Characterization of the Complete Mitochondrial Genome of the Red Alga Ahnfeltiopsis flabelliformis (Rhodophyta, Gigartinales, Phyllophoraceae) and Its Phylogenetic Analysis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Genomic DNA Extraction and Sequencing
2.2. Sequence Assembly, Annotation, and Analysis
2.3. Phylogenetic Analysis
3. Results and Discussion
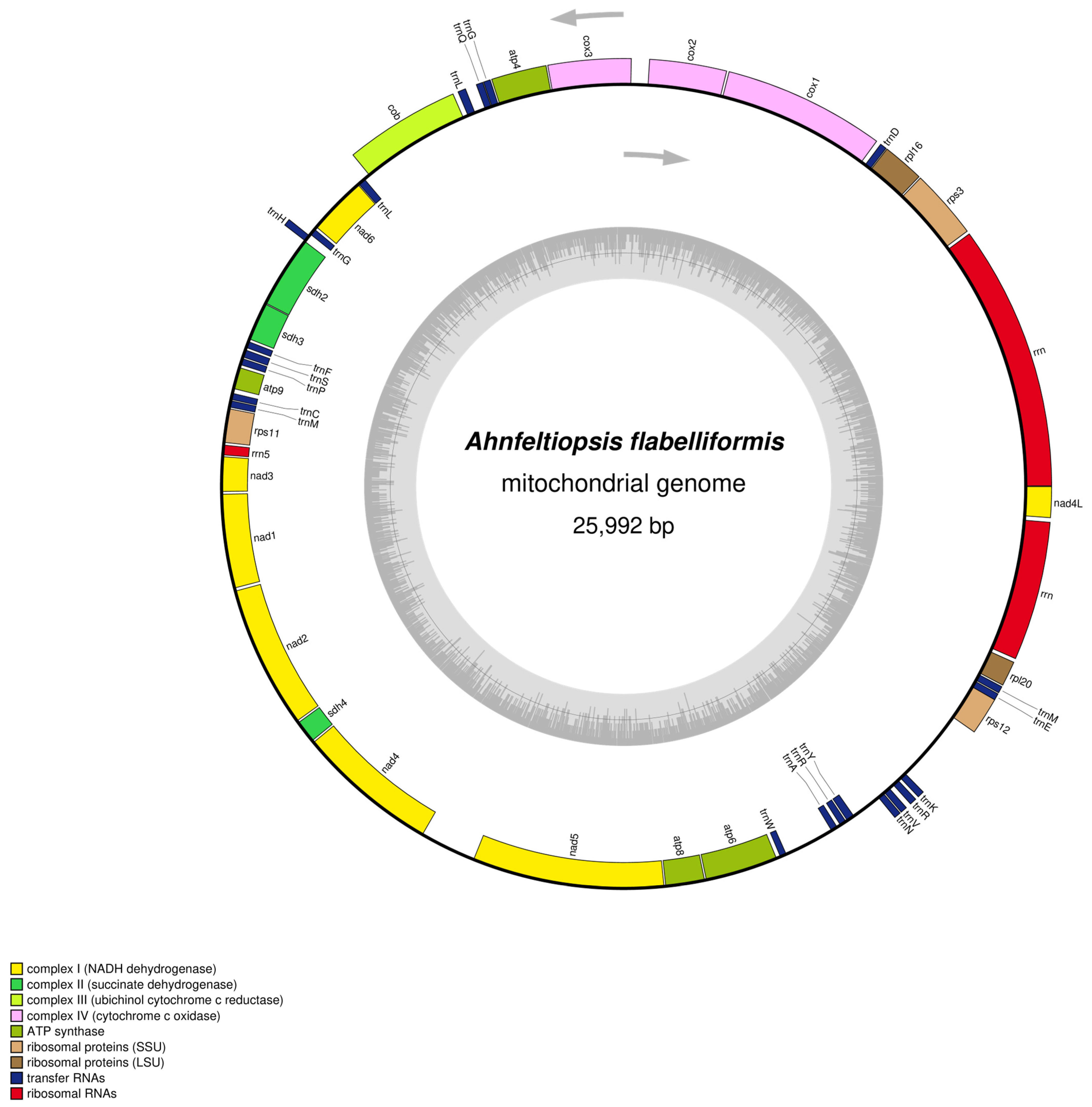
3.1. Mitochondrial Genome Structure and Nucleotide Composition
3.2. Protein-Coding Gene Features
3.3. Ribosomal and Transfer RNA Genes
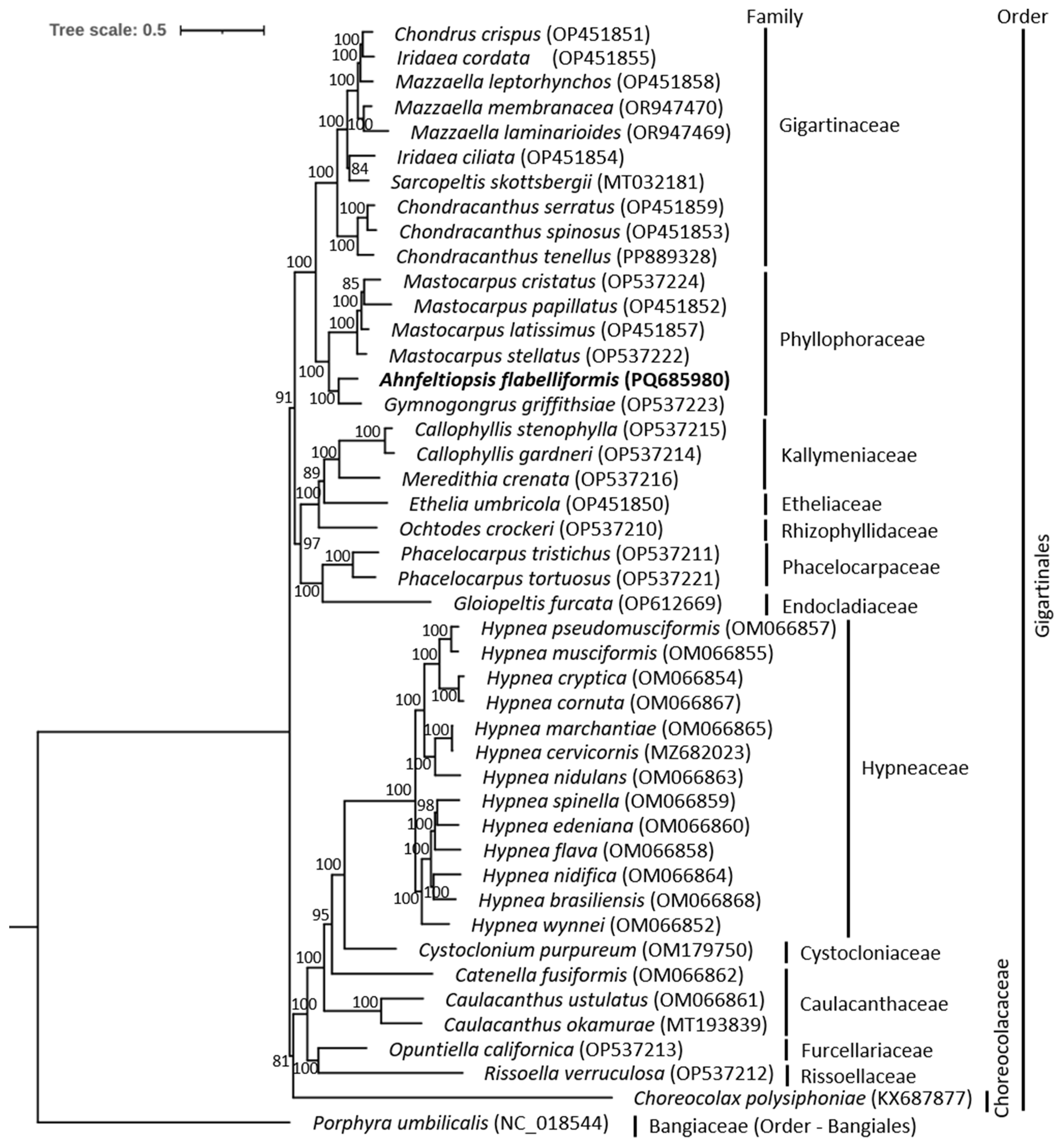
3.4. Phylogenetic Relationship Within Gigartinales
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guiry, G.M. How Many Species of Algae Are There? A Reply. Four Kingdoms, 14 Phyla, 63 Classes and Still Growing. J. Phycol. 2024, 60, 214–228. [Google Scholar] [CrossRef] [PubMed]
- Gurgel, C.F.D.; Lopez-Bautista, J. Red Algae. In Encyclopedia of Life Science; Wiley: Hoboken, NJ, USA, 2007. [Google Scholar] [CrossRef]
- Ohta, K.; Satoh, K.; Matsuda, M.; Enomoto, S.; Kitade, Y.; Mizushina, Y.; Yoshida, H.; Matsukage, A.; Hayakawa, S.; Sakaguchi, K. Sulfoquinovosyldiacylglycerol, KM043, a New Potent Inhibitor of Eukaryotic DNA Polymerases and HIV-1 Reverse Transcriptase from a Marine Red Alga, Gigartina tenella. Chem. Pharm. Bull. 1998, 46, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Michalak, I.; Chojnacka, K. Algae as Production Systems of Bioactive Compounds. Eng. Life Sci. 2015, 15, 160–176. [Google Scholar] [CrossRef]
- Aziz, E.; Naguib, M.M.; Aziz, S.W.; Klimek, J.; Jákói, Z.; Pál, J. An Overview on Red Algae Bioactive Compounds and Their Pharmaceutical Applications. J. Complement. Integr. Med. 2021, 17, 20190203. [Google Scholar] [CrossRef]
- Bixler, H.J.; Porse, H. A Decade of Change in the Seaweed Hydrocolloids Industry. J. Appl. Phycol. 2011, 23, 321–335. [Google Scholar] [CrossRef]
- Guo, Z.; Hu, W.; Wu, G.; Zhu, J.; Yin, Y.; Tang, J.; Zhang, W.; He, P. Carrageenan Oligosaccharides: A Comprehensive Review of Preparation, Isolation, Purification, Structure, Biological Activities, and Applications. Algal Res. 2022, 61, 102593. [Google Scholar] [CrossRef]
- Zhang, J.; Zannie, L.; Scott, W. The Global Carrageenan Industry. In Globalisation and Livelihood Transformations in the Indonesian Seaweed Industry; Routledge: London, UK, 2023; pp. 23–50. [Google Scholar] [CrossRef]
- Kravchenko, A.O.; Menshov, A.S.; Sokolova, E.V.; Ermakova, S.P.; Malyarenko, O.S.; Avilov, S.A.; Zvyagintseva, T.N. Carrageenans and Their Oligosaccharides from Red Seaweeds Ahnfeltiopsis flabelliformis and Mastocarpus pacificus (Phyllophoraceae) and Their Antiproliferative Activity. Int. J. Mol. Sci. 2023, 24, 7657. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, Y.H.; Seo, Y.W.; Park, S. Quorum Sensing Inhibitors from the Red Alga, Ahnfeltiopsis flabelliformis. Biotechnol. Bioprocess Eng. 2007, 12, 308–311. [Google Scholar] [CrossRef]
- Masuda, M. Ahnfeltiopsis (Gigartinales, Rhodophyta) in the Western Pacific. Jpn. J. Phycol. 1993, 41, 1–6. [Google Scholar]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef]
- Lee, Y.P.; Kang, S.Y. A Catalogue of the Seaweeds in Korea; Jeju National University Press: Jeju, Republic of Korea, 2001. [Google Scholar]
- Kim, H.-S.; Boo, S.M.; Lee, I.K.; Sohn, C.H. National List of Species of Korea (Marine Algae); National Institute of Biological Resources, Ministry of Environment: Incheon, Republic of Korea, 2013. [Google Scholar]
- Shibneva, S.Y.; Skriptsova, A.V.; Semenchenko, A.A.; Suzuki, M. Morphological and Molecular Reassessment of Three Species of the Genus Besa (Phyllophoraceae, Rhodophyta) from the Northwest Pacific. Eur. J. Phycol. 2021, 56, 72–84. [Google Scholar] [CrossRef]
- Schneider, C.W.; Wynne, M.J. A Synoptic Review of the Classification of Red Algal Genera a Half Century After Kylin’s Die Gattungen der Rhodophyceen. Bot. Mar. 2007, 50, 197–249. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Beck, N.; Lang, B. MFannot, Organelle Genome Annotation Webserver. Université de Montréal, QC, Canada. Available online: https://megasun.bch.umontreal.ca/apps/mfannot/ (accessed on 1 December 2024).
- Gish, W.; States, D.J. Identification of Protein Coding Regions by Database Similarity Search. Nat. Genet. 1993, 3, 266–272. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar] [CrossRef]
- Lang, B.F.; Laforest, M.J.; Burger, G. Mitochondrial Introns: A Critical View. Trends Genet. 2007, 23, 119–125. [Google Scholar] [CrossRef]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of Nucleotide Composition at Fourfold Degenerate Sites of Animal Mitochondrial Genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Patil, M.P.; Kim, J.O.; Kim, Y.R.; Yoon, S.; Kim, K. Complete Mitogenome Sequencing, Annotation, and Phylogeny of Grateloupia turuturu, a Red Alga with Intronic cox1 Gene. Life 2023, 13, 1642. [Google Scholar] [CrossRef]
- Hughey, J.R.; Leister, G.L.; Gabrielson, P.W.; Hommersand, M.H. Sarcopeltis gen. nov. (Gigartinaceae, Rhodophyta), with S. skottsbergii comb. nov. from Southern South America and S. antarctica sp. nov. from the Antarctic Peninsula. Phytotaxa 2020, 468, 75–88. [Google Scholar] [CrossRef]
- de Jesus, P.B.; de Mattos Lyra, G.; Zhang, H.; Fujii, M.T.; Nauer, F.; de Castro Nunes, J.M.; Davis, C.C.; Oliveira, M.C. Phylogenomics and Taxon-Rich Phylogenies of New and Historical Specimens Shed Light on the Systematics of Hypnea (Cystocloniaceae, Rhodophyta). Mol. Phylogenet. Evol. 2023, 183, 107752. [Google Scholar] [CrossRef]
- Patil, M.P.; Kim, Y.R.; Nakashita, S.; Kim, J.O.; Kim, K. Mitogenome Features and Phylogenetic Analysis of Red Algae, Grateloupia cornea (Rhodophyta, Halymeniales). J. Genet. 2024, 103, 19. [Google Scholar] [CrossRef]
- Sepúlveda-Espinoza, F.; Cofré-Serrano, A.; Veloso-Valeria, T.; Quesada-Calderon, S.; Guillemin, M.L. Characterization of the Organellar Genomes of Mazzaella laminarioides and Mazzaella membranacea (Gigartinaceae, Rhodophyta). J. Phycol. 2024, 60, 797–805. [Google Scholar] [CrossRef]
- Aguilar, A.; Ahumada, T.J.; Amezcua Moreno, N.; Bohn, J.; Bustamante, D.E.; Calderon, M.S.; Cardoso, E.; Carranza, R.; Castillo, M.; Cazares, E.; et al. The Complete Mitochondrial and Plastid Genomes of the Invasive Marine Red Alga Caulacanthus okamurae (Caulacanthaceae, Rhodophyta) from Moss Landing, California, USA. Mitochondrial DNA Part B 2020, 5, 2067–2069. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Liu, P.; Liu, X.; Zhang, J.; Tan, X.; Jia, X.; Jin, Y.; Liu, T.; Hu, Y. Complete Organellar Genomes and Molecular Phylogeny of Hypnea cervicornis (Gigartinales, Florideophyceae) from China. J. Appl. Phycol. 2022, 34, 2705–2717. [Google Scholar] [CrossRef]
- Moeckel, C.; Zaravinos, A.; Georgakopoulos-Soares, I. Strand Asymmetries Across Genomic Processes. Comput. Struct. Biotechnol. J. 2023, 21, 2036–2047. [Google Scholar] [CrossRef]
- Yang, E.C.; Kim, K.M.; Kim, S.Y.; Lee, J.; Boo, G.H.; Lee, J.H.; Nelson, W.A.; Yi, G.; Schmidt, W.E.; Fredericq, S.; et al. Highly Conserved Mitochondrial Genomes Among Multicellular Red Algae of the Florideophyceae. Genome Biol. Evol. 2015, 7, 2394–2406. [Google Scholar] [CrossRef]
- Seo, M.H.; Kang, S.C.; Kim, K.M.; Kwak, M.S.; Jo, J.; Choi, H.G.; Boo, G.H.; Yoon, H.S.; Seo, M.H.; Kang, S.C.; et al. Novel Rearrangements in the Mitochondrial Genomes of the Ceramiales (Rhodophyta) and Evolutionary Implications. Algae 2023, 38, 253–264. [Google Scholar] [CrossRef]
- Butenko, A.; Lukeš, J.; Speijer, D.; Wideman, J.G. Mitochondrial Genomes Revisited: Why Do Different Lineages Retain Different Genes? BMC Biol. 2024, 22, 15. [Google Scholar] [CrossRef]
- Qiu, H.; Price, D.C.; Yang, E.C.; Yoon, H.S.; Bhattacharya, D. Evidence of Ancient Genome Reduction in Red Algae (Rhodophyta). J. Phycol. 2015, 51, 624–636. [Google Scholar] [CrossRef]
- Salomaki, E.D.; Lane, C.E. Red Algal Mitochondrial Genomes Are More Complete Than Previously Reported. Genome Biol. Evol. 2017, 9, 48–63. [Google Scholar] [CrossRef][Green Version]
- Sharp, P.M.; Li, W.H. The Codon Adaptation Index—A Measure of Directional Synonymous Codon Usage Bias, and Its Potential Applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- Lee, Y.; Cho, C.H.; Noh, C.; Yang, J.H.; Park, S.I.; Lee, Y.M.; West, J.A.; Bhattacharya, D.; Jo, K.; Yoon, H.S. Origin of Minicircular Mitochondrial Genomes in Red Algae. Nat. Commun. 2023, 14, 3363. [Google Scholar] [CrossRef]
- Patil, M.P.; Woo, H.E.; Kim, Y.R.; Kim, J.O.; Kim, K. Complete Mitochondrial Genome and Phylogenetic Analysis of the Red Algae Chondracanthus tenellus (Rhodophyta, Gigartinales) from South Korea. Biochem. Genet. 2025, 1–19. [Google Scholar] [CrossRef]
- Beveren, F.V.; Eme, L.; López-García, P.; Ciobanu, M.; Moreira, D. Independent Size Expansions and Intron Proliferation in Red Algal Plastid and Mitochondrial Genomes. Genome Biol. Evol. 2022, 14, evac037. [Google Scholar] [CrossRef] [PubMed]
- Hancock, L.; Goff, L.; Lane, C. Red algae lose key mitochondrial genes in response to becoming parasitic. Genome Biol. Evol. 2010, 2, 897–910. [Google Scholar] [CrossRef]
- Adams, K.L.; Palmer, J.D. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 2003, 29, 380–395. [Google Scholar] [CrossRef]
| Gene | Position | Size (bp) | Coding Strand | IN | Codon | Anti-Codon | Amino Acids | ||
|---|---|---|---|---|---|---|---|---|---|
| Start | End | Start | Stop | ||||||
| rnl | 1 | 2613 | 2613 | H | 0 | - | - | - | - |
| rps3 | 2640 | 3332 | 693 | H | 26 | ATG | TAA | - | 230 |
| rpl16 | 3344 | 3760 | 417 | H | 11 | ATG | TAA | - | 138 |
| trnD | 3765 | 3836 | 72 | H | 4 | - | - | GTC | - |
| cox1 | 3879 | 5471 | 1593 | H | 42 | ATG | TAA | - | 530 |
| cox2 | 5489 | 6250 | 762 | H | 17 | ATG | TAA | - | 253 |
| cox3 | 6421 | 7239 | 819 | H | 170 | ATG | TAA | - | 272 |
| atp4 | 7250 | 7798 | 549 | H | 10 | ATG | TAA | - | 182 |
| trnG | 7808 | 7881 | 74 | H | 9 | - | - | TCC | - |
| trnQ | 7884 | 7955 | 72 | H | 2 | - | - | TTG | - |
| trnL | 8062 | 8145 | 84 | H | 106 | - | - | TAA | - |
| cob | 8192 | 9334 | 1143 | H | 46 | ATG | TAA | - | 380 |
| trnL | 9394 | 9476 | 83 | L | 59 | - | - | TAG | - |
| nad6 | 9481 | 10,098 | 618 | L | 4 | ATG | TAA | - | 205 |
| trnG | 10,123 | 10,195 | 73 | L | 24 | - | - | GCC | - |
| trnH | 10,208 | 10,282 | 75 | H | 12 | - | - | GTG | - |
| sdh2 | 10,283 | 11,041 | 759 | L | 0 | ATG | TAA | - | 252 |
| sdh3 | 11,046 | 11,447 | 402 | L | 4 | ATG | TAA | - | 133 |
| trnF | 11,470 | 11,541 | 72 | L | 22 | - | - | GAA | - |
| trnS | 11,561 | 11,644 | 84 | L | 19 | - | - | TGA | - |
| trnP | 11,655 | 11,727 | 73 | L | 10 | - | - | TGG | - |
| atp9 | 11,753 | 11,983 | 231 | L | 25 | ATG | TAA | - | 76 |
| trnC | 12,027 | 12,098 | 72 | L | 43 | - | - | GCA | - |
| trnM | 12,105 | 12,187 | 83 | L | 6 | - | - | CAT | - |
| rps11 | 12,190 | 12,546 | 357 | L | 2 | ATG | TAA | - | 118 |
| rrn5 | 12,568 | 12,675 | 108 | L | 21 | - | - | - | - |
| nad3 | 12,688 | 13,053 | 366 | L | 12 | ATG | TAA | - | 121 |
| nad1 | 13,075 | 14,055 | 981 | L | 21 | ATG | TAA | - | 326 |
| nad2 | 14,082 | 15,575 | 1494 | L | 26 | ATG | TAA | - | 497 |
| sdh4 | 15,592 | 15,831 | 240 | L | 16 | ATG | TAA | - | 79 |
| nad4 | 15,848 | 17,329 | 1482 | L | 16 | ATG | TAA | - | 493 |
| nad5 | 17,910 | 19,913 | 2004 | L | 580 | ATG | TAA | - | 667 |
| atp8 | 19,925 | 20,335 | 411 | L | 11 | ATG | TAA | - | 136 |
| atp6 | 20,348 | 21,109 | 762 | L | 12 | ATG | TAA | - | 253 |
| trnW | 21,147 | 21,218 | 72 | L | 37 | - | - | TCA | - |
| trnA | 21,739 | 21,812 | 74 | L | 520 | - | - | TGC | - |
| trnR | 21,846 | 21,919 | 74 | L | 33 | - | - | TCT | - |
| trnY | 21,931 | 22,018 | 88 | L | 11 | - | - | GTA | - |
| trnN | 22,330 | 22,402 | 73 | H | 311 | - | - | GTT | - |
| trnV | 22,410 | 22,482 | 73 | H | 7 | - | - | TAC | - |
| trnR | 22,530 | 22,604 | 75 | H | 47 | - | - | ACG | - |
| trnK | 22,632 | 22,705 | 74 | H | 27 | - | - | TTT | - |
| tatC | 22,753 | 23,460 | 708 | H | 47 | ATG | TAA | - | 235 |
| rps12 | 23,461 | 23,844 | 384 | H | 0 | ATG | TAA | - | 127 |
| trnE | 23,830 | 23,901 | 72 | H | −16 | - | - | TTC | - |
| trnM | 23,908 | 23,980 | 73 | H | 6 | - | - | CAT | - |
| rpl20 | 23,992 | 24,243 | 252 | H | 11 | ATG | TAA | - | 83 |
| rns | 24,276 | 25,648 | 1373 | H | 32 | - | - | - | - |
| nad4L | 25,687 | 25,992 | 306 | H | 38 | ATG | TAA | - | 101 |
| Species (Accession No.) | Size (bp) | Nucleotide Composition (%) | AT-Skew | GC-Skew | Number of Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | T | G | C | A + T | G + C | PCG | tRNA | rRNA | ORF* | ||||
| Ahnfeltiopsis flabelliformis (PQ685980) | 25,992 | 37.4 | 33.9 | 14.7 | 14.0 | 71.3 | 28.7 | 0.049 | 0.023 | 24 | 22 | 3 | 0 |
| Gymnogongrus griffithsiae (OP537223) | 25,812 | 37.0 | 33.6 | 15.2 | 14.2 | 70.6 | 29.4 | 0.048 | 0.035 | 24 | 22 | 3 | 0 |
| Mastocarpus cristatus (OP537224) | 25,838 | 35.5 | 31.8 | 16.7 | 16.0 | 67.3 | 32.7 | 0.054 | 0.023 | 24 | 23 | 3 | 1 |
| Mastocarpus latissimus (OP451857) | 26,208 | 36.2 | 32.2 | 16.2 | 15.5 | 68.4 | 31.6 | 0.059 | 0.023 | 25 | 23 | 3 | 1 |
| Mastocarpus papillatus (OP451852) | 26,132 | 34.3 | 30.6 | 18.1 | 17.0 | 64.9 | 35.1 | 0.058 | 0.023 | 25 | 23 | 3 | 1 |
| Mastocarpus stellatus (OP537222) | 25,826 | 36.4 | 32.5 | 15.9 | 15.2 | 68.9 | 31.1 | 0.057 | 0.022 | 23 | 22 | 3 | 0 |
| PCG | Length (bp) | Nucleotide Composition (%) | AT-Skewness | GC-Skewness | |||||
|---|---|---|---|---|---|---|---|---|---|
| A | T | G | C | A + T | G + C | ||||
| rps3 | 693 | 37.2 | 36.9 | 12.3 | 13.6 | 74.2 | 25.8 | 0.004 | −0.050 |
| rpl16 | 417 | 41.2 | 31.4 | 13.9 | 13.4 | 72.7 | 27.3 | 0.135 | 0.018 |
| cox1 | 1593 | 26.9 | 39.5 | 17.0 | 16.6 | 66.4 | 33.6 | −0.189 | 0.013 |
| cox2 | 762 | 31.1 | 36.5 | 17.5 | 15.0 | 67.6 | 32.4 | −0.080 | 0.077 |
| cox3 | 819 | 25.8 | 42.9 | 17.2 | 14.2 | 68.6 | 31.4 | −0.249 | 0.097 |
| atp4 | 549 | 37.5 | 39.2 | 9.3 | 14.0 | 76.7 | 23.3 | −0.021 | −0.203 |
| cob | 1143 | 27.0 | 42.6 | 16.1 | 14.3 | 69.6 | 30.4 | −0.224 | 0.061 |
| nad6 | 618 | 29.1 | 42.9 | 15.0 | 12.9 | 72.0 | 28.0 | −0.191 | 0.075 |
| sdh2 | 759 | 35.7 | 36.2 | 13.7 | 14.4 | 71.9 | 28.1 | −0.007 | −0.023 |
| sdh3 | 402 | 28.6 | 47.5 | 8.2 | 15.7 | 76.1 | 23.9 | −0.248 | −0.313 |
| atp9 | 231 | 26.0 | 40.7 | 20.3 | 13.0 | 66.7 | 33.3 | −0.221 | 0.221 |
| rps11 | 357 | 42.6 | 32.8 | 12.0 | 12.6 | 75.4 | 24.6 | 0.130 | −0.023 |
| nad3 | 366 | 30.3 | 45.9 | 13.7 | 10.1 | 76.2 | 23.8 | −0.204 | 0.149 |
| nad1 | 981 | 28.7 | 40.8 | 16.5 | 14.0 | 69.5 | 30.5 | −0.173 | 0.084 |
| nad2 | 1494 | 28.1 | 46.0 | 13.3 | 12.6 | 74.1 | 25.9 | −0.241 | 0.028 |
| sdh4 | 240 | 35.0 | 43.3 | 10.4 | 11.3 | 78.3 | 21.7 | −0.106 | −0.038 |
| nad4 | 1482 | 27.6 | 44.5 | 14.0 | 13.9 | 72.1 | 27.9 | −0.234 | 0.005 |
| nad5 | 2004 | 26.4 | 44.0 | 16.1 | 13.5 | 70.5 | 29.5 | −0.249 | 0.088 |
| atp8 | 411 | 38.4 | 38.7 | 8.5 | 14.4 | 77.1 | 22.9 | −0.003 | −0.255 |
| atp6 | 762 | 28.7 | 43.8 | 13.0 | 14.4 | 72.6 | 27.4 | −0.208 | −0.053 |
| tatC | 708 | 28.4 | 50.0 | 9.3 | 12.3 | 78.4 | 21.6 | −0.276 | −0.137 |
| rps12 | 384 | 38.3 | 29.9 | 17.2 | 14.6 | 68.2 | 31.8 | 0.122 | 0.082 |
| rpl20 | 252 | 42.9 | 35.3 | 9.9 | 11.9 | 78.2 | 21.8 | 0.096 | −0.091 |
| nad4L | 306 | 32.7 | 41.2 | 14.4 | 11.8 | 73.9 | 26.1 | −0.115 | 0.100 |
| Total | 17,733 | 30.3 | 41.5 | 14.3 | 13.08 | 71.8 | 28.2 | - | - |
| Genes | Length (bp) | Amino Acids | Start Codon | Stop Codon | Coding Strand |
|---|---|---|---|---|---|
| rps3 | 693 | 230 | ATG | TAA | H |
| rpl16 | 417 a,d/435 b/414 c/411 e/420 f | 138 a,d/144 b/137 c/136 e/139 f | ATG | TAA | H |
| cox1 | 1593 a/1596 b/1608 c,d,e,f | 530 a/531 b/535 c,d,e,f | ATG | TAG a,b/TAA c,d,e,f | H |
| cox2 | 762 a/771 b,c,d,e,f | 253 a/256 b,c,d,e,f | ATG | TAA | H |
| cox3 | 819 | 272 | ATG | TAA | H |
| atp4 | 549 a,b/552 c,d,e,f | 182 a,b/183 c,d,e,f | ATG | TAA a,b,c,e,f/TAG d | H |
| cob | 1143 a,b/1146 c,d,e/1152 f | 380 a,b/381 c,d,e/383 f | ATG | TAA a,b,c,e,f/TAG d | H |
| nad6 | 618 a,c,e/609 b/615 d,f | 205 a,c,e/202 b/204 d,f | ATG | TAA a,c,d,e,f/TAG b | L |
| sdh2 | 759 a/762 b/753 c,d,e,f | 252 a/253 b/250 c,d,e,f | ATG | TAA a,b,c/TAG d,e,f | L |
| sdh3 | 402 a,b,d,f/447 c/396 e | 133 a,b,d,f/148 c/131 e | ATG | TAA | L |
| atp9 | 231 | 76 | ATG | TAA | L |
| rps11 | 357 a,b/360 c,d,e,f | 118 a,b/119 c,d,e,f | ATG | TAA a,b,c,d,f/TAG e | L |
| nad3 | 366 | 121 | ATG | TAA | L |
| nad1 | 981 | 326 | ATG | TAA | L |
| nad2 | 1494 | 497 | ATG | TAA | L |
| sdh4 | 240 | 79 | ATG a,b,d,e,f/GTG c | TAA | L |
| nad4 | 1482 a,c/1479 b/1473 d,e,f | 493 a,c/492 b/490 d,e,f | ATG | TAA a,b,c,f/TAG d,e | L |
| nad5 | 2004 | 667 | ATG | TAA | L |
| atp8 | 411 a/414 b,c,d,e,f | 136 a/137 b,c,d,e,f | ATG | TAA | L |
| atp6 | 762 a/771 b,c,d,e,f | 253 a/256 b,c,d,e,f | ATG a/ATT b/ATC c,d,e/ATA f | TAA | L |
| tatC | 708 a/783 b/738 c,d,f/543 e | 235 a/260 b/245 c,d,f/180 e | ATG a/ATA b,c,d,f/GTG e | TAA | H |
| rps12 | 384 | 127 | ATG | TAA | H |
| rpl20 * | 252 a,b,d/264 e | 83 a,b,d/87 e | ATG a,b,d,e | TAA a,b,d/TAG e | H |
| nad4L | 306 | 101 | ATG | TAA | H |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patil, M.P.; Kim, J.-O.; Kim, Y.-R.; Nirmal, N.; Kim, G.-D.; Kim, K. Characterization of the Complete Mitochondrial Genome of the Red Alga Ahnfeltiopsis flabelliformis (Rhodophyta, Gigartinales, Phyllophoraceae) and Its Phylogenetic Analysis. Biology 2025, 14, 638. https://doi.org/10.3390/biology14060638
Patil MP, Kim J-O, Kim Y-R, Nirmal N, Kim G-D, Kim K. Characterization of the Complete Mitochondrial Genome of the Red Alga Ahnfeltiopsis flabelliformis (Rhodophyta, Gigartinales, Phyllophoraceae) and Its Phylogenetic Analysis. Biology. 2025; 14(6):638. https://doi.org/10.3390/biology14060638
Chicago/Turabian StylePatil, Maheshkumar Prakash, Jong-Oh Kim, Young-Ryun Kim, Nilesh Nirmal, Gun-Do Kim, and Kyunghoi Kim. 2025. "Characterization of the Complete Mitochondrial Genome of the Red Alga Ahnfeltiopsis flabelliformis (Rhodophyta, Gigartinales, Phyllophoraceae) and Its Phylogenetic Analysis" Biology 14, no. 6: 638. https://doi.org/10.3390/biology14060638
APA StylePatil, M. P., Kim, J.-O., Kim, Y.-R., Nirmal, N., Kim, G.-D., & Kim, K. (2025). Characterization of the Complete Mitochondrial Genome of the Red Alga Ahnfeltiopsis flabelliformis (Rhodophyta, Gigartinales, Phyllophoraceae) and Its Phylogenetic Analysis. Biology, 14(6), 638. https://doi.org/10.3390/biology14060638








