Simple Summary
To improve cotton architecture for field cultivation, this study employed RNA interference (RNAi) to achieve the graded suppression of GhSP (a key flowering repressor). Field trials revealed the dose-dependent effect of GhSP silencing on the determinate growth and final plant height of transgenic cotton. A mild suppression of GhSP in line GhSPi-#5 led to a semi-dwarf phenotype (70~100 cm) with preserved agronomic traits, which was ideal for cotton production compared to the indeterminate growth of the wild type and excessive dwarfism and compromised fiber quality in severely GhSP-suppressed cotton. These results demonstrate that the precise manipulation of GhSP expression enables targeted improvement in cotton plant architecture.
Abstract
Cotton exhibits indeterminate growth potential at its apical meristem. In field cultivation, it is often necessary to restrict plant height by the foliar application of plant growth regulators or artificial topping. The genetic engineering of cotton architecture offers an efficient, environmentally friendly, and low-cost alternative to current field management. Our study aimed to improve the plant architecture of transgenic cotton by the suppression of GhSP, a key flowering repressor, via the RNA interference method. Sixteen independent transgenic lines were generated and classified as mildly, moderately, and severely suppressed, according to GhSP expression levels. Field evaluation revealed the dose-dependent effects of GhSP silencing on plant height. The mildly suppressed line GhSPi-#5 exhibited a semi-dwarf phenotype of approximately 70~100 cm in height. Negative phenotypes, including excessive dwarf plant architecture and inferior fiber quality and yield traits, were observed in severely GhSP-suppressed transgenic lines. Notably, the mild silencing of GhSP in GhSPi-#5 did not negatively affect leaf and flower organ growth, pollen fertility, major agronomic traits, or fiber quality compared with the wild type. These observations demonstrate the feasibility of manipulating the architecture of transgenic cotton via GhSP silencing.
1. Introduction
Crop architecture comprises plant height, branching patterns, and leaf morphology including shape and size characteristics, with the height and branching features serving as the primary determinants of the architecture formation. Excessively tall plant height increases susceptibility to lodging, which impedes mechanized harvesting, crop yields, and quality formation, whereas extreme dwarfism in the morphological structure frequently diminishes yield potential. An ideal plant architecture is critical for achieving equilibrium between the crop yield potential and lodging resistance in agricultural systems [1,2]. Cotton (Gossypium spp.) constitutes a strategic economic crop, producing the majority of the natural fiber for the global textile industry. Cotton exhibits a monopodial primary axis that maintains indeterminate apical dominance under non-manipulated developmental conditions [3]. Current agronomic practices predominantly employ the foliar application of plant growth regulators (such as mepiquat chloride) and artificial topping to control cotton vertical growth [4,5]. These methods impose significant labor input and the potential risk of xenobiotic contamination from agrochemical leaching. To address the dual imperatives of fiber yield optimization and mechanized harvesting efficiency, breeding programs are increasingly prioritizing the development of semi-dwarf cotton with compact architecture [6,7,8].
The precise regulation and dynamic interplay between florigen and anti-florigen is critical to determine the flowering and apical growth habitat and, consequently, plant architecture [9]. Flowering Locus T (FT) is the first identified florigen from Arabidopsis. FT and its homologs (such as SINGLE FLOWER TRUSS or SFT in tomato), synthesized in leaves, are transported to the apical meristem and promote flowering. Anti-florigens comprise the CEN/TFL1/SP (CETS) gene family, including CENTRORADIALIS (CEN) from Antirrhinum, TERMINAL FLOWER1 (TFL1) from Arabidopsis, SELF-PRUNING (SP) from tomato, and their conserved homologs in taller plants [10,11,12,13]. These anti-florigens primarily function antagonistically with florigens to inhibit the transition to flowering and maintain the indeterminate activity of meristems [14,15]. The SFT (florigen)/SP (anti-florigen) balance model posits that an elevated SFT/SP ratio within meristems drives determinate growth, ultimately leading to apical termination, whereas a reduced ratio sustains indeterminate growth [3,16].
In tomato, mutations in the SP gene induce precocious flowering, progressively reducing leaf numbers in sympodial units and terminating shoots with two consecutive inflorescences, thereby driving the shift from indeterminate to determinate growth [11,17]. The SP mutation has emerged as a pivotal target for crop genetic improvement, revolutionizing tomato breeding. Cultivars harboring SP mutations are recognized as cornerstone genetic traits in modern agriculture due to their controlled growth habit and stable yield [10,11,18]. Combinatorial screening with other floral transition pathway genes has further optimized tomato productivity [19]. This strategy has been successfully extended to crops including soybean [20], grapes [21], barley [22], roses [23], and strawberries [23], demonstrating the universal regulatory role of SP in plant architecture modulation.
In recent years, the cotton ortholog of the SELF-PRUNING (GhSP/GhCEN/GhTFL1) gene has emerged as a research focus for its precise regulation of plant architectural development. The virus-induced gene silencing (VIGS) of GhSP triggers precocious flowering and premature growth cessation, forming terminal flowers on dwarfed main stems and shortened lateral branches [14,15,16,24,25]. The complete knockout of GhSP via CRISPR/Cas9 results in extreme negative traits, precluding their utility in breeding applications [8,26,27]. Remarkably, an L86P substitution in GhTFL1—a targeted mutation weakening (but not abolishing) protein interaction capacity—achieved partial loss of function, generating elite germplasm with optimal plant architecture with moderate height, compact fruiting branches, and a shortened reproductive phase [8].
Notably, cotton GhSP exerts anti-florigen functions with a dose effect because the varied activities and mutant copies in the four homologous alleles result in different phenotypic variations in flowering and plant architecture [4,8]. Nevertheless, there currently lacks detailed analysis of the dose relationship between GhSP expression levels and phenotypic outcomes, and the ideal levels of GhSP expression for balancing plant architecture and yield remain to be elucidated. To address these gaps, this study employed RNA interference (RNAi) technology to generate transgenic cotton lines with gradually lowered GhSP expression levels. We observed that the mildly suppressed GhSP RNAi line demonstrated self-pruning capacity while retaining core agronomic traits comparable to the wild type. These findings underscore that the precise optimization of GhSP expression levels, based on this line’s dose-dependent regulatory mechanism, enables simultaneous improvement in cotton architecture (from semi-dwarf to extreme dwarfing gradients) and coordinated yield traits. This approach establishes a novel molecular breeding strategy for the synergistic enhancement of plant architecture and yield traits in crops.
2. Materials and Methods
2.1. Plant Material and Growth Conditions
The tetraploid upland cotton (G. hirsutum L., AD genome, 2n = 4x = 52) cultivar Jimian No.14 was used for cotton transformation. Cotton seedlings were grown in a climate-controlled greenhouse with a 16 h/8 h (light/dark) schedule and the temperature kept at 26–30 °C. Field-grown cotton plants in a net house in Chongqing, China, were subjected to architectural characterization during the growing season, with the concomitant collection of fiber and ovule specimens from these cultivated populations.
2.2. Vector Construction and Plant Transformation
An intron-containing hairpin RNA construct of the cotton SP gene (GhSP-RNAi) was amplified from cotton genomic DNA as previously described [28]. The 20 µL PCR mixture included 50 ng of cotton genomic DNA, a 400 nM flanking primer (5′-GATTCAGATCTAGGGCAAAC-3′), a 40 nM bridge primer (5′-CCTTGGCATTTC GTAGTTCACAACAGATGCCACATTTGGT-3′), and 10 µL of 2 × PrimeSTAR® Max Premix (Takara, Dalian, China). The PCR thermocycling parameters were as follows: 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s and a final extension of 10 min at 72 °C. The GhSP-RNAi fragment was cloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA) and sequenced and then inserted into a modified pBI121 vector p5 containing selection marker GUS and NPTII genes [29]. The Agrobacterium-mediated transformation of cotton was conducted according to a previously established protocol with minor modifications [29]. Cotton calluses were selected on an MS medium containing 50 mg/L of kanamycin with subculturing every 15 days. The regenerated plants (T0) were verified by PCR amplification of the transgene. The primers are listed in Table S1.
T1 plants generated from the selfing seeds of T0 transformants were selfed to produce T2 seeds, which were sown to form T2 lines. For each transformant, homozygous T2 lines (without non-transgenic segregants) were selected for phenotypic observation and molecular characterization.
2.3. RNA Isolation and RT-qPCR
The shoot tips of transgenic and wild-type seedlings at 14 days after sowing (DAS) were ground in liquid nitrogen, and total RNA was extracted using a Plant Total RNA Extraction kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 1 µg of total RNA using a RT SuperMix Kit (Vazyme, R233-01, Nanjing, China) with a gDNA wiper to eliminate genomic DNA contamination. RT-qPCR amplifications were performed with ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) on the CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA). The thermocycling parameters were as follows: 95 °C for 3 min, followed by 40 cycles of 95 °C for 5 s and 57 °C for 20 s, and a standard melting curve program was added to monitor PCR specificity. Each analysis was repeated with three biological replicates. The primers are listed in Table S1. The GhACT gene served as an internal control.
2.4. Chlorophyll Extraction and Quantification
Chlorophyll extraction and quantification in leaves were performed by a modified protocol [30]. In brief, the target leaf samples were thoroughly rinsed and blotted dry, the leaf vein was removed, and the leaf was cut into strips approximately 2 mm wide. Next, 0.5 g of the prepared leaf strips was transferred to a test tube, 10 mL of 80% acetone was added to fully submerge the tissue, and the tube was sealed tightly. The leaf strips were incubated in a darkroom for a minimum of 24 h, with the tube shaken 2–3 times during the extraction period until the strips became completely bleached. The absorbances at 646 and 663 nm were determined using a spectrophotometer (Varioskan LUX multimode microplate reader, Thermo Fisher Scientific, Waltham, MA, USA). Three biological replicates were used for each sample. The chlorophyll contents were represented by the following formulas:
where V represents the volume of the extraction solution, and W represents the weight of the sample in grams.
2.5. Net Photosynthetic Rates
Net photosynthetic rates were measured in three fully expanded leaves per cotton plant using an LI-6400/XT Portable Photosynthesis System (LI-COR, Lincoln, NE, USA). Measurements were conducted between 10:00 a.m. and 12:00 p.m. under clear-sky conditions, following established protocols as previously described [31].
2.6. Observation of Pollen Morphology
For the purpose of analyzing pollen fertility, pollen grains at the stage of anther dehiscence were liberated and subsequently stained with an I2-KI solution. Following this, photographs were taken using a Zeiss Axio Imager Z2 microscope (Zeiss, Oberkochen, Germany). In addition, the morphological characteristics of the anther and pollen grains were observed and photographed using a scanning electron microscope (SEM) platform (SU3500, Hitachi, Tokyo, Japan), operated at an accelerating voltage of 3 kV.
2.7. Histological Sectioning
Histological sections of the shoot tips were prepared as described previously [32]. In brief, shoot apices were cut from cotton seedings at 14 DAS and immediately immersed in a 50% FAA fixative solution (absolute ethanol: formaldehyde: acetic acid, 10:2:1). Dehydration was performed in a graded ethanol series (30%, 50%, 70%, 95%, and 100%, all v/v), and then the ethanol was gradually replaced with xylene. The tissues were embedded in paraffin wax and sliced into sections with a thickness of 10 µm using a Leica RM2235 Rotary Microtome (Leica, Wetzlar, Germany). Toluidine blue (1% w/v) was used to stain the sections. A Zeiss Axio Imager Z2 microscope (Zeiss, Oberkochen, Germany) was used to image the samples under brightfield illumination.
For the microscopic observation of the mature fibers, naturally opened bolls were harvested on the same day. Only the six cottonseeds in the middle of the locule were collected for microscopic analysis. The fibers from the middle of the cottonseed were fixed for transverse sectioning as described in previous research [33]. Briefly, the 1 cm segments were excised from the mid-region of the bundled fibers, dehydrated in a graded ethanol series, embedded in paraffin, and sectioned into 10 µm-thick slices using a rotary microtome (Leica RM2235, Wetzlar, Germany). The sections were stained with fast green (1% w/v), then observed and photographed under an optical microscope (Zeiss Axio Imager Z2, Zeiss, Oberkochen, Germany). The wall thickness of fiber transverse sections were measured in the collected images using ImageJ 2 (http://imagej.net/Fiji, accessed on 22 May 2025).
2.8. Cotton Fiber Quality Assessment
The naturally matured cotton of each line was harvested in late August for fiber yield trait evaluation. After drying and ginning using a roller gin (SY-20, Jianghe Machinery Plant, Xinxiang, Henan, China), the fibers and cottonseeds were separately weighed to determine lint percentages. To evaluate fiber quality, a minimum of 15 g of fibers was collected per sample. Fiber quality traits (including fiber length, the uniformity index, strength, the micronaire value, and the elongation ratio) were analyzed using a High-Volume Instrument (HVI) test system (HFT 9000, Uster Technologies AG, Uster, Switzerland), in accordance with Chinese National Standard GB/T 20392-2006 [34], at the Center for Cotton Fiber Quality Inspection and Testing at the Chinese Ministry of Agriculture (Anyang, Henan, China). Three biological replicates were analyzed per sample.
3. Results
3.1. Down-Regulation of GhSP in Cotton
To avoid the severe dwarfism phenotype resulting from GhSP knockout in cotton (G. hirsutum) [27], RNA interference (RNAi) was employed to modulate GhSP expression levels and improve the cotton architecture. Accordingly, an shRNA-based interference element was designed using the third intron of the GhSP gene as the loop and the fourth exon as the sense strand (Figure 1A). The interference element was driven by a cauliflower mosaic virus (CaMV) 35S promoter to assemble the RNAi vector CaMV35S:GhSP-RNAi (Figure 1B). A total of 16 independent transgenic CaMV35S:GhSP-RNAi (GhSPi) cotton lines were obtained through cotton genetic transformation and were further confirmed with PCR analysis (Figure 1C). RT-qPCR analysis revealed the different-level suppression of GhSP gene expression in transgenic lines compared to the wild type (Figure 1D). These transgenic cotton lines were categorized into three groups based on the degree of GhSP expression suppression, i.e., mildly suppressed (the GhSP expression level was decreased by less than 30%; GhSPi-#1, -#5, and -#12), moderately suppressed (decreased by 30–60%; GhSPi-#2, -#3, -#4, -#7, -#8, -#9, -#10, and -#16), and severely suppressed (over 60%; GhSPi-#6, -#11, -#13, -#14, and -#15). Representative transformants GhSPi-#5 (mild), GhSPi-#8 (moderate), and GhSPi-#14 (severe) were selected for subsequent phenotypic analyses (Figure 1D).
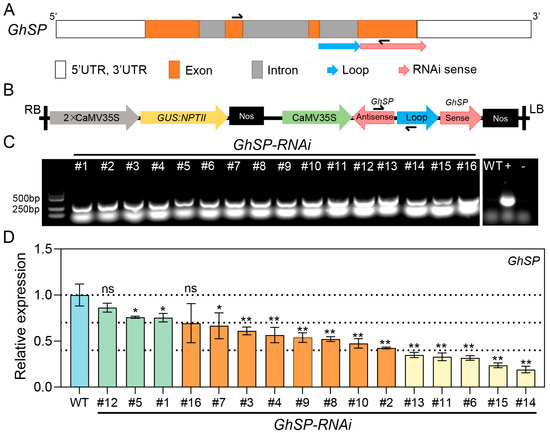
Figure 1.
Suppressing the expression of GhSP in cotton via RNA interference. (A) Schematic representation of the GhSP gene structure. The positions of the GhSP specific primers for RT-qPCR are indicated by the black arrows. (B) Schematic diagram of the T-DNA construct for the RNAi vector targeting the GhSP gene in transgenic cotton. The transgene specific primers are indicated by the black arrows. CaMV35S, cauliflower mosaic virus 35S promoter; 2 × CaMV35S, two CaMV35Ss in series; Nos, the terminator region of A. tumefaciens nopaline synthase gene; NPTII, the neomycin phosphotransferase gene; GUS, the β-glucuronidase gene. (C) PCR amplification validation of transgenic cotton plants (#1~16). Negative controls included WT and ddH2O (−), and the positive control (+) was the RNAi vector CaMV35S:GhSP-RNAi. (D) Relative expression levels of GhSP in T2 transgenic cotton plants. RNA was isolated from the shoot apex (14 DAS) of transgenic cotton plants and the wild type. Statistical significance was determined by Student’s t-test (n = 3). *, p < 0.05, **, p < 0.01; ns, not significant.
3.2. Silencing GhSP Induces Determinate Main Stems in Transgenic Cotton in a Dose-Dependent Manner
To investigate the impact of varying GhSP silencing levels on cotton architecture, the T2 generations of GhSPi-#5, GhSPi-#8, and GhSPi-#14 were sown in a field. All the GhSPi transgenic cotton lines exhibited self-pruning at 90 days after sowing (DAS), showing graded architectural alterations. GhSPi-#5 displayed a semi-dwarf architecture (70~100 cm), GhSPi-#8 showed a dwarf phenotype (50~70 cm), and GhSPi-#14 demonstrated extreme dwarfism (20~40 cm) consistent with the GhSP knockout line (Figure 2A) [27]. At 140 days after sowing (boll maturation stage), GhSPi-#5, GhSPi-#8, and GhSPi-#14 still maintained the self-pruning architecture (Figure 2B,C). Quantitative analyses revealed that GhSPi transgenic cotton lines exhibited significant reductions in plant height, main stem height, and the internode number compared to the wild type. These architectural parameters showed strong positive correlations with GhSP transcript levels (Figure 2D–G). The boll count, a critical determinant of cotton yield, is intricately modulated by plant architecture. Statistical analysis revealed that the number of cotton bolls in GhSP transgenic lines was significantly reduced compared to the wild type, except for an increase observed in GhSPi-#5 at 90 DAS. Like plant height, main stem height, and the internode number, the boll count of GhSPi transgenic cotton displayed a positive correlation with GhSP expression levels (Figure 2H). These results demonstrated that GhSP expression levels exhibited the dose-dependent regulation of cotton architecture. Notably, GhSPi-#5, with the mild down-regulation of GhSP expression, achieved self-pruning architecture while preventing the extreme phenotypic aberrations observed in GhSPi-#14 (Figure 2).
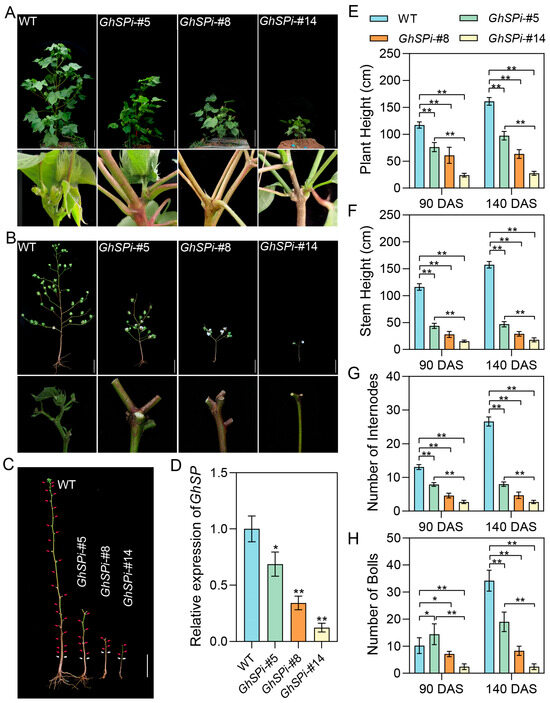
Figure 2.
Plant architecture of GhSPi transgenic cotton. Morphological comparison of plant architecture between GhSPi transgenic cotton and the wild type at 90 DAS (A) and 140 DAS (B). (C) Main stems of GhSPi transgenic and wild-type cotton at 140 DAS. White arrows designate the cotyledon location; red arrows mark the node position. (D) RT-qPCR analysis of GhSP expression in GhSPi transgenic cotton and the wild type at 14 DAS. Statistical significance was determined by Student’s t-test (n = 3). *, p < 0.05, **, p < 0.01. (E–H) Quantitative assessment of plant height (E), stem height (F), internode number (G), and boll number (H) in GhSPi transgenic lines and the wild type. Scale bars, 20 cm. Statistical significance was determined by Student’s t-test (n = 10). *, p < 0.05, **, p < 0.01.
The activity of shoot apical meristems (SAM) is coordinately regulated by complex genetic networks and influenced by internal/external environments, ultimately determining plant architecture and the flowering time. To further investigate the impact of altered GhSP expression levels on apical meristem differentiation, histological analyses were performed on shoot apices at 14 DAS. Comparative observations revealed that GhSPi-#5 exhibited wild type-like apical bud morphology with sustained growth potential, whereas GhSPi-#14 displayed complete conversion to flower bud and the loss of the upward differentiation capacity (Figure S1A). These findings suggested that the suppression of GhSP expression induced apical meristem ablation, leading to determinate growth, while the mild suppression of GhSP levels delayed this developmental switch. GhSP interacts with GhFD to mediate the antagonistic suppression of florigen GhFT activity, thereby establishing the transcriptional repression of downstream inflorescence meristem regulatory genes such as APETALA1 (GhAP1) [6]. RT-qPCR analysis revealed up-regulated GhAP1 expression in the shoot apices of the GhSPi-#5, GhSPi -#8, and GhSPi -#14 lines at 14 DAS, showing a negative correlation with GhSP expression levels (Figure S1B). This transcriptional repressive relationship suggested that GhSP-mediated GhAP1 suppression was also dose-dependent.
3.3. Effects of GhSP Suppression on Leaf and Floral Organ Development
Leaves serve as the primary site for photosynthesis in plants. Compared to the wild type, the GhSPi-#14 line exhibited a smaller leaf size and darker coloration, while GhSPi-#5 displayed leaf dimensions comparable to the wild type (Figure 3A). Chlorophyll content analysis revealed a significant increase in GhSPi transgenic cotton relative to the WT (Figure 3A,B). Furthermore, the net photosynthetic rate was enhanced in GhSPi transgenic lines compared to the wild type, except for GhSPi-#5 (Figure 3D). Observations of the floral organs on the day of anthesis demonstrated that the severely dwarfed GhSPi-#14 line developed smaller flowers and petals but produced larger, darker-colored bracts, while the GhSPi-#5 (semi-dwarf) line showed no significant differences in floral organ development compared to the wild type, indicating that the appropriate suppression of GhSP expression does not critically affect these traits (Figure 3E–H). Pollen fertility and viability are fundamental to plant reproductive success and are critically linked to cotton yield. GhSPi-#14 exhibited shortened stigmas and reduced anther numbers, whereas the GhSPi-#5 and GhSPi-#8 lines showed stigma and anther characteristics comparable to those of the WT (Figure 3I,J). Microscopic analysis revealed no significant differences in pollen fertility among GhSPi-#5, GhSPi-#8, and the wild type, whereas GhSPi-#14 displayed partially shrunken pollen grains (approximately 16%) (Figure 3K). Further scanning electron microscopy (SEM) observation of pollen morphology confirmed these findings. These results imply that the extreme suppression of GhSP (e.g., GhSPi-#14) induces leaf size reduction, floral organ malformation, and partial pollen sterility, whereas the mild down-regulation of GhSP expression (e.g., GhSPi-#5) maintains normal leaf and floral development, preserves pollen fertility, and rescues the associated negative traits caused by the excessive suppression of GhSP expression.
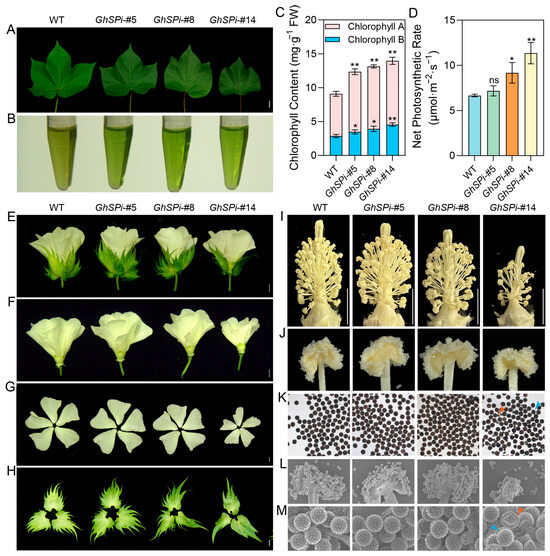
Figure 3.
Leaf, flower, and anther phenotypes of GhSPi transgenic cotton. (A) Leaves of GhSPi transgenic cotton and the wild type. Scale bars, 20 mm. Extraction (B) and measurement (C) of chlorophyll content in the leaves of GhSPi transgenic cotton and the wild type. (D) Net photosynthetic rate of GhSPi transgenic cotton and wild-type leaves. Whole flower (E), bract-peeled flower (F), petals (G), and bracts (H) of GhSPi transgenic cotton and the wild type. Scale bars, 10 mm. (I,J) The anthers from day 0 post-anthesis of GhSPi transgenic cotton and the wild type. Scale bars, 10 mm. Comparative analysis of pollen morphology in GhSPi transgenic cotton and the wild type via I2-KI staining (K) and scanning electron microscopy (L,M). Blue arrows denote normal pollen grains; red arrows highlight shrunken (abortive) pollen grains. Scale bars, 10 mm. Significant differences between GhSPi transgenic plants and the wild-type control in C and D were determined by Student’s t-test. *, p < 0.05; **, p < 0.01; ns, not significant, n = 3.
3.4. Mild Silencing of GhSP Preserves Key Agronomic Traits in Cotton
To identify the effects of plant architecture alterations caused by GhSP down-regulation on cotton yield and the key agronomic traits of the GhSPi lines and the wild type were evaluated. The bolls of the extremely dwarfed line GhSPi-#14 were markedly smaller than those of the WT, whereas the semi-dwarfed line GhSPi-#5 exhibited boll dimensions comparable to those of the WT (Figure 4A,B). Quantitative analysis of boll weight revealed that GhSPi-#5 exhibited no significant difference compared to the WT, while GhSPi-#8 and GhSPi-#14 showed significant reductions (Figure 4C). The primary cause of this outcome likely stems from a low proportion of sterile seeds in GhSPi-#5, comparable to the wild type, whereas GhSPi-#8 and GhSPi-#14 harbored substantially more sterile seeds, thereby limiting their boll weight development (Figure 4D). Additionally, we observed some conjoined bolls in the GhSPi lines, with the frequency of this trait increasing as GhSP expression levels decreased (Figure S2). Fiber length serves as a critical parameter for evaluating cotton fiber quality. Our study demonstrated that fibers from GhSPi-#8 and GhSPi-#14 were significantly shortened compared to the wild type, whereas no statistically significant difference was observed in GhSPi-#5 (Figure 4E,G). Additionally, GhSPi-#14 exhibited a markedly reduced seed size, while both GhSPi-#5 and GhSPi-#8 showed no significant variation relative to the wild type (Figure 4F). Analysis of several yield-related traits revealed a marked increase in lint percentage for GhSPi-#14; however, its seed index and lint index showed significant reductions, collectively reflecting an overall yield loss. In contrast, GhSPi-#5 and GhSPi-#8 exhibited no statistically significant differences in these parameters compared to the wild type (Figure 4H–J). Further fiber quality analysis revealed that GhSPi-#5 displayed no significant differences in fiber properties compared to the wild type, except for a reduced micronaire value.While the excessive suppression of GhSP expression (GhSPi-#8 and GhSPi-#4) led to shortened fiber length, a decreased uniformity index, and reduced micronaire values, which collectively indicate inferior fiber quality (Table S2). Additionally, a cross-sectional analysis of fibers demonstrated that GhSPi transgenic cotton exhibited reduced fiber cell wall thickness compared to wild-type plants, with this phenotype being more pronounced in GhSPi-#8 and GhSPi-#14 (Figure 4K,L).
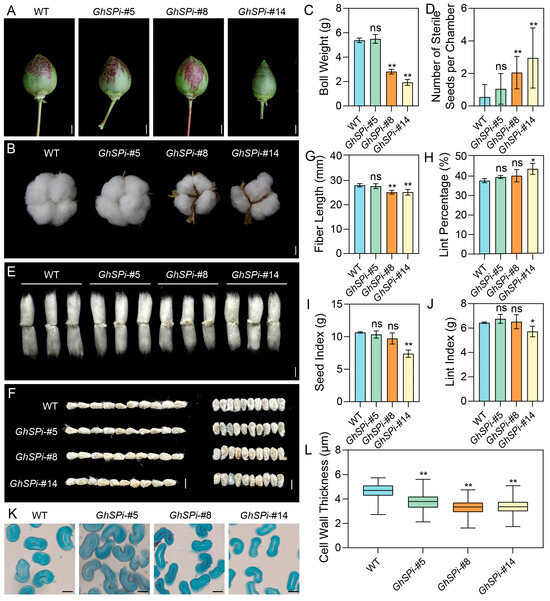
Figure 4.
Several agronomic traits related to yield in GhSPi transgenic cotton. Immature (at 30 DPA, (A)) and mature (B) cotton bolls from transgenic and wild-type plants. Scale bars, 10 mm. (C) Weight of individual mature cotton bolls (n = 18). (D) Number of sterile seeds in individual chambers (n = 20). Mature fibers (E) and seeds (F) of GhSPi transgenic plants and the wild type. Scale bars, 10 mm. Measurement and statistical comparison of fiber length (G), lint percentage (H), the seed index (I), and the lint index (J) were conducted between GhSPi transgenic cotton lines and the wild type (n = 3). (K) Microscopic section observation of mature fibers. Scale bars, 10 µm. (L) Statistical analysis of mature fiber cell wall thickness (n > 150). Student’s t-test was used for statistical analyses, *, p < 0.05, **, p < 0.01; ns, not significant.
Taken together, these results suggest that the mild suppression of GhSP expression (e.g., in GhSPi-#5) improves cotton architecture without significant fiber quality and yield penalties. Conversely, the excessive inhibition of GhSP causes significant declines in both fiber quality and yield.
4. Discussion
The florigen (FT/SFT) and anti-florigen (TFL1/SP) systems collaboratively regulate vegetative and reproductive growth, determining plant architecture. This regulatory module has been harnessed in crop improvement to enhance yields across species. In tomato, SP mutants are recognized as one of the most transformative genetic traits in modern agriculture [11]. By leveraging combinations of selected mutations involving various components of the flowering pathway, tomato productivity has been further optimized [19]. For instance, tomato plants heterozygous for a loss-of-function allele of SFT exhibit up to a 60% increase in yield [35]. In cotton, cotton leaf crumple virus (CLCrV)-mediated GhFT/SFT over-expression decouples flowering from photoperiodic control, inducing premature flowering while retaining the indeterminate growth habit of main stems. Conversely, tobacco rattle virus (TRV)-driven GhFT/SFT silencing delays maturation and amplifies indeterminacy [3,15,16]. The over-expression of GhSP suppresses the vegetative-to-reproductive transition in branches, while its silencing triggers determinate growth, with terminal flowers on the main stem and shorter lateral branches [14,16,24]. These findings underscore the significant potential of the florigen/anti-florigen system in cotton breeding improvement. For example, the GhTFL1L86P mutation, which weakens GhTFL1’s protein interaction capacity, produces semi-dwarf, compact cotton plants [8]. Our research employed RNA interference technology to generate heritable transgenic cotton with stepwise changes in GhSP expression levels. Among these, cotton plants with severely suppressed GhSP expression levels exhibited extreme dwarfism, similar to that of plants with CRISPR/Cas9-knockout GhSP [27]. In contrast, the GhSPi-#5 line with mild GhSP suppression achieved a semi-dwarf phenotype (Figure 1). The expression levels of GhSP exhibited negative correlation with cotton height, the internode number, boll weight, etc., confirming that GhSP exerts its functions in a dose-dependent manner.
Unlike the urgent demand for lodging resistance traits in crops such as rice and wheat, the pursuit of plant architecture in cotton breeding is primarily driven by the need to adapt to high-density planting and mechanical harvesting. Genome-wide association studies (GWASs) have identified multiple quantitative trait loci (QTLs) associated with plant architectural traits [36,37,38,39], demonstrating that cotton architecture regulation involves the interplay of multiple genes. For instance, a major locus (PH1) controlling plant height encoding gibberellin 2-oxidase 1A (GhPH1) suppresses cotton plant height by inactivating bioactive gibberellins (GAs) [5], while elevated endogenous GA levels induce a taller plant [40]. A synergistic regulatory module involving the GA and strigolactone (SL) pathways (GhD53-GhGARF-GhPH1/PAVPH1) coordinates plant height modulation [5]. Additionally, the NAC family gene GhSBI1 was identified as a core regulator of short fruit branches, shortening internodes by repressing GA signaling [41]. The miR164-GhCUC2-GhBRC1-GhNCED1 regulatory module further controls ABA levels to shape aerial plant architecture [42]. Here, we have identified a mild GhSP-suppressed line, GhSPi-#5, with optimal plant height alongside preserved fiber yield and quality. This validates the feasibility of precise GhSP expression manipulation for cotton agronomic trait optimization. Notably, multiple factors, including the chromosome position, the copy number, the genetic background, and even generations, may influence the expression level of a transgene and, consequently, its functions. Considering the dose-dependent manner of GhSP functioning, it is necessary to continuously monitor GhSP expression levels and involve multiple transgenic events to create an optimal cotton germplasm for a certain genetic background or environment.
To address the challenges posed by compressed planting windows due to climatic factors in cotton-growing regions, the development of early-maturing cotton cultivars with shortened growth periods, enabling late sowing and early harvesting, has become a critical breeding objective [6]. Early-maturing varieties typically exhibit a dwarf and compact plant architecture, indicating a strong correlation between early maturity and plant architecture [43]. Recent studies have indicated that the FT-SP module plays a pivotal role in coordinating flowering time and plant architectural development. Silencing anti-florigen SP not only induces dwarfism but also accelerates flowering, a hallmark of early maturity [16,24]. However, early maturity traits are often associated with compromised fiber quality, which makes it difficult to meet the requirements for high-end textile products [6]. Knocking out GhSP [27], or severely suppressing GhSP expression levels (as in our study), results in extremely dwarfed plants with very low boll numbers and reduced fiber quality, making them unsuitable for production. Our study found that mildly suppressing GhSP could maintain plant height within the agronomically suitable range of 70~100 cm while significantly increasing the number of bolls per plant and reducing the rate of sterile seeds compared to extremely dwarfed plants, with fiber quality comparable to that of the wild type (Figure 2 and Figure 4). This phenotypic divergence likely stems from developmental plasticity regulated by the dose-dependent effects of GhSP expression. The severe suppression of GhSP prematurely terminates indeterminate growth in apical meristems, presumably resulting in insufficient photosynthetic source capacity to meet the sink demand of developing bolls, ultimately triggering imbalanced assimilate partitioning. In contrast, the mild suppression of GhSP delays the developmental transition from indeterminate to determinate growth patterns in shoot apices, thereby increasing plant biomass and maintaining the source–sink ratio within an optimized range. This study reveals the nonlinear relationship between GhSP expression levels and plant architecture and yield quality traits, providing new strategies for molecular design breeding in early-maturing cotton. It is proposed that utilizing tissue-specific promoters (such as those activated during the reproductive growth stage) or photoperiod-responsive elements to precisely regulate the spatiotemporal expression of GhSP can not only break through the architectural constraints of traditional early-maturing varieties but also avoid physiological deterioration in fiber quality.
5. Conclusions
In this study, RNA interference was employed to down-regulate GhSP expression in cotton, generating transgenic lines (GhSPi) with varying suppression levels. The results show that GhSP expression regulates cotton architecture in a dose-dependent manner. Mild suppression (such as GhSPi-#5, <30% reduction) resulted in a semi-dwarf phenotype (70~100 cm), with normal leaf/floral organ development and pollen fertility comparable to those of the wild type. In contrast, severe suppression (such as GhSPi-#14, >60% reduction) caused extreme dwarfism (20~40 cm) and the premature termination of apical meristem activity into floral buds, accompanied by reduced leaf size, floral organ malformation, and partial pollen sterility. Agronomic trait analysis revealed that GhSPi-#5 exhibited no significant differences from the wild type in boll size, fiber length, and yield metrics, whereas severely suppressed lines displayed a reduced boll size, compromised fiber quality, and an increased sterile seed ratio. This study demonstrates that mild GhSP suppression (e.g., GhSPi-#5) optimizes plant architecture while avoiding the detrimental effects of severe suppression on photosynthesis, reproduction, and fiber quality, providing an effective strategy for cotton architecture improvement.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology14060601/s1, Figure S1: Stem apex characteristics of GhSPi transgenic cotton; Figure S2: Conjoined bolls appeared in GhSPi transgenic lines; Table S1: Primers used in this study; Table S2: GhSP-RNAi transgenic cotton (#5, #8, #14) and wild-type mature fiber quality analysis.
Author Contributions
Y.X. conceived and designed the research; Y.W., Q.L., W.Y. and J.C. performed the experiments; Y.W., Q.S. and Z.C. analyzed the data; J.Z., A.L. and J.K. participated in the discussion of the results; Y.W. and Q.L. drafted the manuscript; Y.X. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32201859); the Natural Science Foundation of Chongqing, China (CSTB2022NSCQ-BHX0041 and CSTB2023NSCQ-MSX0356); the National Key Research and Development Program of China (2024YFD1200304); the Chongqing Graduate Scientific Research Innovation Project (CYB23154).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hong, J.; Su, S.; Wang, L.; Bai, S.; Xu, J.; Li, Z.; Betts, N.; Liang, W.; Wang, W.; Shi, J.; et al. Combined genome-wide association study and epistasis analysis reveal multifaceted genetic architectures of plant height in Asian cultivated rice. Plant Cell Environ. 2023, 46, 1295–1311. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J. Molecular Basis of Plant Architecture. Annu. Rev. Plant Biol. 2008, 59, 253–279. [Google Scholar] [CrossRef] [PubMed]
- McGarry, R.C.; Ayre, B.G. Cotton architecture: Examining the roles of SINGLE FLOWER TRUSS and SELF-PRUNING in regulating growth habits of a woody perennial crop. Curr. Opin. Plant Biol. 2021, 59, 101968. [Google Scholar] [CrossRef]
- Huang, X.; Liu, H.; Ma, B. The Current Progresses in the Genes and Networks Regulating Cotton Plant Architecture. Front. Plant Sci. 2022, 13, 882583. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Chen, B.; Li, H.; Pei, X.; Sun, Y.; Sun, G.; Pan, Z.; Dai, P.; Gao, X.; Geng, X.; et al. The strigolactone-gibberellin crosstalk mediated by a distant silencer fine-tunes plant height in upland cotton. Mol. Plant 2024, 17, 1539–1557. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Y.; Liu, J.; Wang, Z.; Li, F.; Ge, X. Recent advances and future perspectives in early-maturing cotton research. New Phytol. 2022, 237, 1100–1114. [Google Scholar] [CrossRef]
- Yang, Z.; Gao, C.; Zhang, Y.; Yan, Q.; Hu, W.; Yang, L.; Wang, Z.; Li, F. Recent progression and future perspectives in cotton genomic breeding. J. Integr. Plant Biol. 2023, 65, 548–569. [Google Scholar] [CrossRef]
- Wang, G.; Wang, F.; Xu, Z.; Wang, Y.; Zhang, C.; Zhou, Y.; Hui, F.; Yang, X.; Nie, X.; Zhang, X.; et al. Precise fine-turning of GhTFL1 by base editing tools defines ideal cotton plant architecture. Genome Biol. 2024, 25, 59. [Google Scholar] [CrossRef]
- Eshed, Y.; Lippman, Z.B. Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science 2019, 366, 705. [Google Scholar] [CrossRef]
- Carmel-Goren, L.; Liu, Y.S.; Lifschitz, E.; Zamir, D. The SELF-PRUNING gene family in tomato. Plant Mol. Biol. 2003, 52, 1215–1222. [Google Scholar] [CrossRef]
- Pnueli, L.; Carmel-Goren, L.; Hareven, D.; Gutfinger, T.; Alvarez, J.; Ganal, M.; Zamir, D.; Lifschitz, E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 1998, 125, 1979–1989. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.; Carpenter, R.; Copsey, L.; Vincent, C.; Rothstein, S.; Coen, E. Control of inflorescence architecture in Antirrhinum. Nature 1996, 379, 791–797. [Google Scholar] [CrossRef]
- Bradley, D.; Ratcliffe, O.; Vincent, C.; Carpenter, R.; Coen, E. Inflorescence commitment and architecture in Arabidopsis. Science 1997, 275, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Si, Z.; Liu, H.; Zhu, J.; Chen, J.; Wang, Q.; Fang, L.; Gao, F.; Tian, Y.; Chen, Y.; Chang, L.; et al. Mutation of SELF-PRUNING homologs in cotton promotes short-branching plant architecture. J. Exp. Bot. 2018, 69, 2543–2553. [Google Scholar] [CrossRef]
- McGarry, R.C.; Rao, X.; Li, Q.; van der Knaap, E.; Ayre, B.G.; Sharwood, R. SINGLE FLOWER TRUSS and SELF-PRUNING signal developmental and metabolic networks to guide cotton architectures. J. Exp. Bot. 2020, 71, 5911–5923. [Google Scholar] [CrossRef]
- McGarry, R.C.; Prewitt, S.F.; Culpepper, S.; Eshed, Y.; Lifschitz, E.; Ayre, B.G. Monopodial and sympodial branching architecture in cotton is differentially regulated by the Gossypium hirsutum SINGLE FLOWER TRUSS and SELF-PRUNING orthologs. New Phytol. 2016, 212, 244–258. [Google Scholar] [CrossRef]
- Lifschitz, E.; Ayre, B.G.; Eshed, Y. Florigen and anti-florigen—A systemic mechanism for coordinating growth and termination in flowering plants. Front. Plant Sci. 2014, 5, 465. [Google Scholar] [CrossRef] [PubMed]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2016, 49, 162–168. [Google Scholar] [CrossRef]
- Park, S.J.; Jiang, K.; Tal, L.; Yichie, Y.; Gar, O.; Zamir, D.; Eshed, Y.; Lippman, Z.B. Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 2014, 46, 1337–1342. [Google Scholar] [CrossRef]
- Liu, B.; Watanabe, S.; Uchiyama, T.; Kong, F.; Kanazawa, A.; Xia, Z.; Nagamatsu, A.; Arai, M.; Yamada, T.; Kitamura, K.; et al. The Soybean Stem Growth Habit Gene Dt1 Is an Ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol. 2010, 153, 198–210. [Google Scholar] [CrossRef]
- Fernandez, L.; Torregrosa, L.; Segura, V.; Bouquet, A.; Martinez-Zapater, J.M. Transposon-induced gene activation as a mechanism generating cluster shape somatic variation in grapevine. Plant J. 2010, 61, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Comadran, J.; Kilian, B.; Russell, J.; Ramsay, L.; Stein, N.; Ganal, M.; Shaw, P.; Bayer, M.; Thomas, W.; Marshall, D.; et al. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 2012, 44, 1388–1392. [Google Scholar] [CrossRef] [PubMed]
- Iwata, H.; Gaston, A.; Remay, A.; Thouroude, T.; Jeauffre, J.; Kawamura, K.; Oyant, L.H.S.; Araki, T.; Denoyes, B.; Foucher, F. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 2012, 69, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Teng, Z.; Kong, J.; Liu, X.; Wang, W.; Zhang, X.; Zhai, T.; Deng, X.; Wang, J.; Zeng, J.; et al. Natural variation in a CENTRORADIALIS homolog contributed to cluster fruiting and early maturity in cotton. BMC Plant Biol. 2018, 18, 286. [Google Scholar] [CrossRef]
- Prewitt, S.F.; Ayre, B.G.; McGarry, R.C. Cotton CENTRORADIALIS/TERMINAL FLOWER 1/SELF-PRUNING genes functionally diverged to differentially impact plant architecture. J. Exp. Bot. 2018, 69, 5403–5417. [Google Scholar] [CrossRef]
- Chen, W.; Yao, J.; Li, Y.; Zhao, L.; Liu, J.; Guo, Y.; Wang, J.; Yuan, L.; Liu, Z.; Lu, Y.; et al. Nulliplex-branch, a TERMINAL FLOWER 1 ortholog, controls plant growth habit in cotton. Theor. Appl. Genet. 2019, 132, 97–112. [Google Scholar] [CrossRef]
- Zhang, J.; Si, Z.; Chen, R.; Liu, W.; Shi, Y.; Shi, Z.; Mei, H.; Hu, Y.; Fang, L.; Zhang, T. A new model system for cotton indoor genetic and genomic research. Sci. China Life Sci. 2023, 66, 1444–1446. [Google Scholar] [CrossRef]
- Xiao, Y.H.; Yin, M.H.; Hou, L.; Pei, Y. Direct Amplification of Intron-Containing Hairpin RNA Construct from Genomic DNA. BioTechniques 2006, 41, 548–552. [Google Scholar] [CrossRef]
- Luo, M.; Xiao, Y.; Li, X.; Lu, X.; Deng, W.; Li, D.; Hou, L.; Hu, M.; Li, Y.; Pei, Y. GhDET2, a steroid 5alpha-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J. 2007, 51, 419–430. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, Y.; Liang, F.; Tang, J.; Ma, P.; Tian, J.; Jiang, C.; Zhang, W. The net photosynthetic rate of the cotton boll-leaf system determines boll weight under various plant densities. Eur. J. Agron. 2021, 125, 126251. [Google Scholar] [CrossRef]
- Li, X.; Wu, Y.; Liu, Z.; Wei, H.; Wang, H.; Yu, S. Apical meristem transcriptome analysis identifies a role for the blue light receptor gene GhFKF1 in cotton architecture development. Crop J. 2024, 12, 1126–1136. [Google Scholar] [CrossRef]
- Yan, Q.; Wang, Y.; Li, Q.; Zhang, Z.; Ding, H.; Zhang, Y.; Liu, H.; Luo, M.; Liu, D.; Song, W.; et al. Up-regulation of GhTT2-3A in cotton fibres during secondary wall thickening results in brown fibres with improved quality. Plant Biotechnol. J. 2018, 16, 1735–1747. [Google Scholar] [CrossRef] [PubMed]
- GB/T 20392-2006; Test Method of Properties of Cotton Fibers by High Volume Instruments. Standards Press of China: Beijing, China, 2006.
- Krieger, U.; Lippman, Z.B.; Zamir, D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010, 42, 459–463. [Google Scholar] [CrossRef]
- Su, J.; Li, L.; Zhang, C.; Wang, C.; Gu, L.; Wang, H.; Wei, H.; Liu, Q.; Huang, L.; Yu, S. Genome-wide association study identified genetic variations and candidate genes for plant architecture component traits in Chinese upland cotton. Theor. Appl. Genet. 2018, 131, 1299–1314. [Google Scholar] [CrossRef]
- Wen, T.; Dai, B.; Wang, T.; Liu, X.; You, C.; Lin, Z. Genetic variations in plant architecture traits in cotton (Gossypium hirsutum) revealed by a genome-wide association study. Crop J. 2019, 7, 209–216. [Google Scholar] [CrossRef]
- Ji, G.; Liang, C.; Cai, Y.; Pan, Z.; Meng, Z.; Li, Y.; Jia, Y.; Miao, Y.; Pei, X.; Gong, W.; et al. A copy number variant at the HPDA-D12 locus confers compact plant architecture in cotton. New Phytol. 2020, 229, 2091–2103. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Wang, P.; Zhang, M.; Abbas, M.; Zhang, J.; Liang, C.; Wang, Y.; Wei, Y.; Meng, Z.; Zhang, R. UAV-based time-series phenotyping reveals the genetic basis of plant height in upland cotton. Plant J. 2023, 115, 937–951. [Google Scholar] [CrossRef]
- Xiao, Y.H.; Li, D.M.; Yin, M.H.; Li, X.B.; Zhang, M.; Wang, Y.J.; Dong, J.; Zhao, J.; Luo, M.; Luo, X.Y.; et al. Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J. Plant Physiol. 2010, 167, 829–837. [Google Scholar] [CrossRef]
- Zhong, W.; Wu, L.; Li, Y.; Li, X.; Wang, J.; Pan, J.; Zhu, S.; Fang, S.; Yao, J.; Zhang, Y.; et al. GhSBI1, a CUP-SHAPED COTYLEDON 2 homologue, modulates branch internode elongation in cotton. Plant Biotechnol. J. 2024, 22, 3175–3193. [Google Scholar] [CrossRef]
- Zhan, J.; Chu, Y.; Wang, Y.; Diao, Y.; Zhao, Y.; Liu, L.; Wei, X.; Meng, Y.; Li, F.; Ge, X. The miR164-GhCUC2-GhBRC1 module regulates plant architecture through abscisic acid in cotton. Plant Biotechnol. J. 2021, 19, 1839–1851. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, C.; Huang, J.; Liu, Q.; Wei, H.; Wang, H.; Liu, G.; Gu, L.; Yu, S. Genomic analyses reveal the genetic basis of early maturity and identification of loci and candidate genes in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 2021, 19, 109–123. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).