Simple Summary
Xanthomonas is a large genus of plant-associated bacteria that cause disease on various crops. But their involvement in ornamental plant diseases, especially in the Araceae and Araliaceae families, has been less studied. This review compiles recent information about their taxonomy, phylogenetic relationships, and potential virulence mechanisms of Xanthomonas strains affecting these ornamental hosts. Using advanced tools, researchers have reclassified many of these strains into distinct species and pathovars and revealed their evolutionary relationships. Despite this progress, the placement of several strains remains unresolved, especially those from lesser-studied genera like Epipremnum and Rhaphidophora. The review also discusses genes associated with bacterial pathogenicity, such as those involved in secretion systems, exopolysaccharide production, and host cell wall degradation. These molecular insights, combined with future genomic and functional studies, are essential to better define species boundaries and how Xanthomonas adapts and causes disease in different plant hosts. This knowledge is key to understanding how these bacteria cause disease in ornamental plants and to guiding future research and control efforts.
Abstract
The genus Xanthomonas (family Xanthomonadaceae) comprises 39 validly published species and is associated with a broad host range, infecting hundreds of monocot and dicot plants worldwide. While many Xanthomonas species are notorious for causing leaf spot and blight diseases in major agricultural crops, less attention has been given to their impact on ornamental plants. In Hawaii and other key production regions, xanthomonads have posed persistent threats to popular ornamentals in the Araceae and Araliaceae families. This review synthesizes the evolving phylogenetic and taxonomic framework of Xanthomonas strains isolated from Araceae and Araliaceae, highlighting recent advances enabled by multilocus sequence analysis and whole genome sequencing. We discuss the reclassification of key pathovars, unresolved phylogenetic placements, and the challenges of pathovar delineation within these plant families. Additionally, we examine current knowledge of molecular determinants of pathogenicity, including gene clusters involved in exopolysaccharide and lipopolysaccharide biosynthesis, flagellar assembly, cell-wall-degrading enzymes, and secretion systems (types II, III, and VI). Comparative genomics and functional studies reveal that significant gaps remain in our understanding of the genetic basis of host adaptation and virulence in these xanthomonads. Addressing these knowledge gaps will be crucial for developing effective diagnostics and management strategies for bacterial diseases in ornamental crops.
1. Introduction to Xanthomonas
The genus Xanthomonas (from Greek: xanthos, “yellow”, and monas, “entity”) comprises Gram-negative, rod-shaped, aerobic, polar monotrichous, and xanthomonadin-producing bacteria. Xanthomonas belongs to the family Xanthomonadaceae (syn. Lysobacteraceae), order Xanthomonadales (syn. Lysobacterales), class Gammaproteobacteria, and phylum Pseudomonadota (formerly Proteobacteria) [1,2,3,4]. The genus currently includes 39 validly published species and several others with pending or invalid status [5] (LPSN, accessed 22 June 2025). The type species is Xanthomonas campestris [1].
Xanthomonas is a major group of phytopathogenic and plant-associated bacteria, collectively infecting more than 400 plant species across both monocots and dicots [6,7,8,9,10,11]. Many species and pathovars cause economically significant diseases worldwide. For example, X. oryzae pathovars cause bacterial leaf blight and leaf streak of rice (Oryza sativa), X. campestris pv. campestris causes black rot in crucifers, and X. citri pv. citri and X. citri pv. malvacearum cause citrus canker and cotton bacterial blight, respectively. The bacterial leaf spot of tomatoes and peppers is caused by four xanthomonads: X. euvesicatoria pv. euvesicatoria, X. euvesicatoria pv. perforans, X. hortorum pv. gardneri, and X. vesicatoria. Other notable diseases include the bacterial spot and canker of stone fruits and almonds (X. arboricola pv. pruni) and walnut blights (X. arboricola pv. juglandis) [12,13,14,15].
In addition to food crops, Xanthomonas spp. also cause substantial losses in ornamentals, affecting plants such as anthurium (Araceae), begonia (Begoniaceae), hibiscus (Malvaceae), poinsettia (Euphorbiaceae), pelargonium and geranium (Geraniaceae), English ivy (Araliaceae), and zinnia (Asteraceae) [16,17,18]. This review focuses on bacterial leaf blight and spot diseases caused by Xanthomonas spp. on economically important ornamentals in the Araceae and Araliaceae families, particularly those affecting Anthurium spp. (Araceae) and Polyscias spp. (Araliaceae) in Hawaii, Florida, California, and other major production regions [19,20,21,22,23,24]. Typical leaf blight and spot symptoms on A. andraeanum and P. guilfoylei are shown in Figure 1.

Figure 1.
Leaf blight (white circle) and leaf spot (white arrow) symptoms on infected plants in the Araceae and Araliaceae families. (A) Water-soaked lesions of leaf spot and leaf blight on the leaves of Anthurium andraeanum caused by X. phaseoli pv. dieffenbachiae (syn. X. axonopodis pv. dieffenbachiae and X. campestris pv. dieffenbachiae). (B) Water-soaked lesions surrounding of leaf spot and leaf blight symptoms on the leaves of Polyscias guilfoylei caused by X. euvesicatoria pv. polysciadis (formerly X. hortorum pv. hederae).
2. Taxonomy, Host Range, and Phylogeny of Xanthomonas
Bacterium hyacinthi [25], later reclassified as Xanthomonas hyacinthi [26], was the first recognized xanthomonad plant pathogen. Early plant-pathogenic bacteria were classified as Pseudomonas or Phytomonas based on colony pigmentation and pathogenicity [27]. Dowson [1] established the genus Xanthomonas for Gram-negative, yellow-pigmented, polar-flagellated bacteria, distinguishing them from Bacterium and Pseudomonas.
Later, additional xanthomonads were described as pathogens with specific host ranges, highlighting their genetic and phenotypic diversity. Initially, distinguishing Xanthomonas species was challenging because biochemical and physiological features alone provided insufficient resolution [28]. As a result, early taxonomy relied on the “new host/new species” concept, where isolates from different hosts were classified as separate species [28,29,30]. This host-based approach led to a proliferation of species, with 60 recognized by Burkholder in 1957 and over 200 reported by 1974. Before the Eighth Edition of Bergey’s Manual [31], it was recognized that many of these species lacked sufficient bacteriological and physiological distinction. Consequently, the taxonomy was revised, consolidating the genus into five species: X. campestris, X. albilineans, X. ampelina, X. axonopodis, and X. fragariae. Subsequently, X. ampelina was transferred to the genus Xylophilus as Xylophilus ampelina [32]. To retain meaningful distinctions among plant-pathogenic strains, the pathovar system was introduced, allowing differentiation based on host range within the broader species framework [33].
Over time, more genotypic analyses—including GC content determination, DNA fingerprinting, DNA–DNA hybridization (DDH), ribosomal RNA sequencing, and multilocus sequence analysis (MLSA)—were developed and combined with morphological, biochemical, and physiological characteristics to clarify the taxonomy and nomenclature of Xanthomonas [26,34,35,36]. Based on DDH characterization, Vauterin et al. [26] proposed reclassifying xanthomonads into 20 species. The classification by DDH corresponded with the grouping results obtained by amplified fragment length polymorphism (AFLP) and repetitive-element palindromic polymerase chain reaction (rep-PCR) [34,35]. In contrast, a phylogenetic analysis of the 16S rRNA gene showed limited taxonomic resolution among the 20 Xanthomonas species, grouping 15 species into a large cluster, along with two other clusters (the X. sacchari cluster and the X. albilineans–X. translucens–X. hyacinthi cluster). This lack of resolution was attributed to the highly conserved nature of the 16S rRNA sequence within Xanthomonas [37].
Although the polyphasic approach described above was implemented to achieve better resolution of Xanthomonas spp., debate and controversy have persisted [38,39]. Additional species were described and published using valid and sometimes invalid criteria (https://lpsn.dsmz.de/genus/xanthomonas, accessed on 22 June 2025). In today’s context, the whole genome sequencing technology provides more detailed and comprehensive genomic information, setting a higher standard for species delineation. The current major standards for delineating bacterial species include cut-off values of 70% digital DNA–DNA hybridization (dDDH) and 95% average nucleotide identity (ANI) [40,41,42,43]. Nevertheless, a standard is still lacking for designating subspecific levels based on genomic relatedness, and according to Young et al. [39] and Constantin et al. [14], the resolution of pathovars in phylogenetic trees remains inadequate. The term “pathovar” technically refers only to a group of strains with distinct pathogenicity on a specific host or host range and does not represent a formal taxonomic rank. Such a designation must be supported by data from pathogenicity and host range tests. In this context, we describe the nomenclatural evolution of Xanthomonas strains associated with hosts belonging to the Araceae and Araliaceae (Figure 2).
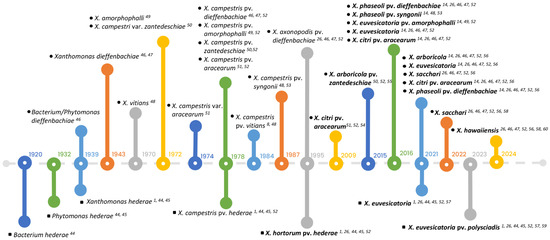
Figure 2.
The history of nomenclatural changes of xanthomonads isolated from Araceae and Araliaceae from 1920 to 2023. The upper part of the timeline shows the identification and reclassification of strains isolated from Araceae (indicated by black circles), while the lower part highlights the changes of strains isolated from Araliaceae (represented by black squares). Recent taxonomic names for strains are highlighted in bold. References for each event related to the strains are indicated by superscript numbers and are listed consecutively: [1,8,14,26,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60].
2.1. Xanthomonads Associated with the Araceae
The first report of a bacterial leaf blight disease affecting the Araceae was a leaf blight of Dieffenbachia picta (syn. Dieffenbachia seguine) in New Jersey (USA), originally attributed to Bacterium dieffenbachiae (syn. Phytomonas dieffenbachiae) [46]. The pathogen was later renamed Xanthomonas dieffenbachiae [47]. In 1970, Wehlburg described another xanthomonad isolated from Syngonium podophyllum and identified the causal agent of leaf blight as X. vitians, a bacterium primarily known for causing leaf spot on lettuce, but which had not yet been reported on Syngonium. In 1972 and 1974, three xanthomonads isolated from other genera of Araceae were described: X. amorphophalli from Amorphophallus campanulatus in India [49], X. campestris var. zantedeschiae from Zantedeschia aethiopica in South Africa [50], and X. campestris var. aracearum from Xanthosoma sagittifolium in Guadeloupe [51]. Furthermore, due to their similar results in bacteriological tests—including physiological and biochemical reactions resembling those of X. campestris—it was recommended that these four xanthomonads be renamed at the subspecific level, using the term “pathovar” rather than “variant.” Thus, they were named X. campestris pv. amorphophalli, X. campestris pv. aracearum, X. campestris pv. dieffenbachiae, and X. campestris pv. zantedeschiae [52].
The host range of the four aforementioned pathovars covers ten species in eight aroid genera, including Aglaonema, Amorphophallus, Anthurium, Dieffenbachia, Philodendron, Syngonium, Xanthosoma, and Zantedeschia. One Syngonium strain was identified as X. campestris pv. vitians, which mainly causes leaf spot on lettuce [8]. Six other strains isolated from Syngonium podophyllum cultivars ‘Cream’ and ‘White Butterfly’ were distinguished from pathovars dieffenbachiae, zantedeschiae, and vitians on aroids and were proposed as X. campestris pv. syngonii [53]. Notably, X. campestris pv. dieffenbachiae was isolated from six araceous species: Aglaonema, Anthurium, Dieffenbachia, and Philodendron, as well as from Dracaena fragrans in the Asparagaceae family [8,33,61]. Subsequently, the extended host range of X. campestris pv. dieffenbachiae included Xanthosoma sagittifolium, an edible aroid commonly called cocoyam [62], as well as Colocasia, Alocasia, Caladium, Epipremnum, Spathiphyllum, and Rhaphidophora [61]. In contrast, X. campestris pv. zantedeschiae and X. campestris pv. syngonii showed host specificity on Zantedeschia aethiopica and Syngonium podophyllum, respectively [50,53].
With the introduction of DDH analyses, the pathovar dieffenbachiae in X. campestris was reclassified into X. axonopodis, while pathovars syngonii and zantedeschiae retained their original classification under X. campestris [26]. Ah-You et al. [54] proposed that some aroid strains, rather than anthurium strains, should be reclassified under X. citri based on DDH, MLSA (atpD, dnaK, and gyrB), and amplified fragment length polymorphism (AFLP) analyses. Further MLSA analyses and whole genome studies were conducted on aroid strains, and the pathovar zantedeschiae, known for its host specificity to Zantedeschia, was placed in X. arboricola [55] whereas pathovar dieffenbachiae infecting Dieffenbachia, Aglaonema, and primarily Anthurium, and pathovar syngonii restricted to Syngonium were reclassified under X. phaseoli [14,56,57,63]; pathovar amorphophalli isolated from Amorphophallus and some strains originally isolated from Philodendron, Dieffenbachia, and Caladium were redesignated as X. euvesicatoria [14,56,57]; pathovar aracearum isolated from various Araceae genera was placed under X. citri, and four other strains isolated from Anthurium, Aglaonema, Colocasia, and Spathiphyllum were grouped with X. sacchari [14,54,56,57,58,63]. In the maximum likelihood phylogenetic tree (Figure 3) presented in Chaung et al. [57], over fifty aroid strains were distributed among six Xanthomonas species groups (X. arboricola, X. campestris, X. citri, X. euvesicatoria, X. phaseoli, and X. sacchari), aligning closely with groupings reported in previous studies. Meanwhile, three additional aroid strains clustered within the Stenotrophomonas spp. group and were subsequently identified as two new species, S. aracearum and S. oahuensis [57,60]. Furthermore, Chuang et al. [60] identified the spathiphyllum and the colocasia strains, formerly in the X. sacchari phylogenetic group, as a novel species, X. hawaiiensis, based on ANI and dDDH values lower than the delineation of new species. Notably, some strains of the phylogenetically related species, X. sacchari, were considered as commensal bacteria because of the absence of type III secretion system (T3SS) and type III effectors (T3Es) [64,65]. The pathogenicity of X. hawaiiensis on Araceae remains uncertain.
2.2. Xanthomonads Associated with the Araliaceae
The causal agent of bacterial leaf spot disease on Hedera helix L. (English ivy), an araliaceous plant, was first identified as Bacterium hederae in France [44] and later renamed Phytomonas hederae by Burkholder and Guterman in 1932 [45]. After Dowson [1] designated the genus Xanthomonas for monotrichous bacteria producing a yellow xanthomonadin pigment (among other bacteriological characteristics), P. hederae was changed to Xanthomonas hederae. With the introduction of pathovar designations for Xanthomonas, the strains isolated from H. helix were designated as the pathovar X. campestris pv. hederae [52]. DNA homology data and Biolog characteristics supported the grouping of pathovar hederae with two other pathovars, pelargonii and vitians. Consequently, all three pathogens were reclassified into a new species, X. hortorum, and named X. hortorum pv. hederae, X. hortorum pv. pelargonii, and X. hortorum pv. vitians [26].
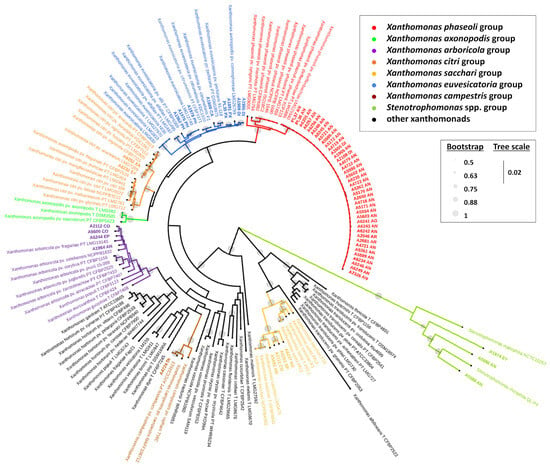
Figure 3.
Phylogenetic tree of Xanthomonas spp. based on the concatenated nucleotide sequences of the partial regions of five housekeeping genes (atpD, dnaA, dnaK, gltA, and gyrB). The abbreviations in the strain names are the following: AG = Aglaonema; AN = Anthurium; AR = Aroid; CA = Caladium; CO = Colocasia; DI = Dieffenbachia; EP = Epipremnum; PH = Philodendron; PX = Polyscias guilfoylei (Panax); SP = Spathiphyllum; SY = Syngonium; XA = Xanthosoma; PT = pathotype strain; and T = type strain. The names of the strains isolated from Araceae and Araliaceae are shown in bold. Reproduced, by permission, from [57,60], Xanthomonas strains isolated from hosts in the Araceae reveal diverse phylogenetic relationships and origins. Phytopathology, 114(8):1963-1974. © The American Phytopathological Society.
Since 1920, leaf spot diseases have been reported worldwide on various Hedera species and cultivars. These include 12 H. helix varieties in the United States [66]; H. helix and H. canariensis in Japan [67]; H. helix in New Zealand, Korea, Greece, Turkey, and Taiwan [68,69,70,71]; H. hibernica in Slovenia [72]; and H. nepalensis var. sinensis in China [73]. The host range of this pathogen has expanded to other members of the Araliaceae family, such as Heptapleurum actinophyllum (syn. Brassaia actinophylla and Schefflera actinophylla; “Heptapleurum actinophyllum (Endl.) [74,75], Fatsia japonica, Plerandra elegantissima (syn. Dizygotheca elegantissima and Schefflera elegantissima); [74], various Polyscias spp. (balfouriana, chinensis, fabian, fruticosa, guilfoylei, and unknown species), and Schefflera arboricola [19,22,24,76]. Additionally, Tolba [76] tested the host range of Xanthomonas strains originally isolated from H. actinophyllum (S. actinophylla), and all eight strains were pathogenic on S. arboricola, P. elegantissima (S. elegantissima), and H. helix, but not on F. japonica. This finding was not supported by an earlier study [19]. These eight strains also failed to infect two araliaceous plants, Aralia nudicaulis and Dendropanax trifidus. The strain originally isolated from H. canariensis did not infect D. trifidus and Tetrapanax papyriferum [67]. Thus, the taxonomy of these pathogens remains uncertain.
Notably, Norman et al. [22] described the heterogeneity of xanthomonads isolated from various genera in the Araliaceae family, including Hedera, Brassaia (syn. Heptapleurum), Polyscias, and Schefflera. Based on metabolic fingerprints, fatty acid methyl ester profiles, and restriction fragment length polymorphism (RFLP) analyses, dendrogram clustering suggested that these strains should be divided into two groups: one primarily comprising Hedera strains and the majority of the others as Polyscias strains. Although advanced genotypic analyses have been recommended to verify the phylogeny and taxonomy of these xanthomonads, strains isolated from Araliaceae hosts other than ivy have not been comprehensively examined using DNA homology or genomic comparison analyses since 1984 [22,76]. Therefore, X. hortorum pv. hederae has been identified as the sole causal agent of bacterial leaf spot and leaf blight affecting six genera in the Araliaceae family: Hedera, Heptapleurum, Fatsia, Plerandra, Polyscias, and Schefflera.
Chuang et al. [56,57] proposed that panax strains PL35 and A1891, isolated from P. guilfoylei in Hawaii, are distinct from X. hortorum phylogenetic clade and belong to a X. euvesicatoria phylogenetic clade, as determined by MLSA concatenating five housekeeping genes (Figure 3). Moreover, the Genome BLAST Distance Phylogeny (GBDP) approach and genomic relatedness values (>98% ANI and >86.7% dDDH) (Figure 4) indicated PL35 should be reclassified as X. euvesicatoria [56]. Additionally, Wang et al. [59] classified strains isolated from P. guilfoylei in Taiwan as X. euvesicatoria based on MLSA. They suggested assigning a novel pathovar, polysciadis, based on host specificity to P. guilfoylei and potentially other Araliaceae members, rather than to Solanum lycopersicum, Euphorbia milii, or Ixora × westii from other families.
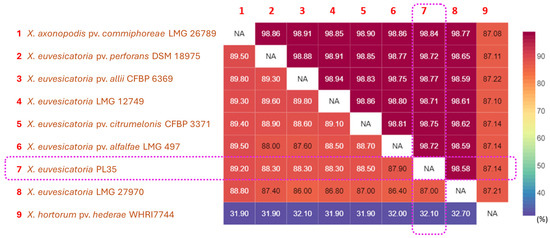
Figure 4.
Pairwise comparison heatmap of ANI and dDDH among whole genome sequences of the panax strain PL35, the type strain of X. hortorum pv. hederae, and other related pathovars within X. euvesicatoria phylogenetic clade. Pairwise ANI values were calculated using CLC Genomics Workbench 22.0.2 (CLC Bio-Qiagen, Arahus, Denmark), while pairwise dDDH values were inferred using the Genome-Genome Distance Calculator (GGDC) v3.0 on the Type Strain Genome Server (TYGS) web server (https://tygs.dsmz.de/, accessed on 22 June 2025) [77,78]. Heatmaps of ANI (%) and dDDH (%) were visualized using displayR (https://www.displayr.com/, accessed on 22 June 2025). The upper triangular portion of the matrix displays ANI values, whereas the lower triangular portion displays dDDH values. The values in the matrix and heatmap bar are shown as percentages (%). The values inside the purple frames indicate that the panax strain PL35 belongs to X. euvesicatoria, based on the species boundary thresholds of 95% for ANI and 70% for DDH.
3. Virulence Mechanisms
Successful infection of a plant by xanthomonads involves the processes of attachment, penetration, and multiplication within the plant; therefore, genes encoding exopolysaccharides (EPS), lipopolysaccharides (LPS), pili, flagella, cell-wall-degrading enzymes (CWDE), and type secretion systems (TSSs) are critical for virulence and pathogenicity [79,80,81,82,83]. The schematic representation of TSSs in xanthomonads is shown in Figure 5. The genetic makeup of pathogenicity determinants in the whole genomes of xanthomonads isolated from the Araceae and Araliaceae families is presented in Table 1. Studies on the pathogenetic mechanisms of Xanthomonas-associated Araceae and Araliaceae remain limited. Therefore, we summarized previous studies on how genes encoding pathogenicity and/or virulence factors influence the interactions between Xanthomonas spp. and their host plants. We did not include the T1SS and T5SS in this review due to the unavailability of recent genomic studies on xanthomonads associated with Araceae and Araliaceae.

Table 1.
Overview of pathogenicity-related gene clusters in Xanthomonas species/pathovars isolated from Araceae and Araliaceae.
3.1. Exopolysaccharides (EPS) and Lipopolysaccharides (LPS)
A defining characteristic of xanthomonads is the production of xanthan, an EPS with pentasaccharide-repeating units which forms a biofilm matrix enabling the pathogen to tolerate environmental stress [84]. A cluster of 12 gum genes, from gumB to gumM, plays an essential role in the assembly of lipid-linked intermediates, polymerization, and secretion of xanthan [85,86]. The gum genetic loci, spanning approximately 15 kb, are highly conserved and exhibit synteny in almost all Xanthomonas species. This includes Araceae strains such as X. citri pv. aracearum, X. euvesicatoria, X. phaseoli pv. dieffenbachiae, and X. phaseoli pv. syngonii [63]. The exceptions are X. albilineans and X. theicola, which lack the entire gum gene cluster [87,88,89,90], while the gumN, gumO, and gumP genes are absent in X. fragariae [91]. The gumD gene, along with gumM, gumH, gumK, and gumI, encodes glycosyltransferases that synthesize a lipid-linked pentasaccharide and is involved in the virulence of X. campestris pv. campestris on broccoli and cabbage [86,92] as well as virulence of X. oryzae pv. oryzae on rice [93]. The gumB, gumC, gumE, and gumJ genes are responsible for xanthan formation and translocation. The defective mutants of these four genes, along with gumM in X. campestris pv. campestris, potentially resulted in cell lethality due to the accumulation of toxic lipid-linked intermediates [86]. Additionally, knockout mutants of gumE, gumI, and gumJ were lethal in X. hortorum pv. vitians [89], and gumJ was also crucial for the survival of X. oryzae pv. oryzae [93].
LPS is a crucial component of the outer membrane (OM) of Gram-negative bacteria, consisting of a hydrophobic lipid A, a hydrophilic core oligosaccharide, and an O-antigen (OA) polysaccharide chain. It protects bacteria from many toxic compounds and is recognized as pathogen-associated molecular patterns (PAMPs) during interactions with plants and animals [80,94]. The LPS gene cluster, i.e., the wxc gene cluster, is located between the highly conserved metB and etfA genes, which encode for cystathionine gamma-synthase and the electron transport flavoprotein subunit alpha proteins, respectively [95,96]. Situated between etfA and metB, the number of genes and their sequence similarity exhibit significant variability across Xanthomonas genomes, with the exception of two conserved genes, wzm and wzt. These genes are involved in the ATP-binding cassette (ABC) transporter pathway [97,98].
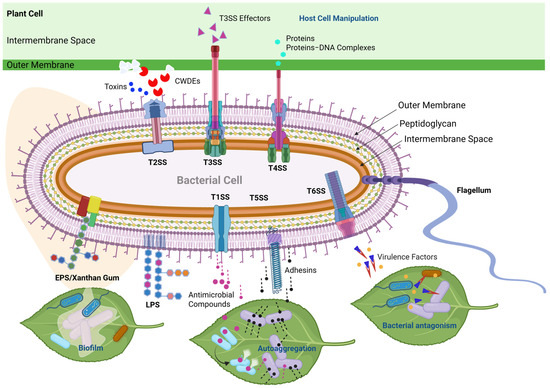
Figure 5.
Schematic representation of type secretion systems (TSSs) and other virulence factors in Xanthomonas species. The T1SS serves primarily to transport various antimicrobial compounds directly from the cytoplasm to the extracellular environment [83,99]. The T2SS is a specialized secretory pathway used to export plant-cell-wall-degrading enzymes (CWDEs) as well as toxins into the extracellular space [100]. The T3SS bridges the inner and outer membranes in order to inject or translocate T3SS effector proteins directly into plant and animal cells [83,101,102,103]. The T4SS is used for bacterial conjugation and the transfer of proteins/effectors, DNA, or protein–DNA complexes from bacteria to prokaryotic or eukaryotic target cells [81,104]. The T5SS is primarily involved in exporting proteins, such as adhesins, which is crucial for host colonization and the establishment of infections in plant tissues [105]. The T6SS has been associated with contact-dependent interbacterial antagonism in Xanthomonas [106,107]. Moreover, the versatile T6SS also secretes virulence factors in eukaryotic hosts [108,109]. EPS/Xanthan Gum is important for the biofilm matrix, enabling the pathogen to tolerate environmental stress and protect against other microbes [84]. LPS protects bacteria from many toxic compounds and is recognized as pathogen-associated molecular patterns (PAMPs) during interactions with plants [80,94].
In aroid strains, variability in LPS was observed, ranging from the shortest length of 19.7 kb in X. citri pv. aracearum LMG 7399 to the longest length of 25.9 kb in X. phaseoli pv. dieffenbachiae LMG 25940 [63]. Both the wzm gene, encoding the O-antigen-permease protein, and the wzt gene, encoding the teichoic ATP-binding protein, were present in X. phaseoli pv. dieffenbachiae LMG 25940, X. phaseoli pv. syngonii LMG 9055, and X. citri pv. aracearum LMG7399. However, only the gene encoding the teichoic ATP-binding protein was found in X. euvesicatoria LMG 12749 [63]. When comparing the type strain X. hortorum pv. hederae CFBP 4925T isolated from ivy (Araliaceae) with other pathovars, X. hortorum pv. cynarae CFBP 4188PT (formerly named X. cynarae) from artichoke and X. hortorum pv. gardneri ATCC 19865PT (formerly named X. gardneri) from tomato, wzm and wzt homologs were identified in all three genomes. Average nucleotide differences of 69.5 in the metB genes and 24 in the etfA genes were observed [110].
The OA precursor gmd (GDP-mannose 4,6-dehydratase encoding gene) and rmd (GDP-4-dehydro-D-rhamnose reductase encoding gene) were reported in X. axonopodis pv. malvacearum, X. campestris pv. campestris ATCC 33913, X. campestris pv. armoraciae 756C, X. euvesicatoria 85–10, X. fuscans subsp. fuscans 4834-R, and three pathovars of X. hortorum (pv. hederae CFBP 4925T, pv. cynarae CFBP4188PT, and pv. gardneri ATCC19865PT) but not in X. citri pv. citri strain 306 or the two X. oryzae pathovars strains BLS 256 and MAFF 311018 [95,110,111,112]. The highly polymorphic LPS loci were attributed to multiple horizontal gene transfer (HGT) and recombination events, especially because several insertion sequence (IS) elements were found in X. oryzae pv. oryzicola BLS 256, X. oryzae pv. oryzae MAFF 311018, and X. campestris pv. campestris ATCC 33913 and 8004 [111,113].
In X. citri pv. citri, which causes citrus canker disease, gene disruption mutants of wxacO (encoding a putative transmembrane protein), rfbC (encoding a truncated O-antigen biosynthesis protein), and nlxA (encoding a putative glycosyltransferase) within the LPS cluster were found to be more sensitive to the antimicrobial peptide polymyxin B, hydrogen peroxide, and other environmental stresses. These mutants also showed impaired swimming and swarming motility and exhibited reduced virulence on grapefruit leaves when spray inoculated [114,115]. Interestingly, mutations in the LPS biosynthesis genes influenced EPS production, as LPS and EPS share some common genetic determinants including precursor molecules at the level of nucleotide sugar metabolism in polysaccharide biosynthesis [95,115,116]. The yield of xanthan produced by the deficient mutants of the wxcB, wxcK, and wxcN genes in X. campestris pv. campestris increased, while the yield of EPS produced by the nlxA mutant of X. citri pv. citri decreased [115,116].
3.2. Type II Secretion System (T2SS)
The Type II secretion system (T2SS) is a highly specialized secretory pathway utilized by Gram-negative bacteria to export plant-cell-wall-degrading enzymes (CWDEs) such as cellulases, pectinases, and xylanases, along with toxins, lipases, proteases, and phospholipases into the extracellular space [100]. The T2SS is comprised of two distinct gene clusters, xcs and xps, which are conserved within the proteobacterial family, including Xanthomonas. However, only the intact xps cluster is present in X. albilineans, X. hyacinthi, X. sacchari, X. theicola, X. fragariae, X. populi, X. bromi, X. axonopodis pv. vasculorum, X. vasicola pathovars vasculorum and musacearum (syn. X. axonopodis pv. musacearum), and X. oryzae pathovars oryzae and oryzicola [83,111,117,118]. The xcs gene cluster contains 12 genes, including the xcsC gene encoding a PDZ-domain-containing protein and genes from xscD/gspD to xscN/gspN. On the other hand, the xps gene cluster is comprised of 11 genes, including xpsE/gspE to xpsK/gspK, the xpsL gene encoding a pilN-domain-containing protein, and genes from xpsM/gspM to xpsN/gspN, along with xpsD/gspD. Both the xcs and xps gene clusters are highly conserved among xanthomonads isolated from the Araceae and Araliaceae families, including X. citri pv. aracearum, X. euvesicatoria, X. phaseoli pv. dieffenbachiae, X. phaseoli pv. syngonii, and X. hortorum pv. hederae [63,118].
The virulence function of xps gene clusters has appeared in several xanthomonads. However, there have been no reports demonstrating the contribution of xcs genes to virulence, except for a study by Szczesny et al. [119], which suggested that xcs genes could partially complement the homologous xpsE and xpsD genes [83]. Hu et al. [120] investigated ten Xps pleiotropic mutants of X. campestris pv. campestris, which were non-pathogenic to cabbage and exhibited reduced accumulation of amylase, endoglucanase, and polygalacturonate lyase. Baptista et al. [121] demonstrated that the absence of the xpsD gene in X. axonopodis pv. citri (syn. X. citri pv. citri), which is crucial for the secretion of various enzymes including cellulases, resulted in the reduced virulence of the pathogen on citrus. Similarly, reduced symptoms on rice were observed in xpsF and xpsD mutants of X. oryzae pv. oryzae due to an export deficiency of xylanase [122,123]. In X. campestris pv. vesicatoria 85-10 (heterotypic syn. X. euvesicatoria), the xpsD gene was found to contribute significantly to virulence, unlike the xcsD genes, while the xpsE gene influenced T3SS-dependent effector protein translocation [119]. Furthermore, defective mutants of T2SS secreted substrates, such as xylanase and protease, which were reported to affect the virulence of X. campestris pv. vesicatoria in tomatoes [124].
3.3. Type III Secretion System (T3SS)
The Type III secretion system (T3SS), consisting of more than 20 proteins encoded by hrp (hypersensitive response and pathogenicity) and hrc (hrp conserved) genes, is a contact-dependent pathway. It forms an apparatus located between the inner and outer membranes to inject or translocate proteins into plant and animal cells [83,101,102,103]. Most T3SS membrane-associated proteins are conserved among different pathogenic bacteria [125], and nine of these proteins, including HrcC, HrcJ, HrcN, and HrcQ to HrcV, are highly conserved hrc-gene-encoded proteins in plant and animal pathogens [126]. Interestingly, genes encoding components of the Type III secretion system (T3SS) share high homology with clustered genes involved in flagellar biosynthesis [103]. Among the Xanthomonas species, the manipulation of pathogenicity by the protein repertoire, termed type III effectors (T3Es,) secreted via T3SS [127], has been studied for more than two decades. White et al. [128] categorized T3Es into 39 classes based on sequence relatedness, identifying various biochemical or structural motifs, including glycerolphosphoryl diester phosphodiesterase, site-specific DNA binding, nuclear localization, M27 zinc protease, and tyrosine phosphatase, among others. A comparison of the genomes from seven Xanthomonas species revealed that eleven T3E groups, such as avirulence Bs2 (AvrBs2), Xanthomonas outer protein F (XopF), XopK-N, XopP-R, XopX, and XopZ, were commonly present. In contrast, XopB, XopD, XopT-U, XopW, XopAC, XopAF, XopAH, XopAJ, and XopAK showed restricted distribution across the seven genomes [128].
The Type III secretion system (T3SS) has been confirmed to play a crucial role as a pathogenicity determinant, with extensive research focused on gene function. Interestingly, it has been reported that some non-pathogenic Xanthomonas strains lack the Hrp T3SS and its associated substrates. These include X. sacchari NCPPB 4393 and R1 strains [64,65], the X. maliensis type strain CFBP 7942 [129], the sugarcane-pathogenic X. albilineans GPE PC73 strain [130], and the cannabis-infecting X. cannabis pv. cannabis strains NCPPB 2877 and NCPPB 3753 [131]. Merda et al. [132] analyzed the T3SS gene cluster and its upstream and downstream genomic contexts (within a 20 kb window) across 82 genomes of various Xanthomonas species/pathovars. They discovered similar hrp gene clusters in the genome of X. hortorum pv. hederae, comprising 20 genes isolated from the Araliaceae family, and in the genomes of X. arboricola pv. zantedeschiae (24 genes) and X. phaseoli pv. dieffenbachiae (22 genes), both isolated from the Araceae family. Genomic rearrangement events were observed on both sides of the 20 kb window in X. arboricola pv. zantedeschiae and X. phaseoli pv. dieffenbachiae. However, only one side of the 20 kb flanking region was analyzed in X. hortorum pv. hederae, which was presumed to be one of the main gene donors in the Maximum Likelihood (ML) phylogeny [132].
Almost all syntenic T3SS, located between the XopAE- and Hpa2-encoding genes, was displayed in four aroid strains. The core hrp gene cluster consisted of 27–30 genes encoding Hrp components, Xop effectors, and hypothetical proteins [63]. Notably, additional mobile elements located in the vicinity of the hrp cluster were observed in the X. euvesicatoria LMG 12749 strain from Philodendron but were absent in the other three aroid strains (LMG 25940 from Anthurium, LMG 7399 from Dieffenbachia, and LMG 9055 from Syngonium) [63]. Similar to X. campestris pv. vesicatoria 85-10, X. oryzae pv. oryzae PXO86, and X. fuscans subsp. fuscans 4834-R strains, mobile genetic elements flanking both sides of the hrp gene cluster were proposed to carry virulence genes and act as a pathogenicity island [112,133]. The highest number of T3Es was predicted in X. euvesicatoria LMG 12749 among the four aroid strains; however, the LMG 12749 strain was very weakly pathogenic to the tested aroids. In contrast, X. citri pv. aracearum LMG 7399 with 24 T3Es, X. phaseoli pv. syngonii LMG 9055 with 22 T3Es, and X. phaseoli pv. dieffenbachiae LMG 25940 with 20 T3Es caused disease on the tested aroids and shared some T3Es (XopE2, XopG, and XopAM) that were lacking in the LMG 12749 strain [63].
From previous studies, the hrp, hrc, hpa (hrp-associated), and effector genes involved in the T3SS have shown various effects on pathogenicity across different Xanthomonas–host pathosystems [134]. For the X. campestris pv. vesicatoria strain 85-10, which contains the avrBs1 avirulence gene, deletion mutants of hrpB1, hrpB2, hrpB4, and hrpB5 in the hrpB operon were unable to infect susceptible pepper plants or to induce a hypersensitive response (HR) in resistant pepper plants [134,135]. With the exception of the hrpF gene, mutants deficient in hrp-hrc genes (including hrpE, hrpD6, hrpD5, hrcS, hrcR, hrcQ, hrcV, hrcU, hrpB1, hrpB2, hrcJ, hrpB4, hrpB5, hrcN, hrpB7, hrcT, hrcC, hrpG, and hrpX) derived from X. oryzae pv. oryzae KACC10859 were unable to cause disease in rice. By contrast, the hrpF null mutant of X. campestris pv. vesicatoria demonstrated that HrpF is required for pathogenicity to pepper and acts as a translocon protein at the plant–pathogen interface [135]. Similar results were observed in the X. campestris pv. vesicatoria and soybean pathosystem [136]. Additionally, the hpaA, hpaF, hpaP, hpa1, and hpa2 disruption mutants of X. oryzae pv. oryzae KACC 10859 exhibited various levels of defective virulence, with the exception of the hpaB disruption mutant, which completely lost its pathogenicity.
3.4. Type IV Secretion System (T4SS)
The Type IV secretion system (T4SS) is a highly complex and versatile multiprotein system for bacterial conjugation and transferring proteins/effectors, DNA, and/or protein–DNA complexes from bacteria to prokaryotic or eukaryotic target cells [81,104]. Three types of T4SS loci are found on the chromosomes and/or plasmids of Gram-negative bacteria, such as the virB, trb, and avhB loci in Agrobacterium tumefaciens. In X. citri pv. citri, there are chromosomal virB loci and plasmid-borne twr/virB loci, while X. campestris pv. campestris possesses only a single virB cluster [113,137,138]. The canonical T4SS was assembled by 12 core proteins including the periplasmic transglycosylase VirB1, a periplasmic VirB7–VirB9–VirB10 repeated core complex, an inner membrane VirB3–VirB6–VirB8 complex, the outer membrane lipoprotein VirB7, VirB2–VirB5-formed extracellular pilus, and three membrane-associated ATPase (VirB4 and VirB11 plus VirD4) [139,140,141]. The ATPase VirD4 is capable of recognizing and interacting with proteins that have C-terminal XVIPCDs (Xanthomonas-VirD4-interacting protein conserved domains) and/or protein–DNA complexes [138]. Furthermore, these toxic XVIPs are secreted by the T4SS to kill other bacterial cells. For instance, virB7 and virD4 gene-defective mutants of X. citri pv. citri lost the ability to lyse Escherichia coli while being co-cultured [141].
In the study by Constantin et al. [63], a virB5-like gene was identified in aroid strains, exhibiting approximately 35% similarity with X. euvesicatoria 85-10. All core protein-encoding genes, with the exception of virB5, were found in X. euvesicatoria LMG 12749 and X. phaseoli pv. dieffenbachiae LMG 25940, sharing 68 to 96% sequence homology. In contrast, only virB10 and virB11 were present in X. phaseoli pv. syngonii LMG 9055, while virB6 and a truncated virD4 were harbored in X. citri pv. aracearum LMG 7399 [63]. Similar to X. hortorum pv. hederae, the virB3-B4, virB8-virB11, and virD4 genes were found in pathovars vitians, pelargonii, and taraxaci [142]. According to subsequent studies, the T4SS of Xanthomonas spp. was shown to indirectly affect pathogenicity on their hosts [143]. The virB7 knockout mutant of X. citri pv. citri developed symptoms similar to the wild type on citrus leaves [144], and a 10-gene deletion mutant of X. campestris pv. campestris, ranging from virD4 to virB4, exhibited unchanged virulence on tested cabbage, Chinese cabbage, Chinese kale, pak choi, and radish [145].
3.5. Type VI Secretion System (T6SS)
In most Gram-negative bacteria, the Type VI secretion system (T6SS) serves as another contact-dependent secretion machinery that translocates virulence factors, including effectors and toxins, into both prokaryotic and eukaryotic cells [108,109]. This contractile injection system consists of three main components: (i) the membrane-spanning complex, formed by proteins encoded by tssJ, tssL, and tssM and (ii) a baseplate, assembled by proteins encoded by tssE-G, tssK, and tssI/vgrG along with PAAR repeats, with the formation of (iii) an extended inner tube facilitated by the Hcp (Haemolysin-Coregulated Protein) encoded by the tssD/hcp gene. This tube is assembled by proteins encoded by tssB and tssC and is coordinated by a TssA-like protein [83,146,147]. Three different subclasses of T6SS loci, comprising 13 core genes, have been reported in Xanthomonas spp. However, none of these were found in X. campestris, X. hyacinthi, X. theicola, X. sacchari, and X. albilineans [83,118,148].
Moreover, the T6SS subclasses (T6SS-I, II, and III) acquired via horizontal gene transfer events exhibited various organization and synteny of core genes among Xanthomonas spp. [149]. Both T6SS-I/T6SS-3 and T6SS-III/T6SS-3 were present in X. phaseoli pv. dieffenbachiae LMG 25940 and X. phaseoli pv. syngonii LMG 9055, as well as in X. euvesicatoria strains including LMG 12749 from aroids, 85-10 from pepper, 91-118 and CFBP 7293 from tomato, and CFBP 3836 (syn. X. alfalfae pv. alfalfae) from alfalfa, and F1 (syn. X. axonopodis pv. citrumelonis) from citrus [63,118,150]. The Xanthomonas citri pv. aracearum LMG 7399 strain possessed only the T6SS-III/T6SS-3 subclass, closely clustered with the T6SS-III of X. phaseoli pv. dieffenbachiae LMG 25940 and X. phaseoli pv. syngonii LMG 9055, while X. hortorum pv. hederae WHRI 7744 from ivy (Araliaceae) contained only the T6SS-II/T6SS-4 subclass, also present in X. oryzae pathovars and X. fragariae [18,63,118,150].
Besides its role in antagonistic activity within the bacterial community, the versatile T6SS has been involved in biofilm formation, antibacterial activity, and virulence in eukaryotic hosts [106,107,151]. The T6SS secretion system is crucial for X. citri pv. citri to defend against soil amoebae, which are bacterial predators that feed in a phagocytic manner [152]. Montenegro Benavides et al. [149] revealed that the vgrG, hcp, and clpV genes of X. phaseoli pv. manihotis, possessing only T6SS-III, were essential for maximizing aggressiveness on cassava, while the clpV gene also significantly impaired motility. The function of the hcp gene in T6SS-I, but not in T6SS-II of X. oryzae pv. oryzae, was demonstrated to play a role in virulence to rice [153]. In contrast, neither the hcp gene in T6SS-I nor the hcp gene in T6SS-II of X. oryzae pv. oryzicola impacted virulence on rice [154]. Furthermore, the tssM mutant in T6SS-III of X. perforans (syn. X. euvesicatoria pv. perforans) [13,14] exhibited increased virulence on tomato leaves during dip inoculation and no significant difference in severity during infiltration infection [155]. Notably, two tssM genes, sharing 65% identity in two T6SS loci (I and III), were found in X. euvesicatoria pv. perforans, and the one in T6SS-III was required for successful epiphytic survival during the asymptomatic phase of tomato infection [155]. Gene expression profiles of X. citri pv. citri, including tssB, clpV, vgrG, and ecfK (a T6SS regulator-encoded gene), were increased during epiphytic growth rather than inside the mesophyll of sweet orange [156].
3.6. Cell-Wall-Degrading Enzymes (CWDEs)
Xanthomonas spp. secrete a diverse array of cellulolytic, hemicellulolytic, and pectinolytic enzymes, collectively termed cell-wall-degrading enzymes (CWDEs), which degrade the plant cell wall matrix. An average of 31 homologs of the CWDE repertoire was retrieved from 26 xanthomonad genomes including complex groups of cellulases, cellobiosidases, polygalacturonase, pectin methylesterases, pectate lyases, rhamnogalacturonases, rhamnogalacturonan acetylesterase, xylanase, and beta-glucosidases xylosidases/arabinosidases [91]. Vieira et al. [157] unraveled a xyloglucan utilization locus comprised of two TonB-dependent transporters, four glycoside hydrolases, and one esterase, which is highly conserved in 34 Xanthomonas species/pathovars that infect monocots and dicots (excluding X. oryzae, X. translucens, and X. albilineans). Previous studies have reported that xanthomonads mainly manipulated the T2SS to secrete CWDEs into extracellular milieu, including the following: xylanases (XCV0965, XCV4358, and XCV4360), protease (XCV3671), and lipase (XCV0536) as T2SS-dependent substrates in X. campestris pv. vesicatoria 85-10 [119,124], cellobiosidase (CbsA), cellulose (ClsA), and lipase/esterase (LipA) via T2SS in X. oryzae pv. oryzae [158,159]. Moreover, an alternative transport route for some T2SS substrates was proposed to attribute to the CWDEs presenting in outer membrane vesicles (OMVs), also called type zero secretion system (T0SS). T2SS-secreted cellulose (Egl) and xylosidase (XynB) were observed in OMVs of X. campestris pv. campestris [160]. Moreover, putative protease (XCV0007), xylanase (XCV4355), and other T2S substrates including lipase (XCV0536), protease (XCV3671), and xylanases (XCV4358 and XCV4360) were detected in OMVs of X. campestris pv. vesicatoria under immune-electron microscopy [124]. Based on a previous genomic study of aroid strains isolated from dieffenbachia, anthurium, philodendron, and syngonium, the two strains possessing the highest number of CWDEs are X. phaseoli pv. dieffenbachiae LMG 25940 (total = 40) with the most pectinolytic and hemicellulolytic enzymes and X. citri pv. aracearum LMG7399 (total = 38) having the most cellulolytic enzymes [63]. Meanwhile, X. euvesicatoria LMG12749 contained the identical number of hemicellulolytic enzymes as LMG 25940 but fewer than the other two groups. Although the X. phaseoli pv. syngonii LMG 9055 strain was the most aggressive on syngonium, it displayed the lowest combination of CWDEs due to truncated or frameshifted sequences and incomplete genome-assembly (232 contigs, N50: 52,766 bp) [63]. The large CWDE repertoire was reported in the genomes of X. hortorum pv. vitians LM 16734 isolated from lettuce and pv. gardneri ATCC 19865 from tomato; however, no details were discussed for X. hortorum pv. hederae from ivy [89,91].
In X. campestris pv. vesicatoria strain 85-10, xylanase (XynC, XCV0965) contributed to bacterial virulence on pepper, and the xynC-deficient mutant showed reduced leaf spot symptoms and lower in planta bacterial multiplication [119]. Two Xyn homologs, XynA and XynB, were found in X. oryzae pv. oryzae, though only XynB displayed dominantly xylanase activity [158]. The pathogenicity of the xynB gene and lipA-xynB double mutants of X. oryzae pv. oryzae was reduced significantly [158]. Moreover, the cbsA disruption mutant and lipA clsA double mutant abolished the virulence of X. oryzae pv. oryzae to rice, whereas clsA- and lipA-deficient mutants partially reduced virulence [158,159]. In comparison with cellulase-, xylanase-, and lipase-encoding genes, protease- and pectin-homogalacturonan (HG)-degrading genes of X. oryzae pv. oryzae showed inefficient virulence to rice. Tayi et al. [161] reported four HG degrading genes including pglA encoding polygalacturonase, pmt encoding pectin methyl esterase, and both pel and pelL encoding pectate lyases, which played minor roles in virulence to rice. Interestingly, ecpA encoding extracellular protease was reported not to affect virulence on rice for X. oryzae pv. oryzae, but the gene is a crucial virulence determinant required for X. oryzae pv. oryzicola to infect rice [162]. Besides CWDEs, non-fimbrial adhesion-like protein A (XadA) promoted the virulence of X. oryzae pv. oryzae to rice [163].
3.7. Flagellum
The flagellum is a complex motility organelle that enables bacteria to swim and swarm. Besides motility, it facilitates adherence to hosts, biofilm formation, protein export, and cellular invasion [164,165,166]. A typical flagellum consists of a cytoplasmic export apparatus, body rings including L, P, MS, and C rings embedded in the cell membrane, tubular axial structures including a hook and rod across the cell membrane, and filaments outside of the membrane [167]. The flagellar structures, composed of ~20,000–30,000 protein subunits of over 30 different proteins, are assembled in a sequential order: basal rings, export apparatus, rod, hook, and then filaments [167,168]. For xanthomonads having a single polar flagellum, four gene clusters are involved in flagellar synthesis and regulation: cluster I contains fliSDC, flgLKJIHGFEDCB, cheV, and flgAM genes. Cluster II consists of fliA, fleN, flhFAB, and fliRQPONMLKJIHGFE genes; cluster III possesses only two flagellar motor protein encoding genes, motA and motB; cluster IV contains chemotaxis protein, methyl-accepting chemotaxis protein, partitioning protein, and motor protein genes [63,82]. The flagellar gene clusters I, III, and IV were conserved in X. citri pv. aracearum, X. euvesicatoria, X. phaseoli pv. dieffenbachiae, and pv. syngonii strains isolated from Araceae, but cluster II showed differences in the gene organization and the number of hypothetical protein-coding genes [63]. No information about flagellar biosynthesis gene clusters in X. hortorum pv. hederae is presently available.
Mutations in either flagellar component genes or regulatory genes of different xanthomonads affect diverse flagellation, motility, and pathogenicity on their hosts. In the study by Darrasse et al. [112] the integrity of the flagellar gene cluster was examined across various Xanthomonas species and pathovars. Among the 38 strained tested, 4.4% including X. albilineans CFBP 2523, X. arboricola pv. corylina CFBP1159, X. cucurbitae CFBP2542, X. fuscans subsp. fuscans CFBP 4885, and X. sacchari CFBP 4641 were non-motile, due to deficiencies in their flagellar gene clusters. Interestingly, the non-flagellated X. fuscans subsp. fuscans CFBP 4885 is highly pathogenic on beans [112]. In X. campestris pv. campestris, the insertion mutants of the flhA gene, which encoded a subunit of an export apparatus, and of the fliA gene, which encoded sigma factor 28 in cluster II, were defective in flagellation and motility, and both mutations caused attenuated virulence on a cabbage leaf [169]. Conversely, flhB- and fleN-deficient mutants of X. campestris pv. campestris displayed identical virulence to parental strains, even though a flhB-deficient mutant was immotile and non-flagellated [169], but a fleN-deficient mutant was hyper-flagellated [170]. Xanthomonas oryzae pv. oryzae mutants with a mutation in the flhF gene, flanking in the downstream of flhB and flhA, showed no interference in flagellation, motility, and pathogenicity to rice using scissor-dipping and spray methods [171]. Whereas the flhF deletion mutant of X. campestris pv. campestris produced one to two lateral flagella like fliA and flgM mutants, the flhF mutant was diminished in pathogenicity to cabbage [172].
Two rpoN homologous regulators, RpoN1 and RpoN2, were revealed in X. citri subsp. citri [173] and X. campestris pv. campestris [169]. Gene rpoN2 is in a large flagellar gene cluster I while gene rpoN1 is harbored in a phosphotransferase system [169,173,174]. Xanthomonas citri subsp. citri rpoN1 and rpoN2 deletion mutants and double mutants were delayed in canker symptom development; however, the distinct regulatory effects on motility were observed in these mutants [173]. Yang et al. [169] and Li et al. [174] reported the mutation of rpoN2 in X. campestris pv. campestris strains ATCC33913 and Xc1, respectively, not only reduced flagellation and motility but also significantly affected the virulence to susceptible cabbage. The direct regulation of RpoN2 with corresponding promoters of genes fliC and fliQ was revealed while analyzing transcriptomic gene expression profiles [174], whereas a knockout mutant of the rpoN1 gene, harbored in a phosphotransferase system, caused similar symptoms to the Xc1 strain. Also, a rpoN1N2 double mutant lost pathogenicity and motility, similar to a rpoN2 gene knockout mutant, and plasmid-born rpoN2 complemented the defective double mutant, but plasmid-born rpoN1 failed to restore swimming ability [174]. Taken together, homologous regulator RpoN2 worked independently and was not interchangeable with RpoN1 [173,174].
4. Conclusions and Future Directions
In conclusion, recent advances in phylogenomic and pathogenicity studies have led to the reclassification of Xanthomonas strains from Araceae into six species (X. arboricola, X. citri, X. euvesicatoria, X. hawaiiensis, X. phaseoli, and X. sacchari) and those from Araliaceae into two species (X. hederae and X. euvesicatoria). Pathogenicity and host range tests identified several pathovars among strains from both the Araceae and Araliaceae families, including X. arboricola pv. zantedeschiae, X. citri pv. aracearum, X. euvesicatoria pv. amorphophalli, X. euvesicatoria pv. polysciadis, X. hortorum pv. hederae, X. phaseoli pv. dieffenbachiae, and X. phaseoli pv. syngonii (Figure 2). This refined taxonomy provides a more robust foundation for disease diagnostics, surveillance, and management in economically important ornamental crops. However, the phylogenetic placement of certain strains from diverse araceous and araliaceous hosts remains unresolved due to limited genomic representation. To address these gaps, future research should prioritize comprehensive genome sequencing of underrepresented strains, especially from genera such as Epipremnum and Rhaphidophora, and apply comparative pan-genome and population genomic analyses to clarify species boundaries and pathovar affiliations. Additionally, functional genomics and reverse genetic studies targeting key pathogenicity determinants—such as secretion systems, exopolysaccharide and lipopolysaccharide biosynthesis, and cell-wall-degrading enzymes—will be essential to unravel the molecular mechanisms underpinning host adaptation and virulence. By integrating high-throughput sequencing with targeted functional assays, the field is poised to resolve outstanding taxonomic ambiguities and accelerate the development of precise diagnostic tools and effective disease management strategies, thereby advancing both the fundamental understanding and practical control of Xanthomonas diseases in Araceae and Araliaceae.
Author Contributions
Conceptualization, M.A.; writing—original draft preparation, S.-C.C.; writing—review and editing, S.-C.C., S.D., L.M.K., A.M.A. and M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the USDA-ARS Agreement no. 58-2040-9-011, Systems Approaches to Improve Production and Quality of Specialty Crops Grown in the U.S. Pacific Basin: sub-project: Genome Informed Next Generation Detection Protocols for Pests and Pathogens of Specialty Crops in Hawaii. This work is also supported by the USDA-NIFA Award No. 2023-67013-39301.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Dowson, W.J. On the systematic position and generic names of the Gram-negative bacterial plant pathogens. Zentralblatt Für Bakteriol. Parasitenkd. Infekt. Und Hyg. 1939, 100, 177–193. [Google Scholar]
- Garrity, G.M.; Brenner, D.J.; Krieg, N.R.; Staley, J.R. Bergey’s Manual of Systematic Bacteriology; Volume 2: The Proteobacteria (Part B); Springer: New York, NY, USA, 2005. [Google Scholar]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Bansal, K.; Patil, P.P.; Patil, P.B. Phylogenomics insights into order and families of Lysobacterales. Access Microbiol. 2019, 1, e000015. [Google Scholar] [CrossRef]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Heiden, N.; Broders, K.A.; Hutin, M.; Castro, M.O.; Roman-Reyna, V.; Toth, H.; Jacobs, J.M. Bacterial Leaf Streak Diseases of plants: Symptom convergence in monocot plants by distant pathogenic Xanthomonas Species. Phytopathology 2023, 113, 2048–2055. [Google Scholar] [CrossRef]
- Bradbury, J.F. Bergey’s Manual of Systematic Bacteriology; Krieg, N.R., Holt, J.G., Eds.; Williams & Wilkins Co.: Philadelphia, PA, USA, 1984; Volume 1. [Google Scholar]
- Leyns, F.; De Cleene, M.; Swings, J.G.; De Ley, J. The host range of the genus Xanthomonas. Bot. Rev. 1984, 50, 308–356. [Google Scholar] [CrossRef]
- Vauterin, L.; Rademaker, J.; Swings, J. Synopsis on the taxonomy of the genus Xanthomonas. Phytopathology 2000, 90, 677–682. [Google Scholar] [CrossRef]
- Ryan, R.P.; Vorhölter, F.-J.; Potnis, N.; Jones, J.B.; Van Sluys, M.-A.; Bogdanove, A.J.; Dow, J.M. Pathogenomics of Xanthomonas: Understanding bacterium–plant interactions. Nat. Rev. Microbiol. 2011, 9, 344–355. [Google Scholar] [CrossRef]
- Mafakheri, H.; Taghavi, S.M.; Zarei, S.; Portier, P.; Dimkić, I.; Koebnik, R.; Kuzmanovic, N.; Osdaghi, E. Xanthomonas bonasiae sp. nov. and Xanthomonas youngii sp. nov., isolated from crown gall tissues. Int. J. Syst. Evol. Microbiol. 2022, 72, 005418. [Google Scholar] [CrossRef]
- Palacio-Bielsa, A.; Roselló, M.; Cambra, M.A.; López, M.M. First report on almond in Europe of bacterial spot disease of stone fruits caused by Xanthomonas arboricola pv. pruni. Plant Dis. 2010, 94, 786. [Google Scholar] [CrossRef]
- Barak, J.D.; Vancheva, T.; Lefeuvre, P.; Jones, J.B.; Timilsina, S.; Minsavage, G.V.; Koebnik, R. Whole-genome sequences of Xanthomonas euvesicatoria strains clarify taxonomy and reveal a stepwise erosion of type 3 effectors. Front. Plant Sci. 2016, 7, 1805. [Google Scholar] [CrossRef] [PubMed]
- Constantin, E.C.; Cleenwerck, I.; Maes, M.; Baeyen, S.; Van Malderghem, C.; De Vos, P.; Cottyn, B. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol. 2016, 65, 792–806. [Google Scholar] [CrossRef]
- Dhakal, U.; Dobhal, S.; Alvarez, A.M. Phylogenetic analyses of xanthomonads causing bacterial leaf spot of tomato and pepper: Xanthomonas euvesicatoria revealed homologous populations despite distant geographical distribution. Microorganisms 2019, 7, 462. [Google Scholar] [CrossRef]
- Daughtrey, M.L.; Wick, R.L.; Peterson, J.L. Compendium of Flowering Potted Plant Diseases; APS Press: St. Paul, MN, USA, 1995; Volume 24. [Google Scholar]
- Mustafa, M.; Yesim, A.; Fulya, B.-G. Bacterial spot and blight diseases of ornamental plants caused by different Xanthomonas species in Turkey. Plant Prot. Sci. 2018, 54, 240–247. [Google Scholar] [CrossRef]
- Dia, N.C.; Morinière, L.; Cottyn, B.; Bernal, E.; Jacobs, J.M.; Koebnik, R.; Pothier, J.F. Xanthomonas hortorum—Beyond gardens: Current taxonomy, genomics, and virulence repertoires. Mol. Plant Pathol. 2022, 23, 597–621. [Google Scholar] [CrossRef]
- Chase, A.R. Xanthomonas campestris pv. hederae causes a leaf spot of five species of Araliaceae. Plant Pathol. 1984, 33, 439–440. [Google Scholar] [CrossRef]
- Nishijima, W.T. Anthurium blight: An overview. Pages 6-8. In Proceedings of the 1st Anthurium Blight Conference; Hawaii Institute of Tropical Agricultural Human Resources (HITAHR), University of Hawaii: Honolulu, HI, USA, 1988. [Google Scholar]
- Sathyanarayana, N.; Reddy, O.R.; Latha, S.; Rajak, R.L. Interception of Xanthomonas campestris pv. dieffenbachiae on Anthurium Plants from the Netherlands. Plant Dis. 1998, 82, 262. [Google Scholar] [CrossRef]
- Norman, D.J.; Chase, A.R.; Stall, R.E.; Jones, J.B. Heterogeneity of Xanthomonas campestris pv. hederae strains from araliaceous hosts. Phytopathology 1999, 89, 646–652. [Google Scholar] [CrossRef]
- EPPO. 2009. PM 7/23 (2): Xanthomonas axonopodis pv. dieffenbachiae. Bull. OEPP 39:393-402. Available online: https://gd.eppo.int/download/doc/276_datasheet_XANTDF.pdf (accessed on 22 June 2025).
- Nelson, S. Bacterial Leaf Blight of Panax (Polyscias guilfoylei). Plant Disease, PD-75; College of Tropical Agriculture and Human Resources, University of Hawai‘i at Mānoa: Honolulu, HI, USA, 2011; Available online: https://www.ctahr.hawaii.edu/oc/freepubs/pdf/PD-75.pdf (accessed on 22 June 2025).
- Wakker, J.H. Vorlaüfige Mittheilungen über Hyacinthenkrankeiten. Bot. Zentralblatt 1883, 14, 1F–6F. [Google Scholar] [CrossRef]
- Vauterin, L.; Hoste, B.; Kersters, K.; Swings, J. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 1995, 45, 472–489. [Google Scholar] [CrossRef]
- Winslow, C.E.; Broadhurst, J.; Buchanan, R.E.; Krumwiede, C.; Rogers, L.A.; Smith, G.H. The families and genera of the bacteria: Final report of the Committee of the Society of American Bacteriologists on characterization and classification of bacterial types. J. Bacteriol. 1920, 5, 191–229. [Google Scholar] [CrossRef] [PubMed]
- Burkholder, W.H.; Starr, M.P. The generic and specific characters of phytopathogenic species of Pseudomonas and Xanthomonas. Phytopathology 1948, 38, 494–502. [Google Scholar]
- Burkholder, W.H. Genus II. Xanthomonas Dowson 1939. In Bergey’s Manual of Determinative Bacteriology, 7th ed.; Breed, R.S., Murray, E.G.D., Smith, N.R., Eds.; Williams & Wilkins: Philadelphia, PA, USA, 1957; pp. 152–183. [Google Scholar]
- Starr, M.P. The genus Xanthomonas. In The Prokaryotes; Starr, M.P., Stolp, H., Trüper, H.G., Balows, A., Schlegel, H.G., Eds.; Springer Verlag: Heidelberg, Germany, 1981. [Google Scholar]
- Bergey, D.H.; Buchanan, R.E.; Gibbons, N.E. Bergey’s Manual of Determinative Bacteriology, 8th ed.; Williams & Wilkins Co.: Philadelphia, PA, USA, 1974. [Google Scholar]
- Willems, A.; Gillis, M.; Kersters, K.; Van Den Broecke, L.; De Ley, J. Transfer of Xanthomonas ampelina Panagopoulos 1969 to a new genus, Xylophilus gen. nov., as Xylophilus ampelinus (Panagopoulos 1969) comb. nov. Int. J. Syst. Bacteriol. 1987, 37, 422–430. [Google Scholar] [CrossRef]
- Dye, D.; Lelliott, R. Genus II. Xanthomonas Dowson 1939; The Williams and Wilkins Co.: Philadelphia, PA, USA, 1974. [Google Scholar]
- Rademaker, J.L.; Hoste, B.; Louws, F.J.; Kersters, K.; Swings, J.; Vauterin, L.; Vauterin, P.; de Bruijn, F.J. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int. J. Syst. Evol. Microbiol. 2000, 50, 665–677. [Google Scholar] [CrossRef]
- Rademaker, J.L.W.; Louws, F.J.; Schultz, M.H.; Rossbach, U.; Vauterin, L.; Swings, J.; de Bruijn, F.J. A comprehensive species to strain taxonomic framework for Xanthomonas. Phytopathology 2005, 95, 1098–1111. [Google Scholar] [CrossRef]
- Young, J.M.; Park, D.C.; Shearman, H.M.; Fargier, E. A multilocus sequence analysis of the genus Xanthomonas. Syst. Appl. Microbiol. 2008, 31, 366–377. [Google Scholar] [CrossRef]
- Hauben, L.; Vauterin, L.; Swings, J.; Moore, E.R. Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int. J. Syst. Bacteriol. 1997, 47, 328–335. [Google Scholar] [CrossRef]
- Schaad, N.W.; Jones, J.B.; Chun, W. Laboratory Guide for Identification of Plant Pathogenic Bacteria, 3rd ed.; American Phytopathological Society: St Paul, MI, USA, 2001. [Google Scholar]
- Young, J.M.; Bull, C.T.; De Boer, S.H.; Firrao, G.; Gardan, L.; Saddler, G.E.; Stead, D.E.; Takikawa, Y. Classification, nomenclature, and plant pathogenic bacteria—A clarification. Phytopathology 2001, 91, 617–620. [Google Scholar] [CrossRef]
- Wayne, L.G.; Moore, W.E.C.; Stackebrandt, E.; Kandler, O.; Colwell, R.R.; Krichevsky, M.I.; Truper, H.G.; Murray, R.G.E.; Grimont, P.A.D.; Brenner, D.J.; et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Evol. Microbiol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57 Pt 1, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, G. Une maladie bactérienne du lierre (Hedera helix L.). CR Acad. Sci. 1920, 171, 121–122. [Google Scholar]
- Burkholder, W.H.; Guterman, C.E.F. Synergism in a bacterial disease of Hedera helix. Int. J. Syst. Evol. Microbiol. 1932, 59 Pt 2, 306–318. [Google Scholar]
- McCulloch, L.; Pirone, P.P. Bacterial leaf spot of dieffenbachia. Phytopathology 1939, 29, 956–962. [Google Scholar]
- Dowson, W.J. On the generic names Pseudomonas, Xanthomonas and Bacterium for certain bacterial plant pathogens. Trans. Br. Mycol. Soc. 1943, 26, 4–14. [Google Scholar] [CrossRef]
- Wehlburg, C. Bacterial leaf blight of Syngonium. Florida Department of Agriculture & Consumer Services, Division of Plant Industry. Plant Pathol. Circ. 1970, 91. Available online: https://www.fdacs.gov/content/download/11098/file/pp91.pdf (accessed on 22 June 2025).
- Jindal, J.K.; Patel, P.N.; Singh, R. Bacterial leaf spot disease on Amorphophallus campanulatus. Indian Phytopathol. 1972, 25, 374–377. [Google Scholar]
- Joubert, J.J.; Truter, S.J. A variety of Xanthomonas campestris pathogenic to Zantedeschia aethiopica. Neth. J. Plant Pathol. 1972, 78, 212–217. [Google Scholar] [CrossRef]
- Berniac, M. Une maladie bacterienne de Xanthosoma sagittifolium (L.) Schott. Ann. De Phytopathol. 1974, 6, 197–202. [Google Scholar]
- Young, J.M.; Dye, D.W.; Bradbury, J.F.; Panagopoulos, C.G.; Robbs, C.F. A proposed nomenclature and classification for plant pathogenic bacteria. N. Z. J. Agric. Res. 1978, 21, 153–177. [Google Scholar] [CrossRef]
- Dickey, R.S.; Zumoff, C.H. Bacterial leaf blight of Syngonium caused by a pathovar of Xanthomonas campestris. Phytopathology 1987, 77, 1257–1262. [Google Scholar] [CrossRef]
- Ah-You, N.; Gagnevin, L.; Grimont, P.A.D.; Brisse, S.; Nesme, X.; Chiroleu, F.; Ngoc, L.; Jouen, E.; Lefeuvre, P.; Vernière, C.; et al. Polyphasic characterization of xanthomonads pathogenic to members of the Anacardiaceae and their relatedness to species of Xanthomonas. Int. J. Syst. Evol. Microbiol. 2009, 59 Pt 2, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Saux, M.F.-L.; Bonneau, S.; Essakhi, S.; Manceau, C.; Jacques, M.-A. Aggressive emerging pathovars of Xanthomonas arboricola represent widespread epidemic clones distinct from poorly pathogenic strains, as revealed by multilocus sequence typing. Appl. Environ. Microbiol. 2015, 81, 4651–4668. [Google Scholar] [CrossRef] [PubMed]
- Chuang, S.-C.; Dobhal, S.; Alvarez, A.; Arif, M. Pathological and molecular biology of Xanthomonas strains causing bacterial leaf blight of Panax (Polyscias guilfoylei) in Hawaii [Oral Presentation]. In Proceedings of the APS Annual Meeting, Virtual, 2–6 August 2021. [Google Scholar]
- Chuang, S.-C.; Dobhal, S.; Pal, K.; Amore, T.D.; Alvarez, A.M.; Arif, M. Xanthomonas strains isolated from hosts in the Araceae reveal diverse phylogenetic relationships and origins. Phytopathology 2024, 114, 1963–1974. [Google Scholar] [CrossRef]
- van der Wolf, J.M.; Krijger, M.C.; Mendes, O.; Brankovics, B.; Bonants, P.J.M.; Didden, L.; Meekes, E. Molecular characterization of Xanthomonas species isolated from Araceae and the development of a triplex TaqMan assay for detection of Xanthomonas phaseoli pv. dieffenbachiae. Eur. J. Plant Pathol. 2022, 163, 167–179. [Google Scholar] [CrossRef]
- Wang, L.H.; Chan, J.J.; Wang, Y.H.; Fang, Z.Q.; Lee, S.; Chu, C.C. Bacterial leaf blight of Polyscias guilfoylei caused by a novel pathovar of Xanthomonas euvesicatoria. Plant Dis. 2023, 107, 298–305. [Google Scholar] [CrossRef]
- Chuang, S.-C.; Dobhal, S.; Alvarez, A.M.; Arif, M. Three new species, Xanthomonas hawaiiensis sp. nov., Stenotrophomonas aracearum sp. nov., and Stenotrophomonas oahuensis sp. nov., isolated from Araceae family. Front. Microbiol. 2024, 15, 1356025. [Google Scholar] [CrossRef]
- Lipp, R.L.; Alvarez, A.M.; Benedict, A.A.; Berestecky, J. Use of monoclonal antibodies and pathogenicity tests to characterize strains of Xanthomonas campestris pv. dieffenbachiae from aroids. Phytopathology 1992, 82, 677–682. [Google Scholar] [CrossRef]
- Pohronezny, K.; Dankers, W.; Schaffer, B.; Valenzuela, H.; Moss, M.A. Marginal necrosis and intercostal leaf spots of cocoyam infected by Xanthomonas campestris pv. dieffenbachiae. Plant Dis. 1990, 74, 573–577. [Google Scholar] [CrossRef]
- Constantin, E.C.; Haegeman, A.; Van Vaerenbergh, J.; Baeyen, S.; Van Malderghem, C.; Maes, M.; Cottyn, B. Pathogenicity and virulence gene content of Xanthomonas strains infecting Araceae, formerly known as Xanthomonas axonopodis pv. dieffenbachiae. Plant Pathol. 2017, 66, 1539–1554. [Google Scholar] [CrossRef]
- Studholme, D.J.; Wasukira, A.; Paszkiewicz, K.; Aritua, V.; Thwaites, R.; Smith, J.; Grant, M. Draft genome sequences of Xanthomonas sacchari and two banana-associated xanthomonads reveal insights into the Xanthomonas Group 1 clade. Genes 2011, 2, 1050–1065. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Lin, H.; Wu, L.; Ren, D.; Ye, W.; Dong, G.; Zhu, L.; Guo, L. Genome sequence of Xanthomonas sacchari R1, a biocontrol bacterium isolated from the rice seed. J. Biotechnol. 2015, 206, 77–78. [Google Scholar] [CrossRef]
- White, R.P.; McCulloch, L. A bacterial disease of Hedera helix. J. Agric. Res. 1934, 48, 807–815. [Google Scholar]
- Suzuki, A.; Kusumoto, S.; Horie, H.; Takikawa, Y. Bacterial leaf spot of ivy caused by Xanthomonas campestris pv. hederae. J. Gen. Plant Pathol. 2002, 68, 398–400. [Google Scholar] [CrossRef]
- Dye, D. Bacterial spot of ivy caused by Xanthomonas hederae (Arnaud, 1920) Dowson 1939, in New Zealand. N. Z. J. Sci. 1967, 10, 481–485. [Google Scholar]
- Lee, S.; Lee, J.; Han, K.; Seo, S.T.; Kim, Y.; Heu, S.; Ra, D.S. Bacterial leaf spot of English ivy caused by Xanthomonas hortorum pv. hederae. Radiat. Prot. Dosim. 2007, 13, 61–65. [Google Scholar]
- Trantas, E.A.; Sarris, P.F.; Mpalantinaki, E.; Papadimitriou, M.; Ververidis, F.; Goumas, D.E. First report of Xanthomonas hortorum pv. hederae causing bacterial leaf spot on ivy in Greece. Plant Dis. 2016, 100, 2158. [Google Scholar] [CrossRef]
- Lu, C.H.; Chiu, Y.H.; Tzeng, J.Y.; Lin, C.Y.; Lin, Y.R.; Deng, W.L.; Chu, C.C. First report of Xanthomonas hortorum pv. hederae causing bacterial leaf spot of Hedera helix in Taiwan. Plant Dis. 2019, 103, 1765. [Google Scholar] [CrossRef]
- Pirc, M.; Dreo, T.; Šuštaršič, M.; Erjavec, J.; Ravnikar, M. First report of Xanthomonas hortorum pv. hederae causing bacterial leaf spot of Hedera hibernica in Slovenia. Plant Dis. 2012, 96, 141. [Google Scholar] [CrossRef]
- Zhang, X.F.; Fu, L.N.; Yang, J.; Li, X.; Ji, G.H. First report of bacterial leaf spot of Hedera nepalensis var. Sinensis caused by Xanthomonas campestris pv. hederae in China. Plant Dis. 2016, 100, 1007. [Google Scholar] [CrossRef]
- Lowry, P.P.; Plunkett, G.M.; Frodin, D.G. Revision of Plerandra (Araliaceae). I. A synopsis of the genus with an expanded circumscription and a new infrageneric classification. Brittonia 2013, 65, 42–61. [Google Scholar] [CrossRef]
- POWO. Plants of the World Online. Royal Botanic Gardens, Kew. 2023. Available online: https://en.wikipedia.org/wiki/Plants_of_the_World_Online (accessed on 22 June 2025).
- Tolba, I.H. Bacterial leaf spot of araliaceous plants caused by Xanthomonas campestris pv. hederae in Egypt. J. Plant Prot. Pathol. 2017, 8, 287–295. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Vojnov, A.A.; Zorreguieta, A.; Dow, J.M.; Daniels, M.J.; Dankert, M.A. Evidence for a role for the gumB and gumCgene products in the formation of xanthan from its pentasaccharide repeating unit by Xanthomonas campestris. Microbiology 1998, 144, 1487–1493. [Google Scholar] [CrossRef]
- Nürnberger, T.; Brunner, F.; Kemmerling, B.; Piater, L. Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 2004, 198, 249–266. [Google Scholar] [CrossRef]
- Guglielmini, J.; Néron, B.; Abby, S.S.; Garcillán-Barcia, M.P.; la Cruz, F.d.; Rocha, E.P.C. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 2014, 42, 5715–5727. [Google Scholar] [CrossRef]
- Moreira, L.M.; Almeida, J.N.F.; Potnis, N.; Digiampietri, L.A.; Adi, S.S.; Bortolossi, J.C.; da Silva, A.C.; da Silva, A.M.; de Moraes, F.E.; de Oliveira, J.C.; et al. Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genom. 2010, 11, 238. [Google Scholar] [CrossRef]
- Alvarez-Martinez, C.E.; Sgro, G.G.; Araujo, G.G.; Paiva, M.R.N.; Matsuyama, B.Y.; Guzzo, C.R.; Andrade, M.O.; Farah, C.S. Secrete or perish: The role of secretion systems in Xanthomonas biology. Comput. Struct. Biotechnol. J. 2021, 19, 279–302. [Google Scholar] [CrossRef]
- Vu, B.; Chen, M.; Crawford, R.J.; Ivanova, E.P. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 2009, 14, 2535–2554. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Katzen, F.; Pühler, A.; Ielpi, L. Xanthan gum biosynthesis and application: A biochemical/genetic perspective. Appl. Microbiol. Biotechnol. 1998, 50, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Katzen, F.; Ferreiro, D.U.; Oddo, C.G.; Ielmini, M.V.; Becker, A.; Puhler, A.; Ielpi, L. Xanthomonas campestris pv. campestris gum mutants: Effects on xanthan biosynthesis and plant virulence. J. Bacteriol. 1998, 180, 1607–1617. [Google Scholar] [CrossRef] [PubMed]
- Pieretti, I.; Pesic, A.; Petras, D.; Royer, M.; Süssmuth, R.D.; Cociancich, S. What makes Xanthomonas albilineans unique amongst xanthomonads? Front. Plant Sci. 2015, 6, 289. [Google Scholar] [CrossRef]
- Bansal, K.; Kumar, S.; Patil, P.B. Phylogenomic insights into diversity and evolution of nonpathogenic Xanthomonas strains associated with Citrus. mSphere 2020, 5, 10-1128. [Google Scholar] [CrossRef]
- Morinière, L.; Mirabel, L.; Gueguen, E.; Bertolla, F. A comprehensive overview of the genes and functions required for lettuce infection by the hemibiotrophic phytopathogen Xanthomonas hortorum pv. vitians. mSystems 2022, 7, e0129021. [Google Scholar] [CrossRef]
- Li, M.; Bao, Y.; Li, Y.; Akbar, S.; Wu, G.; Du, J.; Wen, R.; Chen, B.; Zhang, M. Comparative genome analysis unravels pathogenicity of Xanthomonas albilineans causing sugarcane leaf scald disease. BMC Genom. 2022, 23, 1–671. [Google Scholar] [CrossRef]
- Vandroemme, J.; Cottyn, B.; Baeyen, S.; De Vos, P.; Maes, M. Draft genome sequence of Xanthomonas fragariae reveals reductive evolution and distinct virulence-related gene content. BMC Genom. 2013, 14, 829. [Google Scholar] [CrossRef]
- Chou, F.-L.; Chou, H.-C.; Lin, Y.-S.; Yang, B.-Y.; Lin, N.-T.; Weng, S.-F.; Tseng, Y.-H. The Xanthomonas campestris gumD gene required for synthesis of xanthan gum is involved in normal pigmentation and virulence in causing black rot. Biochem. Biophys. Res. Commun. 1997, 233, 265–269. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Kim, J.-G.; Lee, B.-M.; Cho, J.-Y. Mutational analysis of the gum gene cluster required for xanthan biosynthesis in Xanthomonas oryzae pv. oryzae. Biotechnol. Lett. 2009, 31, 265–270. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef]
- Vorhölter, F.J.; Niehaus, K.; Pühler, A. Lipopolysaccharide biosynthesis in Xanthomonas campestris pv. campestris: A cluster of 15 genes is involved in the biosynthesis of the LPS O-antigen and the LPS core. Mol. Genet. Genom. 2001, 266, 79–95. [Google Scholar] [CrossRef]
- Patil, P.B.; Sonti, R.V. Variation suggestive of horizontal gene transfer at a lipopolysaccharide (lps) biosynthetic locus in Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen of rice. BMC Microbiol. 2004, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Fath, M.J.; Kolter, R. ABC transporters: Bacterial exporters. Microbiol. Rev. 1993, 57, 995–1017. [Google Scholar] [CrossRef] [PubMed]
- Patil, P.B.; Bogdanove, A.J.; Sonti, R.V. The role of horizontal transfer in the evolution of a highly variable lipopolysaccharide biosynthesis locus in xanthomonads that infect rice, citrus and crucifers. BMC Evol. Biol. 2007, 7, 243. [Google Scholar] [CrossRef]
- Smith, T.J.; Sondermann, H.; O’Toole, G.A. Type 1 does the two-step: Type 1 secretion substrates with a functional periplasmic intermediate. J. Bacteriol. 2018, 200, jb.00168-18. [Google Scholar] [CrossRef]
- Sandkvist, M. Biology of type II secretion. Mol. Microbiol. 2001, 40, 271–283. [Google Scholar] [CrossRef]
- Lindgren, P.B.; Peet, R.C.; Panopoulos, N.J. Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J. Bacteriol. 1986, 168, 512–522. [Google Scholar] [CrossRef]
- Alfano, J.R.; Collmer, A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: Trafficking harpins, Avr proteins, and death. J. Bacteriol. 1997, 179, 5655–5662. [Google Scholar] [CrossRef]
- Hueck, C.J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 1998, 62, 379–433. [Google Scholar] [CrossRef]
- Llosa, M.; Gomis-Rüth, F.X.; Coll, M.; Cruz, F.D.L. Bacterial conjugation: A two-step mechanism for DNA transport. Mol. Microbiol. 2002, 45, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gottig, N.; Garavaglia, B.S.; Garofalo, C.G.; Orellano, E.G.; Ottado, J. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS ONE 2009, 4, e4358. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.B.; Peterson, S.B.; Mougous, J.D. Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 2014, 12, 137–148. [Google Scholar] [CrossRef]
- Russell, A.B.; Wexler, A.G.; Harding, B.N.; Whitney, J.C.; Bohn, A.J.; Goo, Y.A.; Tran, B.Q.; Barry, N.A.; Zheng, H.; Peterson, S.B.; et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 2014, 16, 227–236. [Google Scholar] [CrossRef]
- Schwarz, S.; West, T.E.; Boyer, F.; Chiang, W.-C.; Carl, M.A.; Hood, R.D.; Rohmer, L.; Tolker-Nielsen, T.; Skerrett, S.J.; Mougous, J.D. Burkholderia Type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010, 6, e1001068. [Google Scholar] [CrossRef]
- Hachani, A.; Wood, T.E.; Filloux, A. Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 2016, 29, 81–93. [Google Scholar] [CrossRef]
- Timilsina, S.; Kara, S.; Jacques, M.A.; Potnis, N.; Minsavage, G.V.; Vallad, G.E.; Jones, J.B.; Fischer-Le Saux, M. Reclassification of Xanthomonas gardneri (ex Šutić 1957) Jones et al. 2006 as a later heterotypic synonym of Xanthomonas cynarae Trébaol et al. 2000 and description of X. cynarae pv. cynarae and X. cynarae pv. gardneri based on whole genome analyses. Int. J. Syst. Evol. Microbiol. 2019, 69, 343–349. [Google Scholar] [CrossRef]
- Lu, H.; Patil, P.; Van Sluys, M.-A.; White, F.F.; Ryan, R.P.; Dow, J.M.; Rabinowicz, P.; Salzberg, S.L.; Leach, J.E.; Sonti, R.; et al. Acquisition and evolution of plant pathogenesis–associated gene clusters and candidate determinants of tissue-specificity in Xanthomonas. PLoS ONE 2008, 3, e3828. [Google Scholar] [CrossRef]
- Darrasse, A.; Carrère, S.; Barbe, V.; Boureau, T.; Arrieta-Ortiz, M.L.; Bonneau, S.; Briand, M.; Brin, C.; Cociancich, S.; Durand, K.; et al. Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genom. 2013, 14, 1–30. [Google Scholar] [CrossRef]
- da Silva, A.C.R.; Ferro, J.A.; Reinach, F.C.; Farah, C.S.; Furlan, L.R.; Quaggio, R.B.; Monteiro-Vitorello, C.B.; Sluys, M.A.V.; Almeida, N.F.; Alves, L.M.C.; et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 2002, 417, 459–463. [Google Scholar] [CrossRef]
- Li, J.; Wang, N. The wxacO gene of Xanthomonas citri ssp. citri encodes a protein with a role in lipopolysaccharide biosynthesis, biofilm formation, stress tolerance and virulence. Mol. Plant Pathol. 2011, 12, 381–396. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Hu, X.; Wang, N. The novel virulence-related gene nlxA in the lipopolysaccharide cluster of Xanthomonas citri ssp. citri is involved in the production of lipopolysaccharide and extracellular polysaccharide, motility, biofilm formation and stress resistance. Mol. Plant Pathol. 2012, 13, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Steffens, T.; Vorhölter, F.-J.; Giampà, M.; Hublik, G.; Pühler, A.; Niehaus, K. The influence of a modified lipopolysaccharide O-antigen on the biosynthesis of xanthan in Xanthomonas campestris pv. campestris B100. BMC Microbiol. 2016, 16, 93. [Google Scholar] [CrossRef] [PubMed]
- Studholme, D.J.; Wicker, E.; Abrare, S.M.; Aspin, A.; Bogdanove, A.; Broders, K.; Bull, C.T. Transfer of Xanthomonas campestris pv. arecae and X. campestris pv. musacearum to X. vasicola (Vauterin) as X. vasicola pv. arecae comb. nov. and X. vasicola pv. musacearum comb. nov. and description of X. vasicola pv. vasculorum pv. nov. Phytopathology 2020, 110, 1153–1160. [Google Scholar] [CrossRef]
- Timilsina, S.; Potnis, N.; Newberry, E.A.; Liyanapathiranage, P.; Iruegas-Bocardo, F.; White, F.F.; Goss, E.M.; Jones, J.B. Xanthomonas diversity, virulence and plant–pathogen interactions. Nat. Rev. Microbiol. 2020, 18, 415–427. [Google Scholar] [CrossRef]
- Szczesny, R.; Jordan, M.; Schramm, C.; Schulz, S.; Cogez, V.; Bonas, U.; Büttner, D. Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria. New Phytol. 2010, 187, 983–1002. [Google Scholar] [CrossRef]
- Hu, N.T.; Hung, M.N.; Chiou, S.J.; Tang, F.; Chiang, D.C.; Huang, H.Y.; Wu, C.Y. Cloning and characterization of a gene required for the secretion of extracellular enzymes across the outer membrane by Xanthomonas campestris pv. campestris. J. Bacteriol. 1992, 174, 2679–2687. [Google Scholar] [CrossRef]
- Baptista, J.C.; Machado, M.A.; Homem, R.A.; Torres, P.S.; Vojnov, A.A.; do Amaral, A.M. Mutation in the xpsD gene of Xanthomonas axonopodis pv. citri affects cellulose degradation and virulence. Genet. Mol. Biol. 2010, 33, 146–153. [Google Scholar] [CrossRef]
- Ray, S.K.; Rajeshwari, R.; Sonti, R.V. Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Mol. Plant-Microbe Interact. 2000, 13, 394–401. [Google Scholar] [CrossRef]
- Wang, J.C.; So, B.H.; Kim, J.H.; Park, Y.J.; Lee, B.M.; Kang, H.W. Genome-wide identification of pathogenicity genes in Xanthomonas oryzae pv. oryzae by transposon mutagenesis. Plant Pathol. 2008, 57, 1136–1145. [Google Scholar] [CrossRef]
- Solé, M.; Scheibner, F.; Hoffmeister, A.-K.; Hartmann, N.; Hause, G.; Rother, A.; Jordan, M.; Lautier, M.; Arlat, M.; Büttner, D. Xanthomonas campestris pv. vesicatoria secretes proteases and xylanases via the Xps type II secretion system and outer membrane vesicles. J. Bacteriol. 2015, 197, 2879–2893. [Google Scholar] [CrossRef] [PubMed]
- Bonas, U. hrp genes of phytopathogenic bacteria. Bact. Pathog. Plants Anim. 1994, 192, 79–96. [Google Scholar] [CrossRef]
- Bogdanove, A.J.; Beer, S.V.; Bonas, U.; Boucher, C.A.; Collmer, A.; Coplin, D.L.; Cornelis, G.R.; Huang, H.Z.; Hutcheson, S.W.; Panopoulos, N.J.; et al. Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol. Microbiol. 1996, 20, 681–683. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J.T.; Vinatzer, B.A. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr. Opin. Microbiol. 2003, 6, 20–28. [Google Scholar] [CrossRef]
- White, F.F.; Potnis, N.; Jones, J.B.; Koebnik, R. Type III effectors of Xanthomonas. Mol. Plant Pathol. 2009, 10, 749–766. [Google Scholar] [CrossRef]
- Triplett, L.R.; Verdier, V.; Campillo, T.; Van Malderghem, C.; Cleenwerck, I.; Maes, M.; Leach, J.E. Characterization of a novel clade of Xanthomonas isolated from rice leaves in Mali and proposal of Xanthomonas maliensis sp. nov. Antonie Van Leeuwenhoek 2015, 107, 869–881. [Google Scholar] [CrossRef]
- Pieretti, I.; Royer, M.; Barbe, V.; Carrere, S.; Koebnik, R.; Cociancich, S.; Couloux, A.; Darrasse, A.; Gouzy, J.; Jacques, M.-A.; et al. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genom. 2009, 10, 616. [Google Scholar] [CrossRef]
- Jacobs, J.M.; Pesce, C.; Lefeuvre, P.; Koebnik, R. Comparative genomics of a cannabis pathogen reveals insight into the evolution of pathogenicity in Xanthomonas. Front. Plant Sci. 2015, 6, 431. [Google Scholar] [CrossRef]
- Merda, D.; Briand, M.; Bosis, E.; Rousseau, C.; Portier, P.; Barret, M.; Jacques, M.; Fischer-Le Saux, M. Ancestral acquisitions, gene flow and multiple evolutionary trajectories of the type three secretion system and effectors in Xanthomonas plant pathogens. Mol. Ecol. 2017, 26, 5939–5952. [Google Scholar] [CrossRef]
- Noël, L.; Thieme, F.; Nennstiel, D.; Bonas, U. Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 2002, 184, 1340–1348. [Google Scholar] [CrossRef]
- Rossier, O.; Van den Ackerveken, G.; Bonas, U. HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol. Microbiol. 2000, 38, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Büttner, D.; Nennstiel, D.; Klüsener, B.; Bonas, U. Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 2002, 184, 2389–2398. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-G.; Park, B.K.; Yoo, C.-H.; Jeon, E.; Oh, J.; Hwang, I. Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 2003, 185, 3155–3166. [Google Scholar] [CrossRef]
- Chen, L.; Chen, Y.; Wood, D.W.; Nester, E.W. A New Type IV Secretion System Promotes Conjugal Transfer in Agrobacterium tumefaciens. J. Bacteriol. 2002, 184, 4838–4845. [Google Scholar] [CrossRef]
- Alegria, M.C.; Souza, D.P.; Andrade, M.O.; Docena, C.; Khater, L.; Ramos, C.H.I.; Silva, A.C.R.; Farah, C.S. Identification of new protein-protein interactions involving the products of the chromosome- and plasmid-encoded type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri. J. Bacteriol. 2005, 187, 2315–2325. [Google Scholar] [CrossRef]
- Waksman, G.; Orlova, E.V. Structural organisation of the type IV secretion systems. Curr. Opin. Microbiol. 2014, 17, 24–31. [Google Scholar] [CrossRef]
- Low, H.H.; Gubellini, F.; Rivera-Calzada, A.; Braun, N.; Connery, S.; Dujeancourt, A.; Lu, F.; Redzej, A.; Fronzes, R.; Orlova, E.V.; et al. Structure of a type IV secretion system. Nature 2014, 508, 550–553. [Google Scholar] [CrossRef]
- Souza, D.P.; Oka, G.U.; Alvarez-Martinez, C.E.; Bisson-Filho, A.W.; Dunger, G.; Hobeika, L.; Cavalcante, N.S.; Alegria, M.C.; Barbosa, L.R.S.; Salinas, R.K.; et al. Bacterial killing via a type IV secretion system. Nat. Commun. 2015, 6, 6453. [Google Scholar] [CrossRef]
- Rosenthal, E.; Potnis, N.; Bull, C.T. Comparative genomic analysis of the lettuce bacterial leaf spot pathogen, Xanthomonas hortorum pv. vitians, to investigate race specificity. Front. Microbiol. 2022, 13, 840311. [Google Scholar] [CrossRef]
- Sgro, G.G.; Oka, G.U.; Souza, D.P.; Cenens, W.; Bayer-Santos, E.; Matsuyama, B.Y.; Bueno, N.F.; dos Santos, T.R.; Alvarez-Martinez, C.E.; Salinas, R.K.; et al. Bacteria-Killing Type IV Secretion Systems. Front. Microbiol. 2019, 10, 1078. [Google Scholar] [CrossRef]
- Souza, D.P.; Andrade, M.O.; Alvarez-Martinez, C.E.; Arantes, G.M.; Farah, C.S.; Salinas, R.K. A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PLoS Pathog. 2011, 7, e1002031. [Google Scholar] [CrossRef] [PubMed]
- He, Y.-Q.; Zhang, L.; Jiang, B.-L.; Zhang, Z.-C.; Xu, R.-Q.; Tang, D.-J.; Qin, J.; Jiang, W.; Zhang, X.; Liao, J.; et al. Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris. Genome Biol. 2007, 8, R218. [Google Scholar] [CrossRef] [PubMed]
- Brunet, Y.R.; Zoued, A.; Boyer, F.; Douzi, B.; Cascales, E. The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet. 2015, 11, e1005545. [Google Scholar] [CrossRef]
- Wang, J.; Brodmann, M.; Basler, M. Assembly and subcellular localization of bacterial type VI secretion systems. Annu. Rev. Microbiol. 2019, 73, 621–638. [Google Scholar] [CrossRef]
- Potnis, N.; Krasileva, K.; Chow, V.; Almeida, N.F.; Patil, P.B.; Ryan, R.P.; Sharlach, M.; Behlau, F.; Dow, J.M.; Momol, M.; et al. Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genom. 2011, 12, 1–23. [Google Scholar] [CrossRef]
- Montenegro, A.; Alvarez, B.A.; Arrieta-Ortiz, M.L.; Rodriguez-R, L.M.; Botero, D.; Tabima, J.F.; Castiblanco, L.; Trujillo, C.; Restrepo, S.; Bernal, A. The type VI secretion system of Xanthomonas phaseoli pv. manihotis is involved in virulence and in vitro motility. BMC Microbiol. 2021, 21, 14. [Google Scholar] [CrossRef]
- Bayer-Santos, E.; Ceseti, L.d.M.; Farah, C.S.; Alvarez-Martinez, C.E. Distribution, function and regulation of type 6 secretion systems of Xanthomonadales. Front. Microbiol. 2019, 10, 1635. [Google Scholar] [CrossRef]
- Hood, R.D.; Singh, P.; Hsu, F.; Güvener, T.; Carl, M.A.; Trinidad, R.R.S.; Silverman, J.M.; Ohlson, B.B.; Hicks, K.G.; Plemel, R.L.; et al. A Type VI Secretion System of Pseudomonas aeruginosa Targets a Toxin to Bacteria. Cell Host Microbe 2010, 7, 25–37. [Google Scholar] [CrossRef]
- Bayer-Santos, E.; Lima, L.d.P.; Ceseti, L.d.M.; Ratagami, C.Y.; de Santana, E.S.; da Silva, A.M.; Farah, C.S.; Alvarez-Martinez, C.E. Xanthomonas citri T6SS mediates resistance to Dictyostelium predation and is regulated by an ECF σ factor and cognate Ser/Thr kinase. Environ. Microbiol. 2018, 20, 1562–1575. [Google Scholar] [CrossRef]
- Choi, Y.; Kim, N.; Mannaa, M.; Kim, H.; Park, J.; Jung, H.; Han, G.; Lee, H.-H.; Seo, Y.-S. Characterization of Type VI Secretion System in Xanthomonas oryzae pv. oryzae and Its Role in Virulence to Rice. Plant Pathol. J. 2020, 36, 289–296. [Google Scholar] [CrossRef]
- Zhu, P.-C.; Li, Y.-M.; Yang, X.; Zou, H.-F.; Zhu, X.-L.; Niu, X.-N.; Xu, L.-H.; Jiang, W.; Huang, S.; Tang, J.-L.; et al. Type VI secretion system is not required for virulence on rice but for inter-bacterial competition in Xanthomonas oryzae pv. oryzicola. Res. Microbiol. 2019, 171, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Liyanapathiranage, P.; Jones, J.B.; Potnis, N. Mutation of a single core gene, tssM, of type VI secretion system of Xanthomonas perforans influences virulence, epiphytic survival, and transmission during pathogenesis on tomato. Phytopathology 2022, 112, 752–764. [Google Scholar] [CrossRef] [PubMed]
- Ceseti, L.M.; de Santana, E.S.; Ratagami, C.Y.; Barreiros, Y.; Lima, L.D.P.; Dunger, G.; Farah, C.S.; Alvarez-Martinez, C.E. The Xanthomonas citri pv. citri Type VI Secretion System is induced during epiphytic colonization of citrus. Curr. Microbiol. 2019, 76, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Vieira, P.S.; Bonfim, I.M.; Araujo, E.A.; Melo, R.R.; Lima, A.R.; Fessel, M.R.; Murakami, M.T. Xyloglucan processing machinery in Xanthomonas pathogens and its role in the transcriptional activation of virulence factors. Nat. Commun. 2021, 12, 4049. [Google Scholar] [CrossRef]
- Rajeshwari, R.; Jha, G.; Sonti, R.V. Role of an in planta-expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol. Plant-Microbe Interact. 2005, 18, 830–837. [Google Scholar] [CrossRef]
- Jha, G.; Rajeshwari, R.; Sonti, R.V. Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. Mol. Plant-Microbe Interact. 2007, 20, 31–40. [Google Scholar] [CrossRef]
- Sidhu, V.K.; Vorhölter, F.-J.; Niehaus, K.; Watt, S.A. Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris. BMC Microbiol. 2008, 8, 87. [Google Scholar] [CrossRef]
- Tayi, L.; Maku, R.; Patel, H.K.; Sonti, R.V. Action of multiple cell wall–degrading enzymes is required for elicitation of innate immune responses during Xanthomonas oryzae pv. oryzae infection in rice. Mol. Plant-Microbe Interact. 2016, 29, 599–608. [Google Scholar] [CrossRef]
- Zou, H.-S.; Song, X.; Zou, L.-F.; Yuan, L.; Li, Y.-R.; Guo, W.; Che, Y.-Z.; Zhao, W.-X.; Duan, Y.-P.; Chen, G.-Y. EcpA, an extracellular protease, is a specific virulence factor required by Xanthomonas oryzae pv. oryzicola but not by X. oryzae pv. oryzae in rice. Microbiology 2012, 158, 2372–2383. [Google Scholar] [CrossRef]
- Ray, S.K.; Rajeshwari, R.; Sharma, Y.; Sonti, R.V. A high-molecular-weight outer membrane protein of Xanthomonas oryzae pv. exhibits similarity to non-fimbrial adhesins of animal pathogenic bacteria and is required for optimum virulence. Mol. Microbiol. 2002, 46, 637–647. [Google Scholar] [CrossRef]
- Moens, S.; Vanderleyden, J. Functions of bacterial flagella. Crit. Rev. Microbiol. 1996, 22, 67–100. [Google Scholar] [CrossRef] [PubMed]
- Kirov, S.M. Bacteria that express lateral flagella enable dissection of the multifunctional roles of flagella in pathogenesis. FEMS Microbiol. Lett. 2003, 224, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Haiko, J.; Westerlund-Wikström, B. The role of the bacterial flagellum in adhesion and virulence. Biology 2013, 2, 1242–1267. [Google Scholar] [CrossRef] [PubMed]
- Kubori, T.; Shimamoto, N.; Yamaguchi, S.; Namba, K.; Aizawa, S.-I. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 1992, 226, 433–446. [Google Scholar] [CrossRef]
- Morimoto, Y.V.; Minamino, T. Structure and function of the bi-directional bacterial flagellar motor. Biomolecules 2014, 4, 217–234. [Google Scholar] [CrossRef]
- Yang, T.C.; Leu, Y.W.; Chang-Chien, H.C.; Hu, R.M. Flagellar biogenesis of Xanthomonas campestris requires the alternative sigma factors RpoN2 and FliA and is temporally regulated by FlhA, FlhB, and FlgM. J. Bacteriol. 2009, 191, 2266–2275. [Google Scholar] [CrossRef]
- Lu, C.D. Characterization of the fleN gene in Xanthomonas campestris pv. Campestris. Master’s Thesis, Asia University, Taichung City, Taiwan, 2005. Available online: https://hdl.handle.net/11296/bjnz8f (accessed on 22 June 2025).
- Shen, Y.; Chern, M.-S.; Goes Silva, F.; Ronald, P. Isolation of a Xanthomonas oryzae pv. oryzae flagellar operon region and molecular characterization of flhF. Mol. Plant-Microbe Interact. 2001, 14, 204–213. [Google Scholar] [CrossRef]
- Chang-Chien, H.C. Characterization of the fliA, flgM, flhF genes in Xanthomonas campestris pv. campestris. Master’s Thesis, Asia University, Taichung City, Taiwan, 2007. Available online: https://hdl.handle.net/11296/n58trv (accessed on 22 June 2025).
- Gicharu, G.K.; Sun, D.-L.; Hu, X.; Fan, X.-J.; Zhuo, T.; Wu, C.-W.; Zou, H.-S. The sigma 54 genes rpoN1 and rpoN2 of Xanthomonas citri subsp. citri play different roles in virulence, nutrient utilization and cell motility. J. Integr. Agric. 2016, 15, 2032–2039. [Google Scholar] [CrossRef]
- Li, K.; Wu, G.; Liao, Y.; Zeng, Q.; Wang, H.; Liu, F. RpoN1 and RpoN2 play different regulatory roles in virulence traits, flagellar biosynthesis, and basal metabolism in Xanthomonas campestris. Mol. Plant Pathol. 2020, 21, 907–922. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).