Analysis of Multitrophic Biodiversity Patterns in the Irtysh River Basin Based on eDNA Metabarcoding
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sample Collection and Processing
2.3. Determination of Environmental Factors
2.4. Selection of Explanatory Variables and Data Sources
2.5. eDNA Metabarcoding Analysis
2.6. Species Composition and -Diversity Analysis
2.7. Impact Analysis of Community Structure
3. Results
3.1. Community Composition
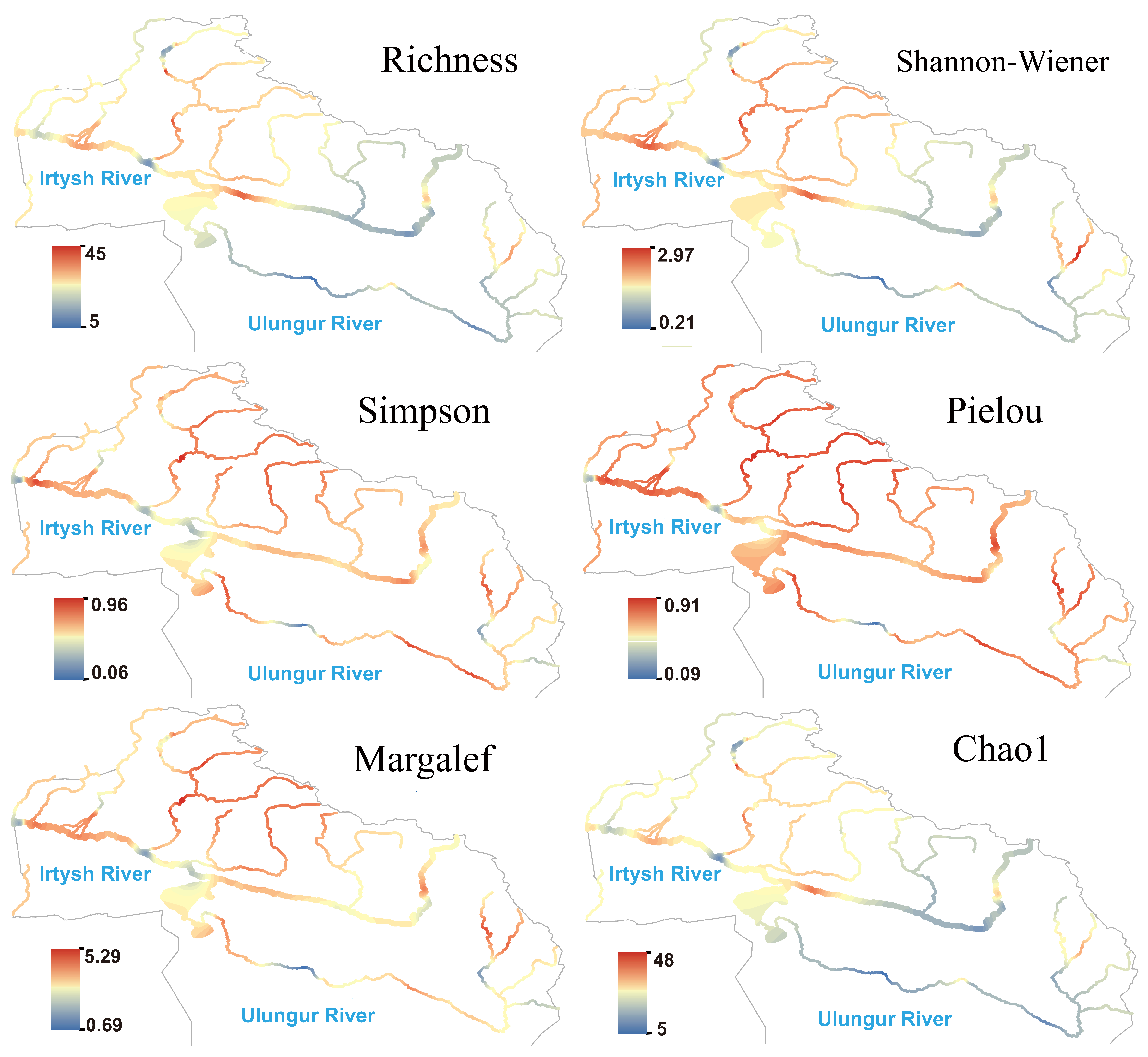
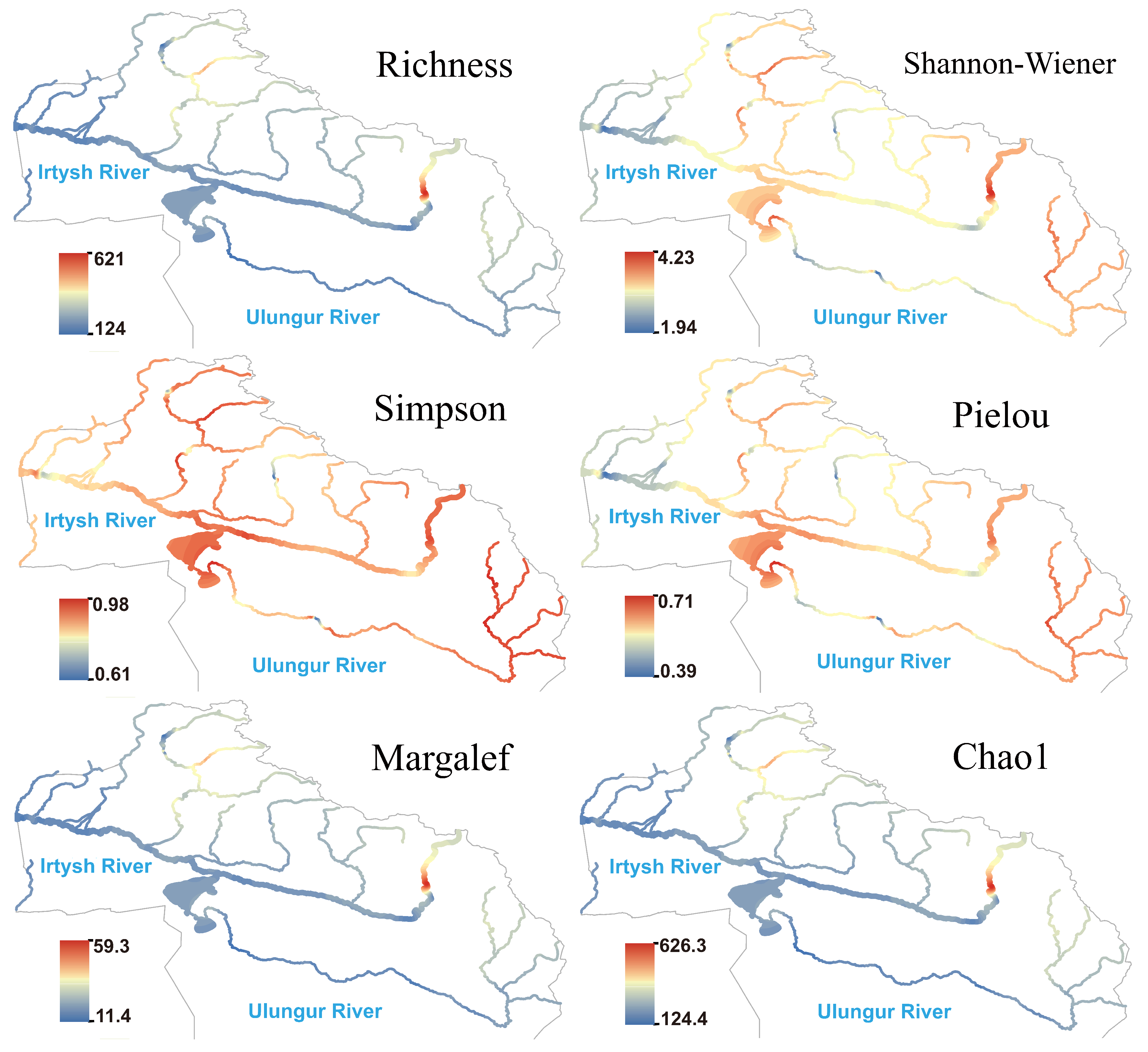
3.2. Patterns of Alpha Diversity
3.3. Contributions of Natural and Anthropogenic Factors to Diversity

3.4. Impacts of Key Environmental Factors on Diversity
3.5. Impacts of Natural and Anthropogenic Factors on Communities
3.5.1. Distance Decay Patterns of Basin Biological Communities
3.5.2. Stressor Affecting Basin Biological Communities
4. Discussion
4.1. Community Composition and Diversity Distribution Patterns
4.2. The Explanation of Climatic and Environmental Factors and Biological Factors on the -Diversity of Plankton
4.3. The Driving Mechanisms of -Diversity Patterns
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomsen, P.F.; Kielgast, J.; Iversen, L.L.; Wiuf, C.; Rasmussen, M.; Gilbert, M.T.P.; Orlando, L.; Willerslev, E. Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 2012, 21, 2565–2573. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Bonin, A.; Zinger, L.; Coissac, E. Environmental DNA: For Biodiversity Research and Monitoring; Oxford University Press: Oxford, UK, 2018. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Eichmiller, J.J.; Miller, L.M.; Sorensen, P.W. Optimizing techniques to capture and extract environmental DNA for detection and quantification of fish. Mol. Ecol. Resour. 2016, 16, 56–68. [Google Scholar] [CrossRef]
- Bae, M.J.; Li, F.; Kwon, Y.S.; Chung, N.; Choi, H.; Hwang, S.J.; Park, Y.S. Concordance of diatom, macroinvertebrate and fish assemblages in streams at nested spatial scales: Implications for ecological integrity. Ecol. Indic. 2014, 47, 89–101. [Google Scholar] [CrossRef]
- Elbrecht, V.; Vamos, E.E.; Meissner, K.; Aroviita, J.; Leese, F. Assessing strengths and weaknesses of DNA metabarcoding-based macroinvertebrate identification for routine stream monitoring. Methods Ecol. Evol. 2017, 8, 1265–1275. [Google Scholar] [CrossRef]
- Huang, X.; Xu, J.; Liu, B.; Guan, X.; Li, J. Assessment of Aquatic Ecosystem Health with Indices of Biotic Integrity (IBIs) in the Ganjiang River System, China. Water 2022, 14, 278. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, P.; Zhang, D.; Chang, J. Evaluation and comparison of the benthic and microbial indices of biotic integrity for urban lakes based on environmental DNA and its management implications. J. Environ. Manag. 2023, 341, 118026. [Google Scholar] [CrossRef]
- Zhu, M.; Yang, N.; Li, Y.; Zhang, W.; Wang, L.; Niu, L.; Wang, L.; Zhang, H. Assessing the effects of cascade dams on river ecological status using multi-species interaction-based index of biotic integrity (Mt-IBI). J. Environ. Manag. 2021, 299, 113585. [Google Scholar] [CrossRef]
- Hu, H.; Wei, X.Y.; Liu, L.; Wang, Y.B.; Jia, H.J.; Bu, L.K.; Pei, D.S. Supervised machine learning improves general applicability of eDNA metabarcoding for reservoir health monitoring. Water Res. 2023, 246, 120686. [Google Scholar] [CrossRef]
- Sims, A.; Zhang, Y.; Gajaraj, S.; Brown, P.B.; Hu, Z. Toward the development of microbial indicators for wetland assessment. Water Res. 2013, 47, 1711–1725. [Google Scholar] [CrossRef]
- David, G.M.; López-García, P.; Moreira, D.; Alric, B.; Deschamps, P.; Bertolino, P.; Restoux, G.; Rochelle-Newall, E.; Thébault, E.; Simon, M.; et al. Small freshwater ecosystems with dissimilar microbial communities exhibit similar temporal patterns. Mol. Ecol. 2021, 30, 2162–2177. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, W.; Li, X.; Lu, W.; Li, J. Strong linkages between dissolved organic matter and the aquatic bacterial community in an urban river. Water Res. 2020, 184, 116089. [Google Scholar] [CrossRef]
- Li, Y.; Wu, H.; Shen, Y.; Wang, C.; Wang, P.; Zhang, W.; Gao, Y.; Niu, L. Statistical determination of crucial taxa indicative of pollution gradients in sediments of Lake Taihu, China. Environ. Pollut. 2019, 246, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Prygiel, E.; Laplace-Treyture, C. Performance of a multimetric index based on phytoplankton to evaluate the ecological quality of French large rivers: The IPHYGE index. Ecol. Indic. 2024, 166, 112303. [Google Scholar] [CrossRef]
- Fontaine, L.; Pin, L.; Savio, D.; Friberg, N.; Kirschner, A.K.T.; Farnleitner, A.H.; Eiler, A. Bacterial bioindicators enable biological status classification along the continental Danube river. Commun. Biol. 2023, 6, 862. [Google Scholar] [CrossRef] [PubMed]
- Tamames, J.; Abellan, J.J.; Pignatelli, M.; Camacho, A.; Moya, A. Environmental distribution of prokaryotic taxa. BMC Microbiol. 2010, 10, 85. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, W.; Zhu, D.; Huang, Q.; Wu, L.; Liu, X. Determining Critical Thresholds of Environmental Flow Restoration Based on Planktonic Index of Biotic Integrity (PIBI): A Case Study in the Typical Tributaries of Poyang Lake. Int. J. Environ. Res. Public Health 2023, 20, 169. [Google Scholar] [CrossRef]
- Zhao, K.; Wang, L.; You, Q.; Zhang, J.; Pang, W.; Wang, Q. Impact of cyanobacterial bloom intensity on plankton ecosystem functioning measured by eukaryotic phytoplankton and zooplankton indicators. Ecol. Indic. 2022, 140, 109028. [Google Scholar] [CrossRef]
- Yang, J.R.; Lv, H.; Isabwe, A.; Liu, L.; Yu, X.; Chen, H.; Yang, J. Disturbance-induced phytoplankton regime shifts and recovery of cyanobacteria dominance in two subtropical reservoirs. Water Res. 2017, 120, 52–63. [Google Scholar] [CrossRef]
- Moyle, P.B.; Leidy, R.A. Loss of Biodiversity in Aquatic Ecosystems: Evidence from Fish Faunas. In Conservation Biology; Springer: Boston, MA, USA, 1992; pp. 127–169. [Google Scholar] [CrossRef]
- Yang, Q.; Weigelt, P.; Fristoe, T.S.; Zhang, Z.; Kreft, H.; Stein, A.; Seebens, H.; Dawson, W.; Essl, F.; König, C.; et al. The global loss of floristic uniqueness. Nat. Commun. 2021, 12, 7290. [Google Scholar] [CrossRef]
- Li, X.; Jia, P.; Niu, C.; Xing, Y.; Li, H.; Liu, H.; Tang, W.; Zhao, Y. Species diversity of freshwater fish and assessment on watershed health in the Irtysh River and Ulungur River basins in Xinjiang, China. Biodivers. Sci. 2020, 28, 422–434. [Google Scholar] [CrossRef]
- Liu, J. Analysis on the water amount flowing into Ulungur Lake. Energy Energy Conserv. 2015, 5, 103–105. [Google Scholar]
- Goldberg, C.S.; Turner, C.R.; Deiner, K.; Klymus, K.E.; Thomsen, P.F.; Murphy, M.A.; Spear, S.F.; McKee, A.; Oyler-McCance, S.J.; Cornman, R.S.; et al. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 2016, 7, 1299–1307. [Google Scholar] [CrossRef]
- Li, H.; Zhang, S.; Zou, K.; Chen, Z.; Chen, X.; Jiang, P.; Cao, Y.; Li, M. Establishment and optimization of environmental DNA extraction method from water of Pearl River Estuary. South China Fish. Sci. 2022, 18, 30–37. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; McCliment, E.A.; Ducklow, H.W.; Huse, S.M. A Method for Studying Protistan Diversity Using Massively Parallel Sequencing of V9 Hypervariable Regions of Small-Subunit Ribosomal RNA Genes. PLoS ONE 2009, 4, e6372. [Google Scholar] [CrossRef]
- Ministry of Environmental Protection of the People’s Republic of China; General Administration of Quality Supervision and of the People’s Republic of China. Environmental Quality Standards for Surface Water. 2002. Available online: https://www.mee.gov.cn/ywgz/fgbz/bz/bzwb/shjbh/shjzlbz/200206/t2002060166497.shtml (accessed on 1 July 2025).
- HJ 828-2017; Water Quality—Determination of the Chemical Oxygen Demand-Dichromate Method. Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2017.
- HJ 636-2012; Water Quality—Determination of Total Nitrogen-Alkaline Potassium Persulfate Digestion UV Spectrophotometric Method. Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2012.
- HJ 535-2009; Water Quality—Determination of Ammonia Nitrogen―Nessler’s Reagent Spectrophotometry. Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2009.
- HJ/T 84-2016; Determination of Water Inorganic Anions (F-, Cl-, NO2-, Br-, NO3-, PO43-, SO32-, SO42-)—Ion Chromatography Method. Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2016.
- HJ 670-2013; Water Quality—Determination of Orthophosphate and Total Phosphorus-Continuous Flow Analysis (CFA) and Ammonium Molybdate Spectrophotometry. Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2013.
- Filipe, A.F.; Markovic, D.; Pletterbauer, F.; Tisseuil, C.; De Wever, A.; Schmutz, S.; Bonada, N.; Freyhof, J. Forecasting fish distribution along stream networks: Brown trout (Salmo trutta) in Europe. Divers. Distrib. 2013, 19, 1059–1071. [Google Scholar] [CrossRef]
- Friedrichs-Manthey, M.; Langhans, S.D.; Hein, T.; Borgwardt, F.; Kling, H.; Jähnig, S.C.; Domisch, S. From topography to hydrology—The modifiable area unit problem impacts freshwater species distribution models. Ecol. Evol. 2020, 10, 2956–2968. [Google Scholar] [CrossRef]
- McRae, S.E.; Allan, J.D.; Burch, J.B. Reach- and catchment-scale determinants of the distribution of freshwater mussels (Bivalvia: Unionidae) in south-eastern Michigan, U.S.A. Freshw. Biol. 2004, 49, 127–142. [Google Scholar] [CrossRef]
- Amoatey, P.; Baawain, M.S. Effects of pollution on freshwater aquatic organisms. Water Environ. Res. 2019, 91, 1272–1287. [Google Scholar] [CrossRef]
- Domisch, S.; Amatulli, G.; Jetz, W. Near-global freshwater-specific environmental variables for biodiversity analyses in 1 km resolution. Sci. Data 2015, 2, 150073. [Google Scholar] [CrossRef]
- Venter, O.; Sanderson, E.W.; Magrach, A.; Allan, J.R.; Beher, J.; Jones, K.R.; Possingham, H.P.; Laurance, W.F.; Wood, P.; Fekete, B.M.; et al. Sixteen years of change in the global terrestrial human footprint and implications for biodiversity conservation. Nat. Commun. 2016, 7, 12558. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Gilmore, S.R.; Sutherland, T.F.; Hajibabaei, M.; Miller, K.M.; Westfall, K.M.; Pawlowski, J.; Abbott, C.L. Biotic signals associated with benthic impacts of salmon farms from eDNA metabarcoding of sediments. Mol. Ecol. 2021, 30, 3158–3174. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Miya, M.; Fukunaga, T.; Sado, T.; Iwasaki, W. MitoFish and MiFish Pipeline: A Mitochondrial Genome Database of Fish with an Analysis Pipeline for Environmental DNA Metabarcoding. Mol. Biol. Evol. 2018, 35, 1553–1555. [Google Scholar] [CrossRef]
- Jeunen, G.J.; Dowle, E.; Edgecombe, J.; von Ammon, U.; Gemmell, N.J.; Cross, H. crabs—A software program to generate curated reference databases for metabarcoding sequencing data. Mol. Ecol. Resour. 2023, 23, 725–738. [Google Scholar] [CrossRef]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.M.; et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 2024, 42, 715–718. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Mingguang, Z.; Guangpeng, F.; Haihua, W.; Jianhua, C.; Chenchen, S.; Yanping, Z.; Haixin, Z.; Yilong, F.; Yuan, Y.; Weikang, X. Dietary analysis of ferocious fishes in Lake Poyang based on stomach content analysis and eDNA Metabarcoding technology. J. Lake Sci. 2025, 37, 532. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Pielou, E.C. Species Diversity and Pattern Diversity in the Study of Ecological Succession. J. Theor. Biol. 1966, 10, 370–383. [Google Scholar] [CrossRef]
- Chao, A. Non-parametric Estimation of the Number of Classes in a Population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Margalef, R. Information Theory in Ecology. Gen. Syst. 1958, 3, 36–71. [Google Scholar]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package. 2025. Available online: https://github.com/vegandevs/vegan (accessed on 4 July 2025).
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- Ziemann, M.; Stirling, M.; Pilling, L. Forestmodel: Forest Plot of Regression Models; CRAN: Vienna, Austria, 2024; Available online: https://cran.r-project.org/web/packages/forestmodel/refman/forestmodel.html (accessed on 1 June 2025).
- Burnham, K.P.; Anderson, D.R. Model selection and multimodel inference: A practical information-theoretic approach. J. Wildl. Manag. 2003, 67, 655. [Google Scholar]
- Nakagawa, S.; Schielzeth, H. A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 2013, 4, 133–142. [Google Scholar] [CrossRef]
- Brooks, M.; Kristensen, K.; Maechler, M.; Bolker, B.M.; Bruun, K.; Skaug, H.J.; Magnusson, A. glmmTMB: Generalized Linear Mixed Models using Template Model Builder, CRAN: Vienna, Austria, 2024. Available online: https://cran.r-project.org/web/packages/glmmTMB/index.html (accessed on 10 June 2025).
- Bartoń, K. MuMIn: Multi-Model Inference, CRAN: Vienna, Austria, 2024. Available online: https://cran.r-project.org/web/packages/MuMIn/index.html (accessed on 1 August 2025).
- Heino, J.; Tolonen, K.T. Ecological drivers of multiple facets of beta diversity in a lentic macroinvertebrate metacommunity. Limnol. Oceanogr. 2017, 62, 2431–2444. [Google Scholar] [CrossRef]
- Petitpierre, B.; Broennimann, O.; Kueffer, C.; Daehler, C.; Guisan, A. Selecting predictors to maximize the transferability of species distribution models: Lessons from cross-continental plant invasions. Glob. Ecol. Biogeogr. 2017, 26, 275–287. [Google Scholar] [CrossRef]
- Liu, C.; Comte, L.; Xian, W.; Chen, Y.; Olden, J.D. Current and projected future risks of freshwater fish invasions in China. Ecography 2019, 42, 2074–2083. [Google Scholar] [CrossRef]
- Krupa, E.; Romanova, S.; Serikova, A.; Shakhvorostova, L. A Comprehensive Assessment of the Ecological State of the Transboundary Irtysh River (Kazakhstan, Central Asia). Water 2024, 16, 973. [Google Scholar] [CrossRef]
- Kłosiński, P.; Kobak, J.; Kakareko, T. Competitive interactions for food resources between invasive Ponto-Caspian gobies and their native competitors in the context of global warming. NeoBiota 2025, 97, 91–119. [Google Scholar] [CrossRef]
- Thompson, R.M.; Dunne, J.A.; Woodward, G. Freshwater food webs: Towards a more fundamental understanding of biodiversity and community dynamics. Freshw. Biol. 2012, 57, 1329–1341. [Google Scholar] [CrossRef]
- Šimek, K.; Grujčić, V.; Mukherjee, I.; Kasalický, V.; Nedoma, J.; Posch, T.; Mehrshad, M.; Salcher, M.M. Cascading effects in freshwater microbial food webs by predatory Cercozoa, Katablepharidacea and ciliates feeding on aplastidic bacterivorous cryptophytes. FEMS Microbiol. Ecol. 2020, 96, fiaa121. [Google Scholar] [CrossRef] [PubMed]
- Pierangelini, M.; Ryšánek, D.; Lang, I.; Adlassnig, W.; Holzinger, A. Terrestrial adaptation of green algae Klebsormidium and Zygnema (Charophyta) involves diversity in photosynthetic traits but not in CO2 acquisition. Planta 2017, 246, 971–986. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, B.; Zhang, Y.; Shen, Y.; Cheng, C.; Yuan, W.; Guo, P. Organic Matter Decomposition in River Ecosystems: The Role of Microbial Interactions Regulated by Environmental Factors. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Saleem, A.; Anwar, S.; Saud, S.; Kamal, T.; Fahad, S.; Nawaz, T. Cyanobacteria diversity and ecological roles: Insights into cyanobacterial adaptations and environmental implications. J. Umm-Qura Univ. Appl. Sci. 2025. [Google Scholar] [CrossRef]
- Sala, M.M.; Steen, A.D.; Herndl, G.J.; Ortega-Retuerta, E.; Osterholz, H.; Niggemann, J. Microbe-Organic Matter Interactions in Aquatic Systems: Advances and Challenges; American Society of Limnology and Oceanography: Waco, TX, USA, 2021. [Google Scholar]
- Gurung, J.K. Review on Fish Diversity and Habitat Relationship with Environmental Variables. Damak Campus J. 2023, 12, 17–23. [Google Scholar] [CrossRef]
- Welbara, D.E.; Gebre-Meskel, D.K.; Hailu, T.F. Morpho-functional traits of phytoplankton functional groups: A review. Biologia 2024, 79, 1983–1998. [Google Scholar] [CrossRef]
- Tsui, C.K.M.; Baschien, C.; Goh, T.K. Biology and Ecology of Freshwater Fungi. In Fungal Biology; Springer: Cham, Switzerland, 2016; pp. 285–313. [Google Scholar] [CrossRef]
- Burnap, R.L. Systems and Photosystems: Cellular Limits of Autotrophic Productivity in Cyanobacteria. Front. Bioeng. Biotechnol. 2015, 3. [Google Scholar] [CrossRef]
- Gallardo, B.; Clavero, M.; Sánchez, M.I.; Vilà, M. Global ecological impacts of invasive species in aquatic ecosystems. Glob. Change Biol. 2015, 22, 151–163. [Google Scholar] [CrossRef]
- Liang, Z.; Gozlan, R.E.; Liu, J.; Jackson, D.; Guo, C. Invasive Fish Reshape Biodiversity Patterns in China’s Freshwater Lakes. Glob. Change Biol. 2025, 31, e70267. [Google Scholar] [CrossRef]
- Zi, F.; Song, T.; Liu, J.; Wang, H.; Serekbol, G.; Yang, L.; Hu, L.; Huo, Q.; Song, Y.; Huo, B.; et al. Environmental and Climatic Drivers of Phytoplankton Communities in Central Asia. Biology 2024, 13, 717. [Google Scholar] [CrossRef]
- Chang, C.; Ren, M.; Wang, H.; Ye, S.; Tang, X.; He, D.; Hu, E.; Li, M.; Pan, B. Riverine network size determined major driving factors of the composition and diversity of aquatic invertebrate communities in a multi-tributary mountain river basin. Water Res. 2025, 276, 123257. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Zi, F.; Huang, Y.; Fang, L.; Zhang, Y.; Liu, Y.; Chang, J.; Li, J. Assessment of Aquatic Ecosystem Health in the Irtysh River Basin Using eDNA Metabarcoding. Water 2025, 17, 246. [Google Scholar] [CrossRef]
- Kazbar, A.; Cogne, G.; Urbain, B.; Marec, H.; Le-Gouic, B.; Tallec, J.; Takache, H.; Ismail, A.; Pruvost, J. Effect of dissolved oxygen concentration on microalgal culture in photobioreactors. Algal Res. 2019, 39, 101432. [Google Scholar] [CrossRef]
- Mai, S.; He, Y.; Li, W.; Zhao, T. Effects of environmental factors on vertical distribution of the eukaryotic plankton community in early summer in Danjiangkou Reservoir, China. Front. Ecol. Evol. 2023, 11, 1324932. [Google Scholar] [CrossRef]
- Jiang, Y.J.; He, W.; Liu, W.X.; Qin, N.; Ouyang, H.L.; Wang, Q.M.; Kong, X.Z.; He, Q.S.; Yang, C.; Yang, B.; et al. The seasonal and spatial variations of phytoplankton community and their correlation with environmental factors in a large eutrophic Chinese lake (Lake Chaohu). Ecol. Indic. 2014, 40, 58–67. [Google Scholar] [CrossRef]
- Hao, Z.; Wang, Y.; Chen, E.; Mu, X.; Li, J.; La, Q.; De, J.; Liu, Y.; Huang, S.; Fang, W.; et al. Climate and biological factors jointly shape microbial community structure in the Yarlung Zangbo River during the dry season. Sci. Total Environ. 2025, 969, 178930. [Google Scholar] [CrossRef]
- Cruaud, P.; Vigneron, A.; Fradette, M.S.; Dorea, C.C.; Culley, A.I.; Rodriguez, M.J.; Charette, S.J. Annual Protist Community Dynamics in a Freshwater Ecosystem Undergoing Contrasted Climatic Conditions: The Saint-Charles River (Canada). Front. Microbiol. 2019, 10, 02359. [Google Scholar] [CrossRef]
- Duarte, S.; Fernandes, I.; Nogueira, M.J.; Cássio, F.; Pascoal, C. Temperature alters interspecific relationships among aquatic fungi. Fungal Ecol. 2013, 6, 187–191. [Google Scholar] [CrossRef]
- Vasconcelos Rissi, D.; Ijaz, M.; Baschien, C. Comparative genome analysis of the freshwater fungus Filosporella fistucella indicates potential for plant-litter degradation at cold temperatures. G3 Genes|Genomes|Genet. 2023, 13, jkad190. [Google Scholar] [CrossRef] [PubMed]
- Fenoy, E.; Pradhan, A.; Pascoal, C.; Rubio-Ríos, J.; Batista, D.; Moyano-López, F.J.; Cássio, F.; Casas, J.J. Elevated temperature may reduce functional but not taxonomic diversity of fungal assemblages on decomposing leaf litter in streams. Glob. Change Biol. 2022, 28, 115–127. [Google Scholar] [CrossRef]
- Van Beek, L.P.H.; Eikelboom, T.; van Vliet, M.T.H.; Bierkens, M.F.P. A physically based model of global freshwater surface temperature. Water Resour. Res. 2012, 48. [Google Scholar] [CrossRef]
- Abdel-raheem, A.; El-Maghraby, O.; Abd-Elaah, G.; Bakhit, M. Interactions of physico-chemical properties of the River Nile water and fungal diversity in River Nile streams in Delta region. J. Environ. Stud. 2018, 18, 33–45. [Google Scholar] [CrossRef]
- Hyde, K.D.; Fryar, S.; Tian, Q.; Bahkali, A.H.; Xu, J. Lignicolous freshwater fungi along a north–south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol. 2016, 19, 190–200. [Google Scholar] [CrossRef]
- Zhang, R.; Wan, N.; Yang, Y.; Ran, Y. Long-term changes of phytoplankton productivity in freshwater ecosystems of the Pearl River Delta as recorded by organic geochemical proxies in sediment cores. Appl. Geochem. 2023, 159, 105850. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, X.; Wu, Y.; Zhang, G.; Dai, C.; Qiao, G.; Ma, Y. A Review on the Driving Mechanism of the Spring Algal Bloom in Lakes Using Freezing and Thawing Processes. Water 2024, 16, 257. [Google Scholar] [CrossRef]
- Sangma, C.B.K.; Sultana, S. Algae as Indicators of Climate Change. In Climate Change and Microbes; Apple Academic Press: Point Pleasant, NJ, USA, 2022. [Google Scholar]
- Biggs, J.; Everingham, Y.; Skocaj, D.; Schroeder, B.; Sexton, J.; Thorburn, P. The potential for refining nitrogen fertiliser management through accounting for climate impacts: An exploratory study for the Tully region. Mar. Pollut. Bull. 2021, 170, 112664. [Google Scholar] [CrossRef]
- Jia, J.; Xia, M.; Zhang, Y.; Tian, S.; Hu, Y.; Zhang, Z.; Zhai, X.; Qu, B.; Hao, L. Effects of Freshwater Restoration on Phytoplankton and Zooplankton Communities in the Yellow River Delta. Water 2025, 17, 2348. [Google Scholar] [CrossRef]
- Kouzuma, A.; Watanabe, K. Exploring the potential of algae/bacteria interactions. Curr. Opin. Biotechnol. 2015, 33, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Mieczan, T.; Płaska, W.; Adamczuk, M.; Toporowska, M.; Bartkowska, A. Effects of the Invasive Fish Species Ameiurus nebulosus on Microbial Communities in Peat Pools. Water 2022, 14, 815. [Google Scholar] [CrossRef]
- Hoque, M.M.; Espinoza-Vergara, G.; McDougald, D. Protozoan predation as a driver of diversity and virulence in bacterial biofilms. FEMS Microbiol. Rev. 2023, 47, fuad040. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Silin, M.; Wenli, C.; Qiaoyun, H.; Yongguan, Z.; Xiuli, H. Review of Protozoa-Pathogen Interactions and Soil Health. J.-Agro-Environ. Sci. 2023, 42, 481–489. [Google Scholar]
- Song, C.; Murata, K.; Suzaki, T. Intracellular symbiosis of algae with possible involvement of mitochondrial dynamics. Sci. Rep. 2017, 7, 1221. [Google Scholar] [CrossRef]
- Ramanan, R.; Kim, B.H.; Cho, D.H.; Oh, H.M.; Kim, H.S. Algae–bacteria interactions: Evolution, ecology and emerging applications. Biotechnol. Adv. 2016, 34, 14–29. [Google Scholar] [CrossRef]
- Xuejin, F.; Dahui, L.; Wei, Y.; Rui, Z.; Nianzhi, J. Interactions Between Marine Cyanobacteria and Heterotrophic Bacteria: A Case Study of Prochlorococcus. Chin. Sci. Bull. 2021, 66, 3839–3848. [Google Scholar] [CrossRef]
- Pintar, M.R.; Resetarits, W.J., Jr. Prey-driven control of predator assemblages: Zooplankton abundance drives aquatic beetle colonization. Ecology 2017, 98, 2201–2215. [Google Scholar] [CrossRef]
- Escalas, A.; Avouac, A.; Belmaker, J.; Bouvier, T.; Clédassou, V.; Ferraton, F.; Rieuvilleneuve, F.; Rilov, G.; Rovirosa Mulet, A.; Shapiro Goldberg, D.; et al. An invasive herbivorous fish (Siganus rivulatus) influences both benthic and planktonic microbes through defecation and nutrient excretion. Sci. Total Environ. 2022, 838, 156207. [Google Scholar] [CrossRef]
- Bui, N.M.N.; Heyse, J.; Delamare-Deboutteville, J.; Defoirdt, T.; Props, R.; Shelley, C. Bacterial and microalgal communities in carp polyculture systems: Composition, affecting factors and further perspectives. Aquaculture 2024, 582, 740505. [Google Scholar] [CrossRef]
- Zeglin, L.H. Stream microbial diversity in response to environmental changes: Review and synthesis of existing research. Front. Microbiol. 2015, 6, 454. [Google Scholar] [CrossRef]
- Sun, H.; Pan, B.; He, H.; Zhao, G.; Hou, Y.; Zhu, P. Assembly processes and co-occurrence relationships in the bacterioplankton communities of a large river system. Ecol. Indic. 2021, 126, 107643. [Google Scholar] [CrossRef]
- Xiaohui, Z. Algal Community Structure and Its Driving Mechanisms in Rivers on the Qinghai-Tibet Plateau. Ph.D. Thesis, Xi’an University of Technology, Xi’an, China, 2024. [Google Scholar]
- Yang, T.; Meng, W.; Zhang, R.; Gao, T.; Cai, L.; Hai, S.; Zhou, Q. DNA barcoding of fishes in Irtysh River China. Russ. J. Genet. 2016, 52, 969–976. [Google Scholar] [CrossRef]
- Voigt, W.; Perner, J.; Davis, A.J.; Eggers, T.; Schumacher, J.; Bährmann, R.; Fabian, B.; Heinrich, W.; Köhler, G.; Lichter, D.; et al. Trophic levels are differentially sensitive to climate. Ecology 2003, 84, 2444–2453. [Google Scholar] [CrossRef]
- Rezende, F.; Antiqueira, P.A.P.; Petchey, O.L.; Velho, L.F.M.; Rodrigues, L.C.; Romero, G.Q. Trophic downgrading decreases species asynchrony and community stability regardless of climate warming. Ecol. Lett. 2021, 24, 2660–2673. [Google Scholar] [CrossRef]
- Liow, L.H.; Fortelius, M.; Bingham, E.; Lintulaakso, K.; Mannila, H.; Flynn, L.; Stenseth, N.C. Higher origination and extinction rates in larger mammals. Proc. Natl. Acad. Sci. USA 2008, 105, 6097–6102. [Google Scholar] [CrossRef]












Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Song, T.; Zhang, Y.; Zi, F.; Huang, Y.; Fang, L.; Liu, Y.; Zhou, H.; Chang, J. Analysis of Multitrophic Biodiversity Patterns in the Irtysh River Basin Based on eDNA Metabarcoding. Biology 2025, 14, 1661. https://doi.org/10.3390/biology14121661
Chen Y, Song T, Zhang Y, Zi F, Huang Y, Fang L, Liu Y, Zhou H, Chang J. Analysis of Multitrophic Biodiversity Patterns in the Irtysh River Basin Based on eDNA Metabarcoding. Biology. 2025; 14(12):1661. https://doi.org/10.3390/biology14121661
Chicago/Turabian StyleChen, Ye, Tianjian Song, Yuna Zhang, Fangze Zi, Yuxin Huang, Lei Fang, Yu Liu, Hongyang Zhou, and Jiang Chang. 2025. "Analysis of Multitrophic Biodiversity Patterns in the Irtysh River Basin Based on eDNA Metabarcoding" Biology 14, no. 12: 1661. https://doi.org/10.3390/biology14121661
APA StyleChen, Y., Song, T., Zhang, Y., Zi, F., Huang, Y., Fang, L., Liu, Y., Zhou, H., & Chang, J. (2025). Analysis of Multitrophic Biodiversity Patterns in the Irtysh River Basin Based on eDNA Metabarcoding. Biology, 14(12), 1661. https://doi.org/10.3390/biology14121661





