Biooxidation of Arsenopyrite by Acidithiobacillus ferriphilus QBS 3 Exhibits Arsenic Resistance Under Extremely Acidic Bioleaching Conditions
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Culture Conditions
2.2. Mineral Sample
2.3. Arsenic Stress Experiment
2.4. Bioleaching Experiment
2.5. Bioinformatics Analysis
2.6. Total RNA Extraction and qRT-PCR
3. Results
3.1. Stress Response of QBS 3
3.2. Bioleaching Performance of QBS 3 on Arsenopyrite
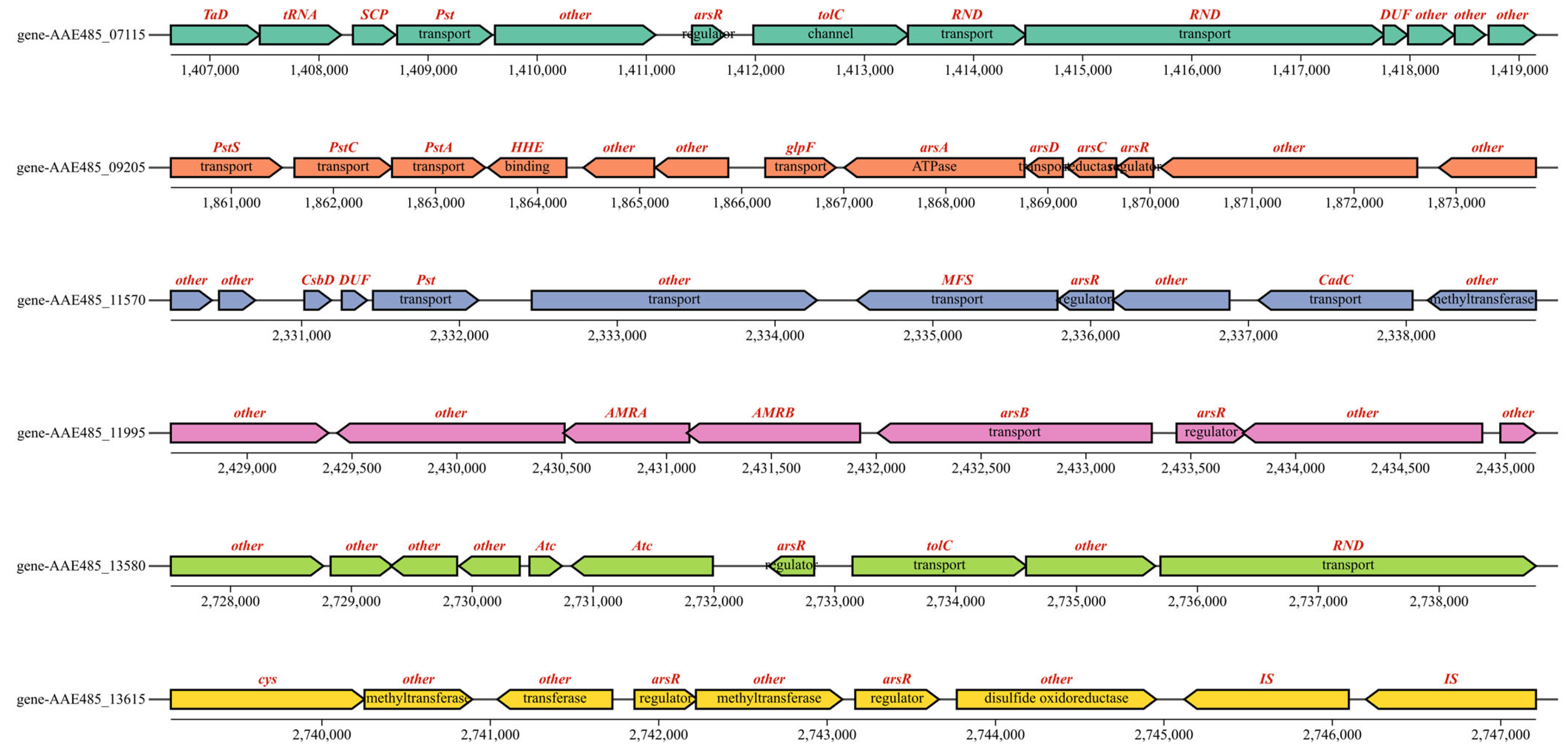
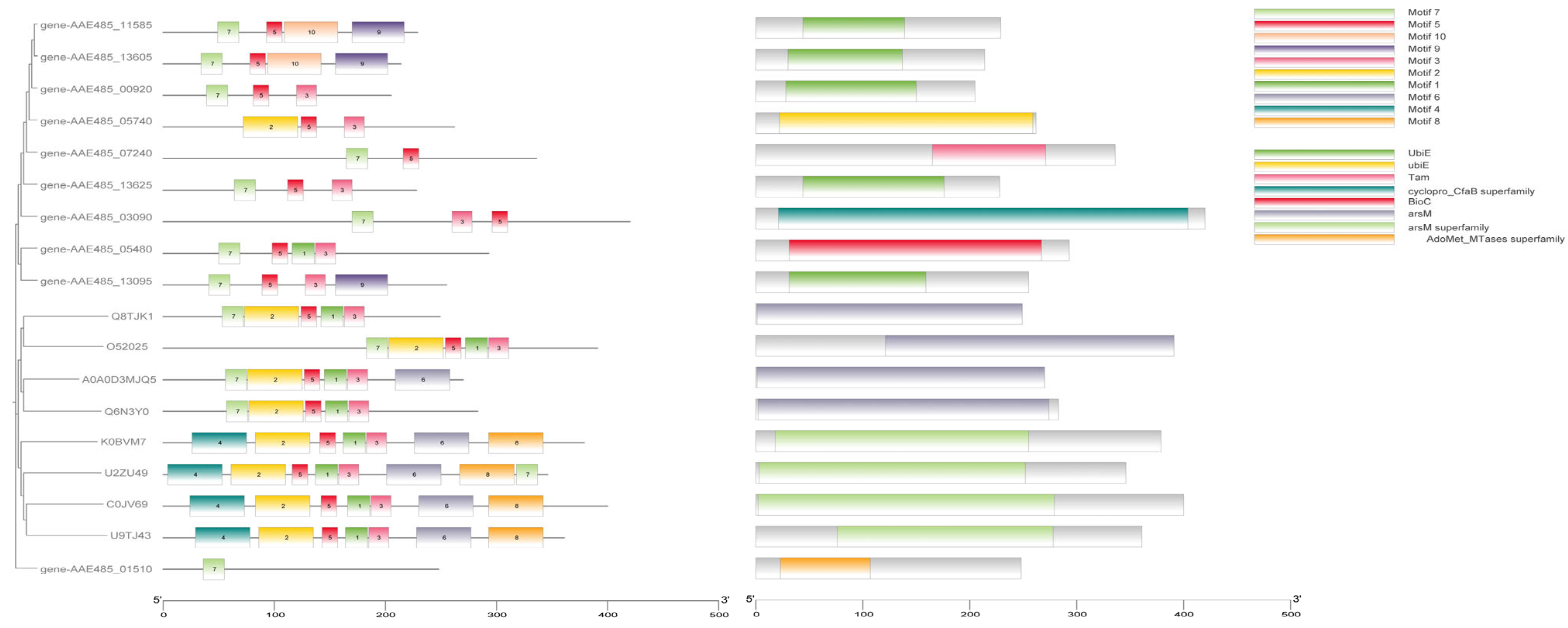
3.3. Mining and Analysis of Arsenic Resistance Genes in QBS 3
3.4. q-RTPCR Experiments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pagnanelli, F.; De Michelis, I.; Di Muzio, S.; Ferella, F.; Veglio, F. Bioassessment of a combined chemical-biological treatment for synthetic acid mine drainage. J. Hazard. Mater. 2008, 159, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, O.; Neculita, C.M.; Yue, X.; Ng, H.Y. Bioelectrochemical treatment of acid mine drainage dominated with iron. J. Hazard. Mater. 2012, 241–242, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Hu, Y.; Luo, J.; Xu, B.; Zhao, J. Geochemical processes controlling fate and transport of arsenic in acid mine drainage (AMD) and natural systems. J. Hazard. Mater. 2009, 165, 13–26. [Google Scholar] [CrossRef]
- Zhang, G.; Chao, X.; Guo, P.; Cao, J.; Yang, C. Catalytic effect of Ag+ on arsenic bioleaching from orpiment (As2S3) in batch tests with Acidithiobacillus ferrooxidans and Sulfobacillus sibiricus. J. Hazard. Mater. 2015, 283, 117–122. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, H.; Abashina, T.; Vainshtein, M. Review on arsenic removal from sulfide minerals: An emphasis on enargite and arsenopyrite. Miner. Eng. 2021, 172, 107133. [Google Scholar] [CrossRef]
- Li, Q.; Li, D.; Qian, F. Pre-oxidation of high-sulfur and high-arsenic refractory gold concentrate by ozone and ferric ions in acidic media. Hydrometallurgy 2009, 97, 61–66. [Google Scholar] [CrossRef]
- Hu, J.; Huang, H.; Xie, H.; Gan, L.; Liu, J.; Long, M. A scaled-up continuous process for biooxidation as pre-treatment of refractory pyrite-arsenopyrite gold-bearing concentrates. Biochem. Eng. J. 2017, 128, 228–234. [Google Scholar] [CrossRef]
- Chen, H.-R.; Li, Q.; Zhao, X.-J.; Zhang, D.-R.; Pakostova, E. Two-step sequential bio-oxidation of arsenopyrite catalyzed by a mesophilic bacterium eliminates hazardous Fe(III)/As-bearing products and enhances mineral dissolution. Chem. Eng. J. 2023, 462, 142259. [Google Scholar] [CrossRef]
- Li, T.; Guo, Z. Mechanisms of arsenic oxidation in the presence of pyrite: An experimental and theoretical study. Sci. Total Environ. 2024, 921, 171072. [Google Scholar] [CrossRef]
- Mandal, B.K.; Suzuki, K.T. Arsenic round the world: A review. Talanta 2002, 58, 201–235. [Google Scholar] [CrossRef]
- Jacobson, T.; Navarrete, C.; Sharma, S.K.; Sideri, T.C.; Ibstedt, S.; Priya, S.; Grant, C.M.; Christen, P.; Goloubinoff, P.; Tamas, M.J. Arsenite interferes with protein folding and triggers formation of protein aggregates in yeast. J. Cell Sci. 2012, 125, 5073–5083. [Google Scholar] [CrossRef] [PubMed]
- Slaughter, D.C.; Macur, R.E.; Inskeep, W.P. Inhibition of microbial arsenate reduction by phosphate. Microbiol. Res. 2012, 167, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Dave, S.R.; Gupta, K.H.; Tipre, D.R. Characterization of arsenic resistant and arsenopyrite oxidizing Acidithiobacillus ferrooxidans from Hutti gold leachate and effluents. Bioresour. Technol. 2008, 99, 7514–7520. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.-X.; Zhang, X.; Tan, W.-S.; Zhu, M.-L. Effect of agitation intensity on the biooxidation process of refractory gold ores by Acidithiobacillus ferrooxidans. Hydrometallurgy 2012, 127–128, 99–103. [Google Scholar] [CrossRef]
- Barragán, C.E.; Márquez, M.A.; Dopson, M.; Montoya Castaño, D. Isolation of Arsenic Resistant and Arsenopyrite Oxidizing Acidithiobacillus Species from pH Neutral Colombian Mine Effluents. Geomicrobiol. J. 2020, 37, 682–689. [Google Scholar] [CrossRef]
- Leng, F.; Li, K.; Zhang, X.; Li, Y.; Zhu, Y.; Lu, J.; Li, H. Comparative study of inorganic arsenic resistance of several strains of Acidithiobacillus thiooxidans and Acidithiobacillus ferrooxidans. Hydrometallurgy 2009, 98, 235–240. [Google Scholar] [CrossRef]
- Zheng, F.; Nie, Z.; Xia, J.; Liu, L.; Yang, H. Domestication of Leptospirillum Ferriphilum Arsenic-tolerant Ability and Mechanism of Domestication. Trans. Nonferrous Met. Soc. China 2019, 29, 617–627. [Google Scholar] [CrossRef]
- Jiang, H.; Liang, Y.; Yin, H.; Xiao, Y.; Guo, X.; Xu, Y.; Hu, Q.; Liu, H.; Liu, X. Effects of Arsenite Resistance on the Growth and Functional Gene Expression of Leptospirillum ferriphilum and Acidithiobacillus thiooxidans in Pure Culture and Coculture. Biomed. Res. Int. 2015, 2015, 203197. [Google Scholar] [CrossRef]
- Flores, A.; Valencia-Marin, M.F.; Chavez-Avila, S.; Ramirez-Diaz, M.I.; de Los Santos-Villalobos, S.; Meza-Carmen, V.; Orozco-Mosqueda, M.D.C.; Santoyo, G. Genome mining, phylogenetic, and functional analysis of arsenic (As) resistance operons in Bacillus strains, isolated from As-rich hot spring microbial mats. Microbiol. Res. 2022, 264, 127158. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, P.; Wei, X.; Peng, H.; Hu, L.; Zhu, X. Migration, transformation of arsenic, and pollution controlling strategies in paddy soil-rice system: A comprehensive review. Sci. Total Environ. 2024, 951, 175500. [Google Scholar] [CrossRef]
- Yachkula, A.; Rozova, O.; Abashina, T.; Vainshtein, M.; Grouzdev, D.; Bulaev, A. Attempts to Stimulate Leaching Activity of Acidithiobacillus ferrooxidans Strain TFBk. Minerals 2022, 12, 1051. [Google Scholar] [CrossRef]
- Falagan, C.; Johnson, D.B. Acidithiobacillus ferriphilus sp. nov., a facultatively anaerobic iron- and sulfur-metabolizing extreme acidophile. Int. J. Syst. Evol. Microbiol. 2016, 66, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Nunez, H.; Moya-Beltran, A.; Covarrubias, P.C.; Issotta, F.; Cardenas, J.P.; Gonzalez, M.; Atavales, J.; Acuna, L.G.; Johnson, D.B.; Quatrini, R. Molecular Systematics of the Genus Acidithiobacillus: Insights into the Phylogenetic Structure and Diversification of the Taxon. Front. Microbiol. 2017, 8, 30. [Google Scholar] [CrossRef]
- Navarro, C.A.; Orellana, L.H.; Mauriaca, C.; Jerez, C.A. Transcriptional and functional studies of Acidithiobacillus ferrooxidans genes related to survival in the presence of copper. Appl. Environ. Microbiol. 2009, 75, 6102–6109. [Google Scholar] [CrossRef]
- Okazaki, T.; Wang, W.; Kuramitz, H.; Hata, N.; Taguchi, S. Molybdenum blue spectrophotometry for trace arsenic in ground water using a soluble membrane filter and calcium carbonate column. Anal. Sci. 2013, 29, 67–72. [Google Scholar] [CrossRef]
- Smith, G.L.; Reutovich, A.A.; Srivastava, A.K.; Reichard, R.E.; Welsh, C.H.; Melman, A.; Bou-Abdallah, F. Complexation of ferrous ions by ferrozine, 2,2’-bipyridine and 1,10-phenanthroline: Implication for the quantification of iron in biological systems. J. Inorg. Biochem. 2021, 220, 111460. [Google Scholar] [CrossRef]
- Amanze, C.; Zheng, X.; Man, M.; Yu, Z.; Ai, C.; Wu, X.; Xiao, S.; Xia, M.; Yu, R.; Wu, X.; et al. Recovery of heavy metals from industrial wastewater using bioelectrochemical system inoculated with novel Castellaniella species. Environ. Res. 2022, 205, 112467. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014, 42, D737–D743. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nkulu, G.; Gaydardzhiev, S.; Mwema, E.; Compere, P. SEM and EDS observations of carrollite bioleaching with a mixed culture of acidophilic bacteria. Miner. Eng. 2015, 75, 70–76. [Google Scholar] [CrossRef]
- Ye, M.; Yan, P.; Sun, S.; Han, D.; Xiao, X.; Zheng, L.; Huang, S.; Chen, Y.; Zhuang, S. Bioleaching combined brine leaching of heavy metals from lead-zinc mine tailings: Transformations during the leaching process. Chemosphere 2017, 168, 1115–1125. [Google Scholar] [CrossRef]
- Corkhill, C.L.; Vaughan, D.J. Arsenopyrite oxidation—A review. Appl. Geochem. 2009, 24, 2342–2361. [Google Scholar] [CrossRef]
- Zhu, T.; Lu, X.; Liu, H.; Li, J.; Zhu, X.; Lu, J.; Wang, R. Quantitative X-ray photoelectron spectroscopy-based depth profiling of bioleached arsenopyrite surface by Acidithiobacillus ferrooxidans. Geochim. Cosmochim. Acta 2014, 127, 120–139. [Google Scholar] [CrossRef]
- Deng, S.; Gu, G.; Xu, B.; Li, L.; Wu, B. Surface characterization of arsenopyrite during chemical and biological oxidation. Sci. Total Environ. 2018, 626, 349–356. [Google Scholar] [CrossRef]
- Cen, L.; Cheng, H.; Liu, Q.; Wang, S.; Wang, X. Arsenic release from arsenopyrite weathering in acid mine drainage: Kinetics, transformation, and effect of biochar. Environ. Int. 2022, 170, 107558. [Google Scholar] [CrossRef]
- Cruz-Hernández, M.; García-Cerón, A.; Maldonado, R.G.S.; Corro-Escorcia, I.A.; Hernández-Ávila, J.; Cerecedo-Sáenz, E.; Flores-Badillo, J.; Toro, N.; Saldana, M.; Gutiérrez-Amador, M.P.; et al. Leveraging Industrial Jarosite Waste for Arsenic(V) and Chromium(III) Adsorption from Water: A Preliminary Study. Appl. Sci. 2025, 15, 31469. [Google Scholar] [CrossRef]
- Eftekhari, N.; Kargar, M.; Zamin, F.; Rastakhiz, N.; Manafi, Z. A Review on Various Aspects of Jarosite and Its Utilization Potentials. Ann. Chim. Sci. Matériaux 2020, 44, 43–52. [Google Scholar] [CrossRef]
- Moinier, D.; Slyemi, D.; Byrne, D.; Lignon, S.; Lebrun, R.; Talla, E.; Bonnefoy, V. An ArsR/SmtB family member is involved in the regulation by arsenic of the arsenite oxidase operon in Thiomonas arsenitoxydans. Appl. Environ. Microbiol. 2014, 80, 6413–6426. [Google Scholar] [CrossRef]
- Rawle, R.; Saley, T.C.; Kang, Y.S.; Wang, Q.; Walk, S.; Bothner, B.; McDermott, T.R. Introducing the ArsR-Regulated Arsenic Stimulon. Front. Microbiol. 2021, 12, 630562. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.; Kaur, M.; Thompson, L.K.; Cox, G. A historical perspective on the multifunctional outer membrane channel protein TolC in Escherichia coli. NPJ Antimicrob. Resist. 2025, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.P.; Liu, Z. Transport pathways for arsenic and selenium: A minireview. Environ. Int. 2009, 35, 512–515. [Google Scholar] [CrossRef]
- Ben Fekih, I.; Zhang, C.; Li, Y.P.; Zhao, Y.; Alwathnani, H.A.; Saquib, Q.; Rensing, C.; Cervantes, C. Distribution of Arsenic Resistance Genes in Prokaryotes. Front. Microbiol. 2018, 9, 2473. [Google Scholar] [CrossRef]
- Yan, G.; Chen, X.; Du, S.; Deng, Z.; Wang, L.; Chen, S. Genetic mechanisms of arsenic detoxification and metabolism in bacteria. Curr. Genet. 2019, 65, 329–338. [Google Scholar] [CrossRef]
- Liang, Y.; Yan, Y.; Shi, L.; Wang, M.; Yuan, X.; Wang, S.; Ye, L.; Yan, Z. Molecular Basis of Thioredoxin-Dependent Arsenic Transformation in Methanogenic archaea. Environ. Sci. Technol. 2025, 59, 443–453. [Google Scholar] [CrossRef]
- Tsai, S.L.; Singh, S.; Chen, W. Arsenic metabolism by microbes in nature and the impact on arsenic remediation. Curr. Opin. Biotechnol. 2009, 20, 659–667. [Google Scholar] [CrossRef]
- Antonelli, R.; Shao, K.; Thomas, D.J.; Sams, R., 2nd; Cowden, J. AS3MT, GSTO, and PNP polymorphisms: Impact on arsenic methylation and implications for disease susceptibility. Environ. Res. 2014, 132, 156–167. [Google Scholar] [CrossRef]
- Jeong, H.; Yoon, C.; Lee, J.S.; Byeon, E. Differential susceptibility to arsenic in glutathione S-transferase omega 2 (GST-O2)-targeted freshwater water flea Daphnia magna mutants. Aquat. Toxicol. 2023, 254, 106364. [Google Scholar] [CrossRef]






| elements | Fe | As | S | O | Zn | Si | Mg |
| (wt.%) | 31.44 | 39.02 | 15.57 | 5.9 | 2.243 | 1.83 | 1.66 |
| Name | Gene Locus | Length (AA) | Dir. | Annotation | BACMAT (%) |
|---|---|---|---|---|---|
| ArsT | 06325 | 322 | ↑ | Thioredoxin disulfide reductase | 42.759 |
| ArsA | 09190 | 589 | ↑ | Arsenical pump-driving ATPase | 68.836 |
| ArsD | 09195 | 121 | ↑ | Arsenical resistance operon trans-acting repressor, metallochaperone | 59 |
| ArsC | 09200 | 162 | ↑ | Arsenate reductase | 36.029 |
| ArsR2 | 09205 | 121 | ↑ | Helix-turn-helix domain-containing protein | |
| ArsB | 11990 | 436 | ↑ | Arsenical pump membrane protein, 11 transmembrane helixes | 61.098 |
| ArsR1 | 11995 | 120 | ↓ | Helix-turn-helix domain-containing protein(down gene overlap 17 bp) | |
| ArsR3 | 13615 | 124 | ↓ | Metalloregulator ArsR/SmtB family transcription factor | |
| ArsM | 13625 | 227 | ↓ | Class I SAM-dependent methyltransferase | 36.842 |
| Name | Gene Locus | Length (AA) | Dir. | Annotation | BACMAT (%) | True |
|---|---|---|---|---|---|---|
| ArsR | 01475 | 122 | ↑ | Helix-turn-helix transcriptional regulator | 42.857 | × |
| CzcR | 07115 | 100 | ↓ | Metalloregulator ArsR/SmtB family transcription factor | 45.455 | × |
| ArsR2 | 09205 | 121 | ↑ | Helix-turn-helix domain-containing protein | √ | |
| ArsR | 11425 | 97 | ↓ | Metalloregulator ArsR/SmtB family transcription factor | 34.848 | × |
| ArsR | 11570 | 118 | ↑ | Metalloregulator ArsR/SmtB family transcription factor | 33.333 | × |
| ArsR1 | 11995 | 120 | ↓ | Helix-turn-helix domain-containing protein | √ | |
| CzcR | 13580 | 125 | ↓ | Metalloregulator ArsR/SmtB family transcription factor | 46.377 | × |
| ArsR3 | 13615 | 124 | ↓ | Metalloregulator ArsR/SmtB family transcription factor | √ | |
| ArsR | 13715 | 232 | ↓ | Winged helix-turn-helix domain-containing protein | × |
| Name | Gene Locus | Length (AA) | Dir. | Annotation | BACMAT (%) | True |
|---|---|---|---|---|---|---|
| ArsM | 00920 | 204 | ↓ | S-adenosylmethionine-dependent methyltransferase activity | 27.48 | × |
| ArsM | 01510 | 248 | ↑ | Class I SAM-dependent methyltransferase | × | |
| ArsM | 03090 | 420 | ↓ | Class I SAM-dependent methyltransferase | × | |
| ArsM | 05480 | 292 | ↓ | Malonyl-CoA methyltransferase activity [Evidence IEA] | 36.28 | × |
| ArsM | 05740 | 261 | ↑ | Methyltransferase activity [Evidence IEA] | 37.03 | × |
| ArsM | 07240 | 336 | ↑ | Class I SAM-dependent methyltransferase | × | |
| ArsM | 11585 | 228 | ↑ | Class I SAM-dependent methyltransferase | 25.87 | × |
| ArsM | 13095 | 254 | ↓ | Class I SAM-dependent methyltransferase | 33.33 | × |
| ArsM | 13605 | 213 | ↓ | Class I SAM-dependent methyltransferase | 31.19 | × |
| ArsM | 13625 | 227 | ↓ | Class I SAM-dependent methyltransferase | 36.84 | √ |
| Name | Gene Locus | Length (AA) | Dir. | Annotation |
|---|---|---|---|---|
| ArsT | 06325 | 322 | ↑ | Thioredoxin disulfide reductase |
| ArsA | 09190 | 589 | ↑ | Arsenical pump-driving ATPase |
| ArsD | 09195 | 121 | ↑ | Arsenical resistance operon trans-acting repressor, metallochaperone |
| ArsC | 09200 | 162 | ↑ | Arsenate reductase |
| ArsR2 | 09205 | 121 | ↑ | Helix-turn-helix domain-containing protein |
| ArsB | 11990 | 436 | ↑ | Arsenical pump membrane protein, 11 transmembrane helixes |
| ArsR1 | 11995 | 120 | ↓ | Helix-turn-helix domain-containing protein |
| ArsR3 | 13615 | 124 | ↓ | Metalloregulator ArsR/SmtB family transcription factor |
| ArsM | 13625 | 227 | ↓ | Class I SAM-dependent methyltransferase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, R.; Liu, S.; Bai, X.; Liu, S.; Liu, Y. Biooxidation of Arsenopyrite by Acidithiobacillus ferriphilus QBS 3 Exhibits Arsenic Resistance Under Extremely Acidic Bioleaching Conditions. Biology 2025, 14, 550. https://doi.org/10.3390/biology14050550
Liu R, Liu S, Bai X, Liu S, Liu Y. Biooxidation of Arsenopyrite by Acidithiobacillus ferriphilus QBS 3 Exhibits Arsenic Resistance Under Extremely Acidic Bioleaching Conditions. Biology. 2025; 14(5):550. https://doi.org/10.3390/biology14050550
Chicago/Turabian StyleLiu, Run, Siyu Liu, Xiaoxuan Bai, Shiping Liu, and Yuandong Liu. 2025. "Biooxidation of Arsenopyrite by Acidithiobacillus ferriphilus QBS 3 Exhibits Arsenic Resistance Under Extremely Acidic Bioleaching Conditions" Biology 14, no. 5: 550. https://doi.org/10.3390/biology14050550
APA StyleLiu, R., Liu, S., Bai, X., Liu, S., & Liu, Y. (2025). Biooxidation of Arsenopyrite by Acidithiobacillus ferriphilus QBS 3 Exhibits Arsenic Resistance Under Extremely Acidic Bioleaching Conditions. Biology, 14(5), 550. https://doi.org/10.3390/biology14050550







