Phylogenomic, Morphological, and Phylogenetic Evidence Reveals Five New Species and Two New Host Records of Nectriaceae (Hypocreales) from China
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Strain Isolation, and Preservation
2.2. DNA Extraction and PCR Amplification
2.3. Whole-Genome Sequencing, Assembly, and Gene Annotation
2.4. Phylogenetic Analyses
2.4.1. Multigene Phylogenetic Analysis
2.4.2. Phylogenomic Analysis
2.5. Genealogical Concordance Phylogenetic Species Recognition Analyses
2.6. Morphological Observations
3. Results
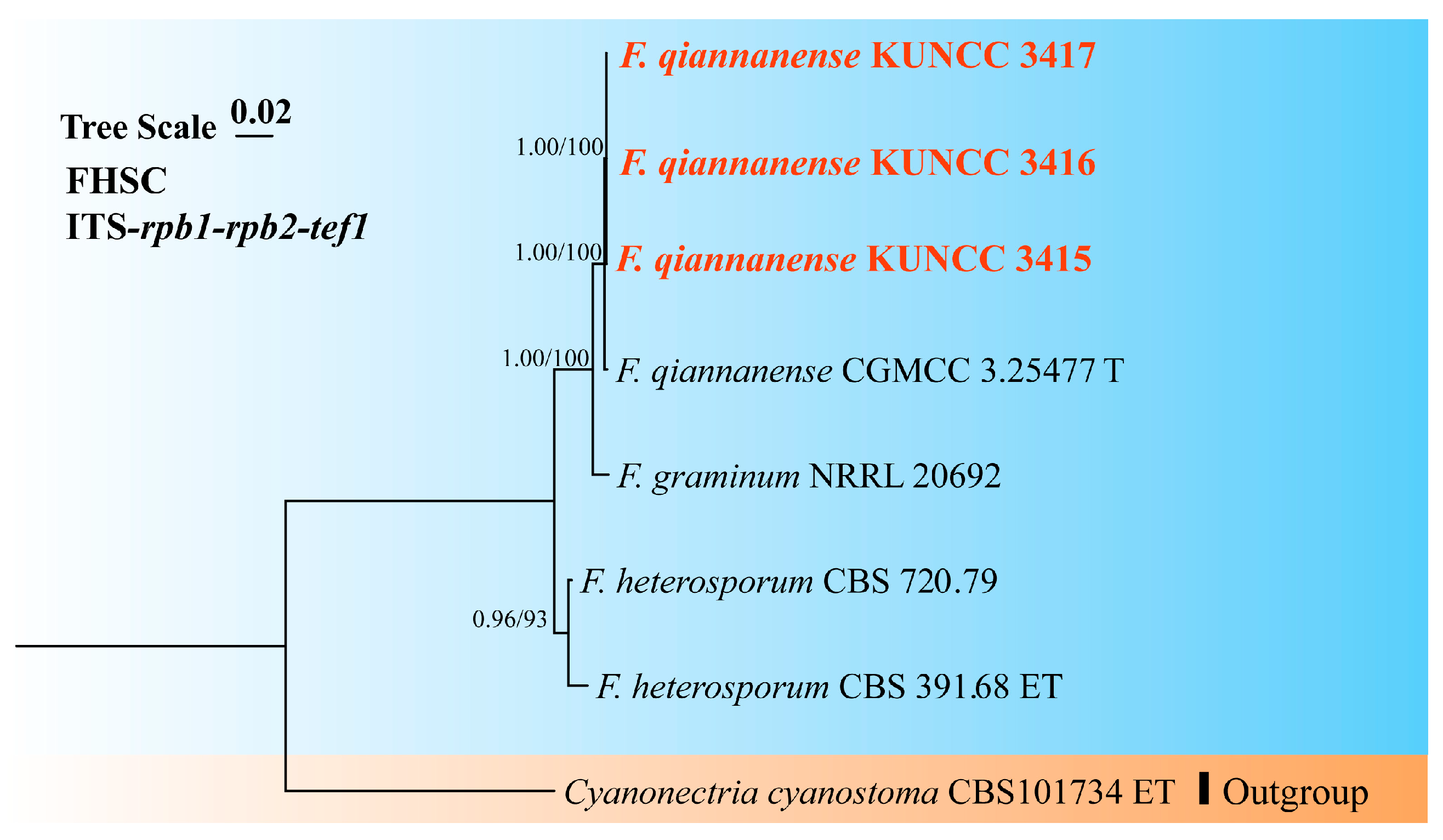
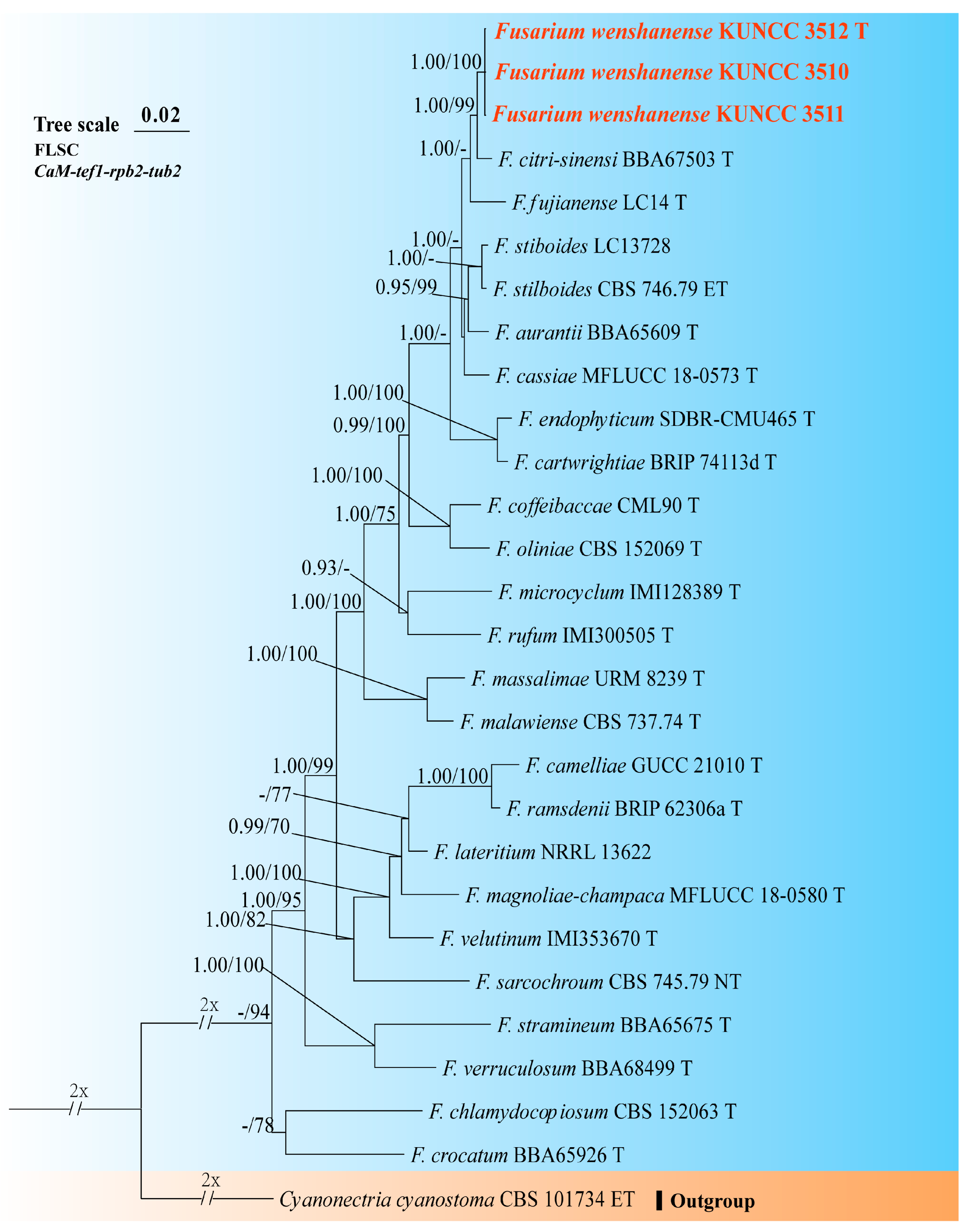
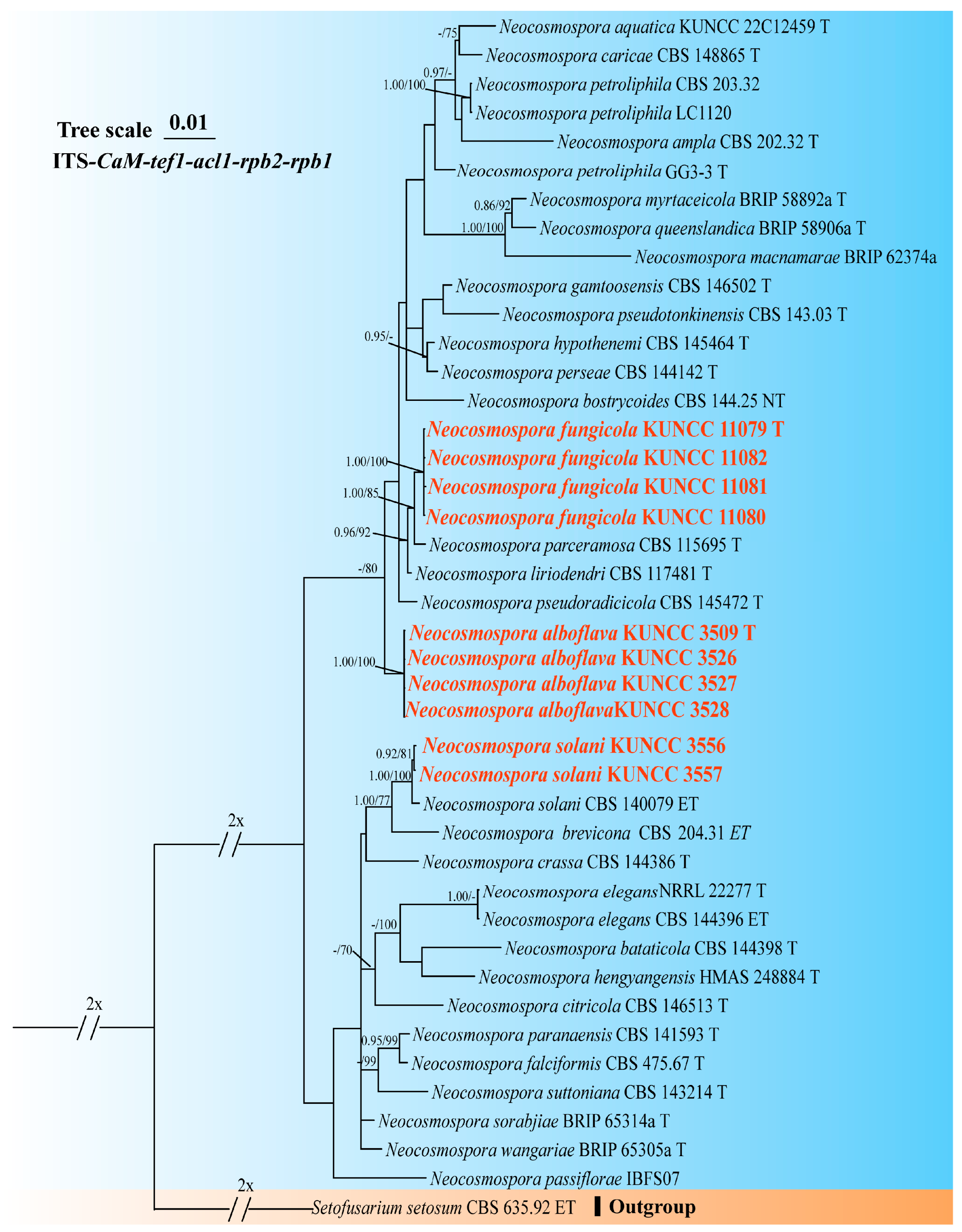
3.1. Molecular Phylogeny
3.2. Genomic Features
3.3. Results of Phylogenomic Analysis
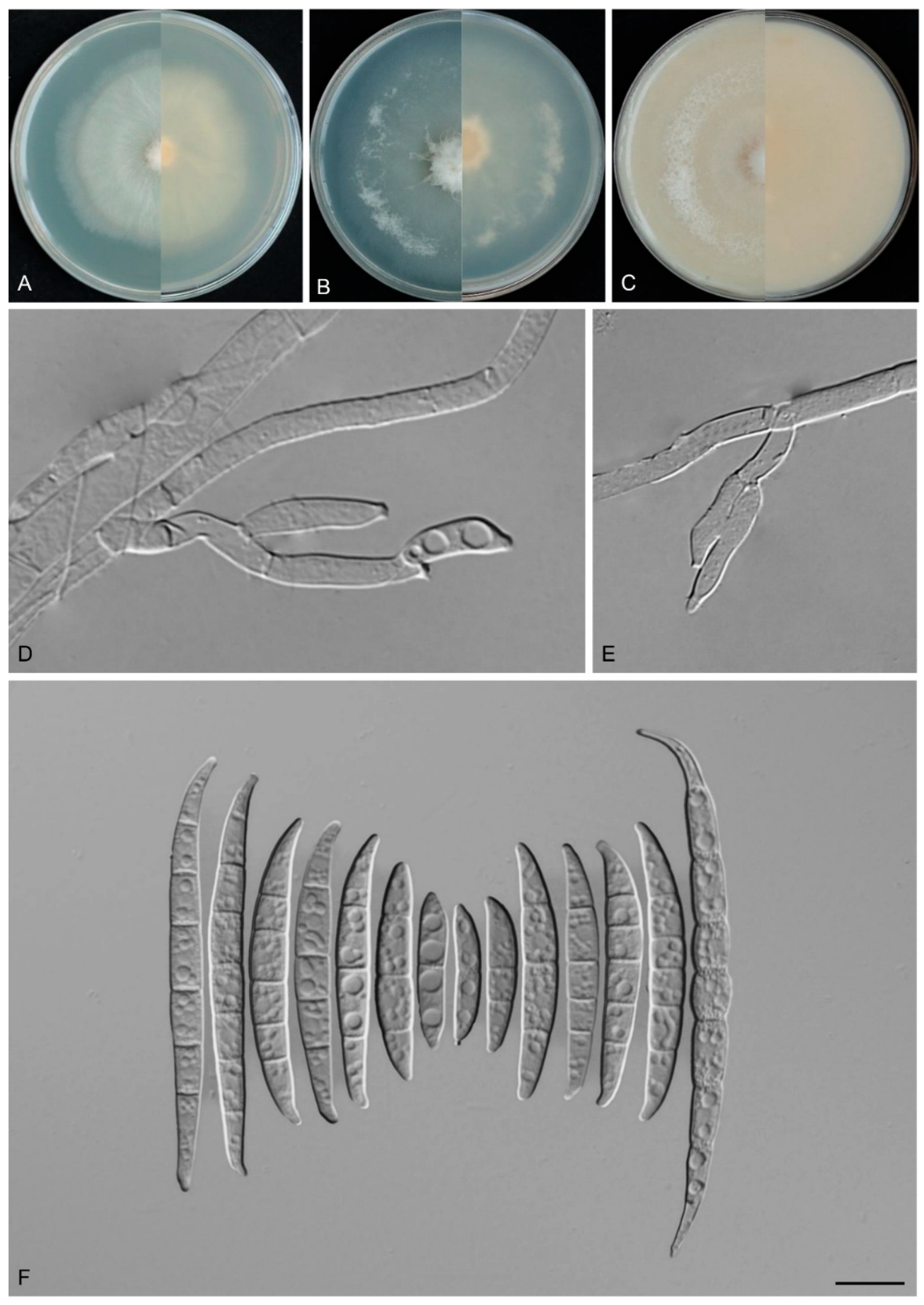
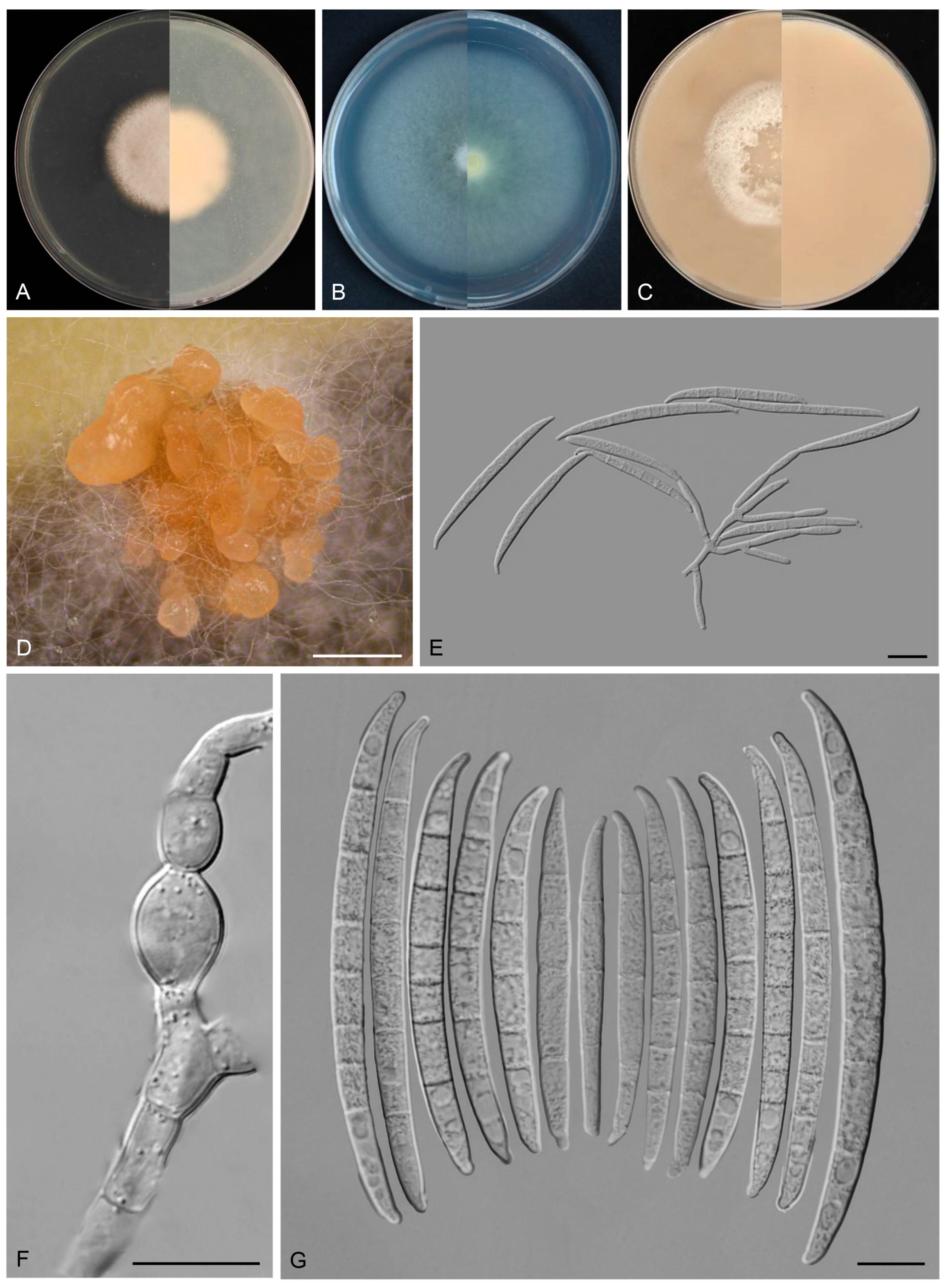
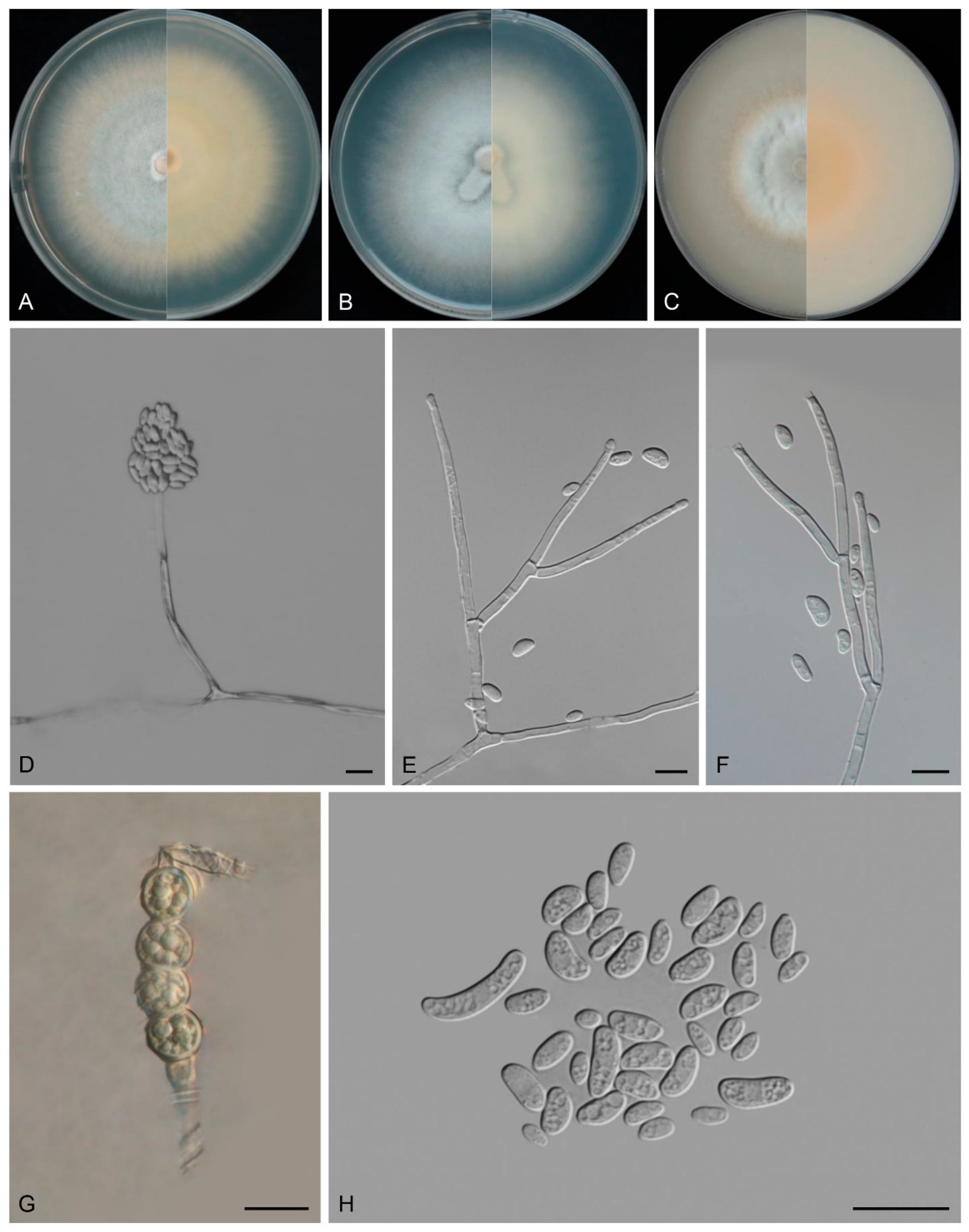
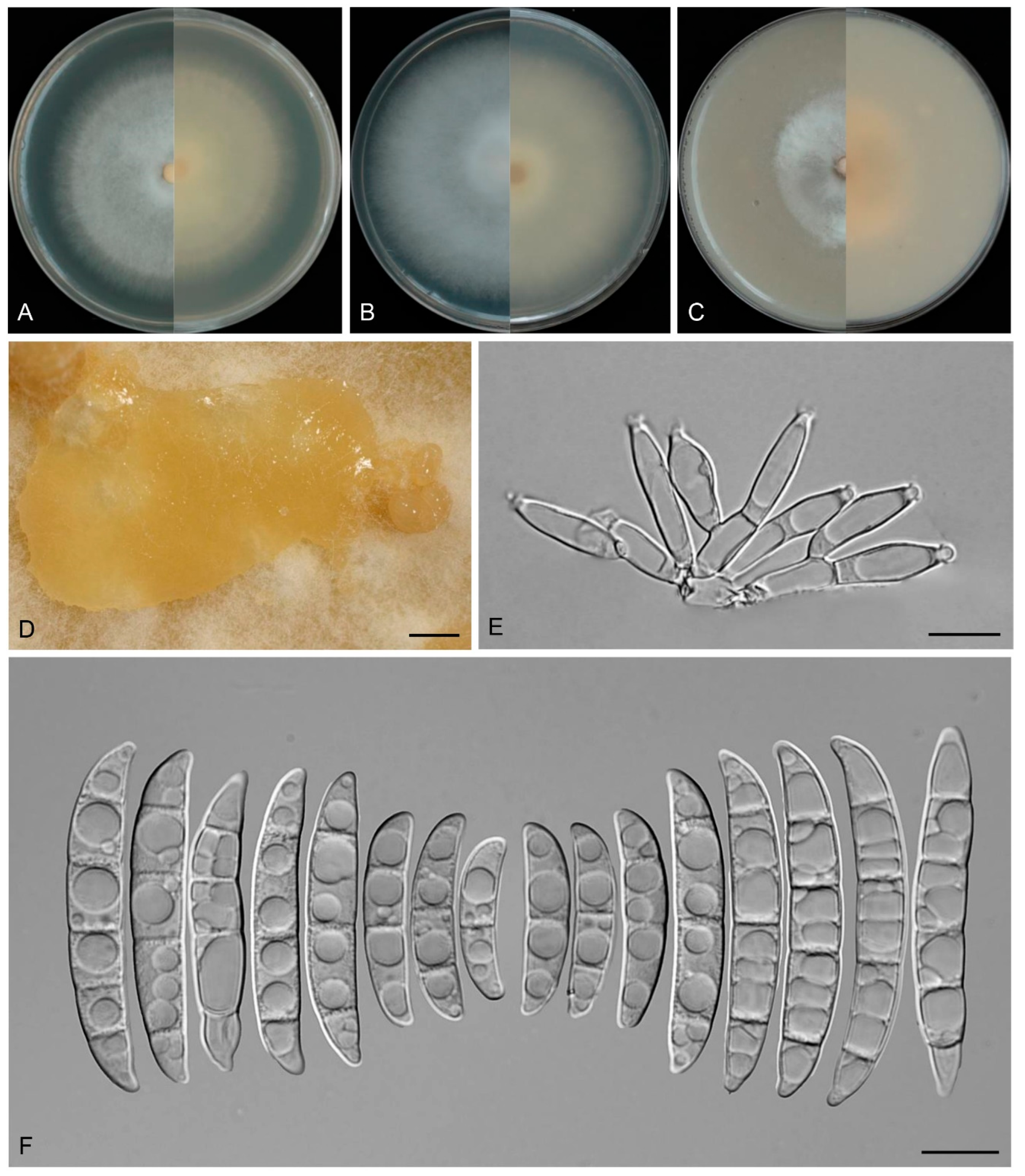
3.4. Taxonomy
4. Discussion
4.1. Species Diversity of Fusarium and Neocosmospora
4.2. Diversity and Potential of Endophytic Fusarioid Fungi
4.3. Evaluation of the Phylogenetic Resolution of Molecular Markers in Fusarium and Related Genera
4.4. Topological Congruence Between Genome-Scale and Multilocus Phylogenetic Trees
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burgess, L. 2011 McAlpine memorial lecture-a love affair with Fusarium. Australas. Plant Pathol. 2014, 43, 359–368. [Google Scholar] [CrossRef]
- Lombard, L.; Van der Merwe, N.; Groenewald, J.; Crous, P. Generic concepts in Nectriaceae. Stud. Mycol. 2015, 80, 189–245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, C.; Nie, L.; Maharachchikumbura, S.S.; Crous, P.; Hyde, K.D.; Xiang, M.; Al-Otibi, F.; Manawasinghe, I.S. Identification and characterization of Albonectria, Fusarium, and Neocosmospora species associated with ornamental plants in Southern China. Mycosphere 2024, 15, 6641–6717. [Google Scholar] [CrossRef]
- Crous, P.W.; Lombard, L.; Sandoval-Denis, M.; Seifert, K.A.; Schroers, H.J.; Chaverri, P.; Gené, J.; Guarro, J.; Hirooka, Y.; Bensch, K.; et al. Fusarium: More than a node or a foot-shaped basal cell. Stud. Mycol. 2021, 98, 100116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zeng, Y.; Wei, T.; Jiang, Y.; Zeng, X. Endophytic Fusarium and allied fungi from Rosa roxburghii in China. Mycosphere 2023, 14, 2092–2207. [Google Scholar] [CrossRef]
- Han, S.L.; Wang, M.M.; Ma, Z.Y.; Raza, M.; Zhao, P.; Liang, J.M.; Gao, M.; Li, Y.J.; Wang, J.W.; Hu, D.M.; et al. Fusarium diversity associated with diseased cereals in China, with an updated phylogenomic assessment of the genus. Stud. Mycol. 2023, 104, 87–148. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Liu, D.; He, X.; Liu, W.; Yu, F. Epidemic identification of fungal diseases in Morchella cultivation across China. J. Fungi 2022, 8, 1107. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Denis, M.; Lombard, L.; Crous, P.W. Back to the roots: A reappraisal of Neocosmospora. Persoonia 2019, 43, 90–185. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.W.; Lee, S.C.; Kim, D.H.; Ko, S.W.; Kim, K.S. Fusarium fruit rot of citrus in Jeju Island. Mycobiology 2000, 28, 158–162. [Google Scholar] [CrossRef]
- Cighir, A.; Mare, A.D.; Vultur, F.; Cighir, T.; Pop, S.D.; Horvath, K.; Man, A. Fusarium spp. in human disease: Exploring the boundaries between commensalism and pathogenesis. Life 2023, 13, 1440. [Google Scholar] [CrossRef] [PubMed]
- Short, D.P.; O’Donnell, K.; Zhang, N.; Juba, J.H.; Geiser, D.M. Widespread occurrence of diverse human pathogenic types of the fungus Fusarium detected in plumbing drains. J. Clin. Microbiol. 2011, 49, 4264–4272. [Google Scholar] [CrossRef] [PubMed]
- Najafzadeh, M.J.; Dolatabadi, S.; de Hoog, S.; Esfahani, M.K.; Haghani, I.; Aghili, S.R.; Ghazvini, R.D.; Rezaei-Matehkolaei, A.; Abastabar, M.; Al-Hatmi, A.M. Phylogenetic analysis of clinically relevant Fusarium species in Iran. Mycopathologia 2020, 185, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Link, H.F. Observationes in ordines plantarum naturales, Dissetatio I. Ges. Naturforschender Freunde Zu Berl. Mag. 1809, 3, 3–42. [Google Scholar]
- Booth, C. The Genus Fusarium; Commonwealth Mycological Institute: Kew, Surrey, UK, 1971. [Google Scholar]
- Nelson, P.E.; Toussoun, T.A.; Marasas, W.F.O. Fusarium Species: An Illustrated Manual for Identification; Pennsylvania State University Press: University Park, PA, USA, 1983. [Google Scholar]
- O’Donnell, K.; Rooney, A.P.; Proctor, R.H.; Brown, D.W.; McCormick, S.P.; Ward, T.J.; Frandsen, R.J.; Lysøe, E.; Rehner, S.A.; Aoki, T. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet. Biol. 2013, 52, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Geiser, D.M.; Aoki, T.; Bacon, C.W.; Baker, S.E.; Bhattacharyya, M.K.; Brandt, M.E.; Brown, D.W.; Burgess, L.W.; Chulze, S.; Coleman, J. One fungus, one name: Defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology 2013, 103, 400–408. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Al-Hatmi, A.M.; Aoki, T.; Brankovics, B.; Cano-Lira, J.F.; Coleman, J.J.; de Hoog, G.S.; Di Pietro, A.; Frandsen, R.J.; Geiser, D.M. No to Neocosmospora: Phylogenomic and practical reasons for continued inclusion of the Fusarium solani species complex in the genus Fusarium. mSphere 2020, 5, e00810-20. [Google Scholar] [CrossRef] [PubMed]
- Geiser, D.M.; Al-Hatmi, A.M.; Aoki, T.; Arie, T.; Balmas, V.; Barnes, I.; Bergstrom, G.C.; Bhattacharyya, M.K.; Blomquist, C.L.; Bowden, R.L. Phylogenomic analysis of a 55.1-kb 19-gene dataset resolves a monophyletic Fusarium that includes the Fusarium solani species complex. Phytopathology 2021, 111, 1064–1079. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.; Amuhenage, T.; Apurillo, C.; Asghari, R.; Aumentado, H.; Bahkali, A.; Bera, I.; Bhunjun, C.; Calabon, M.; Chandrasiri, S. Fungalpedia, an illustrated compendium of the fungi and fungus-like taxa. Mycosphere 2023, 14, 1835–1959. [Google Scholar] [CrossRef]
- Gräfenhan, T.; Schroers, H.J.; Nirenberg, H.; Seifert, K. An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Stud. Mycol. 2011, 68, 79–113. [Google Scholar] [CrossRef] [PubMed]
- Lombard, L.; Sandoval-Denis, M.; Cai, L.; Crous, P.W. Changing the game: Resolving systematic issues in key Fusarium species complexes. Persoonia 2019, 43, i–ii. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.M.; Crous, P.W.; Sandoval-Denis, M.; Han, S.L.; Liu, F.; Liang, J.M.; Duan, W.J.; Cai, L. Fusarium and allied genera from China: Species diversity and distribution. Persoonia 2022, 48, 1–53. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.F. Wilt Disease of Cotton, Watermelon and Cowpea (Neocosmospora nov. gen.); United States Department of Agriculture, Division of Vegetable Physiology and Pathology: Washington, DC, USA, 1899; 17, pp. 1–72.
- Zeng, Z.Q.; Zhuang, W.Y. New species of Neocosmospora (Ascomycota) from China as evidenced by morphological and molecular data. Life 2023, 13, 1515. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhao, W.; Xia, J.; Cai, S.; Huai, W.X.; Zhang, R.B.; Li, B.; Peng, H.; Zhang, S. Fusarium and Neocosmospora Species Associated with the Decline of Metasequoia glyptostroboides in China. Plant Dis. 2024, 109, 1031–1050. [Google Scholar] [CrossRef] [PubMed]
- Klomchit, A.; Calabon, M.S.; Worabandit, S.; Weaver, J.A.; Karima, E.M.; Alberti, F.; Greco, C.; Mahanil, S. Unveiling novel Neocosmospora species from Thai mangroves as potent biocontrol agents against Colletotrichum species. J. Appl. Microbiol. 2024, 135, lxae114. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, R.; Liu, J.; Li, J.; Chen, S. First report of Neocosmospora pisi causing blight on Ammopiptanthus mongolicus in China. Plant Dis. 2024, 108, 1898. [Google Scholar] [CrossRef]
- Riaz, M.; Akhtar, N.; Msimbira, L.A.; Antar, M.; Ashraf, S.; Khan, S.N.; Smith, D.L. Neocosmospora rubicola, a stem rot disease in potato: Characterization, distribution and management. Front. Microbiol. 2022, 13, 953097. [Google Scholar] [CrossRef] [PubMed]
- Kamali-Sarvestani, S.; Mostowfizadeh-Ghalamfarsa, R.; Salmaninezhad, F.; Cacciola, S.O. Fusarium and Neocosmospora species associated with rot of Cactaceae and other succulent plants. J. Fungi 2022, 8, 364. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Ward, T.J.; Robert, V.A.; Crous, P.W.; Geiser, D.M.; Kang, S. DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica 2015, 43, 583–595. [Google Scholar] [CrossRef]
- O’Donnell, K.; Whitaker, B.K.; Laraba, I.; Proctor, R.H.; Brown, D.W.; Broders, K.; Kim, H.-S.; McCormick, S.P.; Busman, M.; Aoki, T. DNA sequence-based identification of Fusarium: A work in progress. Plant Dis. 2022, 106, 1597–1609. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Humber, R.A.; Geiser, D.M.; Kang, S.; Park, B.; Robert, V.A.; Crous, P.W.; Johnston, P.R.; Aoki, T.; Rooney, A.P. Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia 2012, 104, 427–445. [Google Scholar] [CrossRef] [PubMed]
- Torres-Cruz, T.J.; Whitaker, B.K.; Proctor, R.H.; Broders, K.; Laraba, I.; Kim, H.-S.; Brown, D.W.; O’Donnell, K.; Estrada-Rodríguez, T.L.; Lee, Y.-H. FUSARIUM-ID v. 3.0: An updated, downloadable resource for Fusarium species identification. Plant Dis. 2022, 106, 1610–1616. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, N.; Sandoval-Denis, M.; Lombard, L.; Visagie, C.; Wingfield, B.; Crous, P.W. Redefining species limits in the Fusarium fujikuroi species complex. Persoonia 2021, 46, 129–162. [Google Scholar] [CrossRef] [PubMed]
- Jambhulkar, P.P.; Bajpai, R.; Reddy, H.J.; Tripathy, P.S.; Varun, P.; Rout, A.K.; Behera, B.K.; Lakshman, D.K.; Nanjundappa, M. Assessment of Genetic Diversity and the Population Structure of Species from the Fusarium fujikuroi Species Complex Causing Fusarium Stalk Rot of Maize. J. Fungi 2024, 10, 574. [Google Scholar] [CrossRef] [PubMed]
- Achari, S.R.; Kaur, J.; Dinh, Q.; Mann, R.; Sawbridge, T.; Summerell, B.A.; Edwards, J. Phylogenetic relationship between Australian Fusarium oxysporum isolates and resolving the species complex using the multispecies coalescent model. BMC Genom. 2020, 21, 248. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Denis, M.; Costa, M.; Broders, K.; Becker, Y.; Maier, W.; Yurkov, A.; Kermode, A.; Buddie, A.; Ryan, M.; Schumacher, R. An integrative re-evaluation of the Fusarium sambucinum species complex. Stud. Mycol. 2024, 110, 1–110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.X.; Shang, Y.X.; Zhang, M.Y.; Zhang, J.J.; Geng, Y.; Xia, J.W.; Zhang, X.G. Phylogenomics, taxonomy and morphological characters of the Microdochiaceae (Xylariales, Sordariomycetes). MycoKeys 2024, 106, 303. [Google Scholar] [CrossRef] [PubMed]
- Haridas, S.; Albert, R.; Binder, M.; Bloem, J.; LaButti, K.; Salamov, A.; Andreopoulos, B.; Baker, S.; Barry, K.; Bills, G. 101 Dothideomycetes genomes: A test case for predicting lifestyles and emergence of pathogens. Stud. Mycol. 2020, 96, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ma, Z.; Hou, L.; Diao, Y.; Wu, W.; Damm, U.; Song, S.; Cai, L. Updating species diversity of Colletotrichum, with a phylogenomic overview. Stud. Mycol. 2022, 101, 1–56. [Google Scholar] [CrossRef] [PubMed]
- Möller, M.; Stukenbrock, E.H. Evolution and genome architecture in fungal plant pathogens. Nat. Rev. Microbiol. 2017, 15, 756–771. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Chavarria, D.A.; Rua-Giraldo, A.L.; Alzate, J.F. An evolutionary view of the Fusarium core genome. BMC Genom. 2024, 25, 304. [Google Scholar] [CrossRef] [PubMed]
- Lizcano Salas, A.F.; Duitama, J.; Restrepo, S.; Celis Ramírez, A.M. Phylogenomic approaches reveal a robust time-scale phylogeny of the Terminal Fusarium Clade. IMA Fungus 2024, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Cheng, Z.Y.; Xie, L.Y.; Tang, M.; Yang, Z.L.; Shen, P.H.; Wang, Y.B. Molecular phylogeny and morphology of Sporodiniella sinensis sp. nov. (Syzygitaceae, Mucorales), an invertebrate-associated species from Yunnan, China. Int. J. Syst. Evol. Microbiol. 2024, 74, 006315. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.B.; Wang, Y.; Fan, Q.; Duan, D.E.; Zhang, G.D.; Dai, R.Q.; Dai, Y.D.; Zeng, W.B.; Chen, Z.H.; Li, D.D.; et al. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): New taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 2020, 103, 1–46. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Coleman, J.J.; Vinatzer, B.A. Genome Resource: Draft genome of Fusarium avenaceum, strain F156N33, isolated from the atmosphere above Virginia and annotated based on RNA sequencing data. Plant Dis. 2022, 106, 720–722. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Sarver, B.A.; Ward, T.J.; Gale, L.R.; Broz, K.; Kistler, H.C.; Aoki, T.; Nicholson, P.; Carter, J.; O’Donnell, K. Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet. Biol. 2011, 48, 1096–1107. [Google Scholar] [CrossRef] [PubMed]
- Gräfenhan, T.; Johnston, P.R.; Vaughan, M.M.; McCormick, S.P.; Proctor, R.H.; Busman, M.; Ward, T.J.; O’Donnell, K. Fusarium praegraminearum sp. nov., a novel nivalenol mycotoxin-producing pathogen from New Zealand can induce head blight on wheat. Mycologia 2016, 108, 1229–1239. [Google Scholar] [PubMed]
- Laurence, M.; Walsh, J.; Shuttleworth, L.; Robinson, D.; Johansen, R.; Petrovic, T.; Vu, T.; Burgess, L.; Summerell, B.; Liew, E. Six novel species of Fusarium from natural ecosystems in Australia. Fungal Divers. 2016, 77, 349–366. [Google Scholar] [CrossRef]
- Sandoval-Denis, M.; Swart, W.J.; Crous, P.W. New Fusarium species from the Kruger national park, South Africa. MycoKeys 2018, 34, 63–92. [Google Scholar] [CrossRef] [PubMed]
- Lombard, L.; Van Doorn, R.; Crous, P. Neotypification of Fusarium chlamydosporum-a reappraisal of a clinically important species complex. Fungal Syst. Evol. 2019, 4, 183–200. [Google Scholar] [CrossRef] [PubMed]
- Maryani, N.; Sandoval-Denis, M.; Lombard, L.; Crous, P.W.; Kema, G. New endemic Fusarium species hitch-hiking with pathogenic Fusarium strains causing Panama disease in small-holder banana plots in Indonesia. Persoonia 2019, 43, 48–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, Q.; Diao, Y.; Duan, W.; Cai, L. Fusarium incarnatum-equiseti complex from China. Persoonia 2019, 43, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sandoval-Denis, M.; Crous, P.W.; Zhang, X.; Lombard, L. Numbers to names-restyling the Fusarium incarnatum-equiseti species complex. Persoonia 2019, 43, 186–221. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; Von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Erixon, P.; Svennblad, B.; Britton, T.; Oxelman, B. Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst. Biol. 2003, 52, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- dos Reis, M. Dating Microbial Evolution with MCMCtree. In Environmental Microbial Evolution: Methods and Protocols; Luo, H., Ed.; Springer: New York, NY, USA, 2022; pp. 3–22. [Google Scholar]
- Lutzoni, F.; Nowak, M.D.; Alfaro, M.E.; Reeb, V.; Miadlikowska, J.; Krug, M.; Arnold, A.E.; Lewis, L.A.; Swofford, D.L.; Hibbett, D.; et al. Contemporaneous radiations of fungi and plants linked to symbiosis. Nat. Commun. 2018, 9, 5451. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Fisher, N.L.; Burgess, L.; Toussoun, T.; Nelson, P.E. Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology 1982, 72, 151–153. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; Blackwell Publishing Professional: Malden, MA, USA, 2006. [Google Scholar]
- Costa, M.; Sandoval-Denis, M.; Moreira, G.; Kandemir, H.; Kermode, A.; Buddie, A.; Ryan, M.; Becker, Y.; Yurkov, A.; Maier, W. Known from trees and the tropics: New insights into the Fusarium lateritium species complex. Stud. Mycol. 2024, 109, 403–450. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, D.-W.; Cui, W.-L.; Zhu, L.-H.; Huang, L. Morphological and phylogenetic analyses reveal three new species of Fusarium (Hypocreales, Nectriaceae) associated with leaf blight on Cunninghamialanceolata in China. MycoKeys 2024, 101, 45. [Google Scholar] [CrossRef] [PubMed]
- Carroll, G. The biology of endophytism in plants with particular reference to woody perennials. Microbiol. Phyllosphere 1986, 203–222. [Google Scholar]
- Ma, L.J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Denis, M.; Guarnaccia, V.; Polizzi, G.; Crous, P.W. Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 2018, 40, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Kharwar, R.N.; Mishra, A.; Gond, S.K.; Stierle, A.; Stierle, D. Anticancer compounds derived from fungal endophytes: Their importance and future challenges. Nat. Prod. Rep. 2011, 28, 1208–1228. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Calabria, J.; Chen, H.W.; Somssich, M. The Arabidopsis thaliana-Fusarium oxysporum strain 5176 pathosystem: An overview. J. Exp. Bot. 2022, 73, 6052–6067. [Google Scholar] [CrossRef] [PubMed]
- McGrann, G.R.; Steed, A.; Burt, C.; Goddard, R.; Lachaux, C.; Bansal, A.; Corbitt, M.; Gorniak, K.; Nicholson, P.; Brown, J.K. Contribution of the drought tolerance-related Stress-responsive NAC 1 transcription factor to resistance of barley to Ramularia leaf spot. Mol. Plant Pathol. 2015, 16, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.F.; Ma, P.; Wang, X.H.; Zhang, L.Q.; Li, Q.D.; Wang, J.L.; Cheng, Z.Y.; Yang, C.R. The studies of fungal population and relationship between fungi and forming of dragons blood resin in Dracaena cochinchinensis. Plant Divers. 1995, 17, 1. [Google Scholar]
- Wang, X.; Zhang, C.; Yang, L.; Yang, X.H.; Lou, J.D.; Cao, Q.E.; Gomes-Laranjo, J. Enhanced dragon’s blood production in Dracaena cochinchinensis by elicitation of Fusarium oxysporum strains. J. Med. Plants Res. 2010, 4, 2633–2640. [Google Scholar]
- Wang, X.H.; Gong, M.; Tang, L.; Zheng, S.; Lou, J.D.; Ou, L.C.; José, G.L.; Zhang, C.H. Cloning, bioinformatics and the enzyme activity analyses of a phenylalanine ammonia-lyase gene involved in dragon’s blood biosynthesis in Dracaena cambodiana. Mol. Biol. Rep. 2013, 40, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Le, T.N.-G.; Nguyen, T.H. First report of emerging fungal pathogens of Cordyceps militaris in Vietnam. Sci. Rep. 2023, 13, 17669. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B.; List, F.B.C.A.; Bolchacova, E. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed]
- Laurence, M.H.; Summerell, B.A.; Burgess, L.W.; Liew, E.C. Genealogical concordance phylogenetic species recognition in the Fusarium oxysporum species complex. Fungal Biol. 2014, 118, 374–384. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Sutton, D.A.; Rinaldi, M.G.; Sarver, B.A.; Balajee, S.A.; Schroers, H.-J.; Summerbell, R.C.; Robert, V.A.; Crous, P.W.; Zhang, N. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 2010, 48, 3708–3718. [Google Scholar] [CrossRef] [PubMed]
- Geiser, D.M.; del Mar Jiménez-Gasco, M.; Kang, S.; Makalowska, I.; Veeraraghavan, N.; Ward, T.J.; Zhang, N.; Kuldau, G.A.; O’donnell, K. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004, 110, 473–479. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Nirenberg, H.I.; Aoki, T.; Cigelnik, E. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 2000, 41, 61–78. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistlerr, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef] [PubMed]
- Reeb, V.; Lutzoni, F.; Roux, C. Contribution of RPB2 to multilocus phylogenetic studies of the Euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Mol. Phylogenet. Evol. 2004, 32, 1036–1060. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Q.; Su, P.; Xiao, J.; Lou, F.; Huang, X.; Yang, Z.; Chen, B.; Shen, P.; Wang, Y. Phylogenomic, Morphological, and Phylogenetic Evidence Reveals Five New Species and Two New Host Records of Nectriaceae (Hypocreales) from China. Biology 2025, 14, 871. https://doi.org/10.3390/biology14070871
Fan Q, Su P, Xiao J, Lou F, Huang X, Yang Z, Chen B, Shen P, Wang Y. Phylogenomic, Morphological, and Phylogenetic Evidence Reveals Five New Species and Two New Host Records of Nectriaceae (Hypocreales) from China. Biology. 2025; 14(7):871. https://doi.org/10.3390/biology14070871
Chicago/Turabian StyleFan, Qi, Pingping Su, Jiachen Xiao, Fangwei Lou, Xiaoyuan Huang, Zhuliang Yang, Baozheng Chen, Peihong Shen, and Yuanbing Wang. 2025. "Phylogenomic, Morphological, and Phylogenetic Evidence Reveals Five New Species and Two New Host Records of Nectriaceae (Hypocreales) from China" Biology 14, no. 7: 871. https://doi.org/10.3390/biology14070871
APA StyleFan, Q., Su, P., Xiao, J., Lou, F., Huang, X., Yang, Z., Chen, B., Shen, P., & Wang, Y. (2025). Phylogenomic, Morphological, and Phylogenetic Evidence Reveals Five New Species and Two New Host Records of Nectriaceae (Hypocreales) from China. Biology, 14(7), 871. https://doi.org/10.3390/biology14070871




