Study on the Degradation of Aflatoxin B1 by Myroides odoratimimus 3J2MO
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Degradation Matrix and Optimization of Conditions
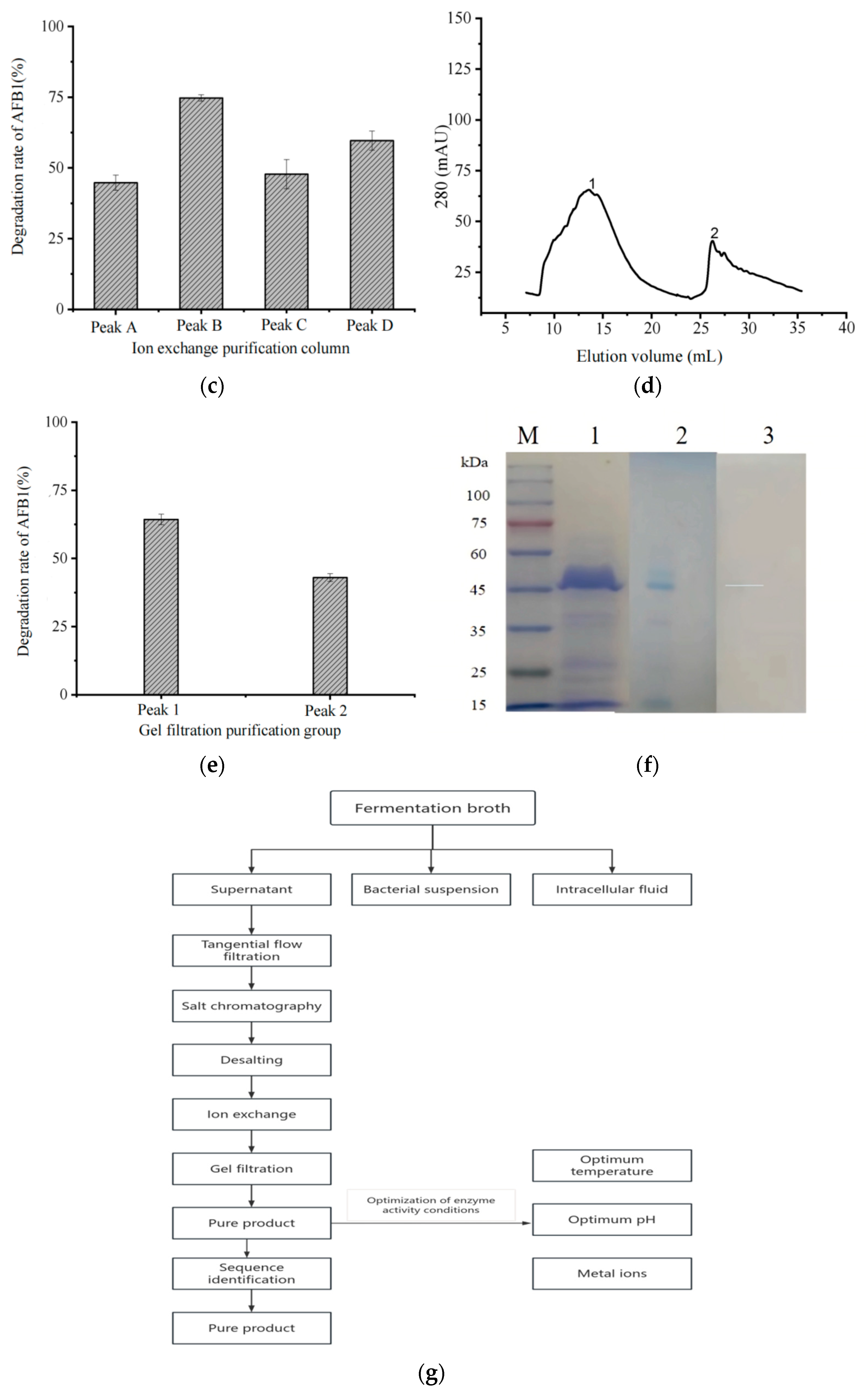
2.2. Purification and Identification of AFB1-Degrading Enzyme
2.3. Bioinformatics Experiment Design
2.4. Data Analysis
3. Results
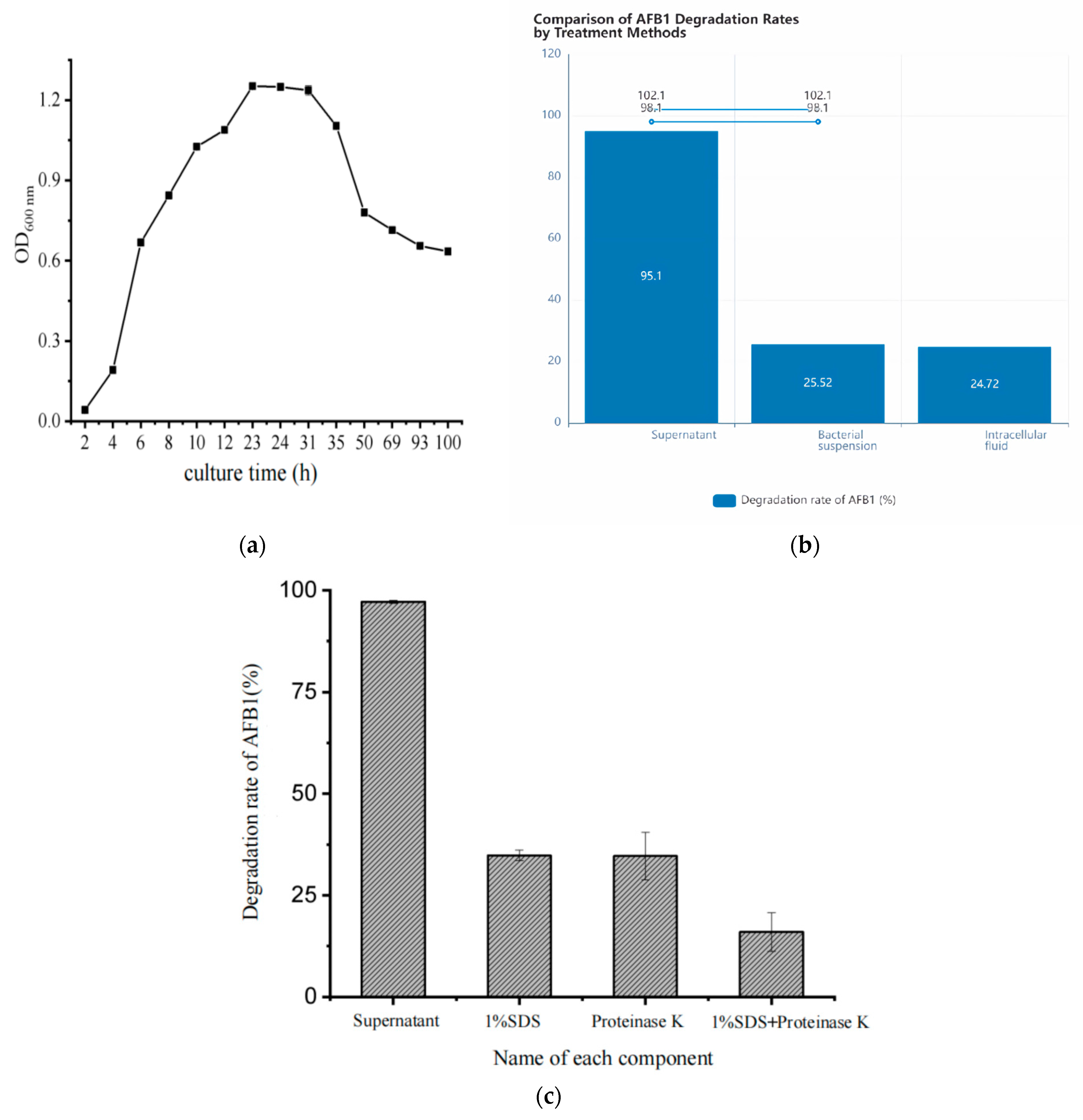
3.1. Determination of Degradation Conditions
3.2. Determination of Degradative Function
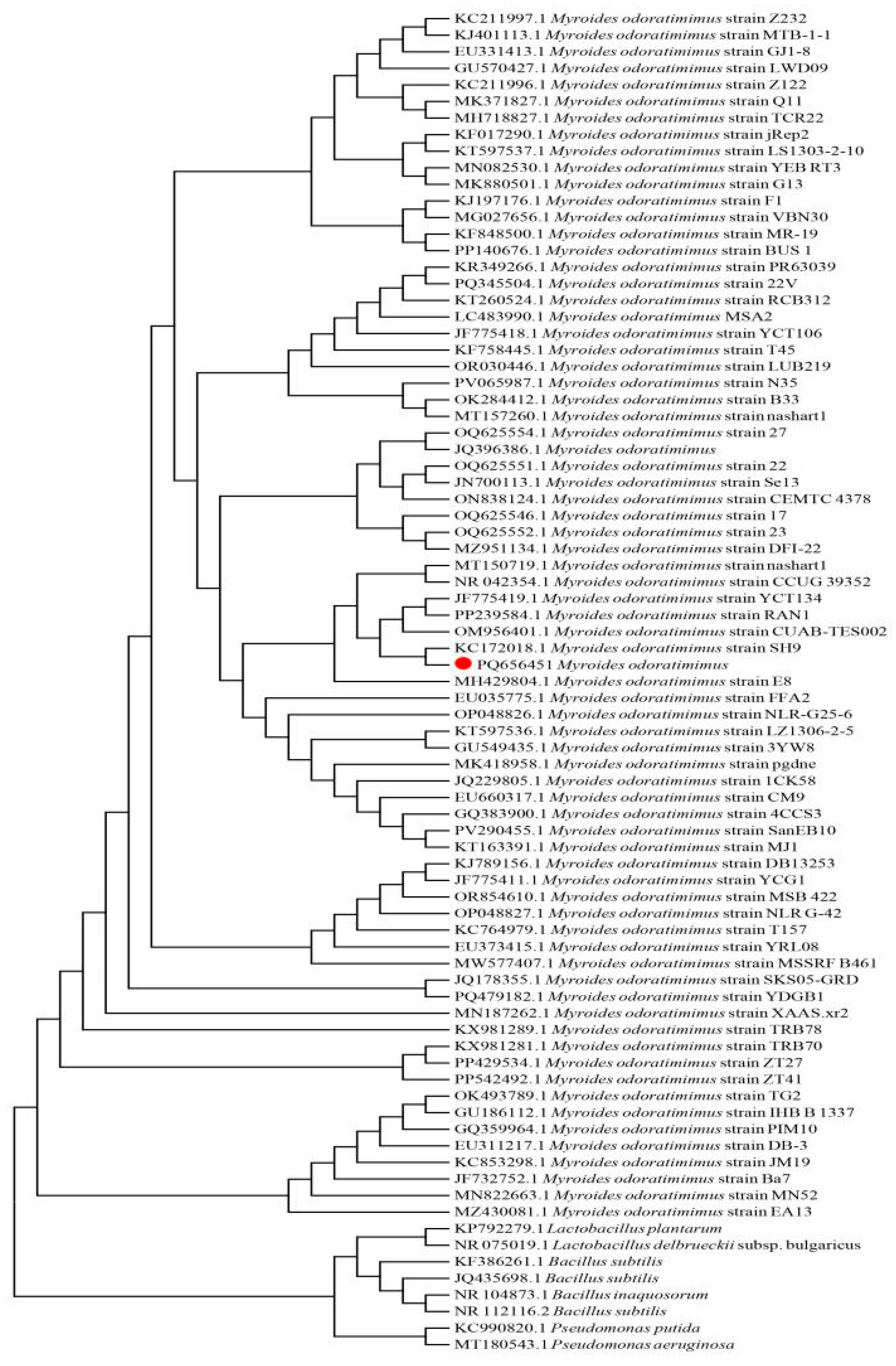
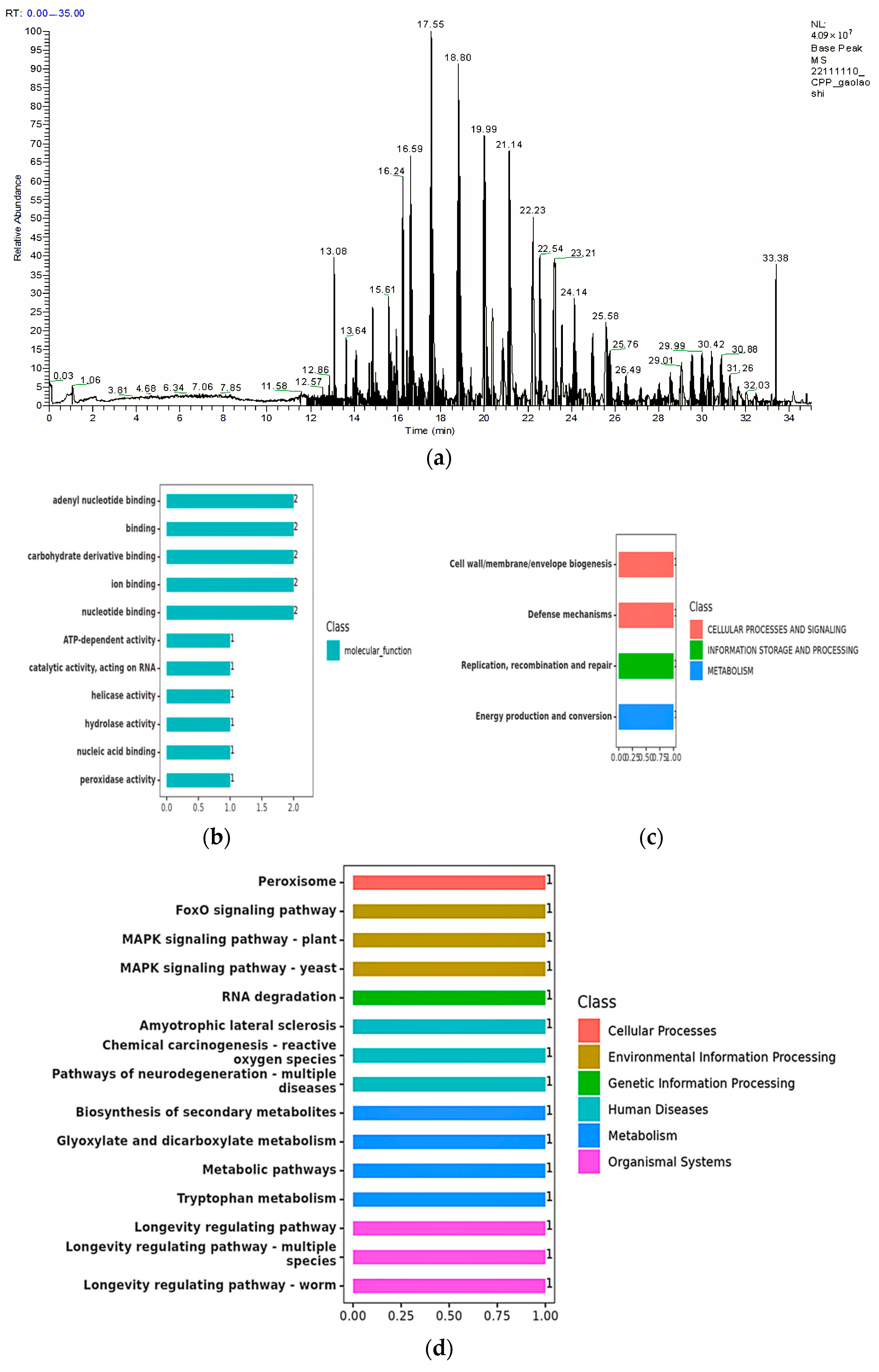
3.3. Bioinformatics Analysis
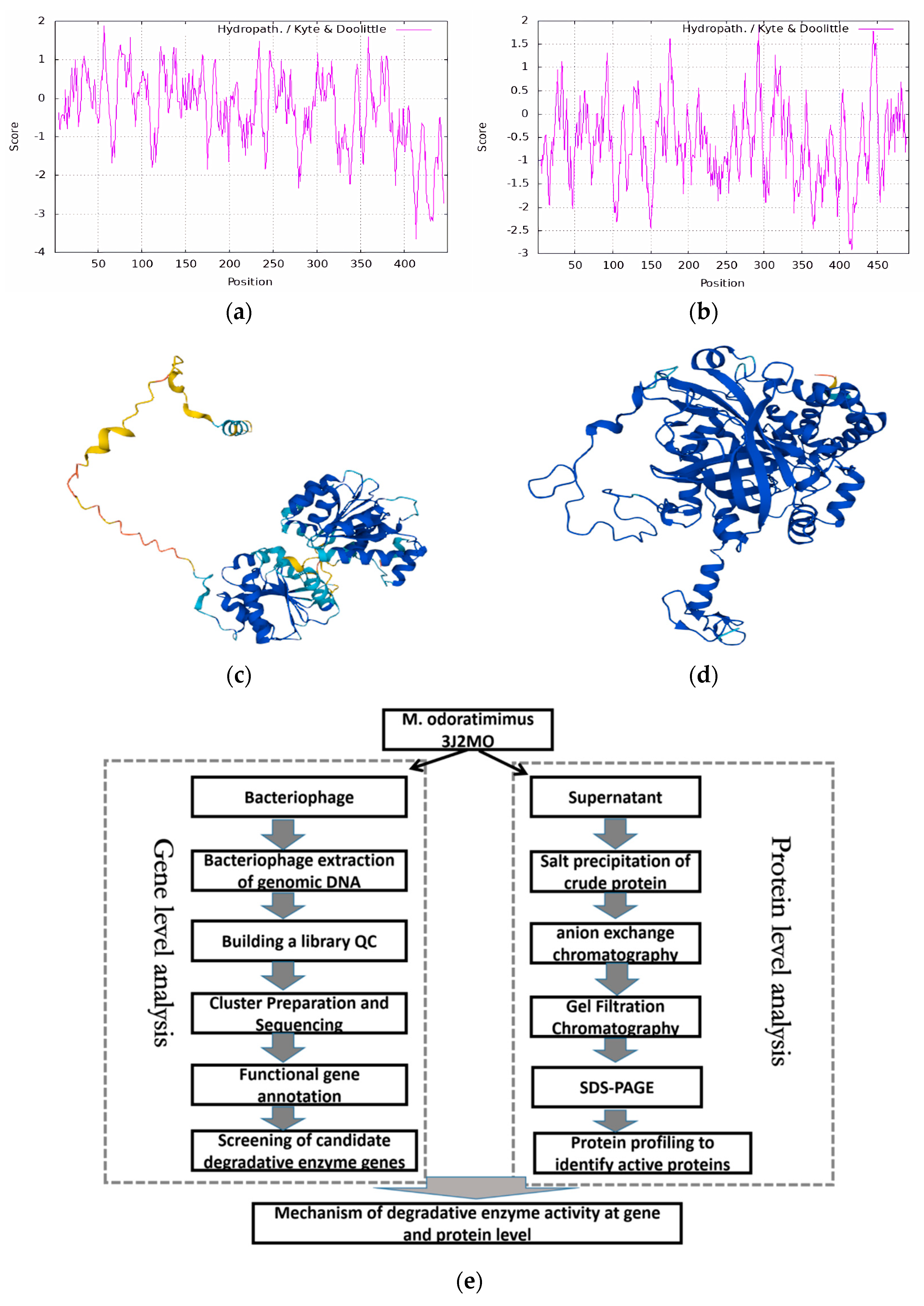
3.4. Functional Analysis of Degrading Enzymes
4. Discussion
5. Conclusions
6. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khoshpey, B.; Farhud, D.D.; Zaini, F. Aflatoxins in Iran: Nature, hazards and carcinogenicity. Iran. J. Public Health 2011, 40, 1. [Google Scholar] [PubMed]
- IARC. IARC Monographs on the Evaluation of Carcinogenic Risks of Chemicals to Humans. Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins; International Agency for Cancer: Lyon, France, 1993; Volume 56, p. 599. [Google Scholar]
- Masood, M.; Iqbal, S.Z.; Asi, M.R.; Malik, N. Natural occurrence of aflatoxins in dry fruits and edible nuts. Food Control. 2015, 55, 62–65. [Google Scholar] [CrossRef]
- Motomura, M.; Toyomasu, T.; Mizuno, K.; Shinozawa, T. Purification and characterization of an aflatoxin degradation enzyme from Pleurotus ostreatus. Microbiol. Res. 2003, 158, 242. [Google Scholar] [CrossRef] [PubMed]
- Gurban, A.-M.; Epure, P.; Oancea, F.; Doni, M. Achievements and Prospects in Electrochemical-Based Biosensing Platforms for Aflatoxin M1 Detection in Milk and Dairy Products. Sensors 2017, 17, 2951. [Google Scholar] [CrossRef]
- Fu, J.; Gu, M.; Tang, X.; Zhang, Q.; Li, P. Early evaluation of aflatoxin occurrence risk using an ultra-sensitive immunoassay for pre-aflatoxin protein biomarker detection. Food Control 2024, 160, 110317. [Google Scholar] [CrossRef]
- Rai, J.P.; Narware, J.; Kumar, R.; Kumar, R.; Pandey, P.; Prakash, N.; Holkar, S.K.; Akhtar, N.; Bhaskar, S.S.; Kumar, S.; et al. Aflatoxin’s Toll on Health: Insights into Human and Animal Impact. Biol. Life Sci. 2024. [Google Scholar] [CrossRef]
- Maneeboon, T.; Roopkham, C.; Mahakarnchanakul, W.; Chuaysrinule, C. Exploration of Pseudomonas knackmussii AD02 for the biological mitigation of post-harvest aflatoxin contamination: Characterization and degradation mechanism. J. Stored Prod. Res. 2024, 109, 102470. [Google Scholar] [CrossRef]
- Campone, L.; Piccinelli, A.L.; Celano, R.; Pagano, I.; Di Sanzo, R.; Carabetta, S.; Russo, M.; Rastrelli, L. Occurrence of aflatoxin M1 in milk samples from Italy analysed by online-SPE UHPLC-MS/MS. Nat. Prod. Res. 2017, 2, 6. [Google Scholar] [CrossRef]
- Chen, C.C.; Yu, Z.P.; Liu, Z.; Yao, Y.; Hagedoorn, P.L.; Schmitz, R.A.; Yang, L.; Yu, L.; Liu, A.; Sheng, X.; et al. Chanoclavine synthase operates by an NADPH-independent superoxide mechanism. Nature 2025, 640, 6. [Google Scholar] [CrossRef]
- Ran, C. Research on the Degradation Technology of Aflatoxin in Peanuts; Chinese Academy of Agricultural Sciences: Beijing, China, 2013. [Google Scholar]
- Amir Ismail, A.I.; Gonçalves, B.L.; Neeff, D.D.; Ponzilacqua, B.; Coppa, C.F.S.C.; Hintzsche, H.; Oliveira, C.A.F. Aflatoxin in foodstuffs: Occurrence and recent advances in decontamination. Food Res. Int. 2018, 113, 74–85. [Google Scholar] [CrossRef]
- Mitchell, N.J.; Bowers, E.; Hurburgh, C.; Wu, F. Potential economic losses to the US corn industry from aflatoxin contamination. Food Addit. Contam. 2016, 33, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Chen, L.; Yue, X.; Bai, Y.; Ding, X.; Li, P.; Zhang, Q.; Zhang, W. Distribution, Aflatoxin production of Aspergillus flavus in soils of typical peanut planting areas in Hubei Province. Chinese J. Oil Crops Sci. 2019, 41, 255–260. [Google Scholar]
- Mahfuz, M.; Gazi, M.A.; Hossain, M.; Islam, M.R.; Fahim, S.M.; Ahmed, T. General and advanced methods for the detection and measurement of aflatoxins and aflatoxin metabolites: A review. Toxin Rev. 2018. [Google Scholar] [CrossRef]
- Li HongBo, L.H.; Wang MiaoYan, W.M.; Hu LiangBin, H.L.; Mo HaiZhen, M.H.; Pan DaoDong, P.D. Isolation and identification of a deep-sea microbacterium that inhibits aflatoxin production. Biotechnol. Prog. 2015, 5, 235–239. [Google Scholar]
- Ji, W.; Li, X.; Yin, X.; Han, H.; Qiao, Z.; Zhang, L.; Miao, R.; Ma, H.; Ming, Y. Research progress on aflatoxin detection methods. J. Henan Univ. Technol. (Nat. Sci. Ed.) 2022, 43, 14–21. [Google Scholar]
- Wambacq, E.; Vanhoutte, I.; Audenaert, K.; De Gelder, L.; Haesaert, G. Occurrence, prevention and remediation of toxigenic fungi and mycotoxins in silage: A review. J. Sci. Food Agric. 2016, 96, 2284–2302. [Google Scholar] [CrossRef]
- Salati, S.; D’Imporzano, G.; Panseri, S.; Pasquale, E.; Adani, F. Degradation of aflatoxin B1 during anaerobic digestion and its effect on process stability. Int. Biodeterior. Biodegrad. 2014, 94, 19–23. [Google Scholar] [CrossRef]
- Xue, M.; Qu, Z.; Moretti, A.; Logrieco, A.F.; Chu, H.; Zhang, Q.; Zhu, Y.G. Aspergillus Mycotoxins: The Major Food Contaminants. Advanced Science 2025, 2412757, 1–24. [Google Scholar] [CrossRef]
- Mwakinyali, S.E.; Ming, Z.; Xie, H.; Zhang, Q.; Li, P. Investigation and characterization of myroides odoratimimus strain 3J2MO aflatoxin B1 degradation. J. Agric. Food Chem. 2019, 67, 4595–4602. [Google Scholar] [CrossRef]
- Fan Sufang, L.P. Comparison on Determination of Aflatoxins in Peanut, Corn and Rice by Liquid Chromatography and Liquid Chromatography Tandem Mass Spectrometr. Food Sci. 2011, 32, 255–258. [Google Scholar]
- Fawole, F.J.; Sahu, N.P.; Shamna, N.; Phulia, V.; Emikpe, B.O.; Adeoye, A.A.; Popoola, O.M. Effects of detoxified Jatropha curcas protein isolate on growth performance, nutrient digestibility and physio-metabolic response of Labeo rohita fingerlings. Aquac. Nutr. 2018, 24, 1223–1233. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, W.; Zhang, S. Purification and identification of an aflatoxin B1 degradation enzyme from Pantoea sp. T6. Toxicon 2019, 157, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Caltabiano, A.M.; Foley, J.P.; Striegel, A.M. Aqueous size-exclusion chromatography of polyelectrolytes on reversed-phase and hydrophilic interaction chromatography columns. J. Chromatogr. A 2018, 1532, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Burgess, R.R. A brief practical review of size exclusion chromatography: Rules of thumb, limitations, and troubleshooting. Protein Expr. Purif. 2018, 150, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Kummari, R.; Bose, K. Gel Filtration Chromatography. In Textbook on Cloning, Expression and Purification of Recombinant Proteins; Bose, K., Ed.; Springer: Singapore, 2022. [Google Scholar] [CrossRef]
- Yang, X. Screening of Aflatoxin B1-Degrading Strains and Purification of Their Active Enzymes; Nanjing University of Science and Technology: Nanjing, China, 2019. [Google Scholar]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1 2006, 2856–2860. [Google Scholar] [CrossRef]
- Kay, H.; Grünewald, E.; Feord, H.K.; Gil, S.; Peak-Chew, S.Y.; Stangherlin, A.; Van Ooijen, G. Deep-coverage spatiotemporal proteome of the picoeukaryote Ostreococcus tauri reveals differential effects of environmental and endogenous 24-hour rhythms. Commun. Biol. 2021, 4, 1147. [Google Scholar] [CrossRef]
- Xu, X.; Geng, F.; Sun, W. Quantitative proteomics and metabolomics analysis reveals the response mechanism of alfalfa (Medicago sativa L.) to o-coumaric acid stress. PLoS ONE 2023, 18, e0295592. [Google Scholar] [CrossRef]
- NY/T 1109-2017; General Biosafety Standard for Microbial Fertilizers. China Agriculture Press: Beijing, China, 2017.
- Haichao, S. Construction of Photochemical Sensor and Detection of Cysteine, Cyromazine and Escherichia coli O157: H7; Shandong Agricultural University: Shandong, China, 2013. [Google Scholar]
- Zhao, Y.; Wang, J.; Chen, J.; Zhang, X.; Guo, M.; Yu, G. A Literature Review of Gene Function Prediction by Modeling Gene Ontology. Front. Genet. 2020, 11, 400. [Google Scholar] [CrossRef]
- Tang, J.; Deng, D.; Song, M.; Liu, Z.; Ma, S.; Wang, Y. The genomic context for aflatoxin B1-degrading Pseudomonas strains. Microbiol. Bull. 2021, 48, 46–56. [Google Scholar] [CrossRef]
- Sun, M.; Zhao, S.; Gilvary, C.; Elemento, O.; Zhou, J.; Wang, F. Graph convolutional networks for computational drug development and discovery. Brief. Bioinform. 2020, 21, 919–935. [Google Scholar] [CrossRef]
- Felicori, L.; Jameson, K.H.; Roblin, P.; Fogg, M.J.; Garcia-Garcia, T.; Ventroux, M.; Cherrier, M.V.; Bazin, A.; Noirot, P.; Wilkinson, A.J. Interdomain flexibility of the replication initiation controller YabA enables simultaneous binding to multiple partners. Nucleic Acids Res. 2016, 44, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.M.; Micolucci, L.; Islam, M.S.; Olivieri, F.; Procopio, A.D. Bioinformatic tools for microRNA dissection. Nucleic Acids Res. 2016, 44, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.X.; Jiang, Y.; Lee, C.-H.; Kharel, P.; Zhang, Y.; van der Zande, A.M.; Huang, P.Y. Achieving sub-0.5-angstrom–resolution ptychography in an uncorrected electron microscope. Science 2024, 383, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Deng, H.; He, J.; Keasling, J.D.; Luo, X. UniKP: A unified framework for the prediction of enzyme kinetic parameters. Nat. Commun. 2023, 640. [Google Scholar] [CrossRef]
- Senghor, L.A.; Ortega-Beltran, A.; Atehnkeng, J.; Callicott, K.A.; Cotty, P.J.; Bandyopadhyay, R. The Atoxigenic Biocontrol Product Aflasafe SN01 Is a Valuable Tool to Mitigate Aflatoxin Contamination of Both Maize and Groundnut Cultivated in Senegal. Plant Disease 2020, 104, 510–520. [Google Scholar] [CrossRef]
- Maxwell, L.A.; Callicott, K.A.; Bandyopadhyay, R.; Mehl, H.L.; Orbach, M.J.; Cotty, P.J. Degradation of Aflatoxins B1 by Atoxigenic Aspergillus flavus Biocontrol Agents. Plant Disease 2021, 105, 2343–2350. [Google Scholar] [CrossRef]
- Fu, C.; Hou, L.; Chen, D.; Huang, T.; Yin, S.; Ding, P.; Li, X.; Huang, X.; Xiong , Y.; Ge, J.; et al. Targeted Detoxification of Aflatoxin B 1 in Edible Oil by an Enzyme–Metal Nanoreactor. J. Agric. Food Chem. 2024, 72, 5966–5974. [Google Scholar] [CrossRef]
- Adegoke, T.V.; Yang, B.; Xing, F.; Tian, X.; Wang, G.; Tai, B.; Si, P.; Hussain, S.; Jahan, I. Microbial Enzymes Involved in the Biotransformation of Major Mycotoxins. J. Agric. Food Chem. 2022, 71, 35–51. [Google Scholar] [CrossRef]
- Liu, S.; Ma, N.; Kong, J.; Zhang, X. Enzyme-mediated controlled polymerization and its application in biomolecular analysis. Microchem. J. 2024, 208, 112581. [Google Scholar] [CrossRef]
- Hu, C.; Yang, O.U.; Li-xia, W.; Bai-xing, Y.; Ying-xin, L.; Da-wei, D. Phosphate rock reduces the bioavailability of heavy metals by influencing the bacterial communities during aerobic composting. J. Integr. Agric. 2021, 20, 1137–1146. [Google Scholar] [CrossRef]
- Battilani, P.; Camardo Leggieri, M.; Rossi, V.; Giorni, P. AFLA-maize, a mechanistic model for Aspergillus flavus infection and aflatoxin B1 contamination in maize. Comput. Electron. Agric. 2013, 94, 38–46. [Google Scholar] [CrossRef]
- Rao, K.R.; Vipin, A.V.; Hariprasad, P.; Appaiah, K.A.; Venkateswaran, G.J.F.C. Biological detoxification of Aflatoxin B1 by Bacillus licheniformis CFR1. Food Control 2017, 71, 234–241. [Google Scholar]
- Petchkongkaew, A.; Taillandier, P.; Gasaluck, P.; Lebrihi, A. Isolation of Bacillus spp. from Thai fermented soybean (Thua-nao): Screening for aflatoxin B1 and ochratoxin A detoxification. J. Applied Microbiol. 2008, 104, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Corassin, C.H.; Bovo, F.; Rosim, R.E.; Oliveira, C.A.F. Efficiency of Saccharomyces cerevisiae and lactic acid bacteria strains to bind aflatoxin M1 in UHT skim milk. Food Control 2013, 31, 80–83. [Google Scholar] [CrossRef]
- Yuqun Yao, Q.X.L. Efficient, fast and robust degradation of chlortetracycline in wastewater catalyzed by recombinant Arthromyces ramosus peroxidase. Sci. Total Environ. 2023, 858. [Google Scholar] [CrossRef]
- Patel, S.S.; Donmez, I. Mechanisms of Helicases. J. Biol. Chem. 2006, 281, 4. [Google Scholar] [CrossRef]
- Loi, M.; Renaud, J.B.; Rosini, E.; Pollegioni, L.; Vignali, E.; Haidukowski, M.; Mulè, G. Enzymatic transformation of aflatoxin B1 by Rh_DypB peroxidase and characterization of the reaction products. Chemosphere 2020, 250. [Google Scholar] [CrossRef]
- Kumar, V.; Bahuguna, A.; Ramalingam, S.; Dhakal, G.; Shim, J.-J.; Kim, M. Recent technological advances in mechanism, toxicity, and food perspectives of enzyme-mediated aflatoxin degradation. Crit. Rev. Food Sci. Nutr. 2022, 62, 5395–5412. [Google Scholar] [CrossRef]
- Wang, J.; Ogata, M.; Hirai, H.; Kawagishi, H. Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 2011, 314, 6. [Google Scholar] [CrossRef]
- Ming, Z. Biodegradation of Aflatoxins by Myroides odoratimimus 3J2MO; Chinese Academy of Agricultural Sciences: Beijing, China, 2019. [Google Scholar]
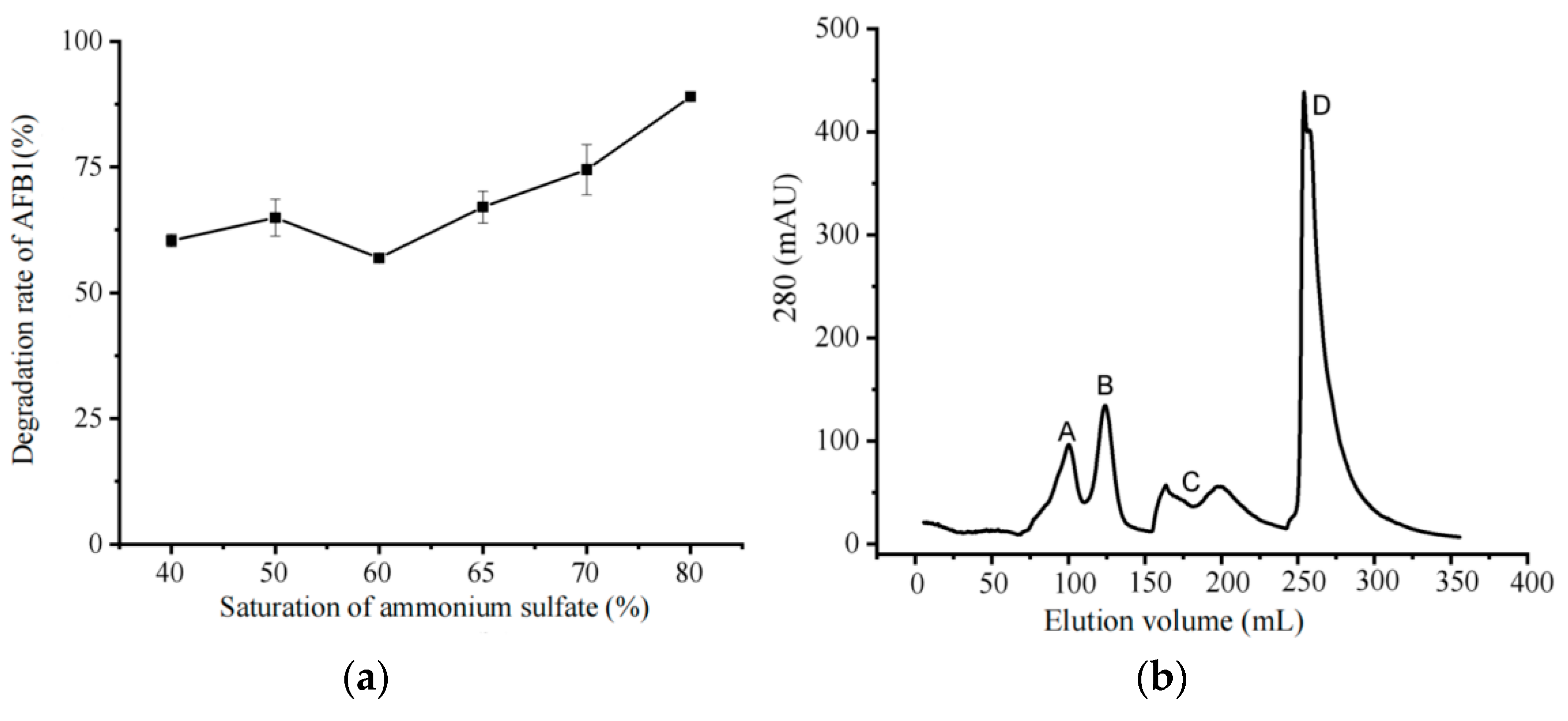
| Ammonium sulfate saturation (%) | 40 | 50 | 60 | 65 | 70 | 80 |
| Protein precipitation (mg) | 0.339 | 0.345 | 0.4205 | 0.4665 | 0.5235 | 0.5925 |
| Analyze Content | Software | Website |
|---|---|---|
| Physical and chemical properties analysis | ProtParam 8.5.6 | http://web.expasy.org/protparam/ accessed on 22 May 2023. |
| Hydrophilicity and hydrophobicity analysis | Protscale | http://web.expasy.org/protscale/ accessed on 11 March 2023. |
| Tertiary structure prediction | AlphaFold 3 | https://alphafold.ebi.ac.uk/ accessed on 11 March 2023. |
| Proteins | Mw (kDa) | Peptides | Coverage | Unique | iBAQ (%) |
|---|---|---|---|---|---|
| 3J2MOGL000150 | 50.839 | 1 | 2.4% | 1 | 83.3995% |
| 3J2MOGL003364 | 55.983 | 1 | 2.2% | 1 | 9.1731% |
| Proteins | Mw (kDa) | Length | GO Description |
|---|---|---|---|
| 3J2MOGL000150 | 50.839 | 449 | helicase activity |
| 3J2MOGL003364 | 55.983 | 492 | peroxidase activity |
| Proteins | Mw (kDa) | Length | COG Description |
|---|---|---|---|
| 3J2MOGL000150 | 50.839 | 449 | helicase activity and hydrolase activity |
| 3J2MOGL003364 | 55.983 | 492 | peroxidase activity |
| Proteins | Mw (kDa) | Length | KEGG Description |
|---|---|---|---|
| 3J2MOGL000150 | 50.839 | 449 | rhlE; ATP-dependent RNA helicase RhlE [EC:3.6.4.13] |
| 3J2MOGL003364 | 55.983 | 492 | katE, CAT, catB, srpA; catalase [EC:1.11.1.6] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Gao, Y.-Y.; Wang, D.; Zhang, Q.; Wang, H.-R.; Zhang, T.-T.; Zhu, M.-J.; Dong, J.; Ling, D.; Feng, P.; et al. Study on the Degradation of Aflatoxin B1 by Myroides odoratimimus 3J2MO. Biology 2025, 14, 724. https://doi.org/10.3390/biology14060724
Wang X, Gao Y-Y, Wang D, Zhang Q, Wang H-R, Zhang T-T, Zhu M-J, Dong J, Ling D, Feng P, et al. Study on the Degradation of Aflatoxin B1 by Myroides odoratimimus 3J2MO. Biology. 2025; 14(6):724. https://doi.org/10.3390/biology14060724
Chicago/Turabian StyleWang, Xue, Yao-Yao Gao, Dun Wang, Qi Zhang, Hao-Ran Wang, Ting-Ting Zhang, Meng-Jie Zhu, Jing Dong, Dong Ling, Peng Feng, and et al. 2025. "Study on the Degradation of Aflatoxin B1 by Myroides odoratimimus 3J2MO" Biology 14, no. 6: 724. https://doi.org/10.3390/biology14060724
APA StyleWang, X., Gao, Y.-Y., Wang, D., Zhang, Q., Wang, H.-R., Zhang, T.-T., Zhu, M.-J., Dong, J., Ling, D., Feng, P., Tang, X.-H., & Li, P.-W. (2025). Study on the Degradation of Aflatoxin B1 by Myroides odoratimimus 3J2MO. Biology, 14(6), 724. https://doi.org/10.3390/biology14060724








