Arabidopsis P4-ATPases ALA1 and ALA7 Enhance Resistance to Verticillium dahliae via Detoxifying Vd-Toxins
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
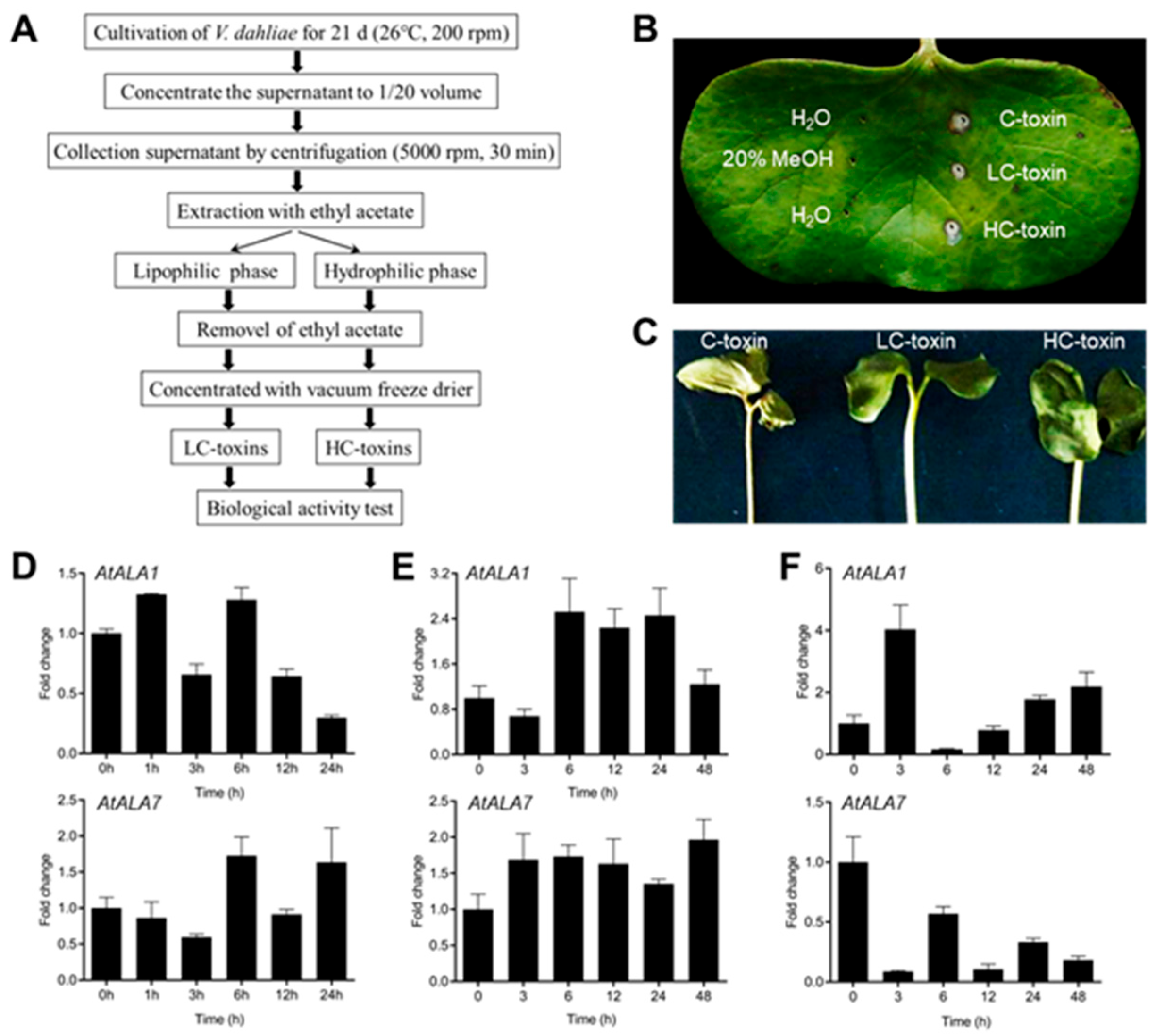
2.2. Vd-Toxins Extraction and Phytotoxicity Assays
2.3. Isolation and Quantitative Real-Time PCR (qRT-PCR)
2.4. RNA Sequencing
2.5. Pathogen Inoculation and Disease Scoring
2.6. Laser Confocal Microscopy Observation
2.7. Statistical Analysis
3. Results
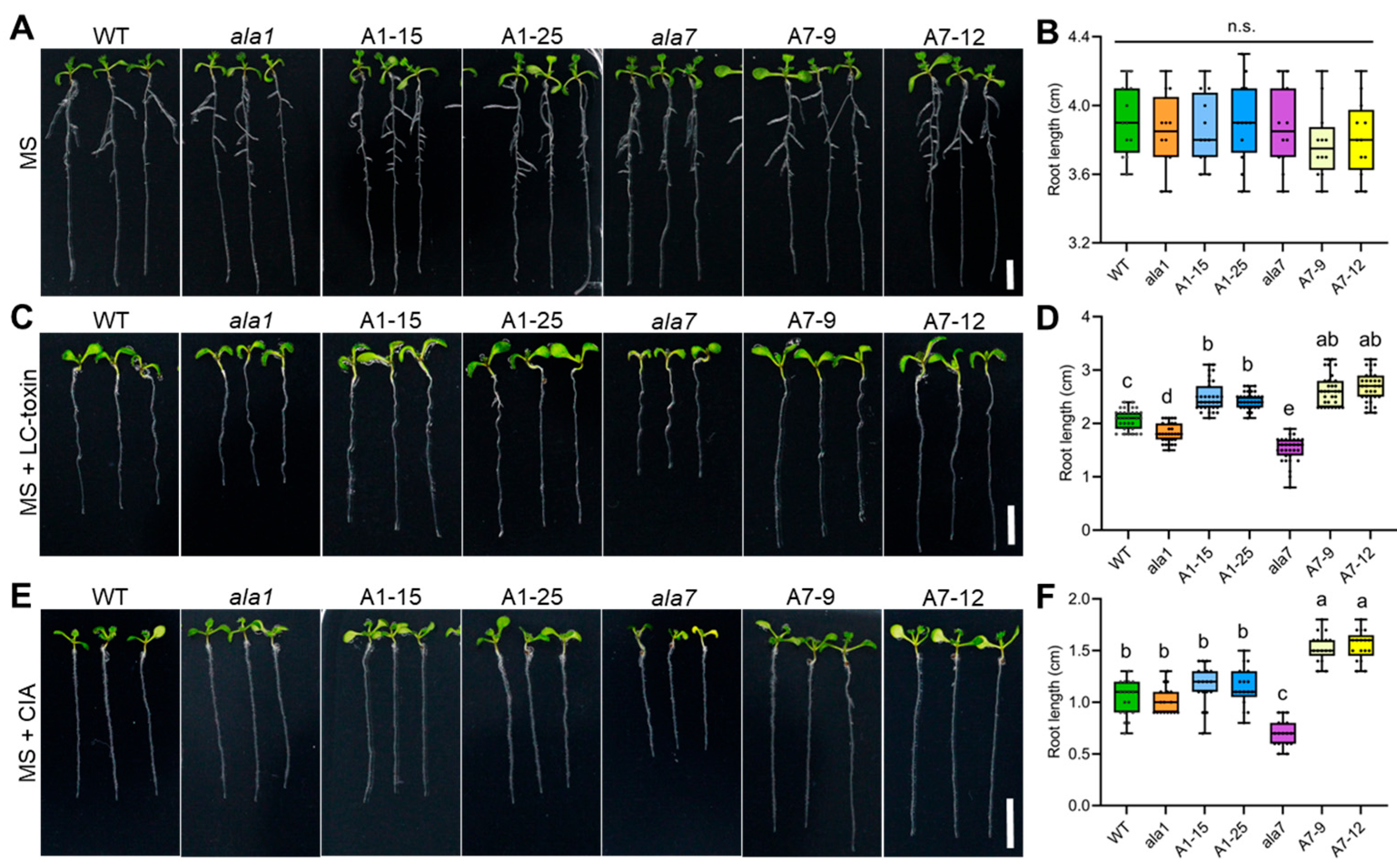
3.1. Both AtALA1 and AtALA7 Contribute to the Resistance of LC-Toxins Secreted by V. dahliae in Arabidopsis
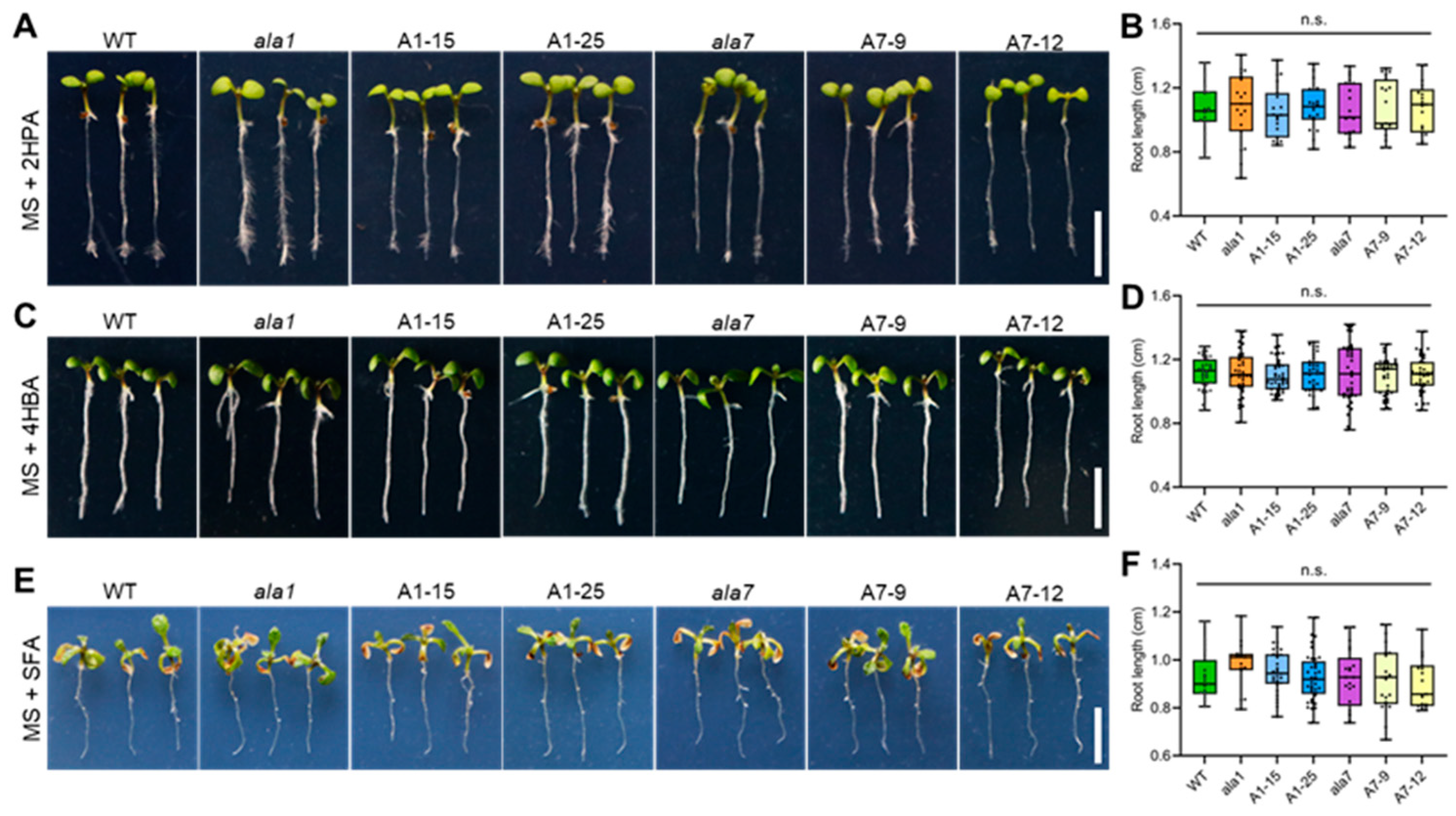
3.2. Overexpression of Either AtALA1 or AtALA7 Enhances the Resistance of Arabidopsis to LC-Toxins Secreted by V. dahliae
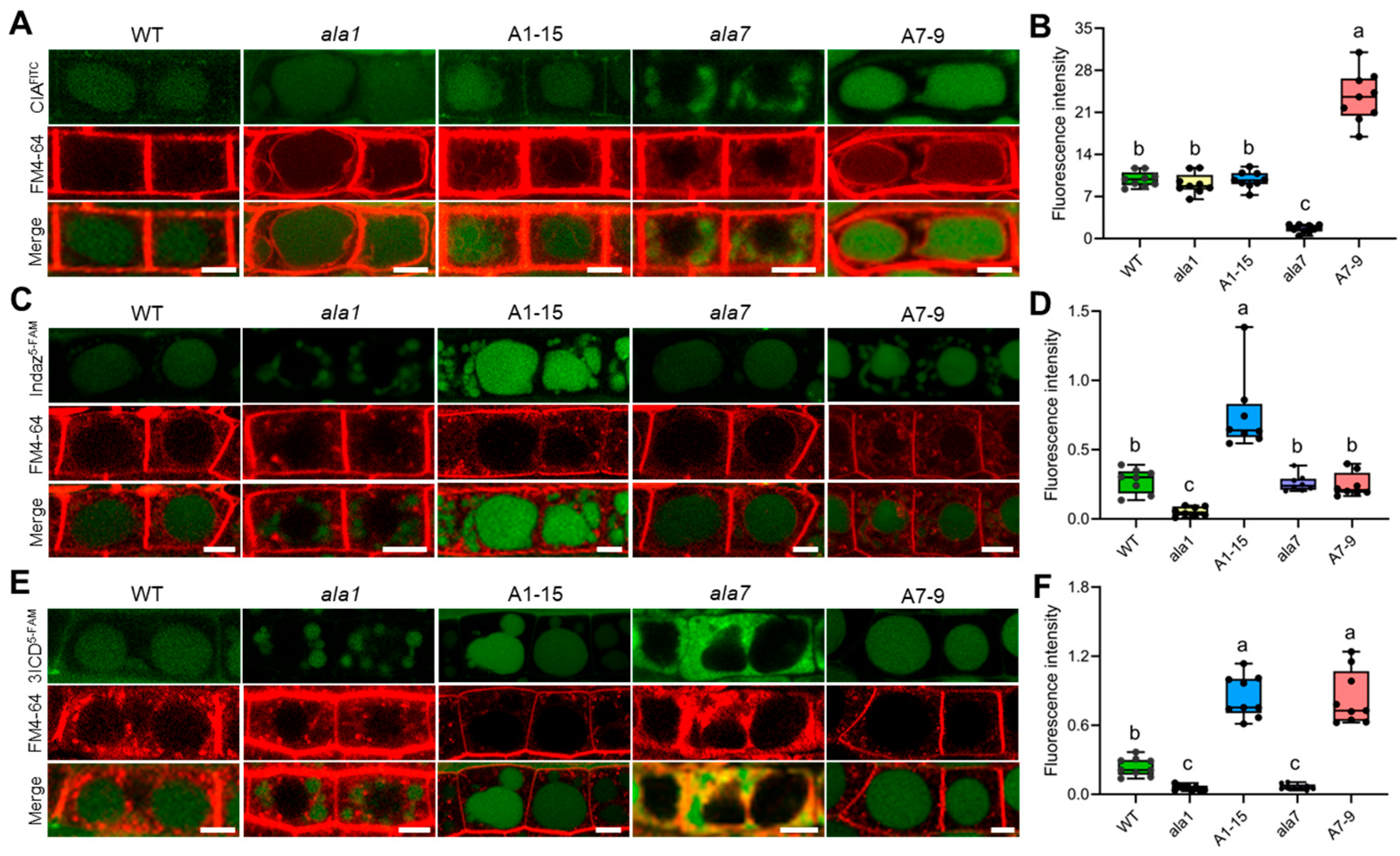
3.3. Overexpression of AtALA1 Promotes Transport Indazole and 3ICD into Vacuoles, While AtALA7 Accumulates CIA and 3ICD to Vacuoles
3.4. AtALA1 or AtALA7 Protect Arabidopsis Plants from ROS Toxicity Triggered by Vd-Toxins
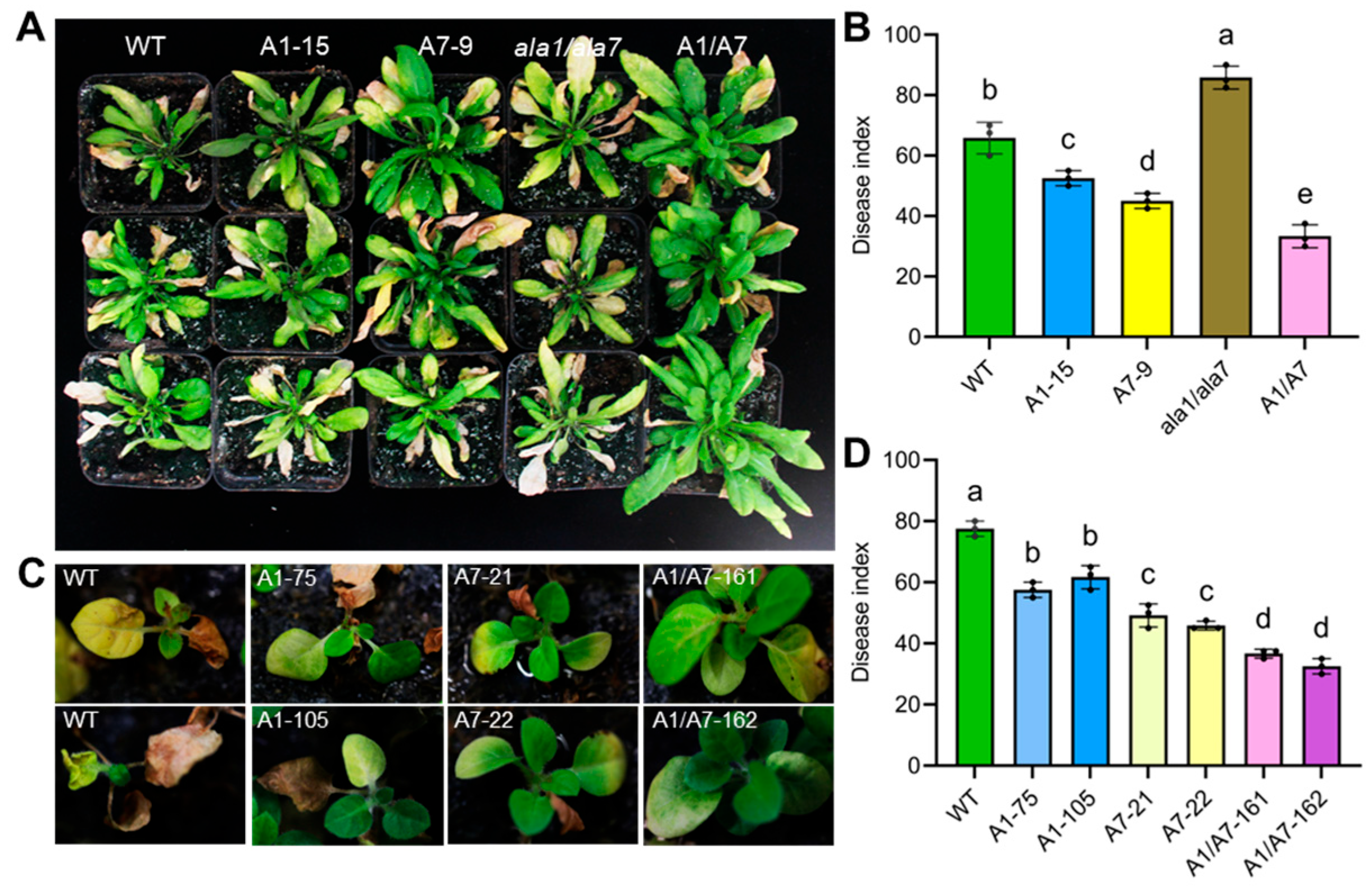
3.5. Aggregation of AtALA1 and AtALA7 Enhances the Plant’s Resistance to V. dahliae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CIA | Cinnamyl acetate |
| cAMP | Cyclic adenosine monophosphate |
| C-toxins | Crude toxin |
| DGEs | Differentially expressed genes |
| HC-toxins | Hydrophilic phase crude toxin |
| H2O2 | Hydrogen peroxide |
| KNGG | Kyoto encyclopedia of genes and genomes |
| LC-toxins | Lipophilic phase crude toxin |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| rpm | Revolutions per minute |
| SFA | Sulfacetamide |
| Vw | Verticillium wilt |
| 2HPA | 2-hydroxypenylacetic acid |
| 3ICD | Indole-3-carboxaldehyde |
| 4HBA | 4-hydroxybenzoic acid |
| 4MBA | 4-methylbenzoic acid |
| 5-FAM | 5-carboxyfluorescein |
References
- Fradin, E.F.; Thomma, B.P. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef]
- Pegg, G.F.; Brady, B.L. Verticillium Wilts; CABI: Wallingford, UK, 2002.
- Bhat, R.G.; Subbarao, K.V. Host range specificity in Verticillium dahliae. Phytopathology 1999, 89, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Díaz, R.M.; Cirulli, M.; Bubici, G.; Del Mar Jiménez-Gasco, M.; Antoniou, P.P.; Tjamos, E.C. Verticillium wilt, a major threat to olive production: Current status and future prospects for its management. Plant Dis. 2012, 96, 304–329. [Google Scholar] [CrossRef] [PubMed]
- Baeza-Montañez, L.; Gómez-Cabrera, R.; García-Pedrajas, M. First report of Verticillium wilt caused by Verticillium dahliae on mango trees (Mangifera indica) in southern Spain. Plant Dis. 2010, 94, 380. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.G.; Zhao, J.H.; Zhang, B.S.; Gao, F.; Wu, X.M.; Yan, Y.S.; Zhang, J.; Guo, H.S. Microbe-induced gene silencing boosts crop protection against soil-borne fungal pathogens. Nat. Plants 2023, 9, 1409–1418. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, J.; Zhao, L.; Feng, Z.; Wei, F.; Bai, H.; Feng, H.; Zhu, H. A review of the pathogenicity mechanism of Verticillium dahliae in cotton. J. Cotton Res. 2022, 5, 3. [Google Scholar] [CrossRef]
- Nachmias, A.; Buchner, V.; Burstein, Y. Biological and immunochemical characterization of a low molecular weight phytotoxin isolated from a protein-lipopolysaccharide complex produced by a potato isolate of Verticillium dahliae Kleb. Physiol. Plant Pathol. 1985, 26, 43–55. [Google Scholar] [CrossRef]
- Palmer, C.S.; Saleeba, J.A.; Lyon, B.R. Phytotoxicity on cotton ex-plants of an 18.5kDa protein from culture filtrates of Verticillium dahliae. Physiol. Mol. Plant Pathol. 2005, 67, 308–318. [Google Scholar] [CrossRef]
- Laouane, H.; Sedra, M.H.; Lazrek, H.B. Isolation and characterization of a new phytotoxic molecule from culture fluids of Verticillium dahliae. Glob. J. Inc. 2013, 13, 37–42. [Google Scholar]
- Jia, Z.Q.; Shi, F.M.; Yuan, H.Y.; Hou, Y.Y.; Li, Y.Z. Comparison of methods of purification and identification of Verticillium dahliae toxins. J. Life Sci. 2007, 1, 67–73. [Google Scholar]
- Buchner, V.; Nachmias, A.; Burstein, Y. Isolation and partial characterization of a phytotoxic glycopeptide from a protein-lipopolysaccharide complex produced by a potato isolate of Verticillium dahliae. FEBS Lett. 1982, 138, 261–264. [Google Scholar] [CrossRef]
- Mansoori, B.; Milton, J.; Smith, C. Isolation and partial purification of a phytotoxin related to pathogenic Verticillium species. J. Phytopathol. 1995, 143, 33–36. [Google Scholar] [CrossRef]
- Cui, X.C.; Luo, D.Q. The study of secondary metabolites of Verticillium dahliae. Asia-Pac. Tradit. Med. 2013, 9, 47–48. [Google Scholar]
- Zhang, D.D.; Wang, X.Y.; Chen, J.Y.; Kong, Z.Q.; Gui, Y.J.; Li, N.Y.; Bao, Y.M.; Dai, X.F. Identification and characterization of a pathogenicity-related gene VdCYP1 from Verticillium dahliae. Sci. Rep. 2016, 6, 27979. [Google Scholar] [CrossRef]
- Xu, F.; Huang, L.; Wang, J.Y.; Ma, C.X.; Tan, Y.Q.; Wang, F.L.; Fan, Y.H.; Luo, M. Sphingolipid synthesis inhibitor fumonisin B1 causes verticillium wilt in cotton. J. Integr. Plant Biol. 2022, 64, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Fan, L.W.; Wu, W.H. Evidences for involvement of endogenous cAMP in Arabidopsis defense responses to Verticillium toxins. Cell Res. 2005, 15, 585–592. [Google Scholar] [CrossRef]
- Yuan, H.Y.; Yao, L.L.; Jia, Z.Q.; Li, Y.; Li, Y.Z. Verticillium dahliae toxin induced alterations of cytoskeletons and nucleoli in Arabidopsis thaliana suspension cells. Protoplasma 2006, 229, 75–82. [Google Scholar] [CrossRef]
- Shi, F.M.; Yao, L.L.; Pei, B.L.; Zhou, Q.; Li, X.L.; Li, Y.; Li, Y.Z. Cortical microtubule as a sensor and target of nitric oxide signal during the defence responses to Verticillium dahliae toxins in Arabidopsis. Plant Cell Environ. 2009, 32, 428–438. [Google Scholar] [CrossRef]
- Yao, L.L.; Zhou, Q.; Pei, B.L.; Li, Y.Z. Hydrogen peroxide modulates the dynamic microtubule cytoskeleton during the defence responses to Verticillium dahliae toxins in Arabidopsis. Plant Cell Environ. 2011, 34, 1586–1598. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, Q.; Zhou, S.; Sun, Y.; Li, X.; Li, Y. H2Bub1 regulates RbohD-dependent hydrogen peroxide signal pathway in the defense responses to Verticillium dahliae toxins. Plant Physiol. 2020, 182, 640–657. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Sun, S.L.; Ge, W.Y.; Zhao, L.F.; Hou, B.Q.; Wang, K.; Lyu, Z.F.; Chen, L.Y.; Xu, S.S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Ren, H.; Wang, F.L.; Chen, J.J.; Ma, L.; Chen, Y.; Li, X.B.; Fan, Y.H.; Jin, D.; Hou, L.; et al. Cell detoxification of secondary metabolites by P4-ATPase-mediated vesicle transport. Elife 2023, 12, e79179. [Google Scholar] [CrossRef]
- Yang, J.; Liang, K.; Ke, H.; Zhang, Y.B.; Meng, Q.; Gao, L.; Fan, J.P.; Li, G.H.; Zhou, H.; Xiao, J.Y.; et al. Enzymatic degradation of deoxynivalenol with the engineered detoxification enzyme Fhb7. JACS Au 2024, 4, 619–634. [Google Scholar] [CrossRef] [PubMed]
- He, W.J.; Yang, P.; Huang, T.; Liu, Y.F.; Zhang, Y.W.; Zhang, W.M.; Zhang, T.T.; Zheng, M.R.; Ma, L.; Zhao, C.X.; et al. Detoxifying bacterial genes for deoxynivalenol epimerization confer durable resistance to Fusarium head blight in wheat. Plant Biotechnol. J. 2024, 22, 619–634. [Google Scholar] [CrossRef]
- Sakuragi, T.; Nagata, S. Regulation of phospholipid distribution in the lipid bilayer by flippases and scramblases. Nat. Rev. Mol. Cell Biol. 2023, 24, 576–596. [Google Scholar] [CrossRef]
- López-Marqués, R.L. Lipid flippases as key players in plant adaptation to their environment. Nat. Plants 2021, 7, 1188–1199. [Google Scholar] [CrossRef]
- Timcenko, M.; Lyons, J.A.; Januliene, D.; Ulstrup, J.J.; Dieudonné, T.; Montigny, C.; Ash, M.R.; Karlsen, J.L.; Boesen, T.; Kühlbrandt, W.; et al. Structure and autoregulation of a P4-ATPase lipid flippase. Nature 2019, 571, 366–370. [Google Scholar] [CrossRef]
- Graham, T.R. Flippases and vesicle-mediated protein transport. Trends Cell Biol. 2004, 14, 670–677. [Google Scholar] [CrossRef]
- Zhou, Y.L.; Yang, Y.; Niu, Y.; Fan, T.T.; Qian, D.; Luo, C.X.; Shi, Y.M.; Li, S.; An, L.Z.; Xiang, Y. The tip-localized phosphatidylserine established by Arabidopsis ALA3 is crucial for Rab GTPase-mediated vesicle trafficking and pollen tube growth. Plant Cell 2020, 32, 3170–3187. [Google Scholar] [CrossRef]
- Zhang, X.X.; Adamowski, M.; Marhava, P.; Tan, S.T.; Zhang, Y.Z.; Rodriguez, L.; Zwiewka, M.; Pukysova, V.; Sanchez, A.S.; Raxwal, V.K.; et al. Arabidopsis flippases cooperate with ARF GTPase exchange factors to regulate the trafficking and polarity of PIN auxin transporters. Plant Cell 2020, 32, 1644–1664. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Niu, Y.; Chen, T.; Zhang, H.K.; Zhang, J.; Qian, D.; Bi, M.M.; Fan, Y.M.; An, L.Z.; Xiang, Y. The phospholipid flippase ALA3 regulates pollen tube growth and guidance in Arabidopsis. Plant Cell 2022, 34, 3718–3736. [Google Scholar] [CrossRef]
- Underwood, W.; Ryan, A.; Somerville, S.C. An Arabidopsis lipid flippase is required for timely recruitment of defenses to the host-pathogen interface at the plant cell surface. Mol. Plant 2017, 10, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Gomès, E.; Jakobsen, M.K.; Axelsen, K.B.; Geisler, M.; Palmgren, M.G. Chilling tolerance in Arabidopsis involves ALA1, a member of a new family of putative aminophospholipid translocases. Plant Cell 2000, 12, 2441–2454. [Google Scholar] [CrossRef]
- Niu, Y.; Qian, D.; Liu, B.; Ma, J.; Wan, D.; Wang, X.; He, W.; Xiang, Y. ALA6, a P4-type ATPase, is involved in heat stress responses in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1732. [Google Scholar] [CrossRef]
- McDowell, S.C.; Lopez-Marques, R.L.; Poulsen, L.R.; Palmgren, M.G.; Harper, J.F. Loss of the Arabidopsis thaliana P4-ATPase ALA3 reduces adaptability to temperature stresses and impairs vegetative, pollen, and ovule development. PLoS ONE 2013, 8, e62577. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Gao, H.; Xu, G.; Wu, D.; Song, S.; Jiang, H.; Zhu, S.; Qi, T.; Xie, D. Arabidopsis ALA1 and ALA2 mediate RNAi-based antiviral immunity. Front. Plant Sci. 2017, 8, 422. [Google Scholar] [CrossRef]
- Guo, Z.; Lu, J.; Wang, X.; Zhan, B.; Li, W.; Ding, S.W. Lipid flippases promote antiviral silencing and the biogenesis of viral and host siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 1377–1382. [Google Scholar] [CrossRef]
- Wang, F.L.; Li, X.B.; Li, Y.J.; Han, J.; Chen, Y.; Zeng, J.Y.; Su, M.; Zhuo, J.X.; Ren, H.; Liu, H.R.; et al. Arabidopsis P4 ATPase-mediated cell detoxification confers resistance to Fusarium graminearum and Verticillium dahliae. Nat. Commun. 2021, 12, 6426. [Google Scholar] [CrossRef]
- Davis, J.A.; Pares, R.B.; Bernstein, T.; McDowell, S.C.; Brown, E.; Stubrich, J.; Rosenberg, A.; Cahoon, E.B.; Cahoon, R.E.; Poulsen, L.R.; et al. The lipid flippases ALA4 and ALA5 play critical roles in cell expansion and plant growth. Plant Physiol. 2020, 182, 2111–2125. [Google Scholar] [CrossRef]
- Botella, C.; Sautron, E.; Boudiere, L.; Michaud, M.; Dubots, E.; Yamaryo-Botte, Y.; Albrieux, C.; Marechal, E.; Block, M.A.; Jouhet, J. ALA10, a phospholipid flippase, controls FAD2/FAD3 desaturation of phosphatidylcholine in the ER and affects chloroplast lipid composition in Arabidopsis thaliana. Plant Physiol. 2016, 170, 1300–1314. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, L.R.; Lopez-Marques, R.L.; McDowell, S.C.; Okkeri, J.; Licht, D.; Schulz, A.; Pomorski, T.; Harper, J.F.; Palmgren, M.G. The Arabidopsis P4-ATPase ALA3 localizes to the Golgi and requires α-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell 2008, 20, 658–676. [Google Scholar] [CrossRef]
- Sanz-Fernández, M.; Rodríguez-Serrano, M.; Sevilla-Perea, A.; Pena, L.; Mingorance, M.D.; Sandalio, L.M.; Romero-Puertas, M.C. Screening Arabidopsis mutants in genes useful for phytoremediation. J. Hazard. Mater. 2017, 335, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Li, X.B.; Niu, G.Q.; Fan, Y.H.; Liu, W.Y.; Wu, Q.; Yu, C.; Wang, J.; Xiao, Y.H.; Hou, L.; Jin, D.; et al. Synthetic dual hormone-responsive promoters enable engineering plants with broad-spectrum resistance. Plant Commun. 2023, 4, 100596. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.Y.; Yan, X.Y.; Bai, W.Q.; Zhang, M.; Chen, Y.; Li, X.B.; Hou, L.; Zhao, J.; Ding, X.Y.; Liu, R.C.; et al. Carpel-specific downregulation of GhCKXs in cotton significantly enhances seed and fiber yield. J. Exp. Bot. 2022, 73, 6758–6772. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, W.W.; He, X.; Liu, M.; Zhang, K.; Shaban, M.; Sun, L.; Zhu, J.C.; Luo, Y.J.; Yuan, D.J.; et al. Functional characterization of cotton genes responsive to Verticillium dahliae through bioinformatics and reverse genetics strategies. J. Exp. Bot. 2014, 65, 6679–6692. [Google Scholar] [CrossRef]
- Cheng, H.Q.; Han, L.B.; Yang, C.L.; Wu, X.M.; Zhong, N.Q.; Wu, J.H.; Wang, F.X.; Wang, H.Y.; Xia, G.X. The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J. Exp. Bot. 2016, 67, 1935–1950. [Google Scholar] [CrossRef]
- Han, L.B.; Li, Y.B.; Wang, F.X.; Wang, W.Y.; Liu, J.; Wu, J.H.; Zhong, N.Q.; Wu, S.J.; Jiao, G.L.; Wang, H.Y.; et al. The cotton apoplastic protein CRR1 stabilizes chitinase 28 to facilitate defense against the fungal pathogen Verticillium dahliae. Plant Cell 2019, 31, 520–536. [Google Scholar] [CrossRef]
- Hua, C.; Zhao, J.H.; Guo, H.S. Trans-kingdom RNA silencing in plant–fungal pathogen interactions. Mol. Plant 2018, 11, 235–244. [Google Scholar] [CrossRef]
- De Coninck, B.; Timmermans, P.; Vos, C.; Cammue, B.P.; Kazan, K. What lies beneath: Belowground defense strategies in plants. Trends Plant Sci. 2015, 20, 91–101. [Google Scholar] [CrossRef]
- Inderbitzin, P.; Subbarao, K.V. Verticillium systematics and evolution: How confusion impedes Verticillium wilt management and how to resolve it. Phytopathology 2014, 104, 564–574. [Google Scholar] [CrossRef]
- Mansoori, B.; Smith, J. Verticillium-toxins their role in pathogenesis. J. Agric. Sci. Technol 2005, 7, 103–114. [Google Scholar]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Kemen, E.; Jones, J.D.G. Obligate biotroph parasitism: Can we link genomes to lifestyles? Trends Plant Sci. 2012, 17, 448–457. [Google Scholar] [CrossRef]
- Vleeshouwers, V.G.A.A.; Oliver, R.P. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant-Microbe Interact. 2014, 27, 196–206. [Google Scholar] [CrossRef]
- Masuda, D.; Ishida, M.; Yamaguchi, K.; Yamaguchi, I.; Kimura, M.; Nishiuchi, T. Phytotoxic effects of trichothecenes on the growth and morphology of Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 1617–1626. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum trichothecene mycotoxins: Biosynthesis, regulation, and management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef]
- Tang, Z.; Wang, H.Q.; Chen, J.; Chang, J.D.; Zhao, F.J. Molecular mechanisms underlying the toxicity and detoxification of trace metals and metalloids in plants. J. Integr. Plant Biol. 2022, 65, 570–593. [Google Scholar] [CrossRef]
- Kistler, H.C.; Broz, K. Cellular compartmentalization of secondary metabolism. Front Microbiol 2015, 6, 68. [Google Scholar] [CrossRef]
- Hathout, A.S.; Aly, S.E. Biological detoxification of mycotoxins: A review. Ann. Microbiol. 2014, 64, 905–919. [Google Scholar] [CrossRef]
- D’Orazio, V.; Stallone, D.; Samer, S.; Loffredo, E.; Cirulli, M.; Bruno, G.L. Phytotoxic metabolites produced by Verticillium dahliae Kleb. in olive wilting: A chemical and spectroscopic approach for their molecular characterisation. Nat. Prod. Res. 2019, 12, 1991–2001. [Google Scholar]
- Chen, J.Y.; Xiao, H.L.; Gui, Y.J.; Zhang, D.D.; Li, L.; Bao, Y.M.; Dai, X.F. Characterization of the Verticillium dahliae exoproteome involves in pathogenicity from cotton-containing medium. Front Microbiol. 2016, 7, 1709. [Google Scholar] [CrossRef]
- Laouane, H.; Lazrek, H. Synthesis and toxicity evaluation of cinnamyl acetate: A new phytotoxin produced by a strain of Verticillium dahliae pathogenic on olive tree. Int. J. Agric. Biol. 2011, 13, 444–446. [Google Scholar]
- Shin, H.W.; Takatsu, H. Phosphatidylserine exposure in living cells. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 166–178. [Google Scholar] [CrossRef]
- Wang, F.L.; Qiu, M.L.; Hou, L.; Li, X.B.; Ren, H.; Zhuo, J.X.; Liu, H.R.; Li, Y.J.; Yang, Y.; Yan, X.Y.; et al. N terminus and C terminus of Arabidopsis P4-ATPase AtALA1 facilitate the detoxification of the mycotoxin deoxynivalenol in wheat. Cell Rep. 2025, 44, 115641. [Google Scholar] [CrossRef]
- You, L.; Zhao, Y.; Kuca, K.; Wang, X.; Oleksak, P.; Chrienova, Z.; Nepovimova, E.; Jaćević, V.; Wu, Q.; Wu, W. Hypoxia, oxidative stress, and immune evasion: A trinity of the trichothecenes T-2 toxin and deoxynivalenol (DON). Arch. Toxicol. 2021, 95, 1899–1915. [Google Scholar] [CrossRef]
- Liu, A.; Sun, Y.; Wang, X.; Ihsan, A.; Tao, Y.; Chen, D.; Peng, D.; Wu, Q.; Wang, X.; Yuan, Z. DNA methylation is involved in pro-inflammatory cytokines expression in T-2 toxin-induced liver injury. Food Chem. Toxicol. 2019, 132, 110661. [Google Scholar] [CrossRef]
- Han, J.; Wang, Q.C.; Zhu, C.C.; Liu, J.; Zhang, Y.; Cui, X.S.; Kim, N.H.; Sun, S.C. Deoxynivalenol exposure induces autophagy/apoptosis and epigenetic modification changes during porcine oocyte maturation. Toxicol. Appl. Pharmacol. 2016, 300, 70–76. [Google Scholar] [CrossRef]
- Zhang, Z.; Fan, K.; Meng, J.; Nie, D.; Zhao, Z.; Han, Z. Deoxynivalenol hijacks the pathway of Janus kinase 2/signal transducers and activators of transcription 3 (JAK2/STAT-3) to drive caspase-3-mediated apoptosis in intestinal porcine epithelial cells. Sci. Total Environ. 2023, 864, 161058. [Google Scholar] [CrossRef]
- Hou, S.; Cheng, Y.; Wang, Z.; Xia, L.; Wang, J.; Wang, H.; Sun, J.; Ma, J.; Yan, Y. DON entry into the nucleus induces DNA damage, apoptosis and cycle arrest in GES-1 cells. Food Chem. Toxicol. 2023, 171, 113531. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Devaiah, S.P.; Narasimhan, R.; Pan, X.; Zhang, Y.; Zhang, W.; Wang, X. Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 2012, 24, 2200–2212. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Qiu, M.; Yao, X.; Li, J.; Ren, H.; Su, M.; Shen, J.; Li, C.; Jiang, Q.; Zhang, Z.; et al. Arabidopsis P4-ATPases ALA1 and ALA7 Enhance Resistance to Verticillium dahliae via Detoxifying Vd-Toxins. Biology 2025, 14, 595. https://doi.org/10.3390/biology14060595
Wang F, Qiu M, Yao X, Li J, Ren H, Su M, Shen J, Li C, Jiang Q, Zhang Z, et al. Arabidopsis P4-ATPases ALA1 and ALA7 Enhance Resistance to Verticillium dahliae via Detoxifying Vd-Toxins. Biology. 2025; 14(6):595. https://doi.org/10.3390/biology14060595
Chicago/Turabian StyleWang, Fanlong, Mingliang Qiu, Xiaoxia Yao, Jiancong Li, Hui Ren, Mei Su, Jiaohuan Shen, Caiwang Li, Qian Jiang, Zixuan Zhang, and et al. 2025. "Arabidopsis P4-ATPases ALA1 and ALA7 Enhance Resistance to Verticillium dahliae via Detoxifying Vd-Toxins" Biology 14, no. 6: 595. https://doi.org/10.3390/biology14060595
APA StyleWang, F., Qiu, M., Yao, X., Li, J., Ren, H., Su, M., Shen, J., Li, C., Jiang, Q., Zhang, Z., Li, Y., Tang, J., Li, X., Fan, Y., & Pei, Y. (2025). Arabidopsis P4-ATPases ALA1 and ALA7 Enhance Resistance to Verticillium dahliae via Detoxifying Vd-Toxins. Biology, 14(6), 595. https://doi.org/10.3390/biology14060595




