Gut Microbiome Engineering for Diabetic Kidney Disease Prevention: A Lactobacillus rhamnosus GG Intervention Study
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Protocol
2.2. Sample Collection and Processing
2.3. Biochemical and Metabolic Assessments
2.4. Metabolic Tolerance Testing
2.5. Histopathological Examination
2.6. Microbiome Analysis
2.6.1. Collection and Preservation of Fecal Samples
2.6.2. DNA Extraction Protocol
2.6.3. Sequence Analysis of the 16S rRNA Gene
2.6.4. Bioinformatics Pipeline
2.7. Alpha Diversity Analysis
- Chao1 index: Estimating species richness with correction for rare taxa;
- According to the Shannon diversity index, which takes into consideration both the richness and evenness of the community’s makeup;
- Faith’s phylogenetic diversity: Incorporating evolutionary relationships between microbial taxa;
- Observed features: Direct count of unique ASVs present in each sample;
- Simpson’s evenness index: Specifically quantifying the equitability of species distribution.
2.8. Beta Diversity and Community Structure
- Weighted UniFrac: Incorporating both phylogenetic relatedness and abundance information;
- Unweighted UniFrac: Focusing on phylogenetic relationships alone (presence/absence);
- Bray–Curtis dissimilarity: Quantifying compositional differences independent of phylogeny;
- Jaccard distance: Assessing community membership overlap.
2.9. Taxonomic Composition Analysis
2.10. Microbiome Ratio Analysis and Functional Prediction
- The Firmicutes to Bacteroidetes (F/B) ratio was estimated as a well-known indicator of the makeup of the gut microbiome. This ratio is often connected with metabolic health status, obesity, and characteristics of metabolic syndrome.
- Prevotella/Bacteroides ratio: Associated with dietary patterns and inflammatory status.
- Beneficial to pathogenic bacteria ratio: Customized metric quantifying the balance of health-promoting versus potentially harmful bacterial populations, with a specific focus on butyrate-producers (Faecalibacterium, Roseburia, Eubacterium) versus uremic toxin producers (Enterobacteriaceae, Clostridioides).
2.11. Statistical Analysis
3. Results
3.1. The Effects of Oral Supplementation with Lactobacillus rhamnosus GG on the Body Weight and Metabolic Indices of Diabetic Rats
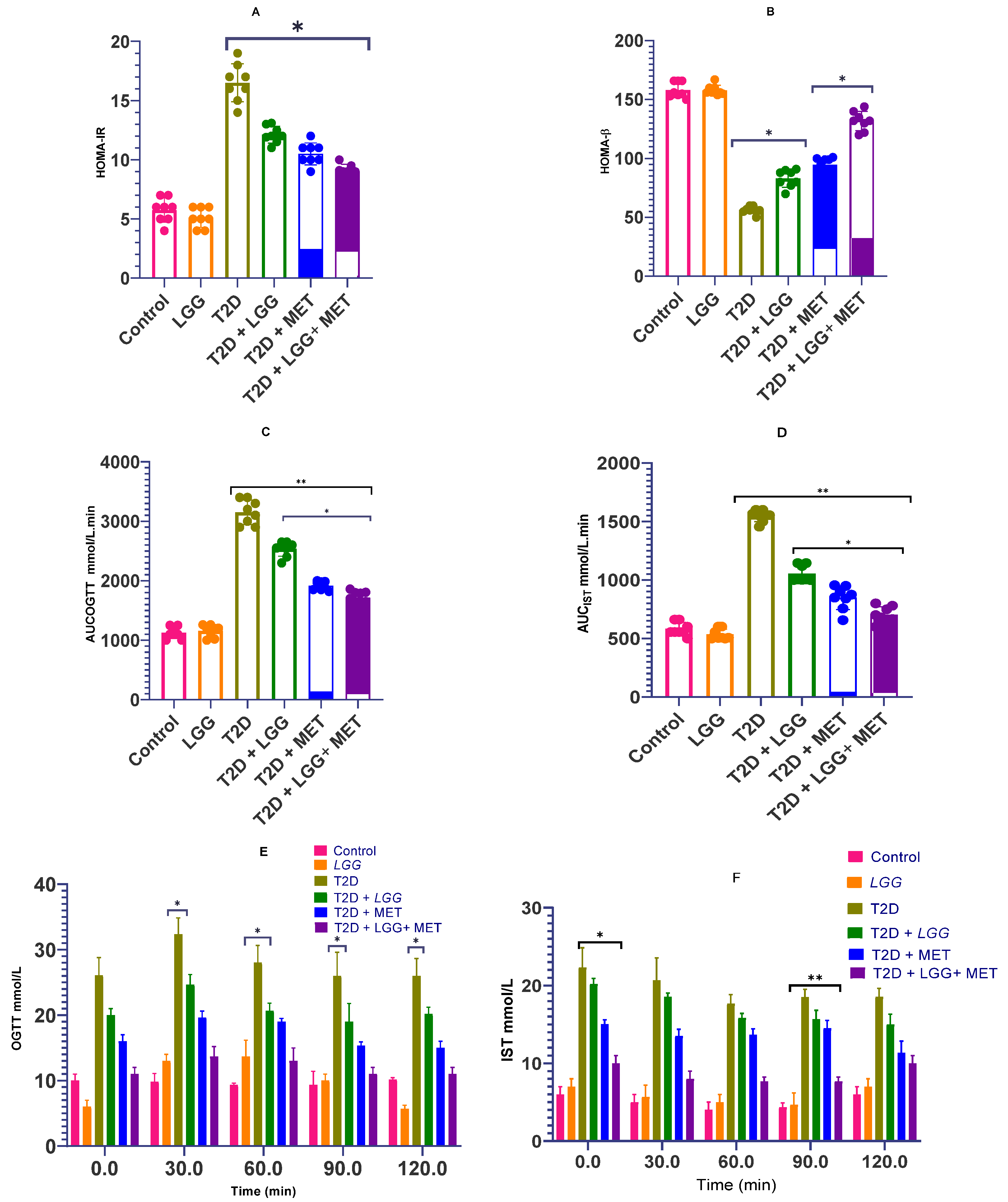
3.2. The Effects of Lactobacillus rhamnosus GG on the Homeostasis of Glucose and Insulin Sensitivity in Rats with Diabetes
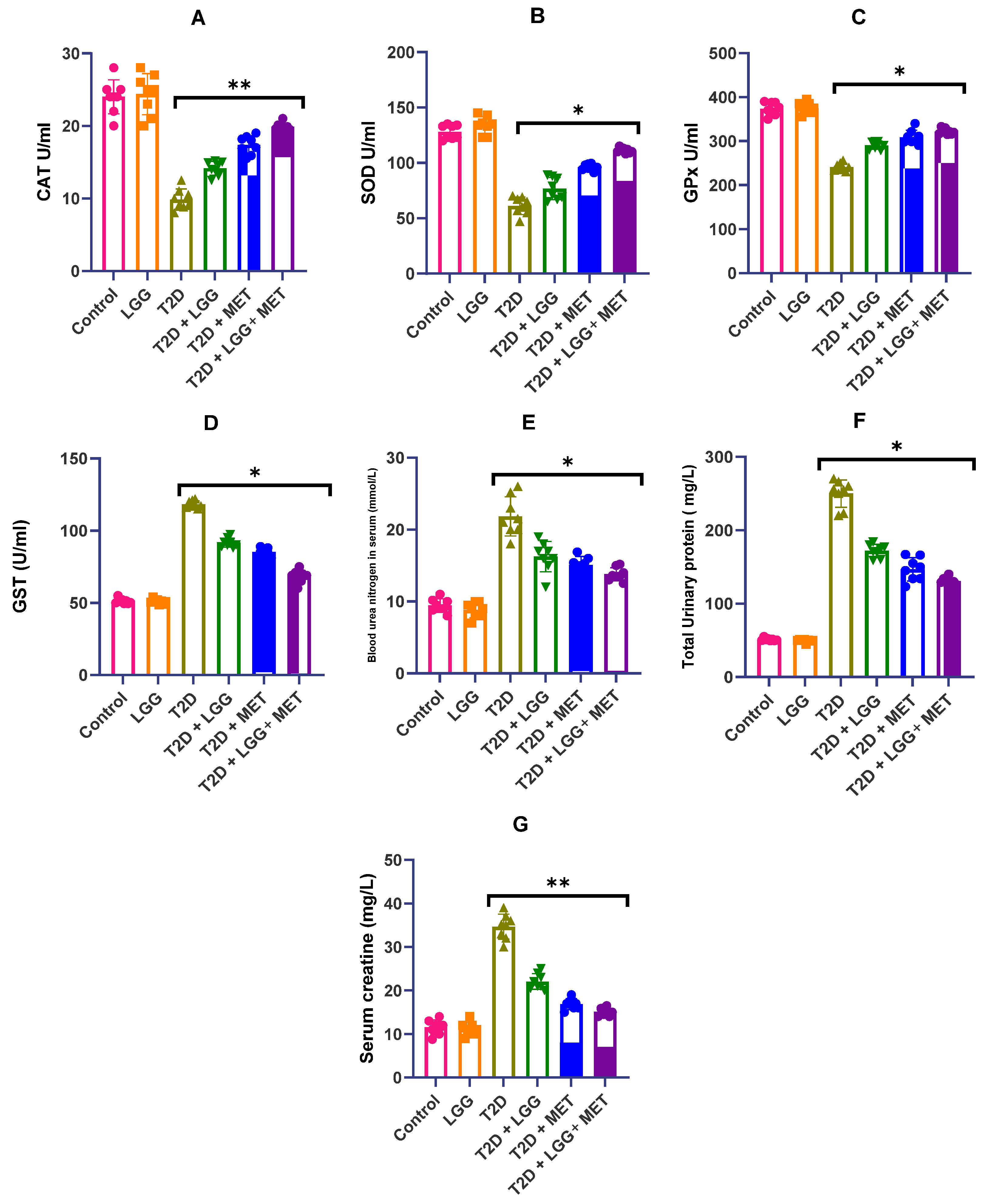
3.3. Analysis of the Influence of Lactobacillus rhamnosus GG on the Activity of Antioxidant Enzymes in Diabetic Rats
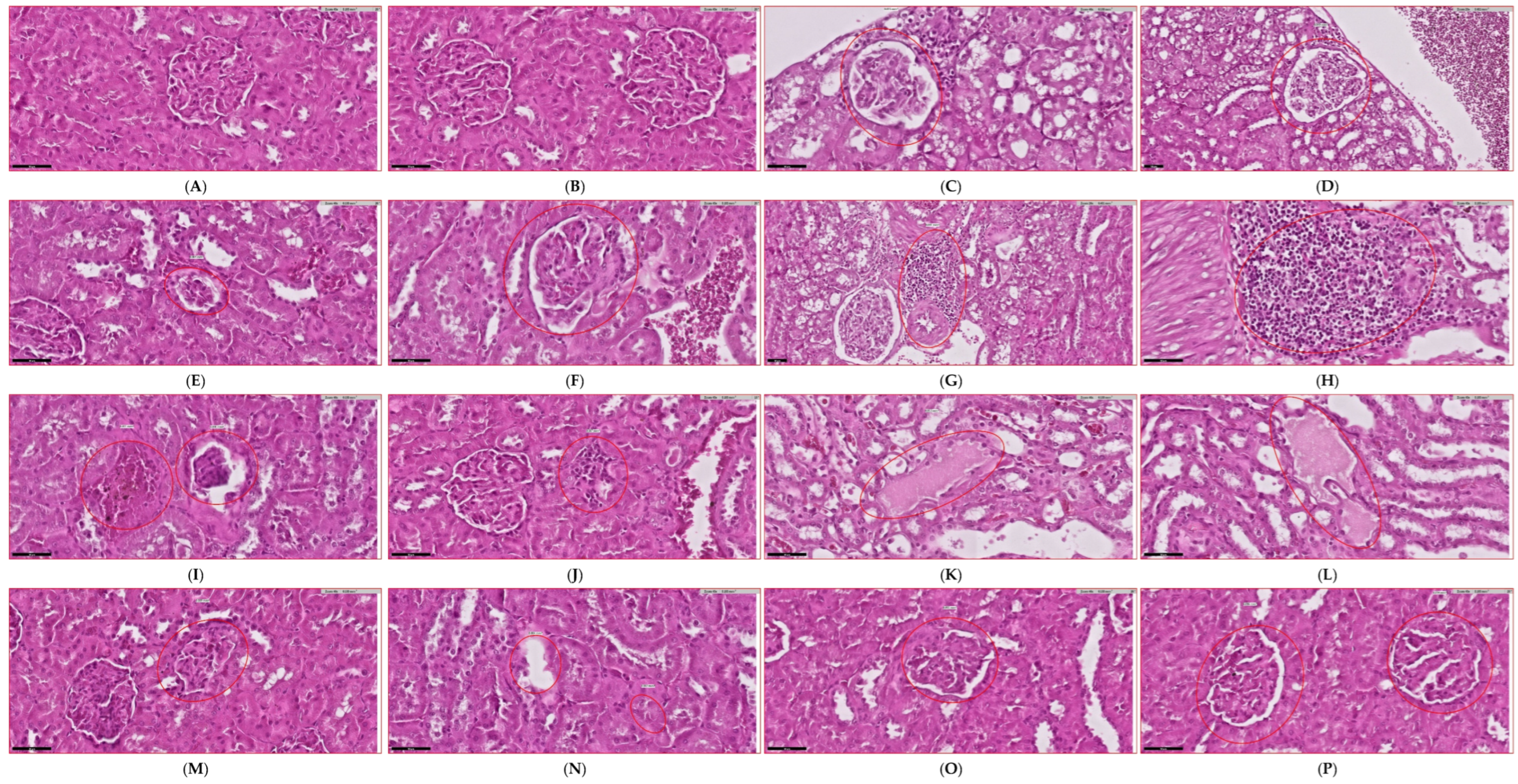
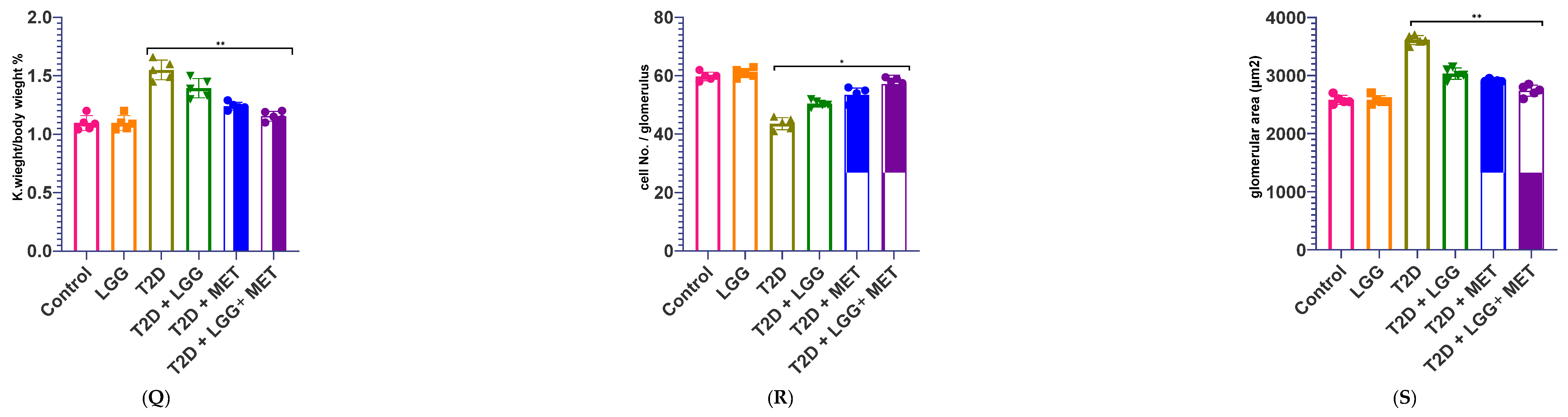
3.4. Evaluation of Renal Functional Parameters
3.5. Gut Microbiome Analysis
3.5.1. Bacterial Phyla Distribution
3.5.2. Alpha Diversity Metrics
3.5.3. Ratio Analysis of Firmicutes and Bacteroidetes
3.5.4. Taxonomic Abundance Profiling
3.5.5. Beta Diversity and Community Structure
3.5.6. Functional Metagenomic Profiling
3.5.7. Phylum-Level Compositional Analysis
4. Discussion
5. Limitations and Future Directions
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gheith, O.; Farouk, N.; Nampoory, N.; Halim, M.; Al-Otaibi, T. Diabetic kidney disease: Worldwide difference of prevalence and risk factors. J. Nephropharmacol. 2015, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, N.D.; Wong, J.; Pahl, M.; Piceno, Y.M.; Yuan, J.; DeSantis, T.Z.; Ni, Z.; Nguyen, T.H.; Andersen, G.L. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013, 83, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef]
- Ishfaq, M.; Hu, W.; Hu, Z.; Guan, Y.; Zhang, R. A review of nutritional implications of bioactive compounds of Ginger (Zingiber officinale Roscoe), their biological activities and nano-formulations. Ital. J. Food Sci. 2022, 34, 1–12. [Google Scholar] [CrossRef]
- Fernández-Pampin, N.; González Plaza, J.J.; García-Gómez, A.; Peña, E.; Rumbo, C.; Barros, R.; Martel-Martín, S.; Aparicio, S.; Tamayo-Ramos, J.A. Toxicology assessment of manganese oxide nanomaterials with enhanced electrochemical properties using human in vitro models representing different exposure routes. Sci. Rep. 2022, 12, 20991. [Google Scholar] [CrossRef]
- Song, Y.; Hu, Z.; Xiong, Z.; Li, S.; Liu, W.; Tian, T.; Yang, X. Comparative transcriptomic and lipidomic analyses indicate that cold stress enhanced the production of the long C18–C22 polyunsaturated fatty acids in Aurantiochytrium sp. Front. Microbiol. 2022, 13, 915773. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, L.; Jin, Z.; Zhang, D. Microbial Cell Factories for Green Production of Vitamins. Front. Bioeng. Biotechnol. 2021, 9, 661562. [Google Scholar] [CrossRef]
- Tan, H.; Chen, J.; Li, Y.; Li, Y.; Zhong, Y.; Li, G.; Liu, L.; Li, Y. Glabridin, a bioactive component of licorice, ameliorates diabetic nephropathy by regulating ferroptosis and the VEGF/Akt/ERK pathways. Mol. Med. 2022, 28, 58. [Google Scholar] [CrossRef]
- Xu, Z.; Zhao, S.J.; Liu, Y. 1,2,3-Triazole-containing hybrids as potential anticancer agents: Current developments, action mechanisms and structure-activity relationships. Eur. J. Med. Chem. 2019, 183, 111700. [Google Scholar] [CrossRef]
- Huang, B.; Song, B.L.; Xu, C. Cholesterol metabolism in cancer: Mechanisms and therapeutic opportunities. Nat. Metab. 2020, 2, 132–141. [Google Scholar] [CrossRef]
- Arcidiacono, M.V.; Carrillo-López, N.; Panizo, S.; Castro-Grattoni, A.L.; Valcheva, P.; Ulloa, C.; Rodríguez-Carrio, J.; Cardús, A.; Quirós-Caso, C.; Martínez-Arias, L.; et al. Barley-ß-glucans reduce systemic inflammation, renal injury and aortic calcification through ADAM17 and neutral-sphingomyelinase2 inhibition. Sci. Rep. 2019, 9, 17810. [Google Scholar] [CrossRef]
- Zhao, Z.; Moghadasian, M.H. Chemistry, natural sources, dietary intake and pharmacokinetic properties of ferulic acid: A review. Food Chem. 2008, 109, 691–702. [Google Scholar] [CrossRef]
- Wang, Y.; Sui, Z.; Wang, M.; Liu, P. Natural products in attenuating renal inflammation via inhibiting the NLRP3 inflammasome in diabetic kidney disease. Front. Immunol. 2023, 14, 1196016. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, W.; Cai, J.; Liu, W.; Akihisa, T.; Li, W.; Kikuchi, T.; Xu, J.; Feng, F.; Zhang, J. The role of metabolites of steviol glycosides and their glucosylated derivatives against diabetes-related metabolic disorders. Food Funct. 2021, 12, 8248–8259. [Google Scholar] [CrossRef] [PubMed]
- Kawano, K.; Mori, S.; Hirashima, T.; Man, Z.W.; Natori, T. Examination of the pathogenesis of diabetic nephropathy in OLETF rats. J. Vet. Med. Sci. 1999, 61, 1219–1222. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.H.; Sweet, D.H.; Lin, S.P.; Yu, C.P.; Chao, P.D.L.; Hou, Y.C. Green tea inhibited the elimination of nephro-cardiovascular toxins and deteriorated the renal function in rats with renal failure. Sci. Rep. 2015, 5, 16226. [Google Scholar] [CrossRef]
- Hu, M.C.; Shi, M.; Zhang, J.; Addo, T.; Cho, H.J.; Barker, S.L.; Ravikumar, P.; Gillings, N.; Bian, A.; Sidhu, S.S.; et al. Renal production, uptake, and handling of circulating αKlotho. J. Am. Soc. Nephrol. 2016, 27, 79–90. [Google Scholar] [CrossRef]
- Gallardo-Becerra, L.; Cornejo-Granados, F.; García-López, R.; Valdez-Lara, A.; Bikel, S.; Canizales-Quinteros, S.; López-Contreras, B.E.; Mendoza-Vargas, A.; Nielsen, H.; Ochoa-Leyva, A. Metatranscriptomic analysis to define the Secrebiome, and 16S rRNA profiling of the gut microbiome in obesity and metabolic syndrome of Mexican children. Microb. Cell Fact. 2020, 19, 61. [Google Scholar] [CrossRef]
- Stevens, L.A.; Coresh, J.; Greene, T.; Levey, A.S. Assessing kidney function—Measured and estimated glomerular filtration rate. N. Engl. J. Med. 2006, 354, 2473–2483. [Google Scholar] [CrossRef]
- Vega Joubert, M.B.; Ingaramo, P.; Oliva, M.E.; D’Alessandro, M.E. Salvia hispanica L. (chia) seed ameliorates liver injury and oxidative stress by modulating NrF2 and NFκB expression in sucrose-rich diet-fed rats. Food Funct. 2022, 13, 7333–7345. [Google Scholar] [CrossRef]
- Abbassi, R.; Pontes, M.C.; Dhibi, S.; Duarte Filho, L.A.; Othmani, S.; Bouzenna, H.; Almeida, J.R.; Hfaiedh, N. Antioxidant properties of date seeds extract (Phoenix dactylifera L.) in alloxan induced damage in rats. Braz. J. Biol. 2023, 83, e274405. [Google Scholar] [CrossRef]
- Sabra, M.S.; Hemida, F.K.; Allam, E.A.H. Adenine model of chronic renal failure in rats to determine whether MCC950, an NLRP3 inflammasome inhibitor, is a renopreventive. BMC Nephrol. 2023, 24, 377. [Google Scholar] [CrossRef]
- Al Za’abi, M.; Ali, H.; Ali, B.H. Effect of flaxseed on systemic inflammation and oxidative stress in diabetic rats with or without chronic kidney disease. PLoS ONE 2021, 16, e0258800. [Google Scholar] [CrossRef]
- Al-Attar, A.M.; Abu Zeid, I.M.; Felemban, L.F.; Omar, A.M.S.; Alkenani, N.A. Protective and antioxidant effects of chia oil and canola oil on testicular injury induced by lead in rats. J. King Saud Univ. Sci. 2023, 35, 102894. [Google Scholar] [CrossRef]
- Hardianto, D. Telaah komprehensif diabetes melitus: Klasifikasi, gejala, diagnosis, pencegahan, dan pengobatan. J. Bioteknol. Biosains Indones. 2020, 7, 304–317. [Google Scholar] [CrossRef]
- Li, D.; Wang, P.; Wang, P.; Hu, X.; Chen, F. Gut microbiota promotes production of aromatic metabolites through degradation of barley leaf fiber. J. Nutr. Biochem. 2018, 58, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, X.; Deng, H.; Huang, H.; Liu, Y.; Zhong, Z.; Shen, L.; Cao, S.; Ma, X.; Zhou, Z.; et al. Integrated transcriptome and metabolomics to reveal the mechanism of adipose mesenchymal stem cells in treating liver fibrosis. Int. J. Mol. Sci. 2023, 24, 16086. [Google Scholar] [CrossRef]
- Ghasemi, A.; Jeddi, S. Streptozotocin as a tool for induction of rat models of diabetes: A practical guide. EXCLI J. 2023, 22, 274–294. [Google Scholar] [CrossRef]
- Sreelatha, G.; Muraleedharan, A.; Sathidevi, P.; Chand, P.; Rajkumar, R. Quantification of DNA damage by the analysis of silver stained comet assay images. IRBM 2015, 36, 306–314. [Google Scholar] [CrossRef]
- Anderson, N.J.; Smith, C.K.; Byrd, J.L. Work-related injury factors and safety climate perception in truck drivers. Am. J. Ind. Med. 2017, 60, 711–723. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Shannon, C.E. A mathematical theory of communication. Bell. Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Illumina. High-throughput sequencing technology for microbial communities. Nat. Methods 2013, 10, 999–1000. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Syst. Rev. 2021, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Vinolo, M.A.; Ferguson, G.J.; Kulkarni, S.; Damoulakis, G.; Anderson, K.; Bohlooly, Y.M.; Stephens, L.; Hawkins, P.T.; Curi, R. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS ONE 2011, 6, e21205. [Google Scholar] [CrossRef]
- Zhong, Y.; Marungruang, N.; Fåk, F.; Nyman, M. Effects of two whole-grain barley varieties on caecal SCFA, gut microbiota and plasma inflammatory markers in rats consuming low- and high-fat diets. Br. J. Nutr. 2015, 113, 1558–1570. [Google Scholar] [CrossRef]
- Dong, N.; Xu, B.; Shi, H. Long noncoding RNA MALAT1 acts as a competing endogenous RNA to regulate Amadori-glycated albumin-induced MCP-1 expression in retinal microglia by a microRNA-124-dependent mechanism. Inflamm. Res. 2018, 67, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Canfora, E.E.; Meex, R.C.R.; Venema, K.; Blaak, E.E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 2019, 15, 261–273. [Google Scholar] [CrossRef]
- Yan, J.K.; Chen, T.T.; Chen, X.; Liu, Y.; Liu, C.; Li, L. Assessing the product quality and biological activities of barley (Hordeum vulgare L.) grasses at different harvest times. Food Biosci. 2022, 46, 101549. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, J.; Liu, J.; Wang, Z.; Chen, M.; Zhou, S. Metagenome of gut microbiota of children with nonalcoholic fatty liver disease. Front. Pediatr. 2019, 7, 518. [Google Scholar] [CrossRef]
- Shokolenko, I.; Venediktova, N.; Bochkareva, A.; Wilson, G.L.; Alexeyev, M.F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009, 37, 2539–2548. [Google Scholar] [CrossRef]
- Jomova, K.; Vondrakova, D.; Lawson, M.; Valko, M. Redox active metal-induced oxidative stress in biological systems. Transit. Met. Chem. 2012, 37, 127–134. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, Y.; Yang, C. Evaluating In Vitro DNA damage using Comet assay. J. Vis. Exp. 2017, 128, e56450. [Google Scholar] [CrossRef]
- Saadatzadeh, A.; Fazeli, M.R.; Jamalifar, H.; Dinarvand, R. Probiotic properties of lyophilized cell-free extract of Lactobacillus casei. Jundishapur J. Nat. Pharm. Prod. 2013, 8, 131–137. [Google Scholar] [CrossRef]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef]
- Gandhi, G.D.; Aamer, W.; Krishnamoorthy, N.; Syed, N.; Aliyev, E.; Al-Maraghi, A.; Kohailan, M.; Alenbawi, J.; Elanbari, M.; Qatar Genome Program Research Consortium (QGPRC); et al. Assessing the genetic burden of familial hypercholesterolemia in a large middle eastern biobank. J. Transl. Med. 2022, 20, 502. [Google Scholar] [CrossRef]
- De Falco, P.; Lazzarino, G.; Felice, F.; Desideri, E.; Castelli, S.; Salvatori, I.; Ciccarone, F.; Ciriolo, M.R. Hindering NAT8L expression in hepatocellular carcinoma increases cytosolic aspartate delivery that fosters pentose phosphate pathway and purine biosynthesis promoting cell proliferation. Redox Biol. 2023, 59, 102585. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lou, H.; Peng, J.; Wang, L.; Li, Z.; Zhao, J.; Li, Y.; Sun, Y.; Zhou, H.; Zhang, X.; et al. Depletion of butyrate-producing bacteria is associated with disease severity in patients with diabetic kidney disease. Gut Microbes 2023, 15, 2166870. [Google Scholar]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Mannerås-Holm, L.; Ståhlman, M.; Olsson, L.M.; Serino, M.; Planas-Fèlix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef]
- He, J.; Zhang, P.; Shen, L.; Niu, L.; Tan, Y.; Chen, L.; Luo, M.; Wang, Z.; Liu, Y.; Zhang, Y.; et al. Short-chain fatty acids mediate the protective effect of gut microbiota on endothelial function and vascular inflammation in hypertension. Front. Pharmacol. 2021, 12, 703486. [Google Scholar]
- Zhang, X.; Zhao, Y.; Xu, J.; Xue, Z.; Zhang, M.; Pang, X.; Zhang, X.; Zhao, L. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 2015, 5, 14405. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Depommier, C.; Derrien, M.; Everard, A. Akkermansia muciniphila: Paradigm for mucosa–microbiota interactions in the human gut. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 691–705. [Google Scholar]
- Davie, J.R. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003, 133 (Suppl. 7), 2485S–2493S. [Google Scholar] [CrossRef]
- Corrêa-Oliveira, R.; Fachi, J.L.; Vieira, A.; Sato, F.T.; Vinolo, M.A. Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 2016, 5, e73. [Google Scholar] [CrossRef]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Lordan, C.; Cryan, J.F.; Dinan, T.G. Gut microbes, diet and mental health: Connections and mechanisms. Nutrients 2019, 11, 1276. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- Wang, B.; Yao, M.; Lv, L.; Ling, Z.; Li, L. The human microbiota in health and disease. Engineering 2017, 3, 71–82. [Google Scholar] [CrossRef]
- Tang, W.H.; Kitai, T.; Hazen, S.L. Gut microbiota in cardiovascular health and disease. Circ Res. 2017, 120, 1183–1196. [Google Scholar] [CrossRef]
- Kitada, M.; Ogura, Y.; Monno, I.; Koya, D. A low-protein diet for diabetic kidney disease: Its effect and molecular mechanism, an approach from animal studies. Nutrients 2018, 10, 544. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Mahurkar, A.; Rahnavard, G.; Crabtree, J.; Orvis, J.; Hall, A.B.; Brady, A.; Creasy, H.H.; McCracken, C.; Giglio, M.G.; et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017, 550, 61–66. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; Deroos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qumsani, A.T. Gut Microbiome Engineering for Diabetic Kidney Disease Prevention: A Lactobacillus rhamnosus GG Intervention Study. Biology 2025, 14, 723. https://doi.org/10.3390/biology14060723
Qumsani AT. Gut Microbiome Engineering for Diabetic Kidney Disease Prevention: A Lactobacillus rhamnosus GG Intervention Study. Biology. 2025; 14(6):723. https://doi.org/10.3390/biology14060723
Chicago/Turabian StyleQumsani, Alaa Talal. 2025. "Gut Microbiome Engineering for Diabetic Kidney Disease Prevention: A Lactobacillus rhamnosus GG Intervention Study" Biology 14, no. 6: 723. https://doi.org/10.3390/biology14060723
APA StyleQumsani, A. T. (2025). Gut Microbiome Engineering for Diabetic Kidney Disease Prevention: A Lactobacillus rhamnosus GG Intervention Study. Biology, 14(6), 723. https://doi.org/10.3390/biology14060723




