20, 224. [Google Scholar] [CrossRef]
- Hage, C.; Michaëlsson, E.; Linde, C.; Donal, E.; Daubert, J.-C.; Gan, L.-M.; Lund, L.H. Inflammatory Biomarkers Predict Heart Failure Severity and Prognosis in Patients with Heart Failure with Preserved Ejection Fraction: A Holistic Proteomic Approach. Circ. Cardiovasc. Genet. 2017, 10, e001633. [Google Scholar] [CrossRef]
- Ouyang, W.; Rutz, S.; Crellin, N.K.; Valdez, P.A.; Hymowitz, S.G. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 2011, 29, 71–109. [Google Scholar] [CrossRef]
- Dumitru, A.V.; Țăpoi, D.A.; Halcu, G.; Munteanu, O.; Dumitrascu, D.-I.; Ceaușu, M.C.; Gheorghișan-Gălățeanu, A.-A. The Polyvalent Role of CD30 for Cancer Diagnosis and Treatment. Cells 2023, 12, 1783. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, A.; Dolcino, M.; Tinazzi, E.; Rigo, A.; Argentino, G.; Patuzzo, G.; Ottria, A.; Beri, R.; Puccetti, A.; Lunardi, C. Characterization of CD30/CD30L(+) Cells in Peripheral Blood and Synovial Fluid of Patients with Rheumatoid Arthritis. J. Immunol. Res. 2015, 2015, 729654. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zhang, L.; Wu, Y.; Zhou, X.; Wang, Y.; Zhang, J.; Liu, Y.; He, Z.; Wang, X. CSF3R as a potential prognostic biomarker and immunotherapy target in glioma. Cent. Eur. J. Immunol. 2024, 49, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Carnevale, S.; Ponzetta, A.; Rigatelli, A.; Carriero, R.; Puccio, S.; Supino, D.; Grieco, G.; Molisso, P.; Di Ceglie, I.; Scavello, F.; et al. Neutrophils Mediate Protection Against Colitis and Carcinogenesis by Controlling Bacterial Invasion and IL22 Production by γδ T Cells. Cancer Immunol. Res. 2024, 12, 413–426. [Google Scholar] [CrossRef]
- Zhang, Q.; Putheti, P.; Zhou, Q.; Liu, Q.; Gao, W. Structures and biological functions of IL-31 and IL-31 receptors. Cytokine Growth Factor Rev. 2008, 19, 347–356. [Google Scholar] [CrossRef]
- Kasraie, S.; Niebuhr, M.; Werfel, T. Interleukin (IL)-31 induces pro-inflammatory cytokines in human monocytes and macrophages following stimulation with staphylococcal exotoxins. Allergy 2010, 65, 712–721. [Google Scholar] [CrossRef]
- Tam, R.C.; Lau, J.Y.; Hong, Z. Mechanisms of action of ribavirin in antiviral therapies. Antivir. Chem. Chemother. 2001, 12, 261–272. [Google Scholar] [CrossRef]
- Ogbomo, H.; Michaelis, M.; Altenbrandt, B.; Doerr, H.W.; Cinatl, J. A novel immunomodulatory mechanism of ribavirin in suppressing natural killer cell function. Biochem. Pharmacol. 2010, 79, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Imran, M.; Rauf, A.; Orhan, I.E.; Shariati, M.A.; Iahtisham-Ul-Haq; IqraYasmin; Shahbaz, M.; Qaisrani, T.B.; Shah, Z.A.; et al. Chrysin: Pharmacological and therapeutic properties. Life Sci. 2019, 235, 116797. [Google Scholar] [CrossRef] [PubMed]
- Schnitzler, P.; Neuner, A.; Nolkemper, S.; Zundel, C.; Nowack, H.; Sensch, K.H.; Reichling, J. Antiviral activity and mode of action of propolis extracts and selected compounds. Phytother. Res. 2010, 24 (Suppl. S1), S20–S28. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-H.; Kwon, B.-E.; Jang, H.; Kang, H.; Cho, S.; Park, K.; Ko, H.-J.; Kim, H. Antiviral Activity of Chrysin Derivatives against Coxsackievirus B3 in vitro and in vivo. Biomol. Ther. 2015, 23, 465–470. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, S.; Chen, Y.; Wang, Y.; Wang, T.; Wo, X.; Dong, Y.; Zhang, J.; Xu, W.; Qu, C.; et al. N-Acetyl cysteine effectively alleviates Coxsackievirus B-Induced myocarditis through suppressing viral replication and inflammatory response. Antivir. Res. 2020, 179, 104699. [Google Scholar] [CrossRef]
- Shimada, K.; Uzui, H.; Ueda, T.; Lee, J.-D.; Kishimoto, C. N-Acetylcysteine Ameliorates Experimental Autoimmune Myocarditis in Rats via Nitric Oxide. J. Cardiovasc. Pharmacol. Ther. 2015, 20, 203–210. [Google Scholar] [CrossRef]



| Primer Sequences | |
|---|---|
| GAPDH | Forward GGTCGGTGTGAACGGATTTG Reverse TGTAGACCATGTAGTTGAGGTCA |
| NT5E | Forward CAGCGATGACTCCACCAAGT Reverse CTCCGGCATCCAAAAACAGC |
| Arg-1 | Forward CATTGGCTTGCGAGACGTAGAC Reverse GCTGAAGGTCTCTTCCATCACC |
| iNOS | Forward TGGAGCCAGTTGTGGATTGTC Reverse GGTCGTAATGTCCAGGAAGTAG |
| Drug | Target | Binding Energy (kcal/mol) |
|---|---|---|
| ICN 1229 | TNFSF10 | −18.793329 |
| ICN 1229 | IL4 | −20.381943 |
| ICN 1229 | ICAM1 | −17.373051 |
| Chrysin | TNFSF10 | −21.043499 |
| Chrysin | IL4 | −17.346796 |
| Chrysin | ICAM1 | −19.609625 |
| Chrysin | NT5E | −23.028145 |
| Simvastatin | TNFSF10 | −20.781334 |
| Simvastatin | IL4 | −19.659033 |
| Simvastatin | ICAM1 | −17.294996 |
| Simvastatin | IL17F | −27.784307 |
| AM-630 | IL4 | −24.125502 |
| AM-630 | ICAM1 | −21.320253 |
| PYRENE | IL4 | −20.845221 |
| PYRENE | ICAM1 | −17.221935 |
| N-Acetyl-L-cysteine | IL10RB | −23.861942 |
| N-Acetyl-L-cysteine | TNFSF10 | −21.000706 |
| N-Acetyl-L-cysteine | IL4 | −22.408953 |
| N-Acetyl-L-cysteine | ICAM1 | −17.972317 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Zhou, P.; Zhu, F.; Wang, Y.; Wang, X.; Chen, J.; Li, Y.; Shao, X. Mechanistic Study of NT5E in Reg3β-Induced Macrophage Polarization and Cooperation with Plasma Proteins in Myocarditis Injury and Repair. Biology 2025, 14, 1017. https://doi.org/10.3390/biology14081017
Zhang S, Zhou P, Zhu F, Wang Y, Wang X, Chen J, Li Y, Shao X. Mechanistic Study of NT5E in Reg3β-Induced Macrophage Polarization and Cooperation with Plasma Proteins in Myocarditis Injury and Repair. Biology. 2025; 14(8):1017. https://doi.org/10.3390/biology14081017
Chicago/Turabian StyleZhang, Shichao, Peirou Zhou, Fanfan Zhu, Yingying Wang, Xuesong Wang, Jingwen Chen, Yumeng Li, and Xiaoyi Shao. 2025. "Mechanistic Study of NT5E in Reg3β-Induced Macrophage Polarization and Cooperation with Plasma Proteins in Myocarditis Injury and Repair" Biology 14, no. 8: 1017. https://doi.org/10.3390/biology14081017
APA StyleZhang, S., Zhou, P., Zhu, F., Wang, Y., Wang, X., Chen, J., Li, Y., & Shao, X. (2025). Mechanistic Study of NT5E in Reg3β-Induced Macrophage Polarization and Cooperation with Plasma Proteins in Myocarditis Injury and Repair. Biology, 14(8), 1017. https://doi.org/10.3390/biology14081017






