Study on the Genomic Basis of Adaptation in Salsk Sheep
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Genotyping
2.2. Quality Control and Data Filtering
2.3. Population Structure and Genetic Diversity Analysis
2.4. Detection of Positive Selection Signatures
2.5. Annotation of Candidate Genes
3. Results and Discussion
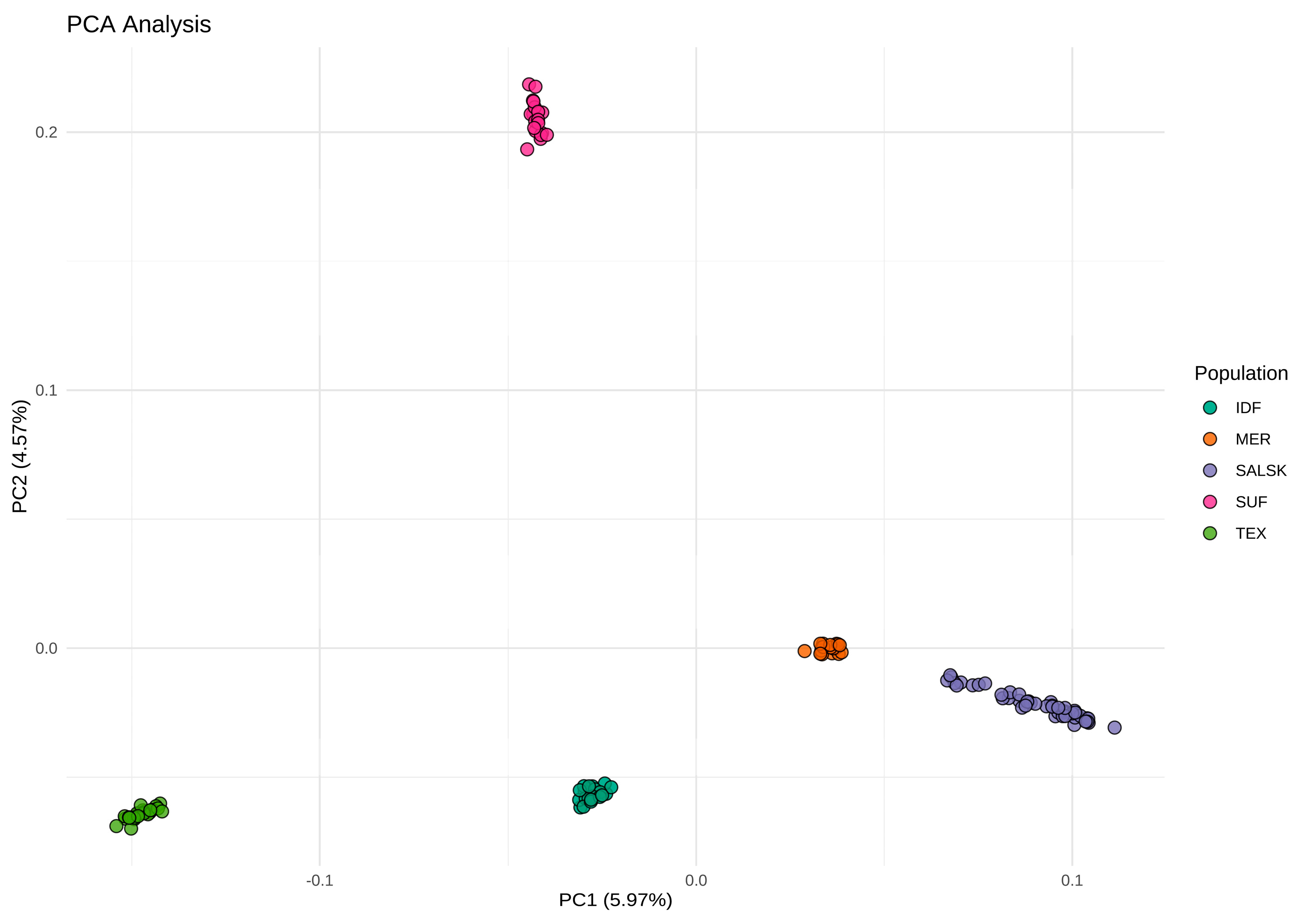
3.1. Principal Component Analysis (PCA)
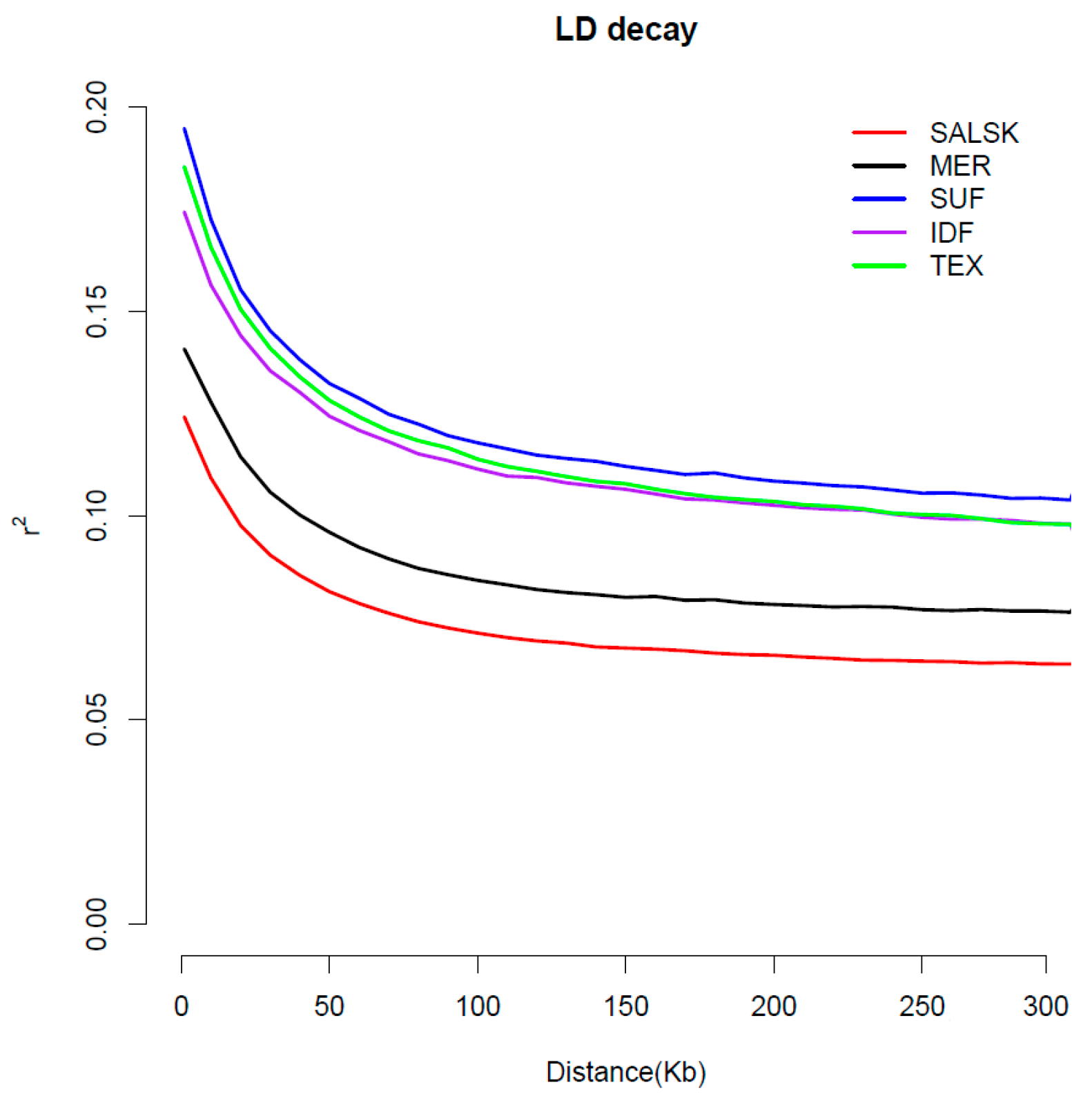
3.2. Linkage Disequilibrium (LD) Analysis
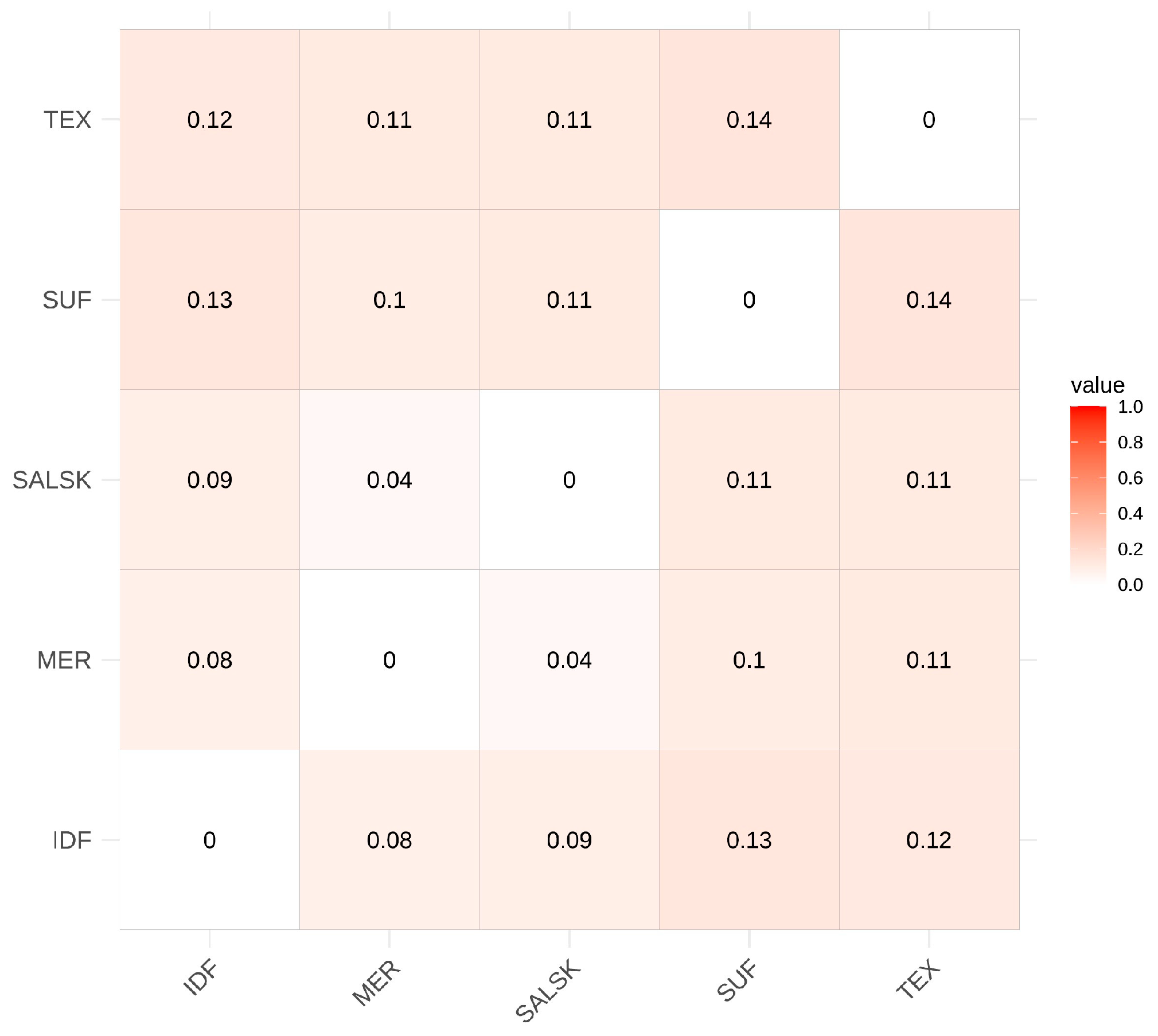
3.3. Genetic Differentiation Between Studied Sheep Breeds (Fst)
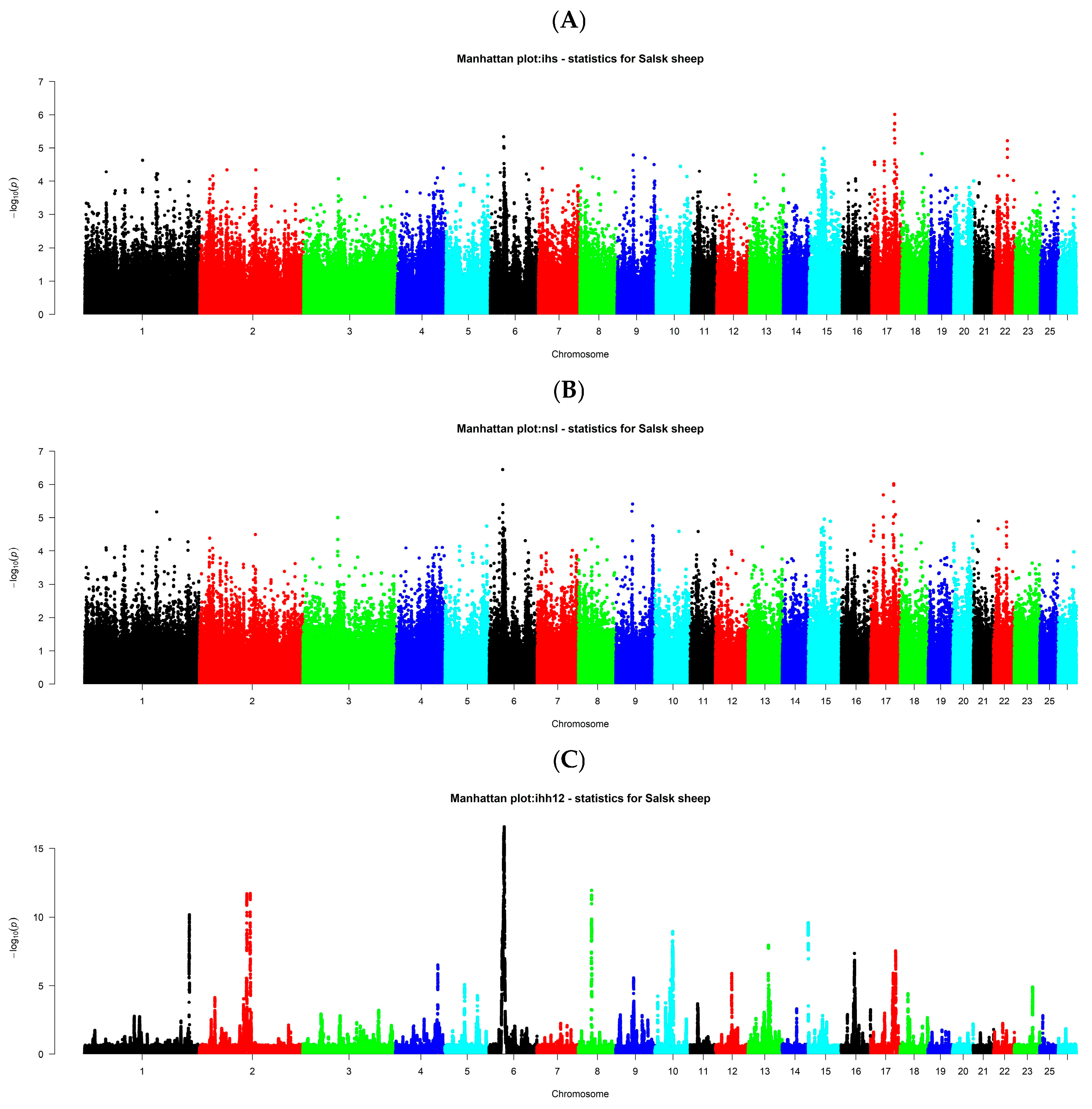
3.4. Intra-Population Selection Signatures (iHS, nSL, iHH12)
3.5. Interpopulation Signatures (XP-EHH)
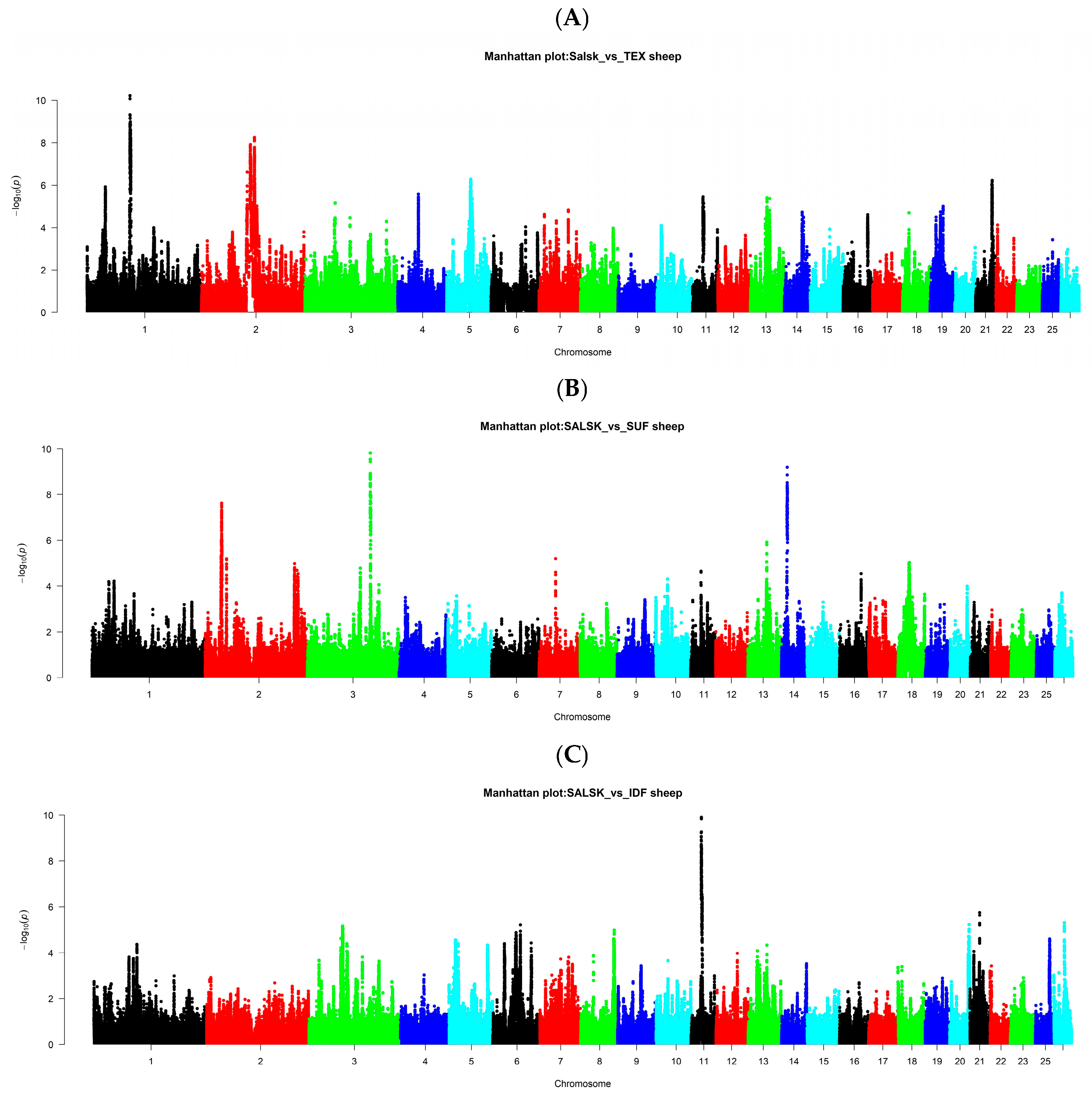
3.6. Salsk vs. Texel
3.7. Salsk vs. Suffolk
3.8. Salsk vs. Île-de-France
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Federal State Budgetary Scientific Institution. All-Russian Research Institute for Animal Breeding. In Sheep and Goat Breeding Yearbook 2025; Federal State Budgetary Scientific Institution: Moscow, Russia, 2025; ISBN 978-5-87958-450-6. Available online: https://vniiplem.ru/l/gisc/sost_plemres/ (accessed on 10 August 2025).
- Kolosov, Y.A.; Aboneev, V.V.; Kulikova, A.Y.; Kolosova, N.N.; Aboneeva, E.V. The influence of selection intensity and selection differential on live weight and wool yield of Salsk sheep Sheep. Goats Wool Bus. 2023, 2, 3–7. [Google Scholar] [CrossRef]
- Chamurliev, N.G.; Kolosov, Y.A.; Degtyar, A.S.; Romanets, T.S. Some biological characteristics of sheep of different origins. News Low. Volga Agrar. Univ. Complex Sci. High. Prof. Educ. 2022, 2, 247–253. [Google Scholar]
- Kolosov, Y.A.; Aboneev, V.V.; Gagloev, A.C.; Lagoda, A.A. Inheritance of the main traits of wool productivity in merino sheep of different origins Sheep. Goats Wool Bus. 2023, 1, 33–36. [Google Scholar] [CrossRef]
- Panigrahi, M.; Rajawat, D.; Nayak, S.S.; Ghildiyal, K.; Sharma, A.; Jain, K.; Lei, C.; Bhushan, B.; Mishra, B.P.; Dutt, T. Landmarks in the history of selective sweeps. Anim. Gen. 2023, 54, 667–688. [Google Scholar] [CrossRef]
- Huber, M.; van Vliet, M.; Giezenberg, M.; Winkens, B.; Heerkens, Y.; Dagnelie, P.C.; Knottnerus, J.A. Towards a ‘patient-centred’ operationalisation of the new dynamic concept of health: A mixed methods study. BMJ Open 2016, 6, e010091. [Google Scholar] [CrossRef]
- Yi, X.; Liang, Y.; Huerta-Sanchez, E.; Jin, X.; Cuo, Z.X.; Pool, J.E.; Xu, X.; Jiang, H.; Vinckenbosch, N.; Korneliussen, T.S.; et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 2010, 329, 75–78. [Google Scholar] [CrossRef]
- Sabeti, P.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; McCarroll, S.A.; Gaudet, R.; et al. Genome-wide detection and characterization of positive selection in human populations. Nature 2007, 449, 913–918. [Google Scholar] [CrossRef]
- Abondio, P.; Cilli, E.; Luiselli, D. Inferring Signatures of Positive Selection in Whole-Genome Sequencing Data: An Overview of Haplotype-Based Methods. Genes 2022, 13, 926. [Google Scholar] [CrossRef]
- Barton, N.H. Genetic hitchhiking. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1553–1562. [Google Scholar] [CrossRef]
- Sabeti, P.; Reich, D.; Higgins, J.; Levine, H.; Richter, D.; Schaffner, S.; Gabriel, S.; Platko, J.; Patterson, N.; McDonald, G.; et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 2002, 419, 832–837. [Google Scholar] [CrossRef]
- Zorc, M.; Horvat, T.; Tanšek, A.; Ferme, T.; Dovč, P. Selection Signatures Reveal Candidate Genes for the Cornish Rex Breed-Specific Phenotype. Genes 2024, 15, 368. [Google Scholar] [CrossRef]
- Lukic, B.; Curik, I.; Drzaic, I.; Ferenčaković, M.; Shihabi, A.; Cubric-Curik, V. Genomic signatures of selection, local adaptation and production type characterisation of East Adriatic sheep breeds. J. Anim. Sci. Biotechnol. 2023, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Ardestani, S.; Aminafshar, M.; Zandi Baghche Maryam, M.B.; Banabazi, M.H.; Sargolzaei, M.; Miar, Y. Signatures of selection analysis using whole-genome sequence data reveals novel candidate genes for pony and light horse types. Genome 2020, 63, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Ceccobelli, S.; Landi, V.; Senczuk, G.; Mastrangelo, S.; Sardina, M.T.; Ben-Jemaa, S.; Persichilli, C.; Karsli, T.; Bâlteanu, V.A.; Raschia, M.A.; et al. A comprehensive analysis of the genetic diversity and environmental adaptability in worldwide Merino and Merino-derived sheep breeds. Genet. Sel. Evol. 2023, 55, 24. [Google Scholar] [CrossRef] [PubMed]
- Rochus, C.M.; Tortereau, F.; Plisson-Petit, F.; Restoux, G.; Moreno-Romieux, C.; Tosser-Klopp, G.; Servin, B. Revealing the selection history of adaptive loci using genome-wide scans for selection: An example from domestic sheep. BMC Genom. 2018, 19, 71. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Szpiech, Z.A.; Hernandez, R.D. Selscan: An Efficient Multithreaded Program to Perform EHH-Based Scans for Positive Selection. Mol. Biol. Evol. 2014, 31, 2824–2827. [Google Scholar] [CrossRef]
- Li, T.; Xing, F.; Zhang, N.; Chen, J.; Zhang, Y.; Yang, H.; Peng, S.; Ma, R.; Liu, Q.; Gan, S.; et al. Genome-Wide Association Analysis of Growth Traits in Hu Sheep. Gene 2024, 15, 1637. [Google Scholar] [CrossRef]
- Khazaei-Koohpar, H.; Gholizadeh, M.; Hafezian, S.H.; Nejati-Javaremi, A. Weighted single-step genome-wide association study for direct and maternal genetic effects associated with birth and weaning weights in sheep. Sci. Rep. 2024, 14, 13120. [Google Scholar] [CrossRef]
- Li, X.; Huang, C.; Liu, M.; Dai, R.; Wu, X.; Ma, X.; Chu, M.; Bao, P.; Pei, J.; Guo, X.; et al. Copy Number Variation of the SOX6 Gene and Its Associations with Growth Traits in Ashidan Yak. Animals 2022, 12, 3074. [Google Scholar] [CrossRef]
- Zhao, R.; Li, J.; Liu, N.; Li, H.; Liu, L.; Yang, F.; Li, L.; Wang, Y.; He, J. Transcriptomic Analysis Reveals the Involvement of lncRNA-miRNA-mRNA Networks in Hair Follicle Induction in Aohan Fine Wool Sheep Skin. Front. Genet 2020, 11, 590. [Google Scholar] [CrossRef]
- Rehman, S.U.; Zhen, Y.; Ding, L.; Saleh, A.A.; Zhang, Y.; Zhang, J.; He, F.; Husien, H.M.; Zhou, P.; Wang, M. Integrative Meta-Analysis: Unveiling Genetic Factors in Meat Sheep Growth and Muscular Development through QTL and Transcriptome Studies. Animals 2024, 14, 1679. [Google Scholar] [CrossRef]
- Li, T.; Jin, M.; Fei, X.; Yuan, Z.; Wang, Y.; Quan, K.; Wang, T.; Yang, J.; He, M.; Wei, C. Transcriptome Comparison Reveals the Difference in Liver Fat Metabolism between Hu and Tibetan Sheep. Animals 2022, 12, 1650. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Huang, S.; Lv, X.; Wang, S.; Cao, X.; Yuan, Z.; Getachew, T.; Mwacharo, J.M.; Haile, A.; Quan, K.; et al. Effect of CUX1 on the Proliferation of Hu Sheep Dermal Papilla Cells and on the Wnt/β-Catenin Signaling Pathway. Genes 2023, 14, 423. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Li, Y.; Chen, Y.; Li, Y.; Yang, K.; Zhao, Y.; Zhang, J.; Liu, J.; Chen, N.; Suolang, Q.; et al. Association between GHR, GHRHR and IGF1 Gene Variants and Milk Traits in Sheep. J. Dairy Sci. 2018, 101, 5972–5982. [Google Scholar]
- Perera, S.; Mankoo, B.; Gautel, M. Developmental regulation of MURF E3 ubiquitin ligases in skeletal muscle. J. Muscle Res. Cell Motil. 2012, 33, 107–122. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Li, M.; Sun, H.; Chen, Q.; Yan, D.; Dong, X.; Pan, Y.; Lu, S. Genome-Wide Association Study of Growth Traits in a Four-Way Crossbred Pig Population. Genes 2022, 13, 1990. [Google Scholar] [CrossRef]
- Guo, D.; Yu, Q.; Tong, Y.; Qian, X.; Meng, Y.; Ye, F.; Jiang, X.; Wu, L.; Yang, Q.; Li, S.; et al. OXCT1 succinylation and activation by SUCLA2 promotes ketolysis and liver tumor growth. Mol. Cell. 2025, 85, 843–856.e6. [Google Scholar] [CrossRef]
- Sun, C.; Kovacs, P.; Guiu-Jurado, E. Genetics of Body Fat Distribution: Comparative Analyses in Populations with European, Asian and African Ancestries. Genes 2021, 12, 841. [Google Scholar] [CrossRef]
- Seasholtz, A.F.; Valverde, R.A.; Denver, R.J. Corticotropin-releasing hormone-binding protein: Biochemistry and function from fishes to mammals. J. Endocrinol. 2002, 175, 89–97. [Google Scholar] [CrossRef]
- Misztal, T.; Młotkowska, P.; Marciniak, E.; Misztal, A. Allopregnanolone reduces neuroendocrine response to acute stressful stimuli in sheep. J. Endocrinol. 2020, 244, 201–211. [Google Scholar] [CrossRef]
- Zhao, F.; Xie, R.; Fang, L.; Xiang, R.; Yuan, Z.; Liu, Y.; Wang, L. Analysis of 206 whole-genome resequencing reveals selection signatures associated with breed-specific traits in Hu sheep. Evol. Appl. 2024, 17, E13697. [Google Scholar] [CrossRef]
- Han, A.; Saijo, K.; Mecklenbräuker, I.; Tarakhovsky, A.; Nussenzweig, M.C. Bam32 links the B cell receptor to ERK and JNK and mediates B cell proliferation but not survival. Immunity 2003, 19, 621–632. [Google Scholar] [CrossRef]
- Amiri Ghanatsaman, Z.; Ayatolahi Mehrgardi, A.; Asadollahpour Nanaei, H.; Esmailizadeh, A. Comparative genomic analysis uncovers candidate genes related with milk production and adaptive traits in goat breeds. Sci. Rep. 2023, 13, 8722. [Google Scholar] [CrossRef]
- Wang, M.D.; Dzama, K.; Hefer, C.A.; Muchadeyi, F.C. Genomic population structure and prevalence of copy number variations in South African Nguni cattle. BMC Genom. 2015, 16, 894. [Google Scholar] [CrossRef]
- Manzari, Z.; Mehrabani-Yeganeh, H.; Nejati-Javaremi, A.; Moradi, M.H.; Gholizadeh, M. Detecting selection signatures in three Iranian sheep breeds. Anim. Genet. 2019, 50, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.E.; Hafez, A.; Ateya, A.; Abdelsabour, T.H.; Mohamed, L.G.; Aboul-Naga, A.M. Single nucleotide polymorphisms, gene expression and evaluation of immunological, antioxidant, and pathological parameters associated with bacterial pneumonia in Barki sheep. Ir. Vet. J. 2025, 78, 11. [Google Scholar] [CrossRef]
- Sonobe, T.; Tsuchimochi, H.; Maeda, H.; Pearson, J.T. Increased contribution of KCa channels to muscle contraction induced vascular and blood flow responses in sedentary and exercise trained ZFDM rats. J. Physiol. 2022, 600, 2919–2938. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Nie, X.; Shi, L.; Shao, F.; Cao, L. A Novel PMVK Variant Associated with Familial Porokeratosis. Hum. Hered. 2023, 88, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Hernandez, L.D.; Galán, J.E.; Janeway, C.A., Jr.; Medzhitov, R.; Flavell, R.A. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 2002, 110, 191–202. [Google Scholar] [CrossRef]
- Yousefi, Z.; Moradi, M.H.; Beige-Nasiri, M.T.; Shirali, M.; Abdollahi-Arpanahi, R. Genomic insights into runs of homozygosity, effective population size and selection signatures in Iranian meat and dairy sheep breeds. PLoS ONE 2025, 20, e0323328. [Google Scholar] [CrossRef]
- Wang, S.; Wu, T.; Sun, J.; Li, Y.; Yuan, Z.; Sun, W. Single-Cell Transcriptomics Reveals the Molecular Anatomy of Sheep Hair Follicle Heterogeneity and Wool Curvature Front. Cell Dev. Biol. 2021, 9, 800157. [Google Scholar]
- Li, S.; Zheng, X.; Nie, Y.; Chen, W.; Liu, Z.; Tao, Y.; Hu, X.; Hu, Y.; Qiao, H.; Qi, Q.; et al. Defining Key Genes Regulating Morphogenesis of Apocrine Sweat Gland in Sheepskin. Front. Genet. 2019, 10, 739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Luo, J.; Shi, C.; Zhu, L.; He, Q.; Tian, H.; Wu, J.; Zhao, J.; Li, C. Genome-wide analysis of the acyl-coenzyme A synthetase family and their association with the formation of goat milk flavor. Front. Genet. 2022, 13, 980463. [Google Scholar]
- Cai, C.; Li, M.; Zhang, Y.; Meng, S.; Yang, Y.; Gao, P.; Guo, X.; Cao, G.; Li, B. Comparative Transcriptome Analyses of Longissimus thoracis Between Pig Breeds Differing in Muscle Characteristics. Front. Genet. 2020, 11, 526309. [Google Scholar] [CrossRef]
- Wang, W.; Ma, C.; Zhang, Q.; Jiang, Y. TMT-labeled quantitative malonylome analysis on the longissimus dorsi muscle of Laiwu pigs reveals the role of ACOT7 in fat deposition. J. Proteom. 2024, 298, 105129. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, M.; Zhou, M.; Wang, Y.; Wu, X.; Zhang, X.; Ding, Y.; Zhao, G.; Yin, Z.; Wang, C. Identification of Signatures of Selection by Whole-Genome Resequencing of a Chinese Native Pig. Front. Genet. 2020, 11, 566255. [Google Scholar] [CrossRef]
- Xu, L.; Yang, L.; Zhu, B.; Zhang, W.; Wang, Z.; Chen, Y.; Zhang, L.; Gao, X.; Gao, H.; Liu, G.E.; et al. Genome-wide scan reveals genetic divergence and diverse adaptive selection in Chinese local cattle. BMC Genom. 2019, 20, 494. [Google Scholar] [CrossRef]
- Ma, X.; Cheng, H.; Liu, Y.; Sun, L.; Chen, N.; Jiang, F.; You, W.; Yang, Z.; Zhang, B.; Song, E.; et al. Assessing Genomic Diversity and Selective Pressures in Bohai Black Cattle Using Whole-Genome Sequencing Data. Animals 2022, 12, 665. [Google Scholar] [CrossRef]
- He, Y.; Huang, Y.; Wang, S.; Zhang, L.; Gao, H.; Zhao, Y.; E, G. Hereditary Basis of Coat Color and Excellent Feed Conversion Rate of Red Angus Cattle by Next-Generation Sequencing Data. Animals 2022, 12, 1509. [Google Scholar] [CrossRef]
- Chen, B.; Yue, Y.; Li, J.; Liu, J.; Yuan, C.; Guo, T.; Zhang, D.; Yang, B.; Lu, Z. Transcriptome-metabolome analysis reveals how sires affect meat quality in hybrid sheep populations. Front. Nutr. 2022, 9, 967985. [Google Scholar] [CrossRef]
- Cheng, J.; Cao, X.; Hanif, Q.; Pi, L.; Hu, L.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Integrating Genome-Wide CNVs Into QTLs and High Confidence GWAScore Regions Identified Positional Candidates for Sheep Economic Traits. Front. Genet. 2020, 11, 569. [Google Scholar] [CrossRef]
- Wu, J.; Fang, S.; Feng, P.; Cai, C.; Zhang, L.; Yang, L. Changes in expression levels of Nod-like receptors in the spleen of ewes. Anim. Reprod. 2023, 20, e20220093. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Y.; Zhao, Z.; Cai, J.; Zhao, S.; Yang, L. Modulation of Nod-like Receptor Expression in the Thymus during Early Pregnancy in Ewes. Vaccines 2022, 10, 2128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cai, J.; Wang, X.; Yang, Z.; Ding, H.; Yang, L. Effects of early pregnancy on NOD-like receptor expression in the ovine endometrium. Front. Vet. Sci. 2024, 11, 1384386. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Li, Y.; Cao, J.; Fang, H.; Zhang, L.; Yang, L. Early Pregnancy Modulates Expression of the Nod-like Receptor Family in Lymph Nodes of Ewes. Animals 2022, 12, 3285. [Google Scholar] [CrossRef]
- Valsdottir, R.; Hashimoto, H.; Ashman, K.; Koda, T.; Storrie, B.; Nilsson, T. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett. 2001, 508, 201–209. [Google Scholar] [CrossRef]
- Satheesan, L.; Kittur, P.M.; Alhussien, M.N.; Karanwal, S.; Madhusoodan, A.P.; Alex, R.; Kamboj, A.; Dang, A.K. Comparative Profiling of Milk Somatic Cells Proteomes Revealed Key Players in Mammary Immune Mechanisms During Mastitis in Tropical Sahiwal (Bos indicus) Cows. Proteom. Clin. Appl. 2024, 18, e202400054. [Google Scholar] [CrossRef]
- Kim, E.; Um, H.; Park, J.; Jung, J.W.; Kim, J.E.; Lee, H.; Shin, E.A.; Pinanga, Y.; Lee, H.; Nam, S.H.; et al. TM4SF5-dependent crosstalk between hepatocytes and macrophages to reprogram the inflammatory environment. Cell Rep. 2021, 37, 110018. [Google Scholar] [CrossRef]
- Sun, L.; Qu, K.; Ma, X.; Hanif, Q.; Zhang, J.; Liu, J.; Chen, N.; Suolang, Q.; Lei, C.; Huang, B. Whole-Genome Analyses Reveal Genomic Characteristics and Selection Signatures of Lincang Humped Cattle at the China-Myanmar Border. Front. Genet. 2022, 13, 833503. [Google Scholar] [CrossRef]
| Gene | Functions | iHS | nSL | iHH12 |
|---|---|---|---|---|
| CCSER1 | Cell cycle, division; growth, fertility | ✓ | ✓ | ✓ |
| ACSL5 | Lipid metabolism, energy balance | ✓ | ✓ | |
| CUX2 | Nervous system development, skin/wool | ✓ | ✓ | |
| SOX6 | Muscle and cartilage differentiation; skeletal development | ✓ | ✓ | |
| LRFN5 | Synaptic function, nervous system | ✓ | ||
| RASSF10 | Cell cycle, growth regulation (Ras pathway) | ✓ | ||
| WNT11 | Wnt signaling; embryogenesis, reproduction | ✓ | ||
| DAPP1 | Immune response (B-cells) | ✓ | ||
| ATP6V0D2 | Bone metabolism, osteoclasts | ✓ | ||
| ARHGEF4 | Wnt/adhesion (RhoGEF); cell migration | ✓ | ||
| TRIM24 | Transcriptional regulation, differentiation | ✓ | ||
| MMRN1 | Extracellular matrix, hemostasis; growth, fattening | ✓ | ||
| CRH | Stress hormone, neuroendocrine regulation | ✓ | ||
| TRIM55 | Muscle protein (MuRF2); muscle mass regulation | ✓ | ||
| OXCT1 | Ketone metabolism; energy adaptation | ✓ | ||
| GHR | Growth hormone receptor; growth, lactation | ✓ | ||
| CIT | Cytokinesis, neurodevelopment | ✓ | ||
| HECTD4 | Ubiquitin ligase; lipid metabolism | ✓ | ||
| SLC8B1 | Ion transport; heart rhythm | ✓ |
| Gene | Salsk Breed—Texel | Salsk Breed—Suffolk | Salsk Breed—Île-De-France |
|---|---|---|---|
| Immune Response and Disease Resistance | |||
| IL6R | ✓ | ||
| IRAK3 | ✓ | ||
| DOCK5 | ✓ | ||
| NLRP1 | ✓ | ||
| RABEP1 | ✓ | ||
| TM4SF5 | ✓ | ||
| FANCA | ✓ | ||
| MINK1 | ✓ | ||
| KIF1C | ✓ | ||
| Metabolism and Energy Exchange | |||
| IL6R | ✓ | ||
| PMVK | ✓ | ||
| WIF1 | ✓ | ||
| ACSF3 | ✓ | ||
| Growth, Tissue Development, and Productivity | |||
| KCNN3 | ✓ | ||
| MYLK3 | ✓ | ||
| Wool development and pigmentation | |||
| WIF1 | ✓ | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lukonina, O.; Bakoev, S.; Kolosov, Y.; Akhmedli, V.; Bakoeva, I.; Kolosova, M.; Usatov, A.; Kolosov, A.; Getmantseva, L. Study on the Genomic Basis of Adaptation in Salsk Sheep. Biology 2025, 14, 1620. https://doi.org/10.3390/biology14111620
Lukonina O, Bakoev S, Kolosov Y, Akhmedli V, Bakoeva I, Kolosova M, Usatov A, Kolosov A, Getmantseva L. Study on the Genomic Basis of Adaptation in Salsk Sheep. Biology. 2025; 14(11):1620. https://doi.org/10.3390/biology14111620
Chicago/Turabian StyleLukonina, Olga, Siroj Bakoev, Yury Kolosov, Vagif Akhmedli, Ilona Bakoeva, Maria Kolosova, Alexandr Usatov, Anatoliy Kolosov, and Lyubov Getmantseva. 2025. "Study on the Genomic Basis of Adaptation in Salsk Sheep" Biology 14, no. 11: 1620. https://doi.org/10.3390/biology14111620
APA StyleLukonina, O., Bakoev, S., Kolosov, Y., Akhmedli, V., Bakoeva, I., Kolosova, M., Usatov, A., Kolosov, A., & Getmantseva, L. (2025). Study on the Genomic Basis of Adaptation in Salsk Sheep. Biology, 14(11), 1620. https://doi.org/10.3390/biology14111620





