Origin of Polyploidy, Phylogenetic Relationships, and Biogeography of Botiid Fishes (Teleostei: Cypriniformes)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling, DNA Extraction, PCR, Library Preparation, and Sequencing
2.2. Mitogenome and Nuclear Gene Assembling
2.3. Datasets and Phylogenetic Analyses
2.3.1. Mitogenome Dataset
2.3.2. Individual Nuclear Gene Datasets
2.3.3. Concatenated Nuclear Gene Dataset
2.3.4. All-Gene Dataset
2.3.5. Phylogenetic Analyses
2.4. Whole Genome Sequencing and Genome Profiling
2.5. Timetree Analyses
2.6. Ancestral Range Reconstruction
3. Results
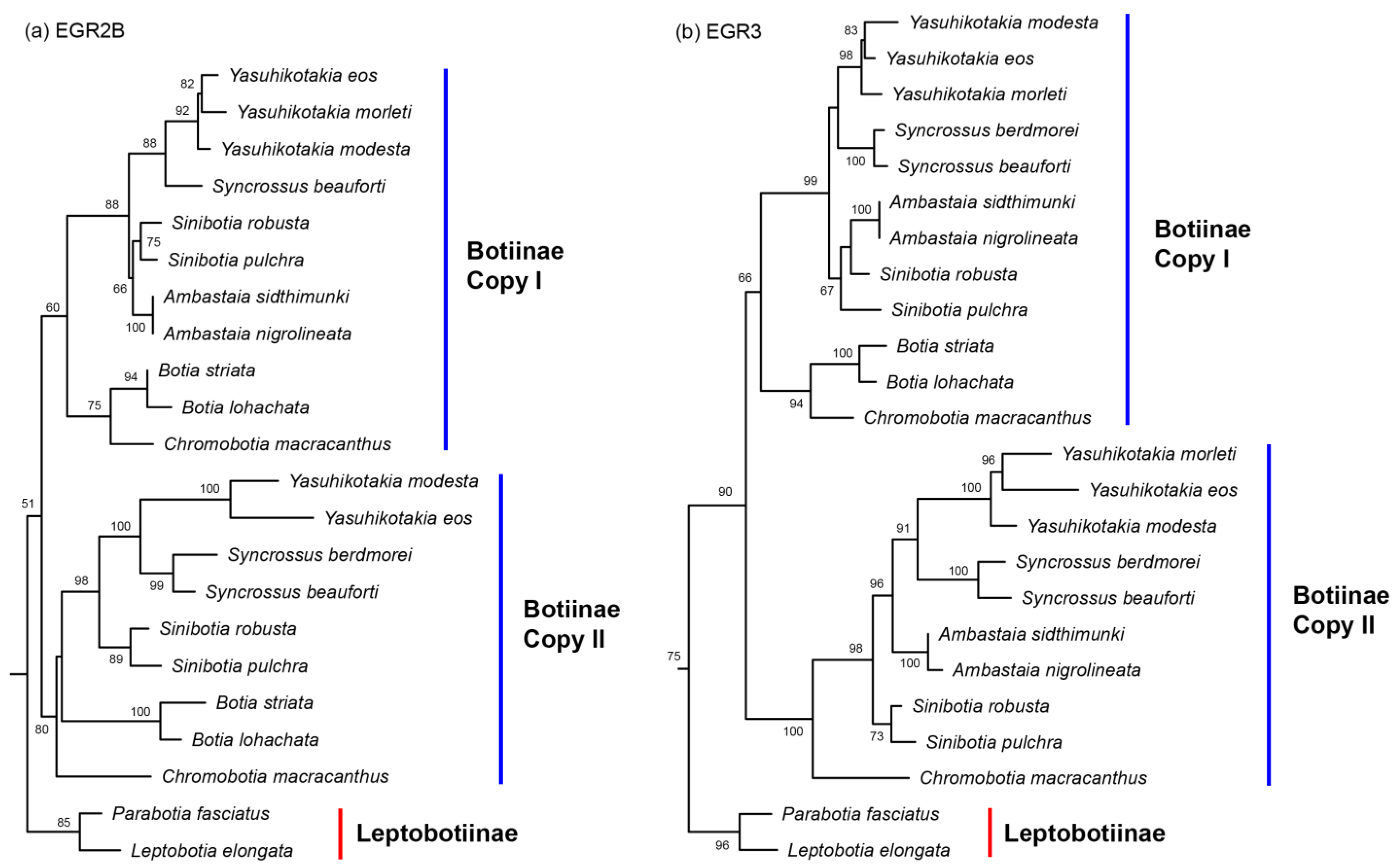
3.1. Nuclear Gene Copies
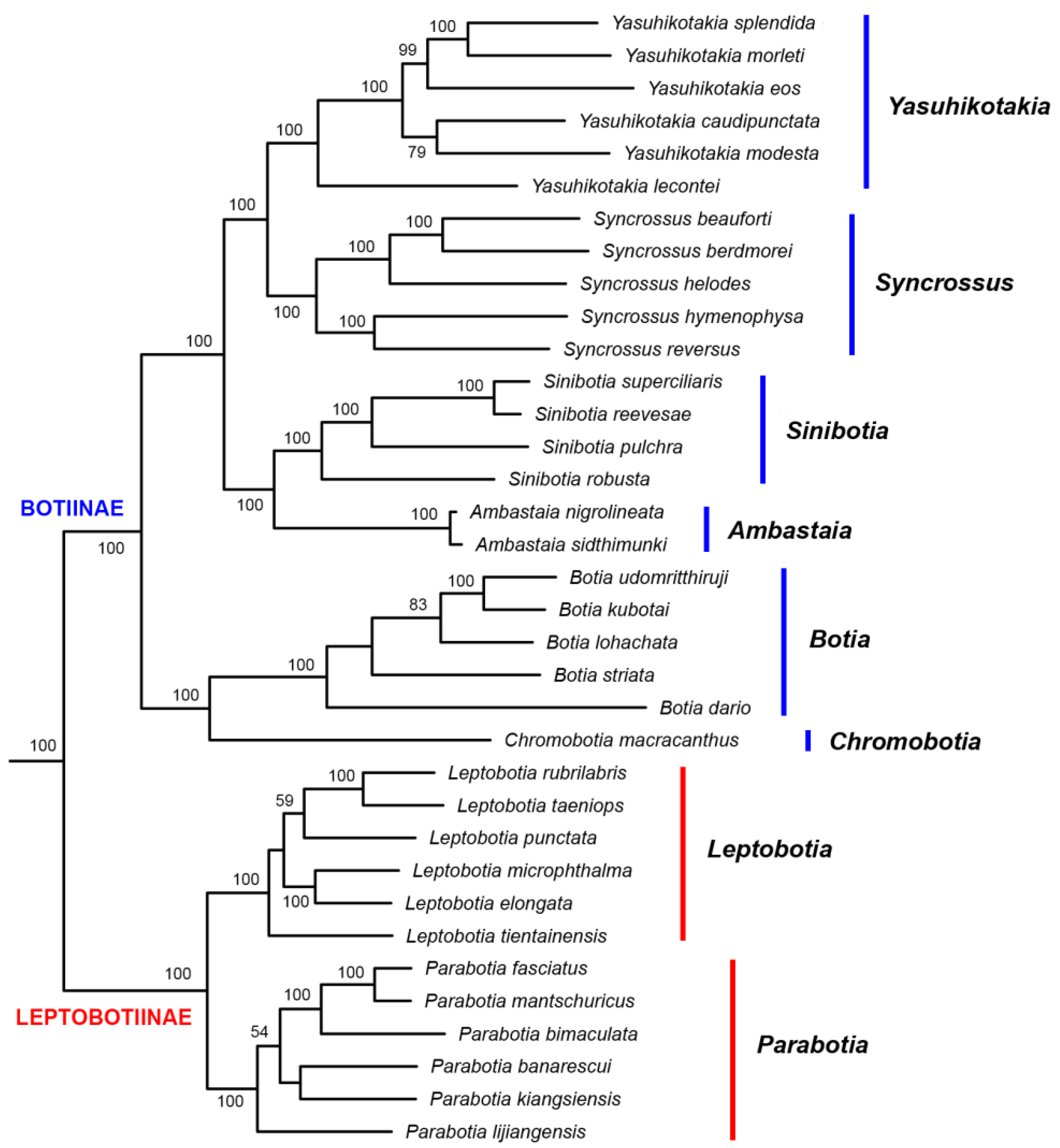
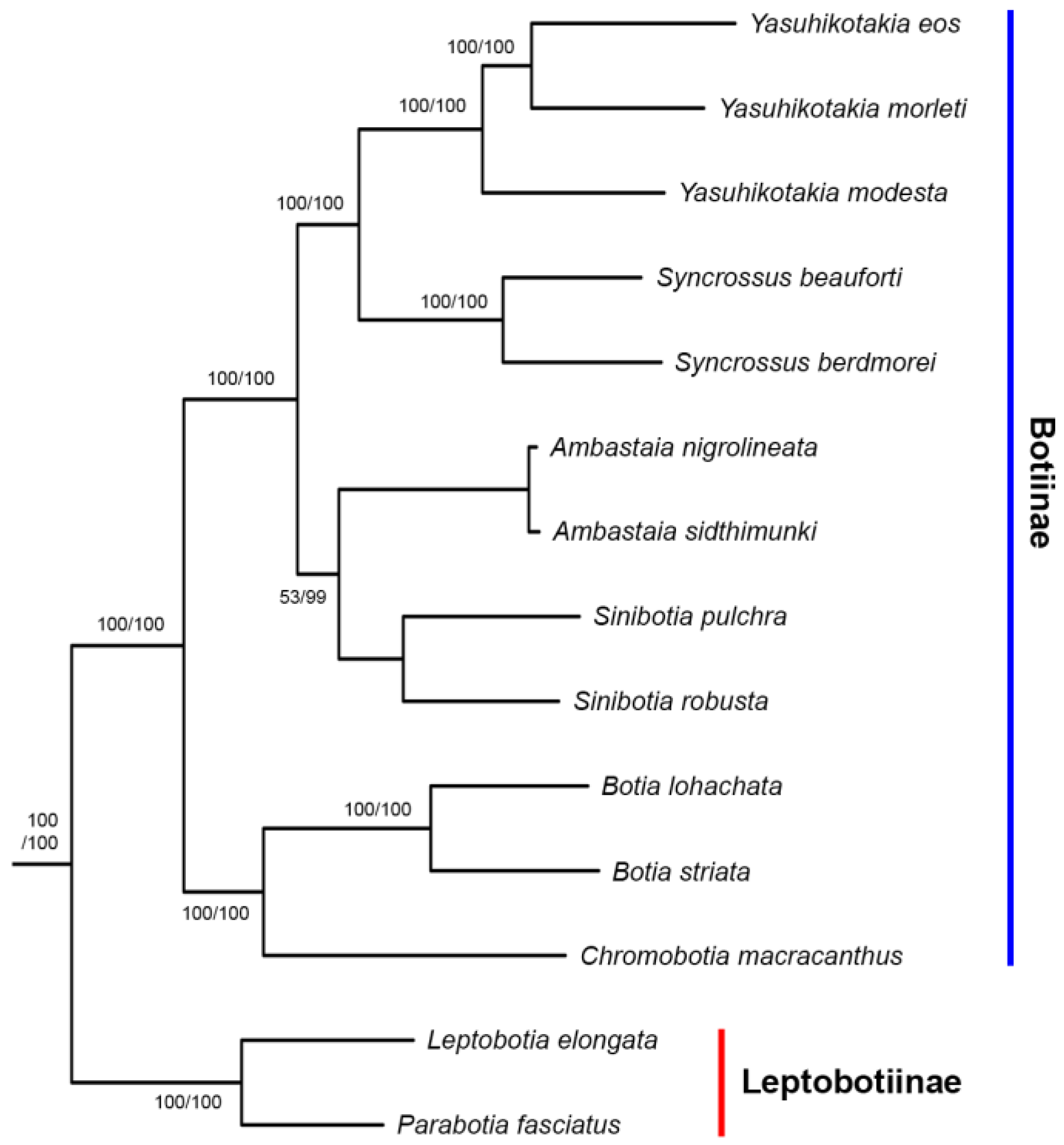
3.2. Phylogenetic Relationships
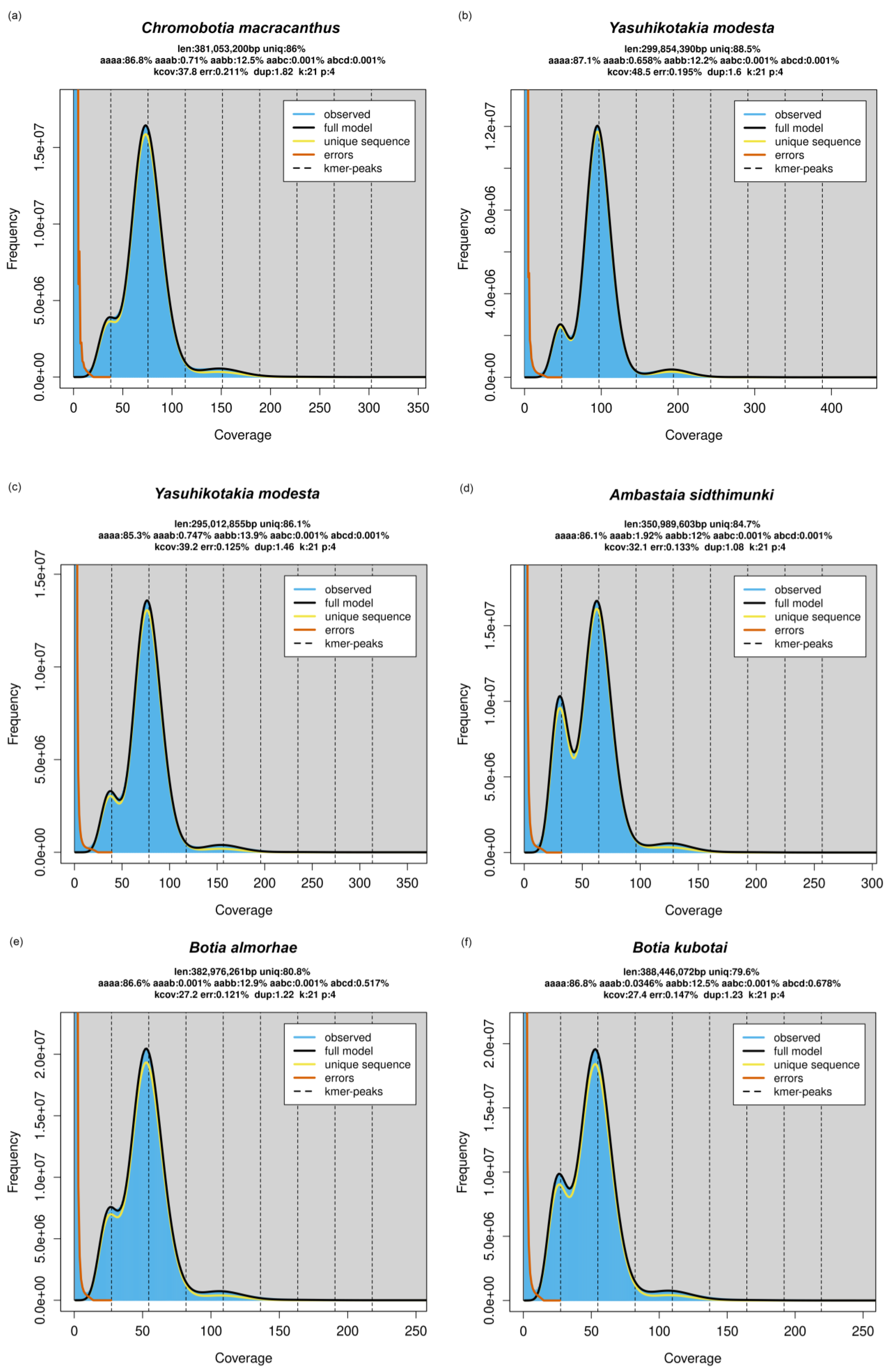
3.3. Whole Genome Sequencing and GenomeScope Profile
3.4. Divergence Time Estimation
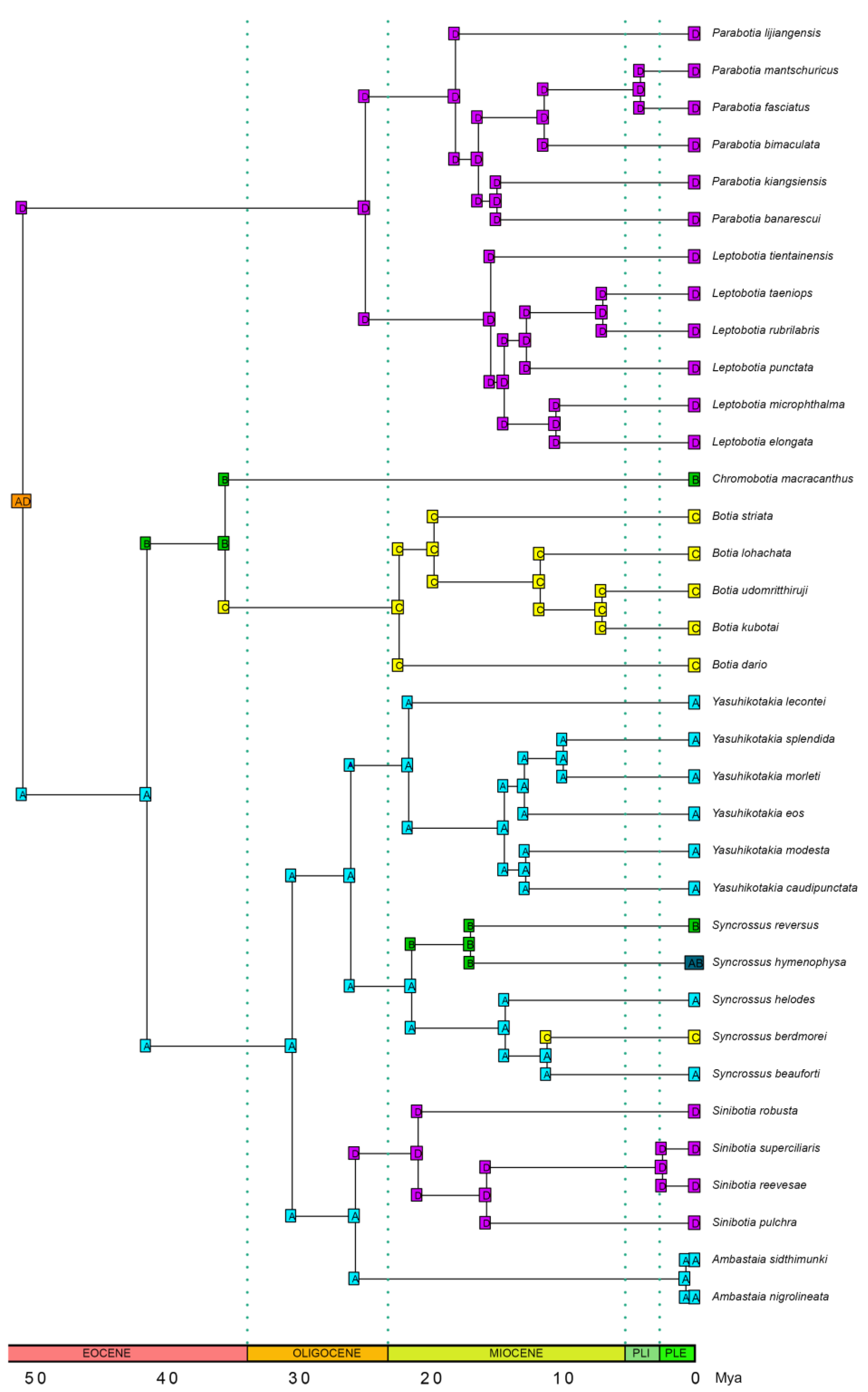
3.5. Ancestral Range Reconstruction Results
4. Discussion
4.1. The Origin of Polyploidy in Botiinae
4.2. Phylogenetic Relationships
4.3. Biogeographical History
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rice, A.; Šmarda, P.; Novosolov, M.; Drori, M.; Glick, L.; Sabath, N.; Meiri, S.; Belmaker, J.; Mayrose, I. The global biogeography of polyploid plants. Nat. Ecol. Evol. 2019, 3, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Ashman, T.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Rothfels, C.J. Polyploid phylogenetics. New Phytol. 2021, 230, 66–72. [Google Scholar] [CrossRef]
- Morris, J.P.; Baslan, T.; Soltis, D.E.; Soltis, P.S.; Fox, D.T. Integrating the study of polyploidy across organisms, tissues, and disease. Annu. Rev. Genet. 2024, 58, 297–318. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.P.; Whitton, J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000, 34, 401–437. [Google Scholar] [CrossRef]
- Mable, B.K.; Alexandrou, M.A.; Taylor, M.I. Genome duplication in amphibians and fish: An extended synthesis. J. Zool. 2011, 284, 151–282. [Google Scholar] [CrossRef]
- Yang, L.; Sado, T.; Hirt, M.; Pasco-Viel, E.; Arunachalam, M.; Li, J.; Wang, X.; Freyhof, J.; Saitoh, K.; Simons, A.M.; et al. Phylogeny and polyploidy: Resolving the classification of cyprinine fishes (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 2015, 85, 97–116. [Google Scholar] [CrossRef]
- Yang, L.; Naylor, G.J.P.; Mayden, R.L. Deciphering reticulate evolution of the largest group of polyploid vertebrates, the subfamily Cyprininae (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 2022, 166, 107323. [Google Scholar] [CrossRef]
- Fricke, R.; Eschmeyer, W.N.; van der Laan, R. (Eds.) Eschmeyer’s Catalog of Fishes: Genera, Species, References, Electronic Version; California Academy of Sciences: San Francisco, CA, USA, 2025; Available online: https://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 15 February 2025).
- Ojima, Y.; Yamamoto, K. Cellular DNA contents of fishes determined by flow cytometry. La Kromosomo II 1990, 57, 1871–1888. [Google Scholar]
- Suzuki, A.; Taki, Y. Tetraplodization in the cobitid subfamily Botinae (Pisces, Cypriniformes). Cytobios 1996, 85, 229–245. [Google Scholar]
- Donsakul, T.; Magtoon, W. Karyotypes of Five Cobitid Fishes (Family Cobitidae): Botia lecontei, B. berdmorei, B. kubotai, Yasuhikotakia sidthimunki and Y. eos in Thailand. In Proceedings of the 48th Kasetsart University Annual Conference: Fisheries, Bangkok, Thailand, 3–5 March 2010; pp. 235–242. [Google Scholar]
- Arai, R. (Ed.) Fish Karyotypes: A Check List; Springer: Tokyo, Japan, 2011. [Google Scholar]
- Kaewmad, P.; Monthatong, M.; Supiwong, W.; Saowakoon, S.; Tanomtong, A. Natural autotetraploid and chromosomal characteristics in the subfamily Botiinae (Cypriniformes, Cobitinae) from Northeast Thailand. Cytologia 2014, 79, 299–313. [Google Scholar] [CrossRef]
- Šlechtová, V.; Bohlen, J.; Freyhof, J.; Ráb, P. Molecular phylogeny of the Southeast Asian freshwater fish family Botiidae (Teleostei: Cobitoidea) and the origin of polyploidy in their evolution. Mol. Phylogenet. Evol. 2006, 39, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Sember, A.; Bohlen, J.; Šlechtová, V.; Altmanová, M.; Pelikánová, Š.; Ráb, P. Dynamics of tandemly repeated DNA sequences during evolution of diploid and tetraploid botiid loaches (Teleostei: Cobitoidea: Botiidae). PLoS ONE 2018, 13, e0195054. [Google Scholar] [CrossRef] [PubMed]
- Bitsikas, V.; Cubizolles, F.; Schier, A.F. A vertebrate family without a functional Hypocretin/Orexin arousal system. Curr. Biol. 2024, 34, 1532–1540. [Google Scholar] [CrossRef]
- Lv, Y.; Li, Y.; Huang, Y.; Wang, J.; Tian, Z.; He, Y.; Shi, J.; Huang, Z.; Wen, Z.; Shi, Q.; et al. Deciphering genome-wide molecular pathways for exogenous Aeromonas hydrophila infection in wide-bodied sand loach (Sinibotia reevesae). Aquac. Rep. 2024, 35, 102033. [Google Scholar] [CrossRef]
- Stebbins, G.L. Variation and Evolution in Plants; Oxford University Press: London, UK, 1950. [Google Scholar]
- Lv, Z.; Nyarko, C.A.; Ramtekey, V.; Behn, H.; Mason, A.S. Defining autopolyploidy: Cytology, genetics, and taxonomy. Am. J. Bot. 2024, 111, e16292. [Google Scholar] [CrossRef]
- Evans, B.J.; Kelley, D.B.; Melnick, D.J.; Cannatella, D.C. Evolution of RAG-1 in polyploid clawed frogs. Mol. Biol. Evol. 2005, 22, 1193–1207. [Google Scholar] [CrossRef][Green Version]
- Gu, H.; Wang, S.; Yang, C.; Tao, M.; Wang, Z.; Liu, S. Global cooling and hot springs may have induced autotetraploidy and autohexaploidy in Schizothorax ancestors, and its implications for polyploid breeding. Aquaculture 2024, 584, 740659. [Google Scholar] [CrossRef]
- Ranallo-Benavidez, T.R.; Jaron, K.S.; Schatz, M.C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 2020, 11, 1432. [Google Scholar] [CrossRef]
- Tang, Q.; Yu, D.; Liu, H. Leptobotia zebra should be revised as Sinibotia zebra (Cypriniformes: Botiidae). Zool. Res. 2008, 29, 1–9. [Google Scholar] [CrossRef]
- Bohlen, J.; Šlechtová, V.; Šlechta, V.; Šlechtová, V.; Sember, A.; Ráb, P. A ploidy difference represents an impassable barrier for hybridisation in animals. Is there an exception among botiid loaches (Teleostei: Botiidae)? PLoS ONE 2016, 11, e0159311. [Google Scholar] [CrossRef]
- Li, C.; Hofreiter, M.; Straube, N.; Corrigan, S.; Naylor, G.J. Capturing protein-coding genes across highly divergent species. BioTechniques 2013, 54, 321–326. [Google Scholar] [CrossRef] [PubMed]
- White, W.T.; Corrigan, S.; Yang, L.; Henderson, A.C.; Bazinet, A.L.; Swofford, D.L.; Naylor, G.J.P. Phylogeny of the manta and devilrays (Chondrichthyes: Mobulidae), with an updated taxonomic arrangement for the family. Zool. J. Linn. Soc. 2018, 182, 50–75. [Google Scholar] [CrossRef]
- Yang, L.; Mayden, R.L.; Naylor, G.J.P. Phylogeny and polyploidy evolution of the suckers (Teleostei: Catostomidae). Biology 2024, 13, 1072. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-J.; Miya, M.; Saitoh, K.; Mayden, R.L. Phylogenetic utility of two existing and four novel nuclear gene loci in reconstructing tree of life of ray- finned fishes: The order Cypriniformes (Ostariophysi) as a case study. Gene 2008, 423, 125–134. [Google Scholar] [CrossRef]
- Lovejoy, N.R.; Collette, B.B. Phylogenetic relationships of New World needlefishes (Teleostei: Belonidae) and the biogeography of transitions between marine and freshwater habitats. Copeia 2001, 2001, 324–338. [Google Scholar] [CrossRef]
- Krueger, F. Trim Galore! 2012. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 1 March 2024).
- Saitoh, K.; Sado, T.; Mayden, R.L.; Hanzawa, N.; Nakamura, K.; Nishida, M.; Miya, M. Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): The first evidence toward resolution of higher-level relationships of the World’s largest freshwater fish clade based on 59 whole mitogenome sequences. J. Mol. Evol. 2006, 63, 826–841. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum Likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods), version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2022. [Google Scholar]
- Marçais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef]
- Saitoh, K.; Sado, T.; Doosey, M.H.; Bart, H.L., Jr.; Inoue, G.; Nishida, M.; Mayden, R.L.; Miya, M. Evidence from mitochondrial genomics supports the lower Mesozoic of South Asia as the time and place of basal divergence of cypriniform fishes (Actinopterygii: Ostariophysi). Zool. J. Linn. Soc. 2011, 161, 633–662. [Google Scholar] [CrossRef]
- Chen, W.-J.; Lavoué, S.; Mayden, R.L. Evolutionary origin and early biogeography of otophysan fishes (Ostariophysi: Teleostei). Evolution 2013, 67, 2218–2239. [Google Scholar] [CrossRef]
- Hirt, M.V.; Arratia, G.; Chen, W.J.; Mayden, R.L.; Tang, K.L.; Wood, R.M.; Simons, A.M. Effects of gene choice, base composition and rate heterogeneity on inference and estimates of divergence times in cypriniform fishes. Biol. J. Linn. Soc. 2017, 121, 319–339. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.R-project.org/ (accessed on 8 May 2025).
- Smith, S.A.; O’Meara, B.C. treePL: Divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 2012, 28, 2689–2690. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Matzke, N.J. Probabilistic historical biogeography: New models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Front. Biogeogr. 2013, 5, 242–248. [Google Scholar] [CrossRef]
- Mason, A.S.; Wendel, J.F. Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution. Front. Genet. 2020, 11, 1014. [Google Scholar] [CrossRef]
- Robertson, F.M.; Gundappa, M.K.; Grammes, F.; Hvidsten, T.R.; Redmond, A.K.; Lien, S.; Martin, S.A.M.; Holland, P.W.H.; Sandve, S.R.; Macqueen, D.J. Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol. 2017, 18, 111. [Google Scholar] [CrossRef]
- Martin, K.J.; Holland, P.W. Enigmatic orthology relationships between Hox clusters of the African butterfly fish and other teleosts following ancient whole-genome duplication. Mol. Biol. Evol. 2014, 31, 2592–2611. [Google Scholar] [CrossRef]
- Macqueen, D.J.; Johnston, I.A. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc. Biol. Sci. 2014, 281, 20132881. [Google Scholar] [CrossRef]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A.; et al. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef]
- Doyle, J.J.; Sherman-Broyles, S. Double trouble: Taxonomy and definitions of polyploidy. New Phytol. 2017, 213, 487–493. [Google Scholar] [CrossRef]
- Du, K.; Stöck, M.; Kneitz, S.; Klopp, C.; Woltering, J.M.; Adolfi, M.C.; Feron, R.; Prokopov, D.; Makunin, A.; Kichigin, I.; et al. The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat. Ecol. Evol. 2020, 4, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, M.; Zoo, M.; Guan, X.; Xu, S.; Chen, W.; Wang, C.; Chen, Y.; He, S.; Guo, B. Recent and recurrent autopolyploidization fueled diversification of snow carp on the Tibetan Plateau. Mol. Biol. Evol. 2024, 41, msae221. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wu, B.; Liu, X.; Hu, Y.; Ming, Y.; Bai, M.; Liu, J.; Xiao, K.; Zeng, Q.; Yang, J.; et al. Whole-genome sequencing reveals autooctoploidy in Chinese sturgeon and its evolutionary trajectories. Genom. Proteom. Bioinform. 2024, 22, qzad002. [Google Scholar] [CrossRef] [PubMed]
- Krabbenhoft, T.J.; MacGuigan, D.J.; Backenstose, N.J.; Waterman, H.; Lan, T.; Pelosi, J.A.; Tan, M.; Sandve, S.R. Chromosome-level genome assembly of Chinese sucker (Myxocyprinus asiaticus) reveals strongly conserved synteny following a catostomid-specific whole-genome duplication. Genome Biol. Evol. 2021, 13, evab190. [Google Scholar] [CrossRef]
- Fontana, F.; Congiu, L.; Mudrak, V.A.; Quattro, J.M.; Smith, T.I.J.; Ware, K.; Doroshov, S.I. Evidence of hexaploid karyotype in shortnose sturgeon. Genome 2008, 51, 113–119. [Google Scholar] [CrossRef]
- Barker, M.S.; Arrigo, N.; Baniaga, A.E.; Li, Z.; Levin, D.A. On the relative abundance of autopolyploids and allopolyploids. New Phytol. 2016, 210, 391–398. [Google Scholar] [CrossRef]
- Spoelhof, J.P.; Soltis, P.S.; Soltis, D.E. Pure polyploidy: Closing the gaps in autopolyploid research. J. Syst. Evol. 2017, 55, 340–352. [Google Scholar] [CrossRef]
- Bagley, J.C.; Mayden, R.L.; Harris, P.M. Phylogeny and divergence times of suckers (Cypriniformes: Catostomidae) inferred from Bayesian total-evidence analyses of molecules, morphology, and fossils. PeerJ 2018, 6, e5168. [Google Scholar] [CrossRef] [PubMed]
- Najman, Y.; Appel, E.; Boudagher-Fadel, M.; Bown, P.; Carter, A.; Garzanti, E.; Godin, L.; Han, J.; Liebke, U.; Oliver, G.; et al. Timing of India- Asia collision: Geological, biostratigraphic, and palaeomagnetic constraints. J. Geophys. Res. 2010, 115, B12416. [Google Scholar] [CrossRef]
- Zachos, J.C.; Dickens, G.R.; Zeebe, R.E. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 2008, 451, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Hall, R. Cenozoic geological and plate tectonic evolution of SE Asia and the SW Pacific: Computer-based reconstructions, model and animations. J. Asian Earth Sci. 2002, 20, 353–434. [Google Scholar] [CrossRef]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Tapponnier, P.; Xu, Z.; Roger, F.; Meyer, B.; Arnaud, N.; Wittlinger, G.; Yang, J. Oblique stepwise rise and growth of the Tibet Plateau. Science 2001, 294, 1671–1677. [Google Scholar] [CrossRef]
- Harrison, T.M.; Copeland, P.; Kidd, W.S.F.; Yin, A. Raising Tibet. Science 1992, 255, 1663–1670. [Google Scholar] [CrossRef]
- Clift, P.D.; Hodges, K.V.; Heslop, D.; Hannigan, R.; Long, H.; Calves, G. Correlated evolution of Himalayan and Tibetan uplift and Asian monsoon intensity. Nat. Geosci. 2008, 1, 875–880. [Google Scholar] [CrossRef]

| Mitogenome | EGR2B | EGR3 | RAG1 | RAG2 | IRBP2 | 7-Nuclear | All-Gene | |
|---|---|---|---|---|---|---|---|---|
| Botiidae | 35 | 14 | 14 | 14 | 13 | 17 | 14 | 14 |
| Outgroup | 85 | 18 | 17 | 18 | 10 | 17 | 18 | 18 |
| Total species | 120 | 32 | 31 | 32 | 23 | 34 | 32 | 32 |
| Total sequences | 120 | 40 | 41 | 32 | 23 | 34 | 32 | 32 |
| Nucleotides (bp) | 14,888 | 828 | 953 | 1497 | 1314 | 864 | 7240 | 22,128 |
| Variable characters (bp) | 8372 | 348 | 393 | 650 | 613 | 446 | 3070 | 10,266 |
| Parsimony-informative characters (bp) | 7188 | 246 | 288 | 513 | 396 | 320 | 2155 | 7909 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Mayden, R.L.; Naylor, G.J.P. Origin of Polyploidy, Phylogenetic Relationships, and Biogeography of Botiid Fishes (Teleostei: Cypriniformes). Biology 2025, 14, 531. https://doi.org/10.3390/biology14050531
Yang L, Mayden RL, Naylor GJP. Origin of Polyploidy, Phylogenetic Relationships, and Biogeography of Botiid Fishes (Teleostei: Cypriniformes). Biology. 2025; 14(5):531. https://doi.org/10.3390/biology14050531
Chicago/Turabian StyleYang, Lei, Richard L. Mayden, and Gavin J. P. Naylor. 2025. "Origin of Polyploidy, Phylogenetic Relationships, and Biogeography of Botiid Fishes (Teleostei: Cypriniformes)" Biology 14, no. 5: 531. https://doi.org/10.3390/biology14050531
APA StyleYang, L., Mayden, R. L., & Naylor, G. J. P. (2025). Origin of Polyploidy, Phylogenetic Relationships, and Biogeography of Botiid Fishes (Teleostei: Cypriniformes). Biology, 14(5), 531. https://doi.org/10.3390/biology14050531






