Genome-Wide Identification and Heat Stress-Induced Expression Profiling of the Hsp70 Gene Family in Phoebe bournei
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Sample Collection
2.2. Characterization and Functional Analysis of Hsp70 Gene Family
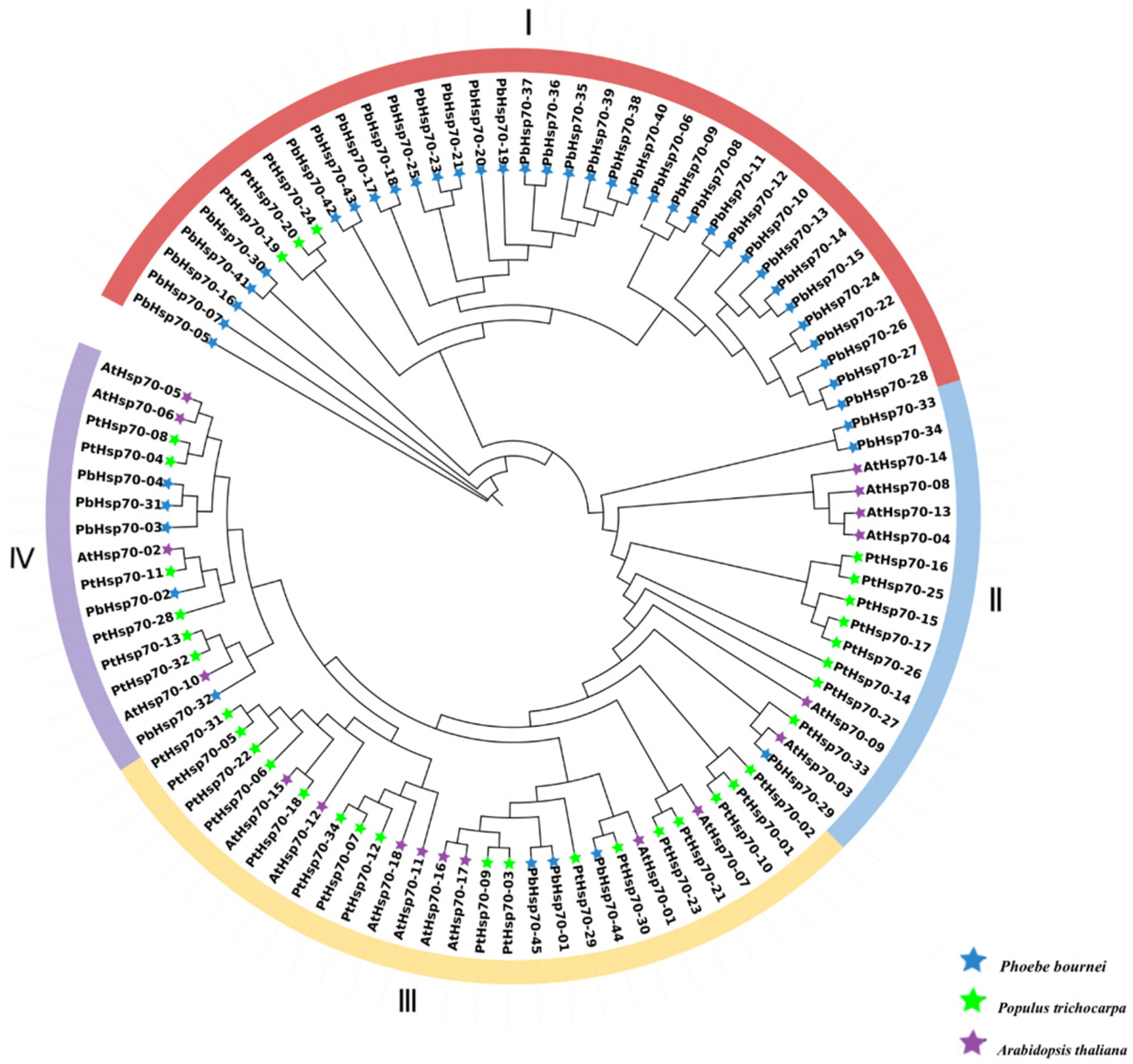
2.3. Phylogenetic Analysis of Hsp70
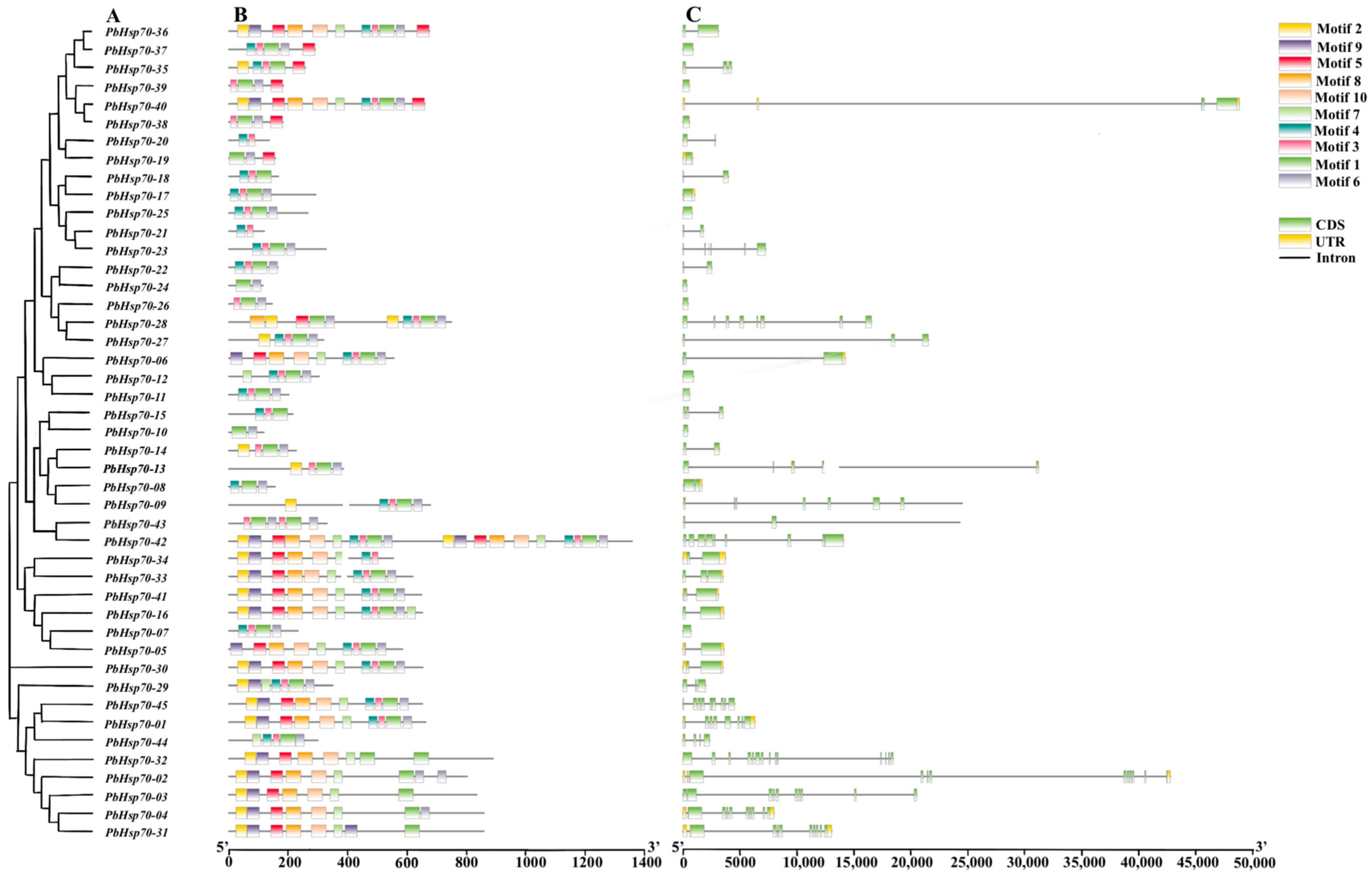
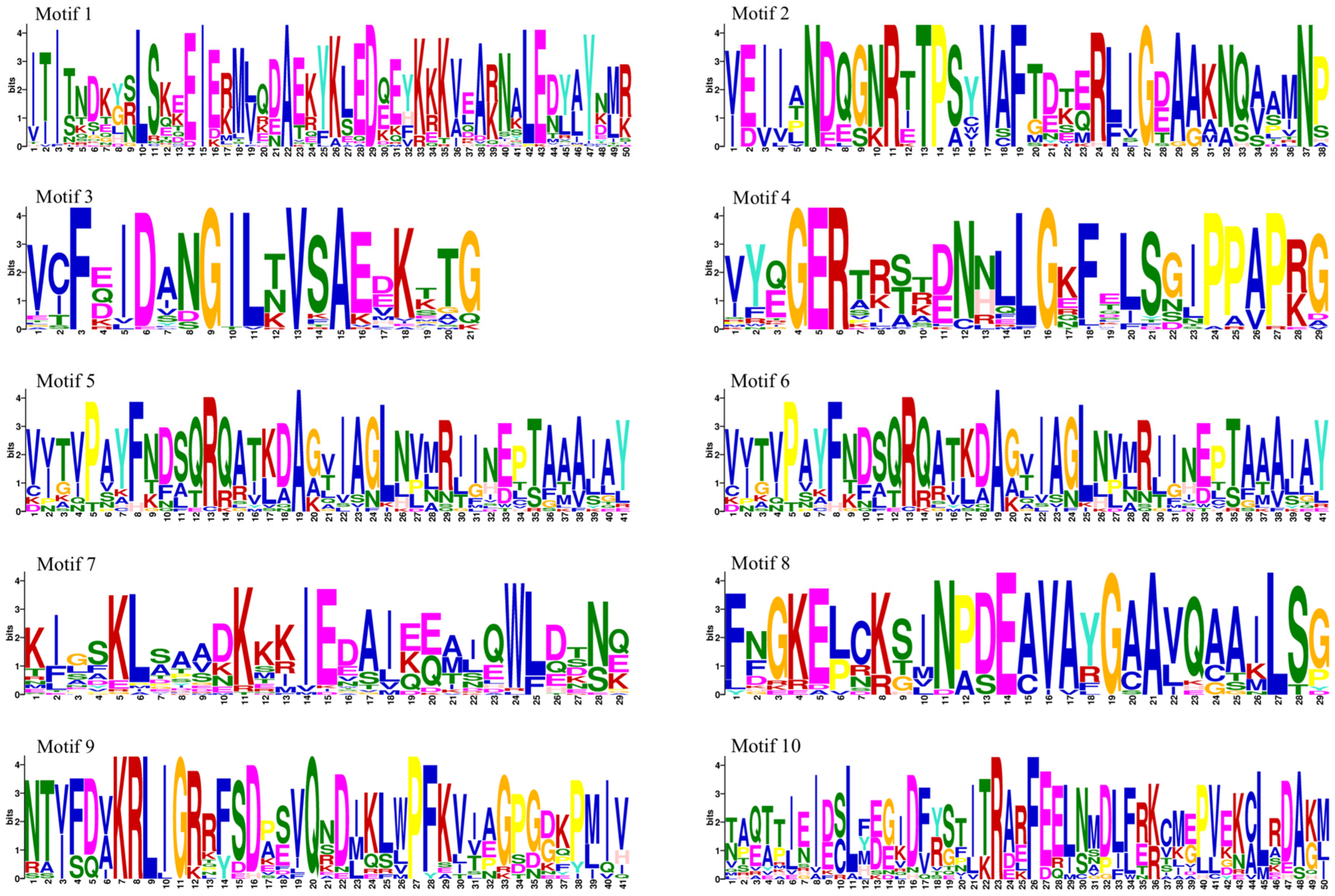
2.4. Motifs and Gene Structures Analysis
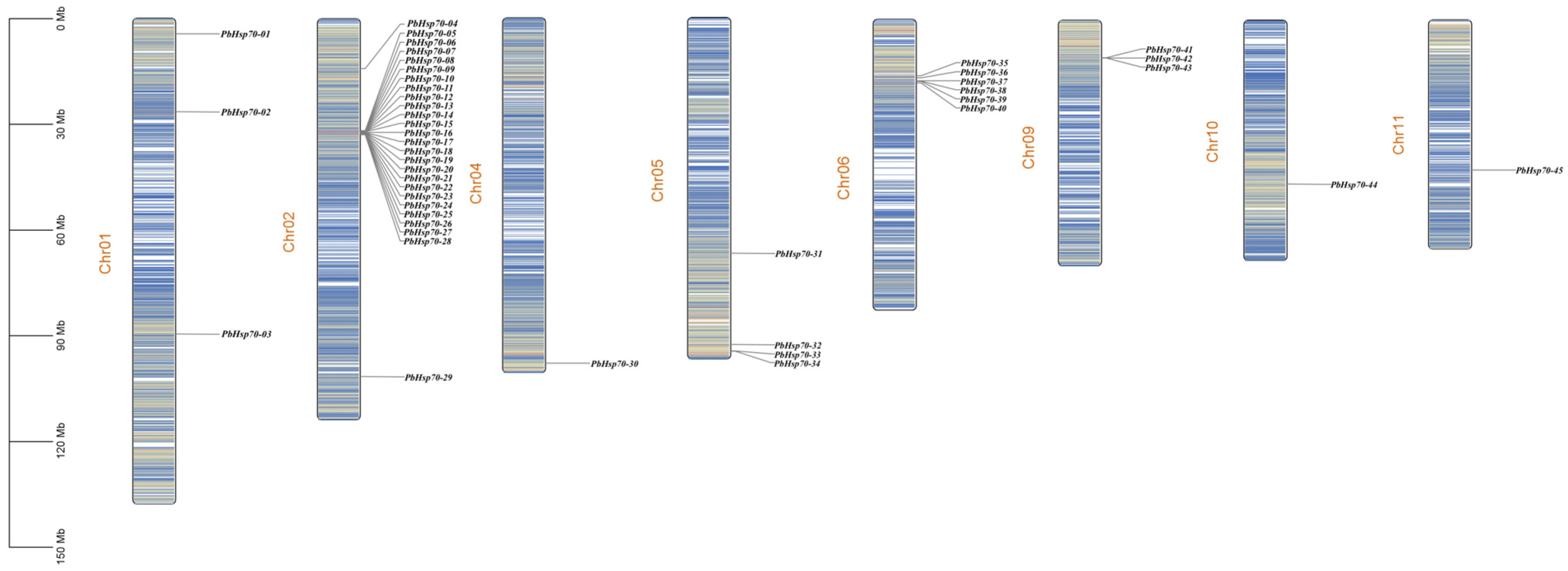
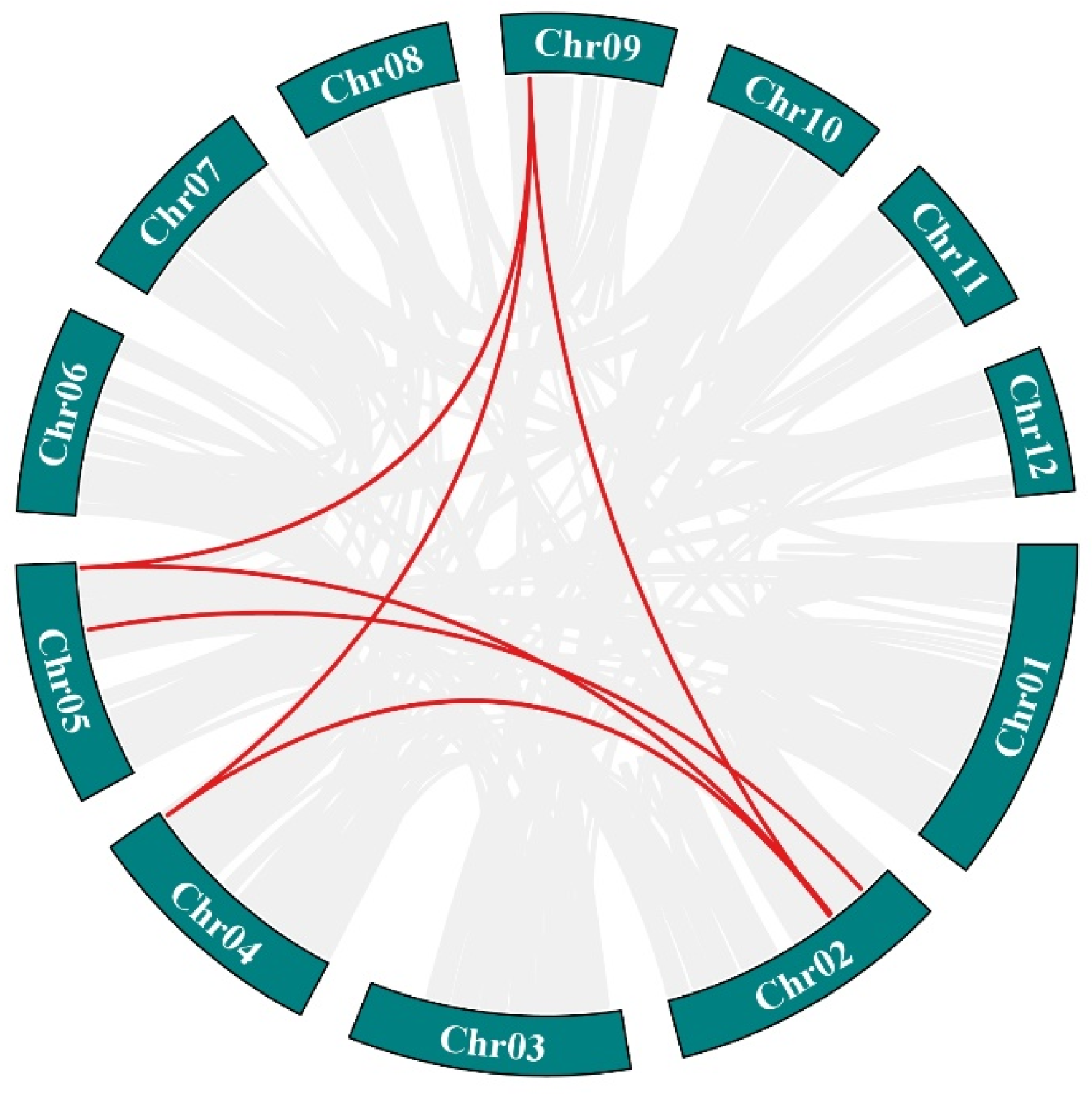
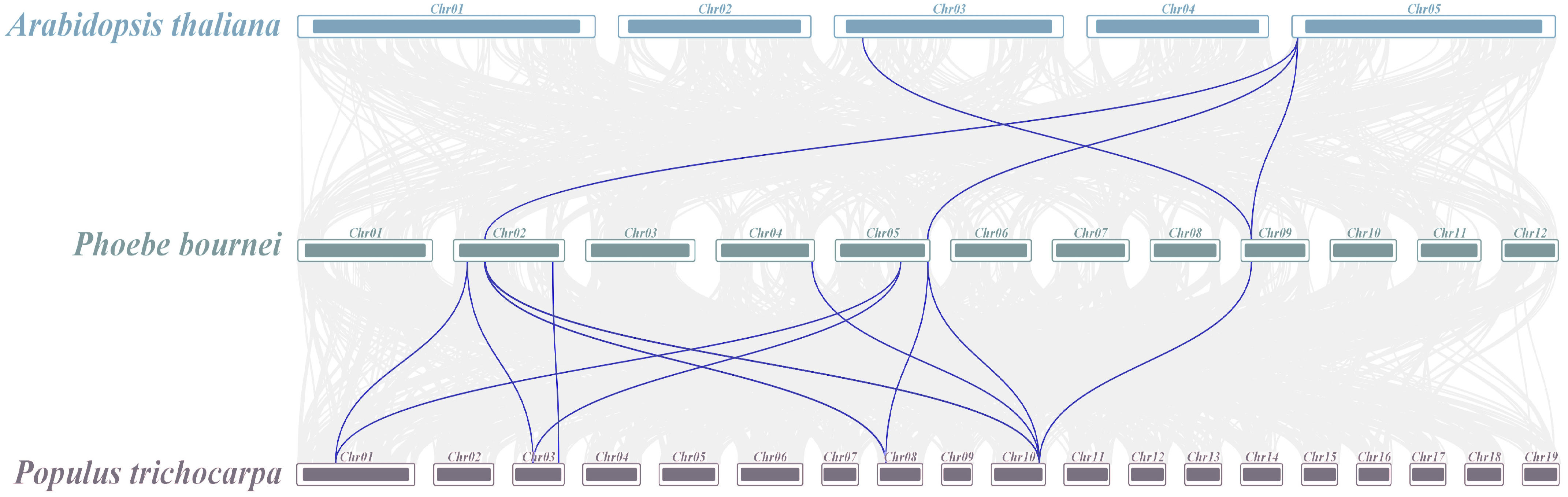
2.5. Chromosomes Localization and Collinearity Analysis of Hsp70s
2.6. Cis-Acting Elements Analysis of Hsp70
2.7. Analysis of Expression, RNA Extraction and RT-qPCR
3. Results
3.1. Identification and Analysis of Hsp70 Proteins
3.2. Phylogeny and Classification of Hsp70s
3.3. Hsp70 Structure and Motif Analysis
3.4. Chromosomal Localization, Collinearity Analysis, and Promoter Analysis of Hsp70s
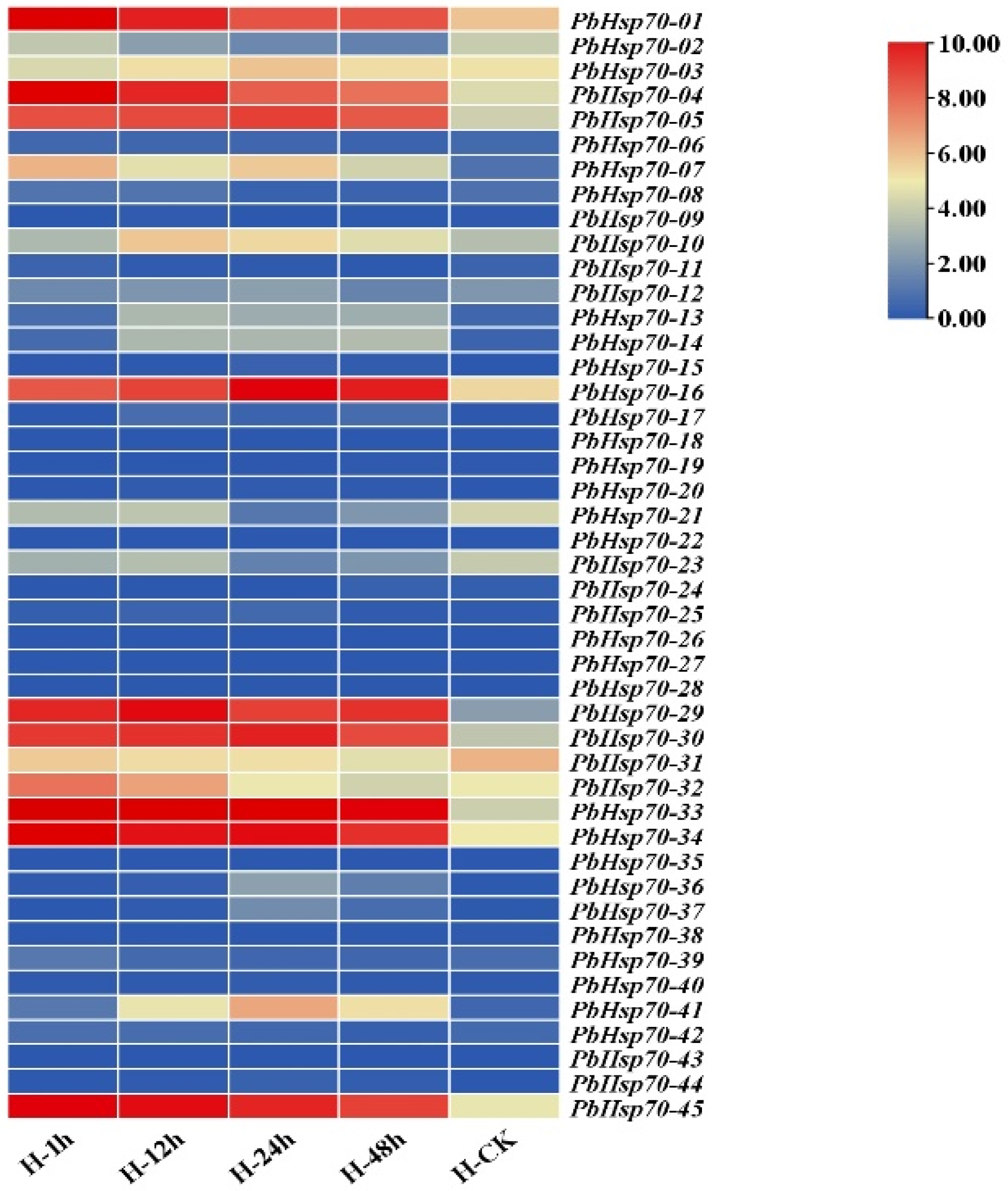
3.5. Expression Pattern of Hsp70s in P. bournei
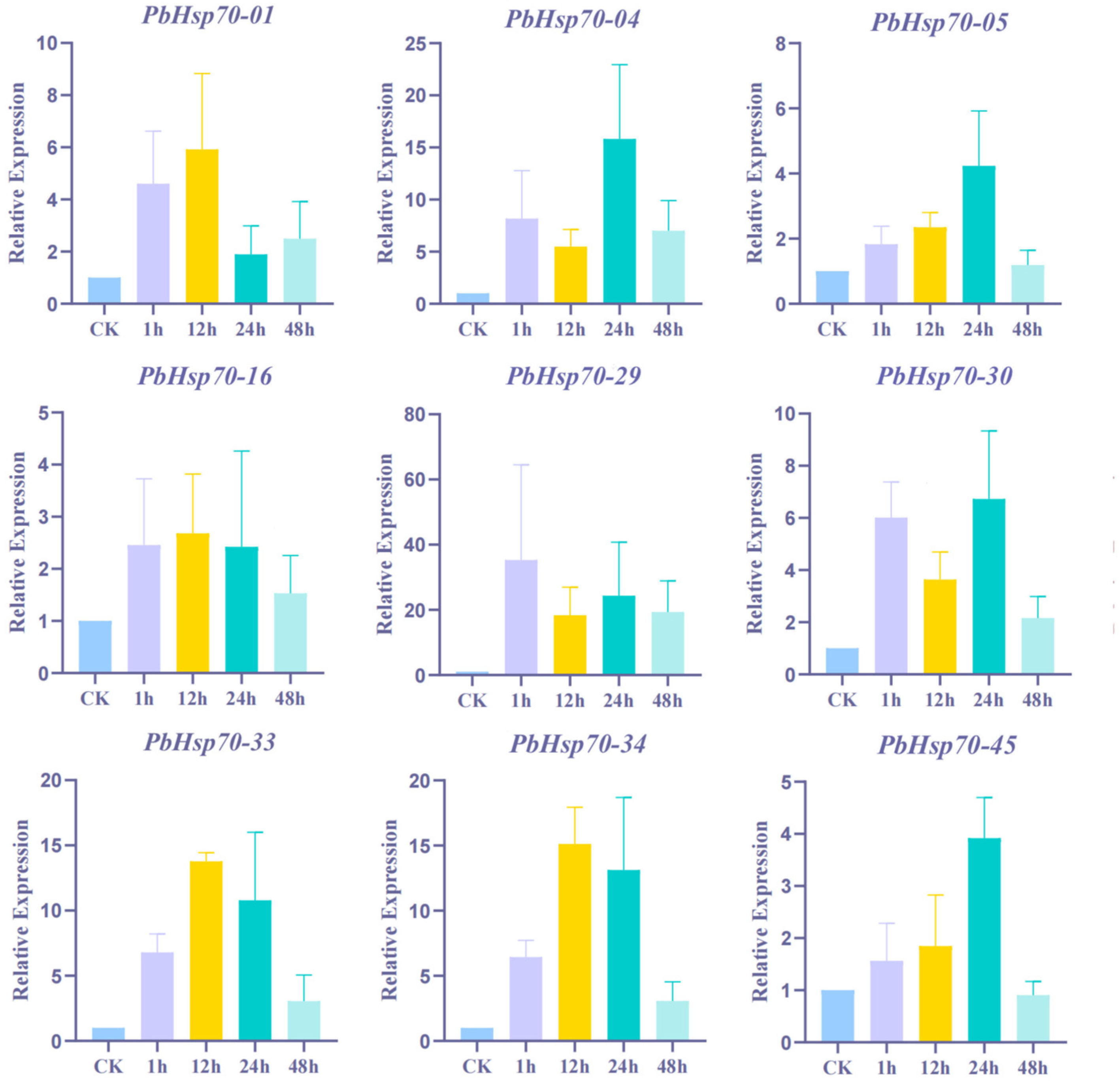
3.6. RT-qPCR Analysis of Hsp70s in P. bournei
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, M.; Liu, J.H.; Ma, X.; Zhai, Y.F.; Gong, Z.H.; Lu, M.H. Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 2016, 252, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Maimbo, M.; Ohnishi, K.; Hikichi, Y.; Yoshioka, H.; Kiba, A. Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol. 2007, 145, 1588–1599. [Google Scholar] [CrossRef]
- Miemyk, J. The 70 kDa stress-related proteins as molecular chaperones. Trends Plant Sci. 1997, 2, 180–187. [Google Scholar] [CrossRef]
- Burdon, R.H. The heat shock proteins. Endeavour 1988, 12, 133–138. [Google Scholar] [CrossRef]
- Radons, J. The human HSP70 family of chaperones: Where do we stand? Cell Stress Chaperones 2016, 21, 379–404. [Google Scholar] [CrossRef]
- Seguí-Simarro, J.M.; Testillano, P.S.; Risueño, M.C. Hsp70 and Hsp90 change their expression and subcellular localization after microspore embryogenesis induction in Brassica napus L. J. Struct. Biol. 2003, 142, 379–391. [Google Scholar] [CrossRef]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef]
- Hartl, F.U. Molecular chaperones in cellular protein folding. Nature 1996, 381, 571–579. [Google Scholar] [CrossRef]
- Sun, X.T.; Li, B.; Zhou, G.M.; Tang, W.Q.; Bai, J.; Sun, D.Y.; Zhou, R.G. Binding of the maize cytosolic Hsp70 to calmodulin, and identification of calmodulin-binding site in Hsp70. Plant Cell Physiol. 2000, 41, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; Gho, H.J.; Nguyen, M.X.; Kim, S.R.; An, G. Genome-wide expression analysis of HSP70 family genes in rice and identification of a cytosolic HSP70 gene highly induced under heat stress. Funct. Integr. Genom. 2013, 13, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Jungkunz, I.; Link, K.; Vogel, F.; Voll, L.M.; Sonnewald, S.; Sonnewald, U. AtHsp70-15-deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. Plant J. 2011, 66, 983–995. [Google Scholar] [CrossRef]
- Su, P.-H.; Li, H.-M. Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 2008, 146, 1231–1241. [Google Scholar] [CrossRef]
- Leng, L.; Liang, Q.; Jiang, J.; Zhang, C.; Hao, Y.; Wang, X.; Su, W. A subclass of HSP70s regulate development and abiotic stress responses in Arabidopsis thaliana. J. Plant Res. 2017, 130, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.S.; DeRocher, A.E.; Keegstra, K.; Vierling, E. Identification of heat shock protein hsp70 homologues in chloroplasts. Proc. Natl. Acad. Sci. USA 1990, 87, 374–378. [Google Scholar] [CrossRef]
- Ge, Y.J.; He, X.Y.; Wang, J.F.; Jiang, B.; Ye, R.H.; Lin, X.C. Physiological and biochemical responses of Phoebe bournei seedlings to water stress and recovery. Acta Physiol. Plant. 2014, 36, 1241–1250. [Google Scholar] [CrossRef]
- Li, X.; Liu, L.L.; Sun, S.X.; Li, Y.M.; Jia, L.; Ye, S.L.; Yu, Y.X.; Dossa, K.; Luan, Y.P. Leaf-transcriptome profiles of Phoebe bournei provide insights into temporal drought stress responses. Front. Plant Sci. 2022, 13, 1010314. [Google Scholar] [CrossRef]
- Wang, F.; Dong, Q.; Jiang, H.; Zhu, S.; Chen, B.; Xiang, Y. Genome-wide analysis of the heat shock transcription factors in Populus trichocarpa and Medicago truncatula. Mol. Biol. Rep. 2012, 39, 1877–1886. [Google Scholar] [CrossRef]
- Tang, X.L.; Jiang, J.; Jin, H.P.; Zhou, C.; Liu, G.Z.; Yang, H. Effects of shading on chlorophyll content and photosynthetic characteristics in leaves of Phoebe bournei. Yingyong Shengtai Xuebao 2019, 30, 2941–2948. [Google Scholar] [CrossRef]
- Han, X.; Zhang, J.; Han, S.; Chong, S.L.; Meng, G.; Song, M.; Wang, Y.; Zhou, S.; Liu, C.; Lou, L. Chromosome-Scale Genome Phoebe bournei reveals contrasting fates of terpene synthase (TPS)-a and TPS-b subfamilies. Plant Commun. 2022, 3, 100410. [Google Scholar] [CrossRef] [PubMed]
- Bukau, B.; Horwich, A.L. The Hsp70 and Hsp60 chaperone machines. Cell 1998, 92, 351–366. [Google Scholar] [CrossRef]
- He, X.; Zhao, X.W.; Zheng, Q.Y.; Zhang, M.M.; Huang, Y.; Liu, Z.J.; Lan, S.R. Whole-Genome Analysis of ZF-HD Genes Among Three Dendrobium Species and Expression Patterns in Dendrobium chrysotoxum. Horticulturae 2024, 10, 610. [Google Scholar] [CrossRef]
- Bennett, J.; Hondred, D.; Register, J.C. Keeping qRT-PCR rigorous and biologically relevant. Plant Cell Rep. 2015, 34, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.Z.; You, L.J.; Yang, F.; Wang, L.N.; Guo, X.Q.; Gao, F.; Hua, C.; Tan, C.; Fang, L.; Shan, R.Q.; et al. CNGBdb: China National GeneBank DataBase. Hereditas 2020, 42, 799–809. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.H.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.Q.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Cox, E.; Tsuchiya, M.T.N.; Ciufo, S.; Torcivia, J.; Falk, R.; Anderson, W.R.; Holmes, J.B.; Hem, V.; Breen, L.; Davis, E.; et al. NCBI taxonomy: Enhanced access via NCBI datasets. Nucleic Acids Res. 2024, 53, D1711–D1715. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Chasovnikova, L.V.; Matveeva, N.A.; Lavrent’ev, V.V. Relation between surface denaturation of immunoglobulin G in monolayers and the pH of the solution. Biofizika 1980, 25, 984–988. [Google Scholar]
- Alabbas, A.B. Integrativesubtractive proteomics, immunoinformatics, docking, and simulation approaches reveal candidate vaccine against Sin Nombre orthohantavirus. Front. Immunol. 2022, 13, 1022159. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Bacon, D.J.; Anderson, W.F. Multiple sequence alignment. J. Mol. Biol. 1986, 191, 153–161. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Zhou, T.; Xu, K.D.; Zhao, F.; Liu, W.Y.; Li, L.Z.; Hua, Z.Y.; Zhou, X. Itol.toolkit accelerates working with iTOL (Interactive Tree of Life) by an automated generation of annotation files. Bioinformatics 2023, 39, btad339. [Google Scholar] [CrossRef]
- Song, J.S.; Gonzales, N.R.; Yamashita, R.A.; Marchler-Bauer, A.; Bryant, S.H. Evolutionary, structural, and functional insights into the seven-transmembrane GPCR superfamily through NCBI’s Conserved Domain Database. Cancer Res. 2015, 75, 1090. [Google Scholar] [CrossRef]
- Li, X.; Ma, L.N.; Mei, X.Y.; Liu, Y.X.; Huang, H.C. ggmotif: An R Package for the extraction and visualization of motifs from MEME software. PLoS ONE 2022, 17, e0276979. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Rombauts, S.; Dehais, P.; Van Montagu, M.; Rouze, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Lotia, S.; Montojo, J.; Dong, Y.; Bader, G.D.; Pico, A.R. Cytoscape App Store. Bioinformatics 2013, 29, 1350–1351. [Google Scholar] [CrossRef]
- Cheung, D.K.B.; Brunke, A.J.; Akkari, N.; Souza, C.M.; Pape, T. Rotational Scanning Electron Micrographs (rSEM): A novel and accessible tool to visualize and communicate complex morphology. Zookeys 2013, 328, 47–57. [Google Scholar] [CrossRef][Green Version]
- Fu, N.N.; Wang, L.; Sun, Q.L.; Wang, Q.G.; Zhang, Y.T.; Han, X.; Yang, Q.; Ma, W.J.; Tong, Z.K.; Zhang, J.H. Genome-wide identification of the bHLH transcription factor family and the regulatory roles of PbbHLH74 in response to drought stress in Phoebe bournei. Int. J. Biol. Macromol. 2024, 283, 137760. [Google Scholar] [CrossRef]
- Nakagawa, T.; Kawaguchi, M. Shoot-applied MeJA suppresses root nodulation in Lotus japonicus. Plant Cell Physiol. 2006, 47, 176–180. [Google Scholar] [CrossRef]
- Cao, S.F.; Cai, Y.T.; Yang, Z.F.; Zheng, Y.H. MeJA induces chilling tolerance in loquat fruit by regulating proline and γ-aminobutyric acid contents. Food Chem. 2012, 133, 1466–1470. [Google Scholar] [CrossRef]
- Fung, R.W.M.; Wang, C.Y.; Smith, D.L.; Gross, K.C.; Tian, M.S. MeSA and MeJA increase steady-state transcript levels of alternative oxidase and resistance against chilling injury in sweet peppers (Capsicum annuum L.). Plant Sci. 2004, 166, 711–719. [Google Scholar] [CrossRef]
- Bowers, J.E.; Chapman, B.A.; Rong, J.K.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-P.; Sun, W.-H.; Xiong, Y.-F.; Jiang, Y.-T.; Liu, X.-D.; Liao, X.-Y.; Zhang, D.-Y.; Jiang, S.-Z.; Li, Y.; Liu, B.; et al. The Phoebe genome sheds light on the evolution of magnoliids. Hortic. Res. 2020, 7, 146. [Google Scholar] [CrossRef] [PubMed]
- Maher, C.; Stein, L.; Ware, D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006, 16, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Danilova, T.; Zhang, J.; Birchler, J.; Peterson, T. Constructing Defined Chromosome Segmental Duplications in Maize. Cytogenet. Genome Res. 2010, 129, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ke, T.; Tehrim, S.; Sun, F.; Liao, B.; Hua, W. PTGBase: An integrated database to study tandem duplicated genes in plants. Database J. Biol. Databases Curation 2015, 2015, bav017. [Google Scholar] [CrossRef]
- Rakoczy-Trojanowska, M.; Bolibok, H. Characteristics and a comparison of three classes of microsatellite-based markers and their application in plants. Cell. Mol. Biol. Lett. 2004, 9, 221–238. [Google Scholar] [CrossRef]
- Panzade, K.P.; Kale, S.S.; Chavan, N.R.; Hatzade, B. Genome-wide analysis of Hsp70 and Hsp100 gene families in Ziziphus jujuba. Cell Stress. Chaperones 2021, 26, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Khatiwada, A.; Yilmaz, A.S.; Wolf, B.J.; Pietrzak, M.; Chung, D.J.; Li, M.Y. multi-GPA-Tree: Statistical approach for pleiotropy informed and functional annotation tree guided prioritization of GWAS results. PLoS Comput. Biol. 2023, 19, e1011686. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol. Plant. 2006, 126, 62–71. [Google Scholar] [CrossRef]
- Colleaux, L.; Michel-Wolwertz, M.R.; Matagne, R.F.; Dujon, B. The apocytochrome b gene of Chlamydomonas smithii contains a mobile intron related to both Saccharomyces and Neurospora introns. Mol. Gen. Genet. 1990, 223, 288–296. [Google Scholar] [CrossRef]
- Foury, F.; Roganti, T.; Lecrenier, N.; Purnelle, B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998, 440, 325–331. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217, 109–119. [Google Scholar] [CrossRef]
- Jeong, H.J.; Jung, K.H. Rice tissue-specific promoters and condition-dependent promoters for effective translational application. J. Integr. Plant Biol. 2015, 57, 913–924. [Google Scholar] [CrossRef]
- Arguello-Astorga, G.; Herrera-Estrella, L. Evolution of light-regulated plant promoters. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 525–555. [Google Scholar] [CrossRef]
- Guiltinan, M.J.; Marcotte, W.R., Jr.; Quatrano, R.S. A plant leucine zipper protein that recognizes an abscisic acid response element. Science 1990, 250, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Hattori, T.; Totsuka, M.; Hobo, T.; Kagaya, Y.; Yamamoto-Toyoda, A. Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol. 2002, 43, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Rohwer, C.L.; Erwin, J.E. Irradiance and photoperiod effects on methyl jasmonate synthesis in Artemisia spp. In Proceedings of the 5th International Symposium on Artificial Lighting in Horticulture, Lillehammer, Norway, 21–24 June 2005; pp. 375–380. [Google Scholar]
- Avdiushko, S.; Croft, K.P.; Brown, G.C.; Jackson, D.M.; Hamilton-Kemp, T.R.; Hildebrand, D. Effect of volatile methyl jasmonate on the oxylipin pathway in tobacco, cucumber, and arabidopsis. Plant Physiol. 1995, 109, 1227–1230. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nicolaides, N.C.; Gualdi, R.; Casadevall, C.; Manzella, L.; Calabretta, B. Positive autoregulation of c-myb expression via Myb binding sites in the 5’ flanking region of the human c-myb gene. Mol. Cell. Biol. 1991, 11, 6166–6176. [Google Scholar] [CrossRef][Green Version]
- Ku, D.H.; Wen, S.C.; Engelhard, A.; Nicolaides, N.C.; Lipson, K.E.; Marino, T.A.; Calabretta, B. c-myb transactivates cdc2 expression via Myb binding sites in the 5’-flanking region of the human cdc2 gene. J. Biol. Chem. 1993, 268, 2255–2259. [Google Scholar] [CrossRef]
- Xu, Y.M.; Liu, Y.; Yi, Y.J.; Liu, J.J. Genome-Wide Identification and Characterization of HSP70 Gene Family in Tausch’s Goatgrass (Aegilops tauschii). Genes 2025, 16, 19. [Google Scholar] [CrossRef]
- Alam, P.; Al Balawi, T.; Manzoor, M.A.; Sabir, I.A. Genome-wide analysis of HSP70 gene family in Beta vulgaris and in-silico expression under environmental stress. BMC Plant Biol. 2025, 25, 214. [Google Scholar] [CrossRef]
- Harrison, R.; Papp, B.; Pál, C.; Oliver, S.G.; Delneri, D. Plasticity of genetic interactions in metabolic networks of yeast. Proc. Natl. Acad. Sci. USA 2007, 104, 2307–2312. [Google Scholar] [CrossRef]
- Henderson, S.T.; Johnson, T.E. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 2001, 11, 1975–1980. [Google Scholar] [CrossRef] [PubMed]
- Desalvo, M.K.; Voolstra, C.R.; Sunagawa, S.; Schwarz, J.A.; Stillman, J.H.; Coffroth, M.A.; Szmant, A.M.; Medina, M. Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata. Mol. Ecol. 2008, 17, 3952–3971. [Google Scholar] [CrossRef] [PubMed]

| Member | Amino Acid (aa) | Grand Average of Hydrophobicity | Theoretical pI | Molecular Weight | Aliphatic Index | Instability Index | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| PbHsp70-01 | 663 | −0.456 | 5.12 | 73,265.1 | 87.95 | 27.01 | E.R. |
| PbHsp70-02 | 803 | −0.428 | 5.6 | 89,802.35 | 79.44 | 51.6 | nucl |
| PbHsp70-03 | 835 | −0.441 | 5.24 | 92,764.05 | 78.12 | 49.06 | cyto |
| PbHsp70-04 | 859 | −0.454 | 5.33 | 95,252.16 | 77.53 | 44.57 | chlo |
| PbHsp70-05 | 585 | −0.416 | 5.11 | 64,506.31 | 84.72 | 35.09 | cyto |
| PbHsp70-06 | 555 | −0.326 | 5.67 | 62,187.26 | 93.64 | 36.96 | cyto |
| PbHsp70-07 | 232 | −0.782 | 4.74 | 25,677.78 | 74.48 | 32.83 | mito |
| PbHsp70-08 | 155 | −0.794 | 5.29 | 17,910.22 | 80.58 | 44.47 | nucl |
| PbHsp70-09 | 678 | −0.592 | 8.97 | 75,778.14 | 80.74 | 53.8 | nucl |
| PbHsp70-10 | 117 | −1.119 | 6.19 | 13,829.69 | 73.5 | 49.48 | cyto |
| PbHsp70-11 | 201 | −0.791 | 6.86 | 22,829.8 | 76.27 | 56.09 | cyto |
| PbHsp70-12 | 304 | −0.436 | 5.96 | 34,209.25 | 90.76 | 30.39 | cyto |
| PbHsp70-13 | 407 | −0.379 | 8.73 | 46,087.08 | 83.69 | 34.91 | chlo |
| PbHsp70-14 | 226 | −0.648 | 5.08 | 25,399.72 | 82.92 | 31.63 | cysk |
| PbHsp70-15 | 215 | −0.396 | 8.56 | 24,200.87 | 83.49 | 32.97 | chlo |
| PbHsp70-16 | 651 | −0.414 | 5.05 | 71,339.71 | 83.01 | 32.89 | cyto |
| PbHsp70-17 | 292 | −0.446 | 6.98 | 32,504.12 | 90.86 | 35.27 | cyto |
| PbHsp70-18 | 166 | −0.743 | 9.06 | 18,816.27 | 78.8 | 36.09 | nucl |
| PbHsp70-19 | 156 | −0.401 | 6.16 | 18,000.74 | 103.21 | 30.24 | cyto/nucl |
| PbHsp70-20 | 135 | −0.457 | 5.87 | 15,406.6 | 90.96 | 43.72 | cyto/nucl |
| PbHsp70-21 | 119 | −0.306 | 9.25 | 12,926.8 | 87.56 | 29.35 | cyto |
| PbHsp70-22 | 165 | −0.520 | 9.49 | 18,538.57 | 88.79 | 40.07 | mito |
| PbHsp70-23 | 327 | −0.385 | 7.69 | 36,460.54 | 87.68 | 30.26 | chlo |
| PbHsp70-24 | 114 | −1.011 | 9.71 | 13,319.34 | 67.72 | 46.18 | nucl |
| PbHsp70-25 | 580 | −0.481 | 5.39 | 65,468.48 | 88.93 | 27.88 | golg |
| PbHsp70-26 | 146 | −0.546 | 6.18 | 16,641.12 | 91.58 | 42.02 | cyto |
| PbHsp70-27 | 319 | −0.609 | 9.56 | 35,633.63 | 75.86 | 35.61 | chlo |
| PbHsp70-28 | 750 | −0.308 | 9.18 | 85,255.63 | 89.36 | 42 | vacu |
| PbHsp70-29 | 366 | −0.613 | 4.95 | 39,992.95 | 75.96 | 32.29 | cyto |
| PbHsp70-30 | 653 | −0.397 | 5.13 | 71,554.09 | 83.52 | 32.98 | cyto |
| PbHsp70-31 | 858 | −0.414 | 5.23 | 94,470.99 | 79.69 | 42.68 | chlo |
| PbHsp70-32 | 889 | −0.417 | 5.48 | 98,969.28 | 87.53 | 38.38 | E.R. |
| PbHsp70-33 | 620 | −0.384 | 4.9 | 67,770.73 | 84.18 | 33.81 | cyto |
| PbHsp70-34 | 554 | −0.392 | 8.78 | 61,513.15 | 83.59 | 39.59 | cyto |
| PbHsp70-35 | 257 | −0.526 | 6.36 | 28,915.68 | 83.19 | 23.18 | cyto |
| PbHsp70-36 | 676 | −0.365 | 9.25 | 76,069.63 | 92.72 | 32.83 | cyto |
| PbHsp70-37 | 291 | −0.477 | 9.75 | 33,607.19 | 99.14 | 27.06 | cyto |
| PbHsp70-38 | 182 | −0.417 | 9.3 | 21,032.39 | 101.87 | 25.20 | cyto |
| PbHsp70-39 | 183 | −0.455 | 9.19 | 21,087.38 | 100.77 | 23.37 | cyto |
| PbHsp70-40 | 660 | −0.321 | 7.18 | 73,827.46 | 93.95 | 32.84 | cyto |
| PbHsp70-41 | 649 | −0.406 | 5.14 | 71,262.93 | 82.97 | 33.84 | cyto |
| PbHsp70-42 | 1358 | −0.308 | 7.94 | 150,806.92 | 86.48 | 38.61 | chlo |
| PbHsp70-43 | 515 | −0.752 | 4.65 | 57,613.73 | 74.08 | 42.83 | cyto |
| PbHsp70-44 | 425 | −0.770 | 4.81 | 47,137.06 | 76.66 | 41.63 | cyto |
| PbHsp70-45 | 652 | −0.494 | 5.26 | 71,897.49 | 86.01 | 27.7 | golg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.; Jiang, Y.; Li, Z.; Niu, Y.; Gong, C.; He, X.; Chen, S.; Cao, S. Genome-Wide Identification and Heat Stress-Induced Expression Profiling of the Hsp70 Gene Family in Phoebe bournei. Biology 2025, 14, 602. https://doi.org/10.3390/biology14060602
Lin Y, Jiang Y, Li Z, Niu Y, Gong C, He X, Chen S, Cao S. Genome-Wide Identification and Heat Stress-Induced Expression Profiling of the Hsp70 Gene Family in Phoebe bournei. Biology. 2025; 14(6):602. https://doi.org/10.3390/biology14060602
Chicago/Turabian StyleLin, Yiming, Yan Jiang, Zhuoqun Li, Yuewang Niu, Chenyu Gong, Xin He, Shipin Chen, and Shijiang Cao. 2025. "Genome-Wide Identification and Heat Stress-Induced Expression Profiling of the Hsp70 Gene Family in Phoebe bournei" Biology 14, no. 6: 602. https://doi.org/10.3390/biology14060602
APA StyleLin, Y., Jiang, Y., Li, Z., Niu, Y., Gong, C., He, X., Chen, S., & Cao, S. (2025). Genome-Wide Identification and Heat Stress-Induced Expression Profiling of the Hsp70 Gene Family in Phoebe bournei. Biology, 14(6), 602. https://doi.org/10.3390/biology14060602






