Combined Multi-Omics and Co-Expression Network Analyses Uncover the Pigment Accumulation Mechanism of Orange-Red Petals in Brassica napus L.
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation and Sampling of Plant Materials
2.2. Phenotypic Observation
2.3. Analysis of Pigment Content
2.4. Metabolomic Analysis
2.5. Transcriptomic Analysis
2.6. Co-Expression Network Analysis of Metabolome and Transcriptome
2.7. Quantitative Real-Time PCR Validation
2.8. Statistical Analysis
3. Results
3.1. Anthocyanins and Carotenoids Contribute to the Color of ‘OrP’ Petals
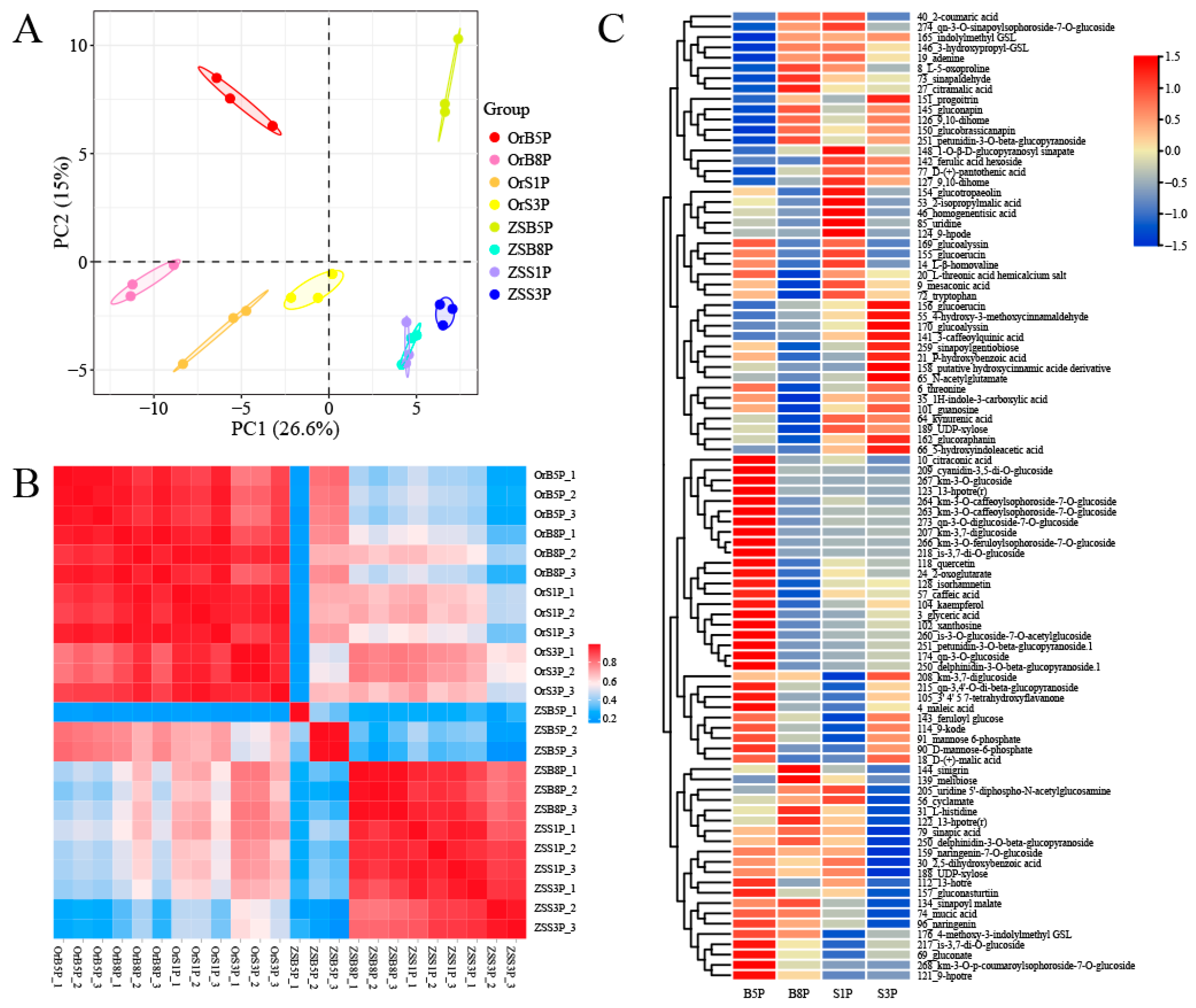
3.2. Pigment-Associated Metabolomic Profiling of ‘OrP’ and ‘ZS11’ Petals
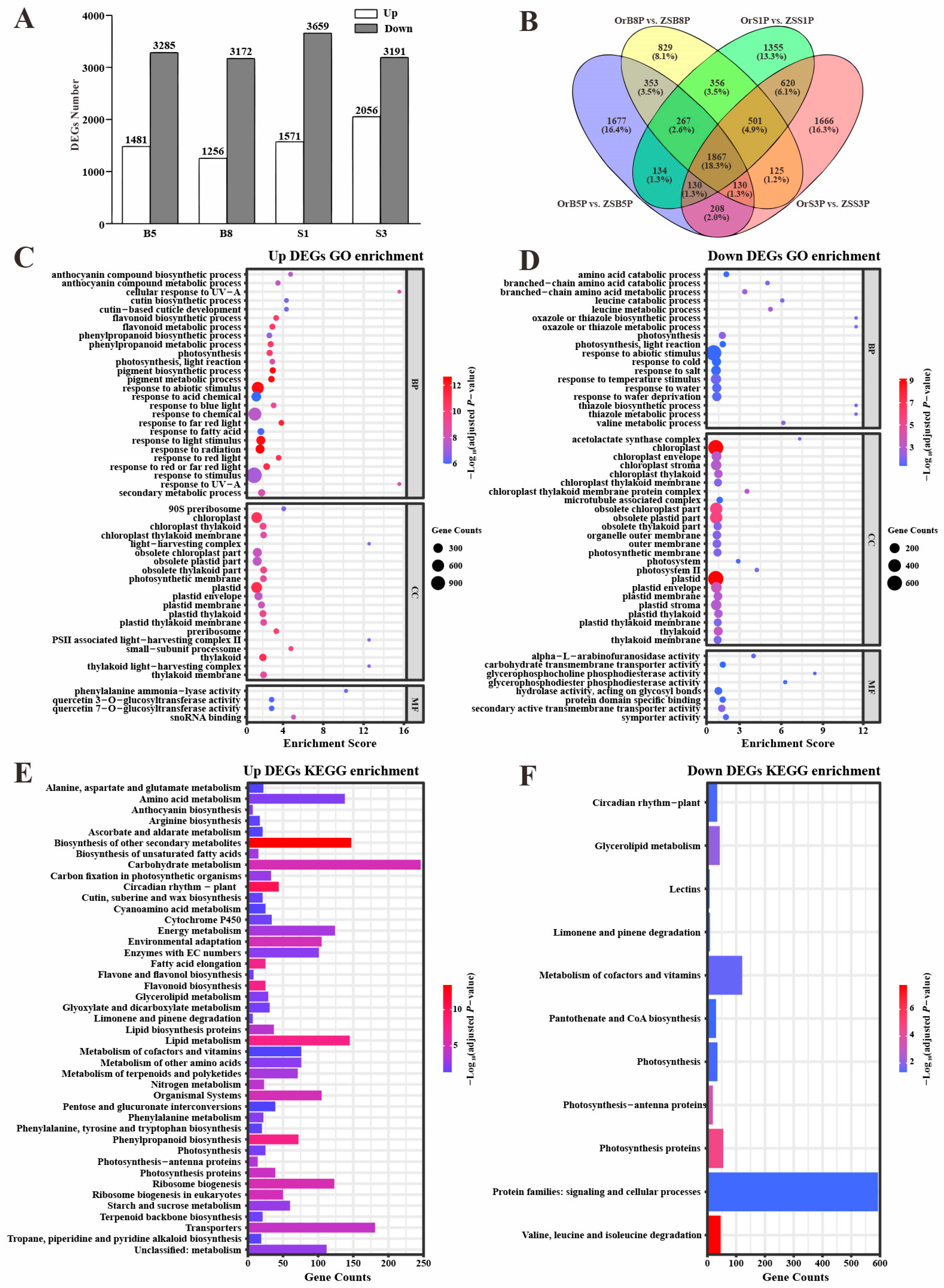
3.3. Pigment-Associated Transcriptomic Profiling of ‘OrP’ and ‘ZS11’ Petals
3.4. WGCNA Reveals Key Genes Associated with Anthocyanin Metabolism in Rapeseed
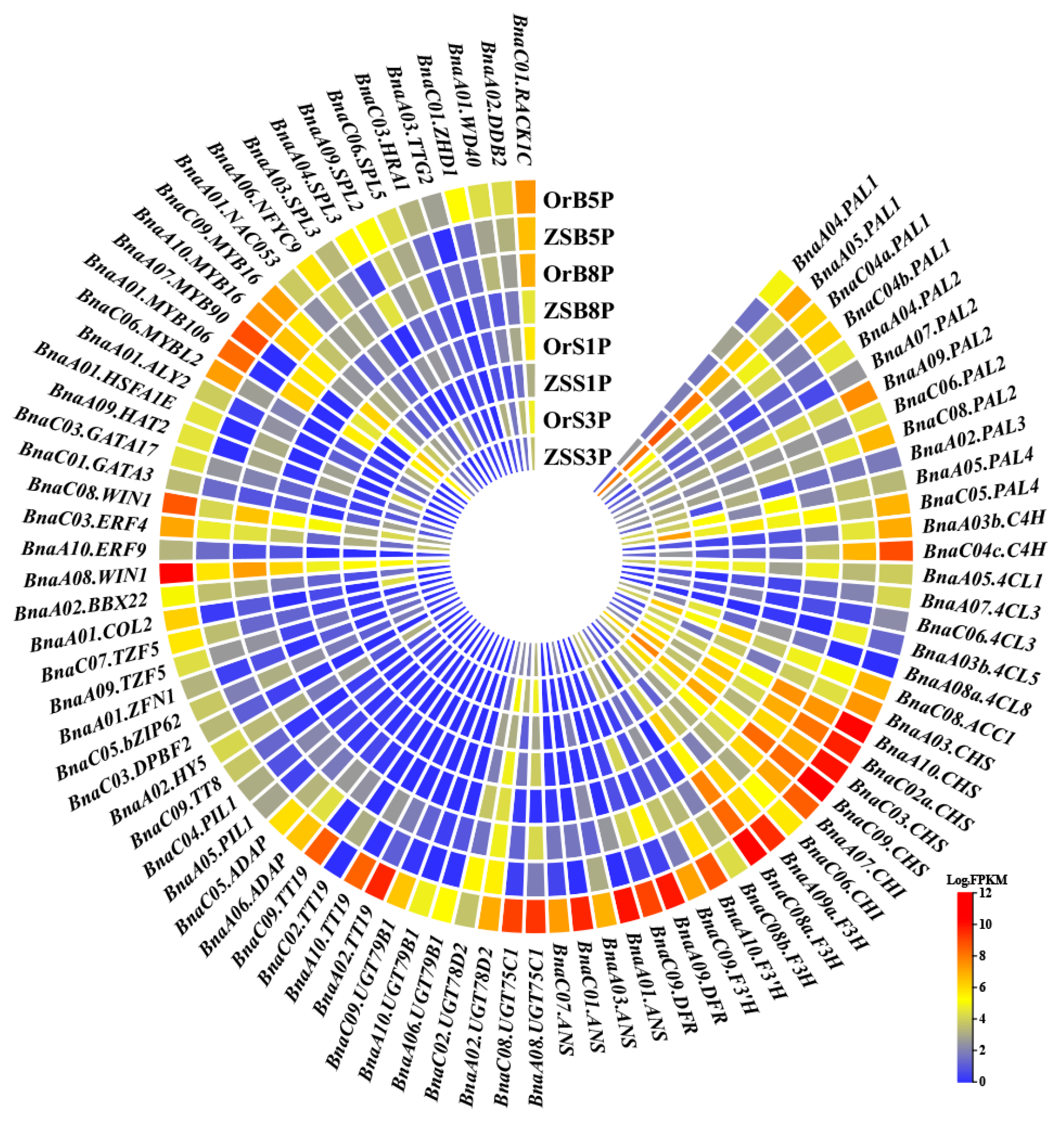
3.5. Exploration of Anthocyanin Metabolism in B. napus
3.6. Transcription Factors Contributing to Anthocyanin Metabolism in Rapeseed
3.7. Quantitative Real-Time PCR Validation of Anthocyanin Metabolism Genes
4. Discussion
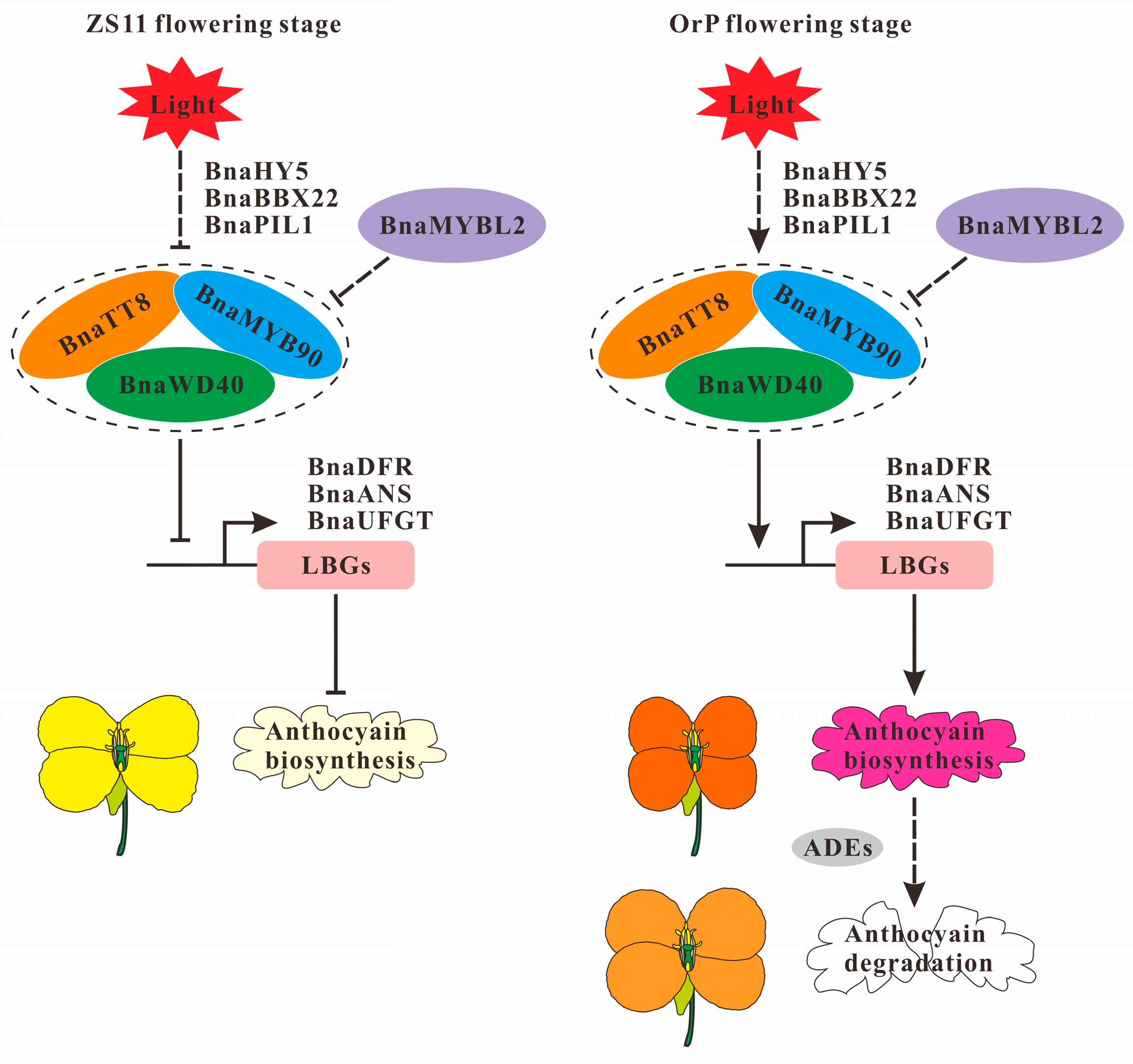
4.1. Formation of Orange-Red Petal Color in ‘OrP’ Rapeseed
4.2. Mechanism of Petals Fading in ‘OrP’ Rapeseed
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DEG | Differentially expressed gene |
| WGCNA | Weighted Gene Co-expression Network Analysis |
| TF | Transcription factor |
| TE | Transposable element |
| CCD | Carotenoid cleavage dioxygenase |
| ZEP | Zeaxanthin epoxidase |
| PAP | Production of anthocyanin pigment |
| RNA-Seq | Transcriptome sequencing |
| FPKM | Fragments Per Kilobase Million |
| FC | Fold change |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| EBGs | Early biosynthetic genes |
| LBGs | Late biosynthetic genes |
| MBW | MYB-bHLH-WD40 |
| ADEs | Anthocyanin degradation enzymes |
| BGLU | β-glucosidase |
| PRX, POD, or PER | Peroxidase |
| PPO | Polyphenol oxidase |
| LAC | Laccase |
References
- Xiao, M.; Wang, H.; Li, X.; Mason, A.S.; Fu, D. Rapeseed as an Ornamental. Horticulturae 2021, 8, 27. [Google Scholar] [CrossRef]
- Zhong, C.; Tang, Y.; Pang, B.; Li, X.; Yang, Y.; Deng, J.; Feng, C.; Li, L.; Ren, G.; Wang, Y.; et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic. Res. 2020, 7, 78. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, C.; Gong, X.; Luan, X.; Wu, Z.; Li, H.; Liu, Q.; Xu, M.; Yu, F. Transcriptome and metabolome analyses reveal a key role of the anthocyanin biosynthetic pathway cascade in the pigmentation of a Cinnamomum camphora red bark mutant (‘Gantong 1’). Ind. Crop. Prod. 2022, 175, 31470. [Google Scholar] [CrossRef]
- Yin, N.W.; Wang, S.X.; Jia, L.D.; Zhu, M.C.; Yang, J.; Zhou, B.J.; Yin, J.M.; Lu, K.; Wang, R.; Li, J.N.; et al. Identification and Characterization of Major Constituents in Different-Colored Rapeseed Petals by UPLC-HESI-MS/MS. J. Agric. Food Chem. 2019, 67, 11053–11065. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, C.; Wang, Y.; Yao, X.; Wang, F.; Wu, J.; King, G.J.; Liu, K. Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol. 2015, 206, 1513–1526. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Chen, D.; Pan, Q.; Li, F.; Zhao, Z.; Ge, X.; Li, Z. Production of red-flowered oilseed rape via the ectopic expression of Orychophragmus violaceus OvPAP2. Plant Biotechnol. J. 2018, 16, 367–380. [Google Scholar] [CrossRef]
- Jia, L.; Wang, J.; Wang, R.; Duan, M.; Qiao, C.; Chen, X.; Ma, G.; Zhou, X.; Zhu, M.; Jing, F.; et al. Comparative transcriptomic and metabolomic analyses of carotenoid biosynthesis reveal the basis of white petal color in Brassica napus. Planta 2021, 253, 8. [Google Scholar] [CrossRef]
- Li, F.; Gong, Y.; Mason, A.S.; Liu, Q.; Huang, J.; Ma, M.; Xiao, M.; Wang, H.; Fu, D. Research progress and applications of colorful Brassica crops. Planta 2023, 258, 45. [Google Scholar] [CrossRef]
- Zheng, X.; Lan, J.; Yu, H.; Zhang, J.; Zhang, Y.; Qin, Y.; Su, X.-D.; Qin, G. Arabidopsis transcription factor TCP4 represses chlorophyll biosynthesis to prevent petal greening. Plant Commun. 2022, 3, 100309. [Google Scholar] [CrossRef]
- Ye, S.; Hua, S.; Ma, T.; Ma, X.; Chen, Y.; Wu, L.; Zhao, L.; Yi, B.; Ma, C.; Tu, J.; et al. Genetic and multi-omics analyses reveal BnaA07.PAP2In-184-317 as the key gene conferring anthocyanin-based color in Brassica napus flowers. J. Exp. Bot. 2022, 73, 6630–6645. [Google Scholar] [CrossRef]
- Liu, Y.; Ye, S.; Yuan, G.; Ma, X.; Heng, S.; Yi, B.; Ma, C.; Shen, J.; Tu, J.; Fu, T.; et al. Gene silencing of BnaA09.ZEP and BnaC09.ZEP confers orange color in Brassica napus flowers. Plant J. 2020, 104, 932–949. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lv, J.; Liu, Z.; Wang, J.; Yang, B.; Chen, W.; Ou, L.; Dai, X.; Zhang, Z.; Zou, X. Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in pepper fruit (Capsicum annuum L.). Food Chem. 2020, 306, 125629. [Google Scholar] [CrossRef] [PubMed]
- Bordignon-Luiz, M.T.; Gauche, C.; Gris, E.F.; Falcão, L.D. Colour stability of anthocyanins from Isabel grapes (Vitis labrusca L.) in model systems. LWT—Food Sci. Technol. 2007, 40, 594–599. [Google Scholar] [CrossRef]
- Zhao, X.; Sheng, F.; Zheng, J.; Liu, R. Composition and Stability of Anthocyanins from Purple Solanum tuberosum and Their Protective Influence on Cr(VI) Targeted to Bovine Serum Albumin. J. Agric. Food Chem. 2011, 59, 7902–7909. [Google Scholar] [CrossRef]
- RABINO, I.; MANCINELLI, A.L. Light, temperature, and anthocyanin production. Plant Physiol. 1986, 81, 922–924. [Google Scholar] [CrossRef] [PubMed]
- Misra, P.; Pandey, A.; Tiwari, M.; Chandrashekar, K.; Sidhu, O.P.; Asif, M.H.; Chakrabarty, D.; Singh, P.K.; Trivedi, P.K.; Nath, P.; et al. Modulation of Transcriptome and Metabolome of Tobacco by Arabidopsis Transcription Factor, AtMYB12, Leads to Insect Resistance. Plant Physiol. 2010, 152, 2258–2268. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Chen, C.; Wang, C.; Liu, Z.; Liu, X.; Zou, L.; Shi, J.; Chen, S.; Chen, J.; Tan, M. Variations in Physiology and Multiple Bioactive Constituents under Salt Stress Provide Insight into the Quality Evaluation of Apocyni Veneti Folium. Int. J. Mol. Sci. 2018, 19, 3042. [Google Scholar] [CrossRef]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinf. 2021, 19, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Bao, Y.; Zhang, Z.; Zhao, W.; Xiao, J.; He, S.; Zhang, G.; Li, Y.; Zhao, G.; Chen, R.; et al. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022, 50, D27–D38. [Google Scholar]
- Song, J.-M.; Guan, Z.; Hu, J.; Guo, C.; Yang, Z.; Wang, S.; Liu, D.; Wang, B.; Lu, S.; Zhou, R.; et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 2020, 6, 34–45. [Google Scholar] [CrossRef]
- Song, J.M.; Liu, D.X.; Xie, W.Z.; Yang, Z.; Guo, L.; Liu, K.; Yang, Q.Y.; Chen, L.L. BnPIR: Brassica napus pan-genome information resource for 1689 accessions. Plant Biotechnol. J. 2021, 19, 412–414. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014, 15, 550. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Sharma, N.; Rahman, M.H.; Strelkov, S.; Thiagarajah, M.; Bansal, V.K.; Kav, N.N.V. Proteome-level changes in two Brassica napus lines exhibiting differential responses to the fungal pathogen Alternaria brassicae. Plant Sci. 2007, 172, 95–110. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, S.; Wei, L.; Huang, Y.; Liu, D.; Jia, Y.; Luo, C.; Lin, Y.; Liang, C.; Hu, Y.; et al. BnIR: A multi-omics database with various tools for Brassica napus research and breeding. Mol. Plant. 2023, 16, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Schwinn, K.E.; Ngo, H.; Kenel, F.; Brummell, D.A.; Albert, N.W.; McCallum, J.A.; Pither-Joyce, M.; Crowhurst, R.N.; Eady, C.; Davies, K.M. The Onion (Allium cepa L.) R2R3-MYB Gene MYB1 Regulates Anthocyanin Biosynthesis. Front. Plant Sci. 2016, 7, 1865. [Google Scholar] [CrossRef] [PubMed]
- Maier, A.; Schrader, A.; Kokkelink, L.; Falke, C.; Welter, B.; Iniesto, E.; Rubio, V.; Uhrig, J.F.; Hulskamp, M.; Hoecker, U. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013, 74, 638–651. [Google Scholar] [CrossRef]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008, 55, 954–967. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, Q.; Pedmale, U.V.; Nito, K.; Fu, W.; Lin, L.; Hazen, S.P.; Chory, J. PIL1 Participates in a Negative Feedback Loop that Regulates Its Own Gene Expression in Response to Shade. Mol. Plant. 2014, 7, 1582–1585. [Google Scholar] [CrossRef]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 Gene Encodes a Basic Helix-Loop-Helix Domain Protein Required for Expression of DFR and BAN Genes in Arabidopsis Siliques. Plant Cell 2000, 12, 1863–1878. [Google Scholar] [CrossRef]
- Zhou, L.-L.; Shi, M.-Z.; Xie, D.-Y. Regulation of anthocyanin biosynthesis by nitrogen in TTG1–GL3/TT8–PAP1-programmed red cells of Arabidopsis thaliana. Planta 2012, 236, 825–837. [Google Scholar] [CrossRef]
- Lee, J.-H.; Terzaghi, W.; Gusmaroli, G.; Charron, J.-B.F.; Yoon, H.-J.; Chen, H.; He, Y.J.; Xiong, Y.; Deng, X.W. Characterization of Arabidopsis and Rice DWD Proteins and Their Roles as Substrate Receptors for CUL4-RING E3 Ubiquitin Ligases. Plant Cell 2008, 20, 152–167. [Google Scholar] [CrossRef]
- Castells, E.; Molinier, J.; Benvenuto, G.; Bourbousse, C.; Zabulon, G.; Zalc, A.; Cazzaniga, S.; Genschik, P.; Barneche, F.; Bowler, C. The conserved factor DE-ETIOLATED 1 cooperates with CUL4-DDB1DDB2 to maintain genome integrity upon UV stress. EMBO J. 2011, 30, 1162–1172. [Google Scholar] [CrossRef]
- Su, J.; Xu, J.; Zhang, S. RACK1, scaffolding a heterotrimeric G protein and a MAPK cascade. Trends Plant Sci. 2015, 20, 405–407. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Habricot, Y.; Condiff, A.S.; Maldiney, R.; Straeten, D.V.D.; Ahmad, M. HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. 2007, 49, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Ma, X.; Gao, X.; Wu, W.; Zhou, B. Light Induced Regulation Pathway of Anthocyanin Biosynthesis in Plants. Int. J. Mol. Sci. 2021, 22, 11116. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX Protein BBX25 Interacts with HY5, Negatively Regulating BBX22 Expression to Suppress Seedling Photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef]
- Liu, Y.; Tang, L.; Wang, Y.; Zhang, L.; Xu, S.; Wang, X.; He, W.; Zhang, Y.; Lin, Y.; Wang, Y.; et al. The blue light signal transduction module FaCRY1-FaCOP1-FaHY5 regulates anthocyanin accumulation in cultivated strawberry. Front. Plant Sci. 2023, 14, 1144273. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Brown, M.; Hatlestad, G.; Akhavan, N.; Smith, T.; Hembd, A.; Moore, J.; Montes, D.; Mosley, T.; Resendez, J.; et al. TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev. Biol. 2016, 419, 54–63. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB–bHLH–WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Sato, F. The function of ETHYLENE RESPONSE FACTOR genes in the light-induced anthocyanin production of Arabidopsis thaliana leaves. Plant Biotechnol. 2018, 35, 87–91. [Google Scholar] [CrossRef]
- Sun, H.; Hu, K.; Wei, S.; Yao, G.; Zhang, H. ETHYLENE RESPONSE FACTORS 4.1/4.2 with an EAR motif repress anthocyanin biosynthesis in red-skinned pears. Plant Physiol. 2023, 192, 1892–1912. [Google Scholar] [CrossRef]
- Ni, J.; Wang, S.; Yu, W.; Liao, Y.; Pan, C.; Zhang, M.; Tao, R.; Wei, J.; Gao, Y.; Wang, D.; et al. The ethylene-responsive transcription factor PpERF9 represses PpRAP2.4 and PpMYB114 via histone deacetylation to inhibit anthocyanin biosynthesis in pear. Plant Cell 2023, 35, 2271–2292. [Google Scholar] [CrossRef]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Tanaka, Y.; Brugliera, F. Flower colour and cytochromes P450. Philos. Trans. R. Soc. B 2013, 368, 20120432. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, G.; Dong, T.; Pan, Y.; Zhao, Z.; Tian, S.; Hu, Z. Anthocyanin Accumulation and Transcriptional Regulation of Anthocyanin Biosynthesis in Purple Bok Choy (Brassica rapa var. chinensis). J. Agric. Food Chem. 2014, 62, 12366–12376. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zhang, Z.; Zhang, L. Anthocyanin Accumulation, Antioxidant Ability and Stability, and a Transcriptional Analysis of Anthocyanin Biosynthesis in Purple Heading Chinese Cabbage (Brassica rapa L. ssp. pekinensis). J. Agric. Food Chem. 2016, 64, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Bak, S.; Beisson, F.; Bishop, G.; Hamberger, B.; Höfer, R.; Paquette, S.; Werck-Reichhart, D. Cytochromes P450. Arab. Book 2011, 9, 1–56. [Google Scholar] [CrossRef]
- Barbagallo, R.N.; Palmeri, R.; Fabiano, S.; Rapisarda, P.; Spagna, G. Characteristic of β-glucosidase from Sicilian blood oranges in relation to anthocyanin degradation. Enzyme Microb. Technol. 2007, 41, 570–575. [Google Scholar] [CrossRef]
- Zhang, Z.; Pang, X.; Duan, X.; Ji, Z.; Jiang, Y. Role of peroxidase in anthocyanin degradation in litchi fruit pericarp. Food Chem. 2005, 90, 47–52. [Google Scholar] [CrossRef]
- Zipor, G.; Duarte, P.; Carqueijeiro, I.; Shahar, L.; Ovadia, R.; Teper-Bamnolker, P.; Eshel, D.; Levin, Y.; Doron-Faigenboim, A.; Sottomayor, M.; et al. In planta anthocyanin degradation by a vacuolar class III peroxidase in Brunfelsia calycina flowers. New Phytol. 2014, 205, 653–665. [Google Scholar] [CrossRef]
- Luo, H.; Deng, S.; Fu, W.; Zhang, X.; Zhang, X.; Zhang, Z.; Pang, X. Characterization of Active Anthocyanin Degradation in the Petals of Rosa chinensis and Brunfelsia calycina Reveals the Effect of Gallated Catechins on Pigment Maintenance. Int. J. Mol. Sci. 2017, 18, 699. [Google Scholar] [CrossRef]
- Zhang, Z.; Pang, X.; Jia, Z.; Jiang, Y. Role of anthocyanin degradation in litchi pericarp browning. Food Chem. 2001, 75, 217–221. [Google Scholar] [CrossRef]
- Tran, L.T.; Taylor, J.S.; Constabel, C.P. The polyphenol oxidase gene family in land plants Lineage-specific duplication and expansion. BMC Genom. 2012, 13, 395. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Davis, E.J.; Ballif, J.; Liang, M.; Bushman, E.; Haroldsen, V.; Torabinejad, J.; Wu, Y. Mutant identification and characterization of the laccase gene family in Arabidopsis. J. Exp. Bot. 2006, 57, 2563–2569. [Google Scholar] [CrossRef] [PubMed]
- Sunil, L.; Shetty, N.P. Biosynthesis and regulation of anthocyanin pathway genes. Appl. Microbiol. Biotechnol. 2022, 106, 1783–1798. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, L.; Li, S.; Zhang, C.; Zeng, L.; Shen, S.; Yin, N.; Zhao, H.; Tang, Z.; Qu, C.; Li, J.; et al. Combined Multi-Omics and Co-Expression Network Analyses Uncover the Pigment Accumulation Mechanism of Orange-Red Petals in Brassica napus L. Biology 2025, 14, 693. https://doi.org/10.3390/biology14060693
Jia L, Li S, Zhang C, Zeng L, Shen S, Yin N, Zhao H, Tang Z, Qu C, Li J, et al. Combined Multi-Omics and Co-Expression Network Analyses Uncover the Pigment Accumulation Mechanism of Orange-Red Petals in Brassica napus L. Biology. 2025; 14(6):693. https://doi.org/10.3390/biology14060693
Chicago/Turabian StyleJia, Ledong, Shengting Li, Chao Zhang, Lijun Zeng, Shulin Shen, Nengwen Yin, Huiyan Zhao, Zhanglin Tang, Cunmin Qu, Jiana Li, and et al. 2025. "Combined Multi-Omics and Co-Expression Network Analyses Uncover the Pigment Accumulation Mechanism of Orange-Red Petals in Brassica napus L." Biology 14, no. 6: 693. https://doi.org/10.3390/biology14060693
APA StyleJia, L., Li, S., Zhang, C., Zeng, L., Shen, S., Yin, N., Zhao, H., Tang, Z., Qu, C., Li, J., & Chen, Z. (2025). Combined Multi-Omics and Co-Expression Network Analyses Uncover the Pigment Accumulation Mechanism of Orange-Red Petals in Brassica napus L. Biology, 14(6), 693. https://doi.org/10.3390/biology14060693






