ITS Sequencing Reveals the Changing Characteristics of Fungal Communities in Different Rice-Growing Substrates Under Salt Stress
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Materials
2.2. Experimental Design
2.3. Substrate Parameters
2.4. ITS Sequencing
2.4.1. Sample Collection
2.4.2. PCR Amplification and Sequencing Library Construction
2.5. Statistical Analysis
3. Results
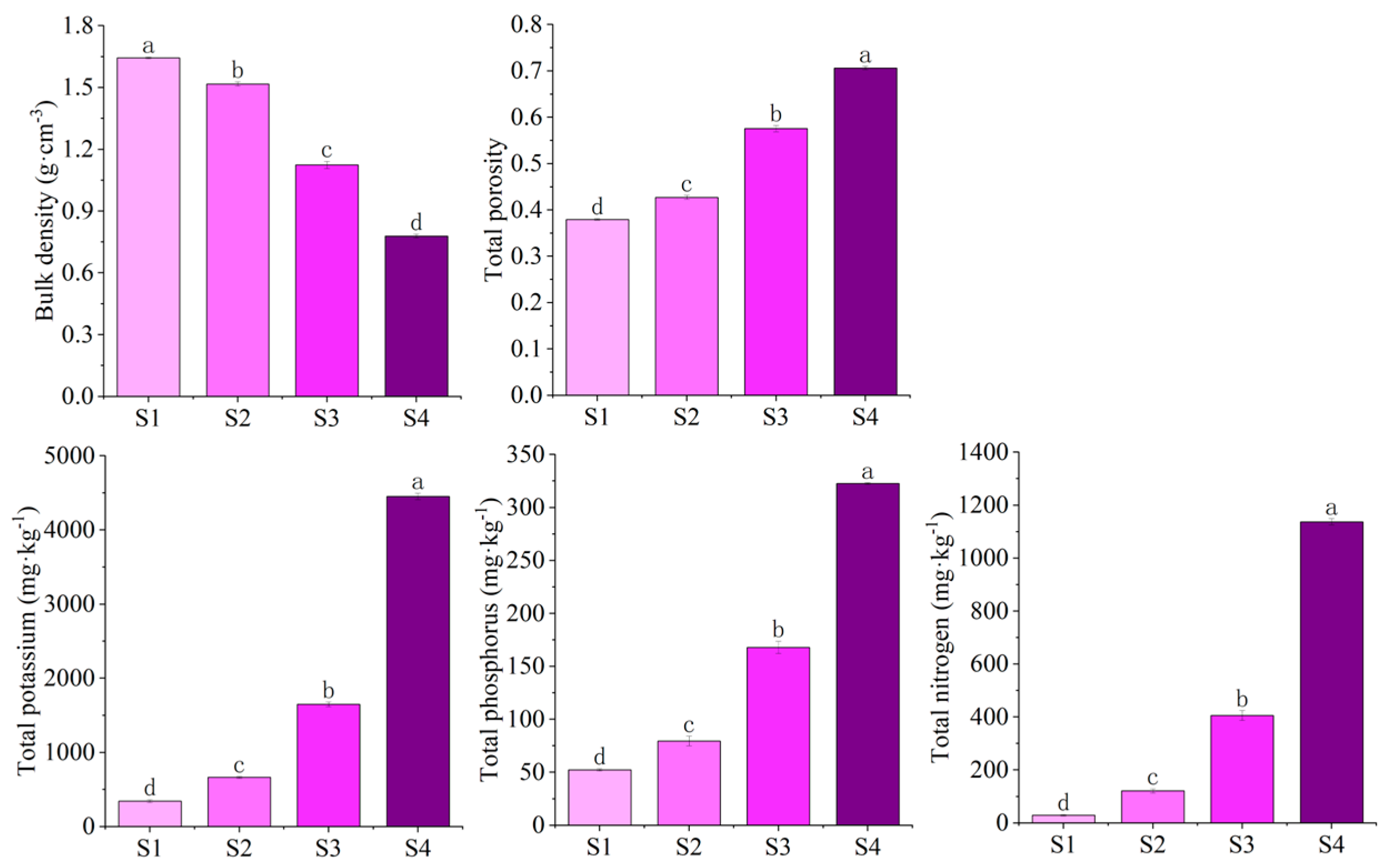
3.1. Bulk Density, Total Porosity, and Nutrient Content of Different Substrates
3.2. Dilution Curve Analysis
3.3. Alpha Diversity Analysis
3.3.1. Alpha Diversity Analysis of Different Comparison Groups
3.3.2. Alpha Diversity Analysis of Different Non-Salt Treatments
3.3.3. Alpha Diversity Analysis of Different Salt Treatments
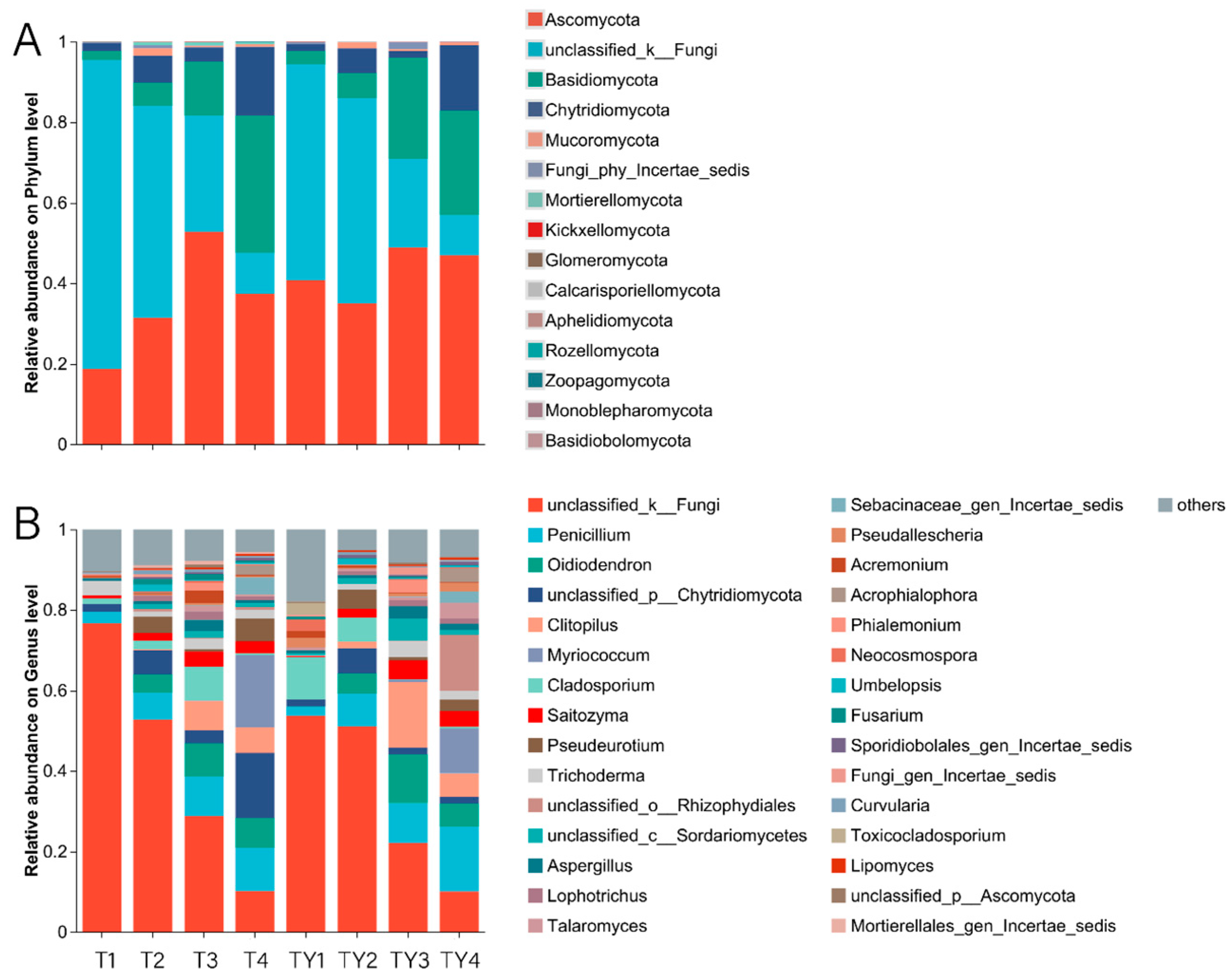
3.4. Species Composition Analysis
3.4.1. Analysis of Community Composition of Different Comparison Groups
3.4.2. Analysis of Community Composition of Different Non-Salt Treatments
3.4.3. Analysis of Community Composition of Different Salt Treatments
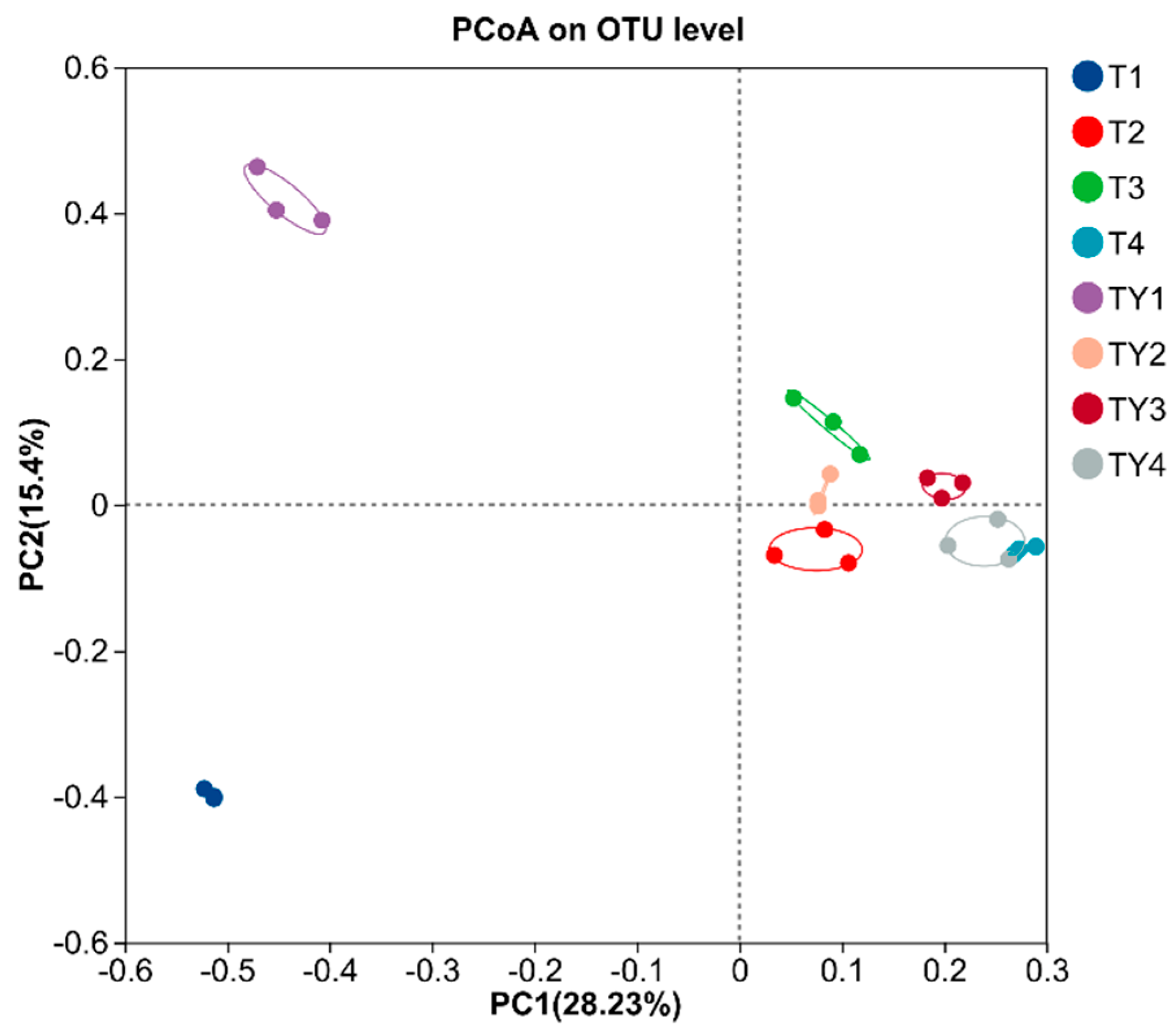
3.5. Beta Diversity Analysis
3.6. Lefse Multi-Level Species Difference Discriminant Analysis
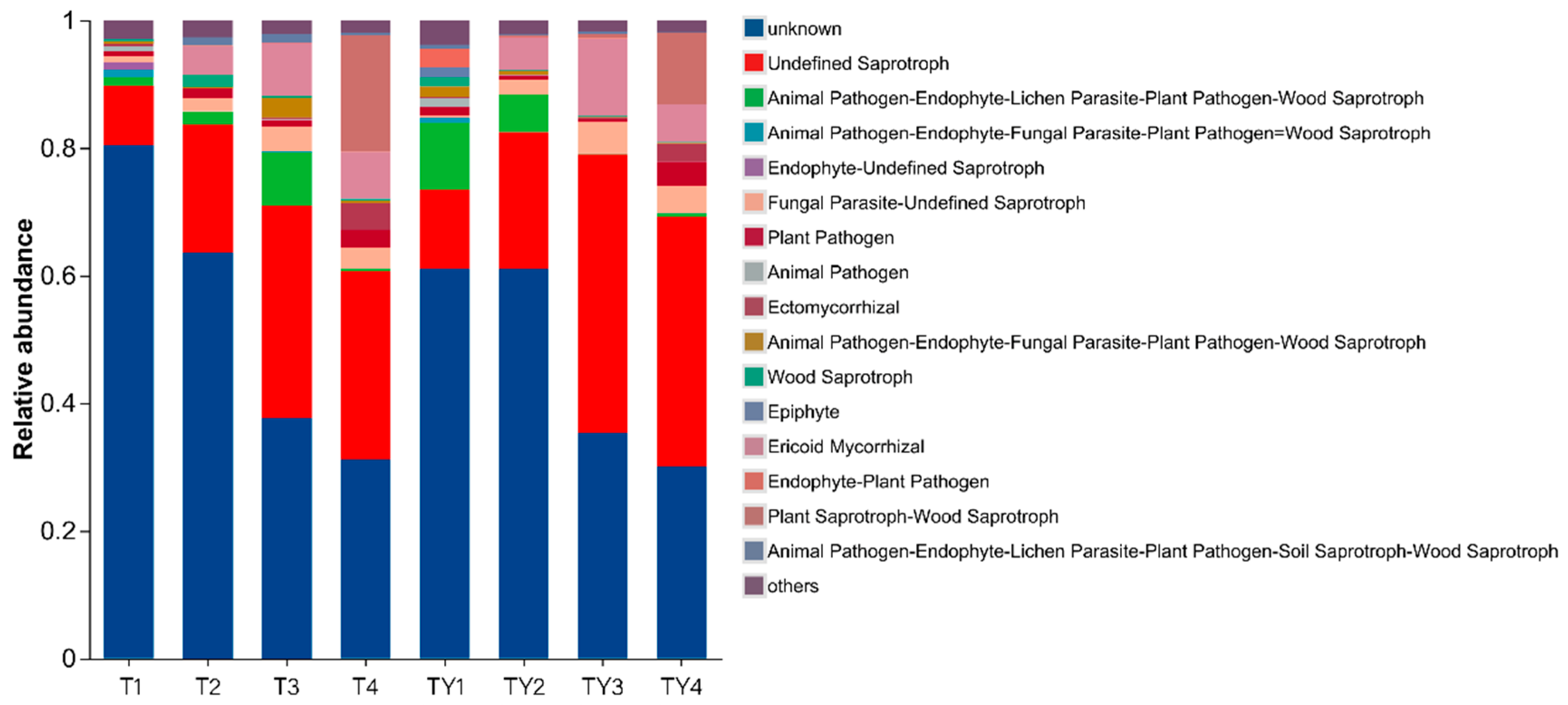
3.7. FUNGuild Function Prediction
3.7.1. FUNGuild Function Prediction of Different Comparison Groups
3.7.2. FUNGuild Function Prediction of Different Non-Salt Treatments
3.7.3. FUNGuild Function Prediction of Different Salt Treatments
3.8. Combined Analysis of Fungal Colonies and Environmental Factors
3.8.1. Canonical Correspondence Analysis (CCA)
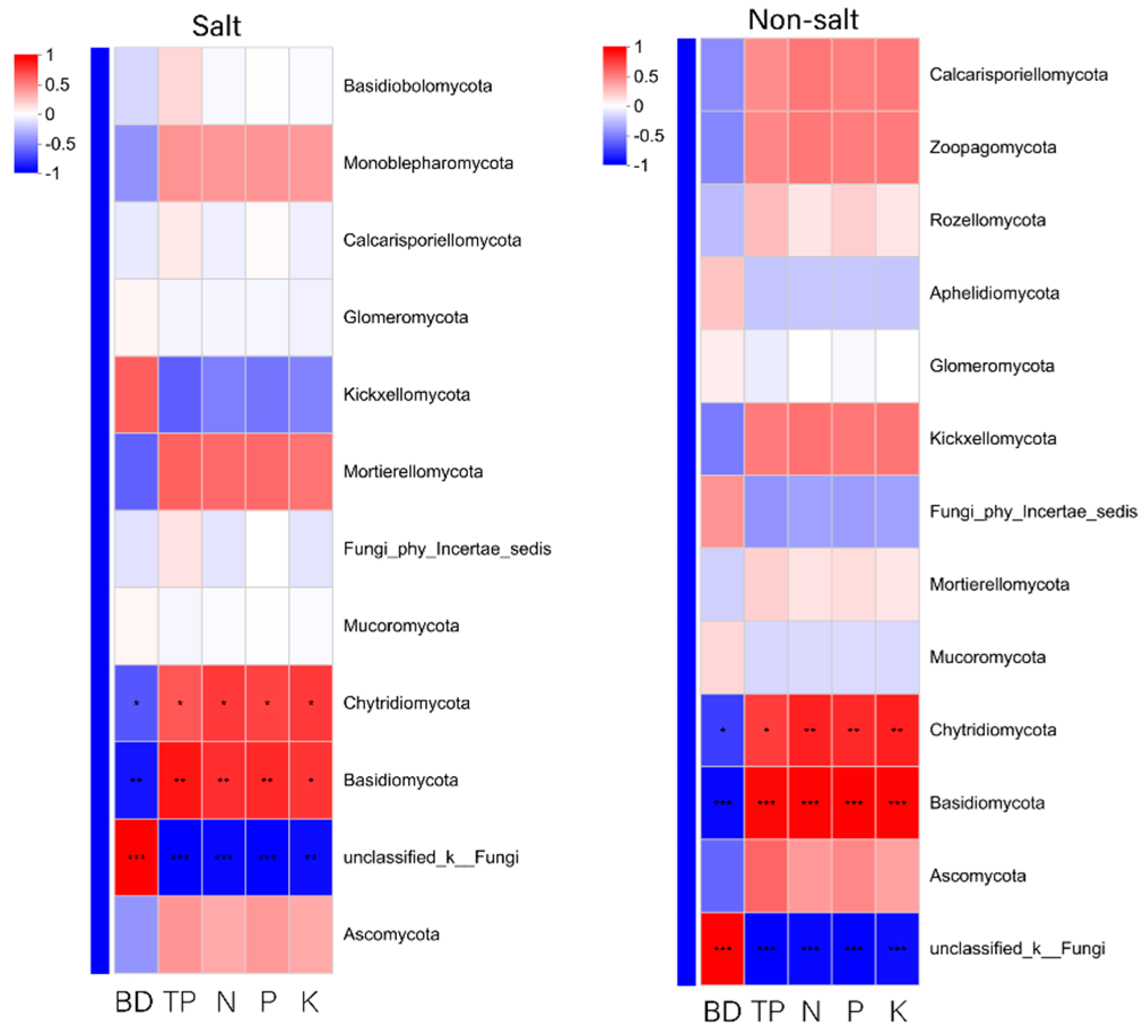
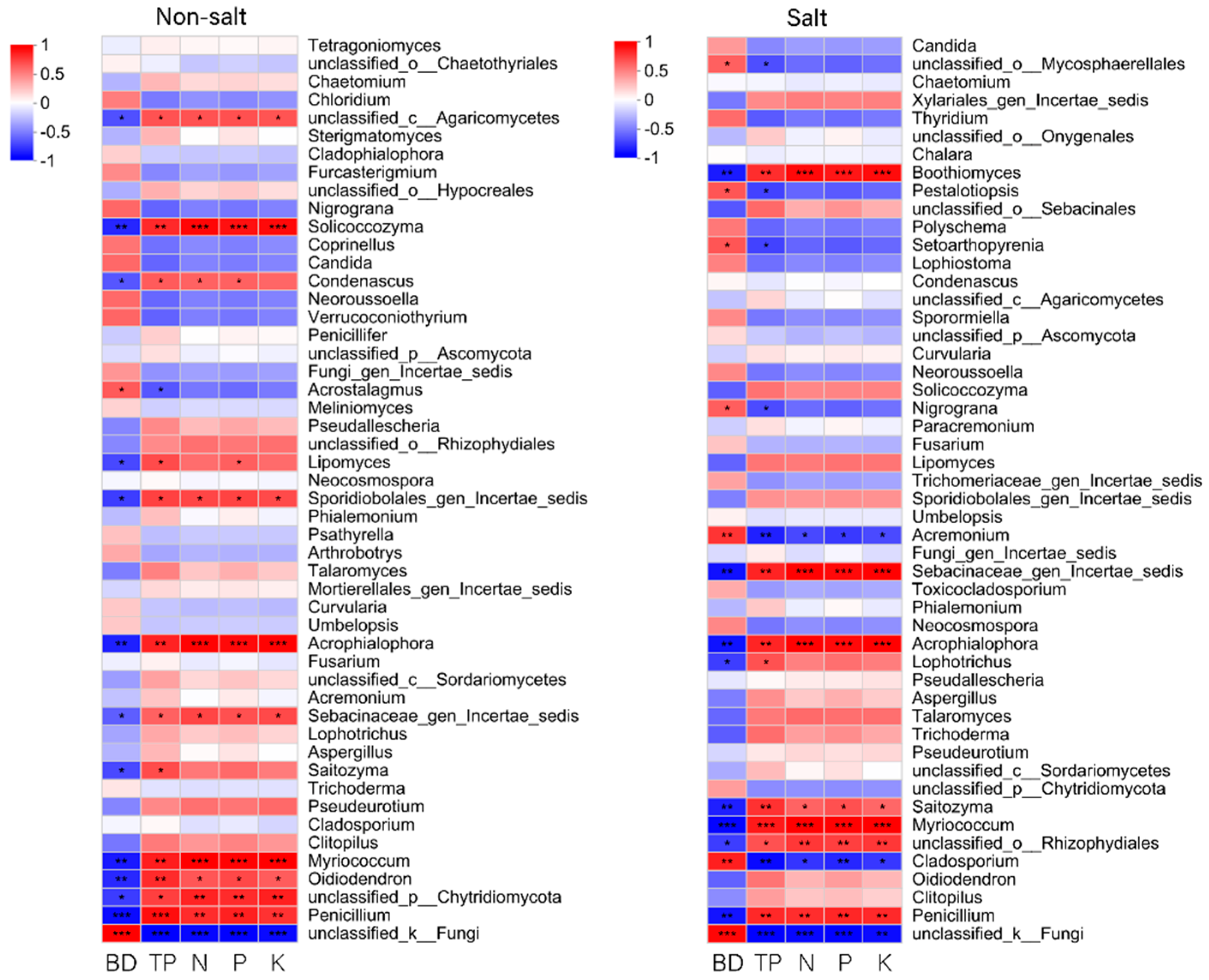
3.8.2. Correlation Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Q.; Song, Y.; An, Y.; Lu, Y.; Zhong, G. Soil Microorganisms: Their Role in Enhancing Crop Nutrition and Health. Diversity 2024, 16, 734. [Google Scholar] [CrossRef]
- Frąc, M.; Hannula, S.E.; Bełka, M.; Jędryczka, M. Fungal biodiversity and their role in soil health. Front. Microbiol. 2018, 9, 707. [Google Scholar] [CrossRef]
- Frąc, M.; Jezierska-Tys, S.; Yaguchi, T. Occurrence, detection, and molecular and metabolic characterization of heat-resistant fungi in soils and plants and their risk to human health. Adv. Agron. 2015, 132, 161–204. [Google Scholar]
- Metternicht, G.I.; Zinck, J. Remote sensing of soil salinity: Potentials and constraints. Remote Sens. Environ. 2003, 85, 1–20. [Google Scholar] [CrossRef]
- Ma, Y.; Tashpolat, N. Current Status and Development Trend of Soil Salinity Monitoring Research in China. Sustainability 2023, 15, 5874. [Google Scholar] [CrossRef]
- Yan, N.; Marschner, P.; Cao, W.; Zuo, C.; Qin, W. Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef]
- Rousk, J.; Elyaagubi, F.K.; Jones, D.L.; Godbold, D.L. Bacterial salt tolerance is unrelated to soil salinity across an arid agroecosystem salinity gradient. Soil Biol. Biochem. 2011, 43, 1881–1887. [Google Scholar] [CrossRef]
- Wei, Y.; Chen, L.; Yin, Z.; Feng, Q.; Xi, H.; Zhang, C.; Gan, K.; Yong, T. Differences in soil fungal communities under salinity gradients in arid and semiarid regions. Glob. Planet. Change 2024, 236, 104425. [Google Scholar] [CrossRef]
- Boumaaza, B.; Gacemi, A.; Benzohra, I.E.; Benada, M.H.; Boudalia, S.; Belaidi, H.; Khaladi, O. Impact of salinity on the behavior of fungi. Int. J. Agric. Biosci. 2022, 11, 139–147. [Google Scholar] [CrossRef]
- Yang, C.; Wang, X.; Miao, F.; Li, Z.; Tang, W.; Sun, J. Assessing the effect of soil salinization on soil microbial respiration and diversities under incubation conditions. Appl. Soil Ecol. 2020, 155, 103671. [Google Scholar] [CrossRef]
- Leitão, A.L.; Enguita, F.J. Gibberellins in Penicillium strains: Challenges for endophyte-plant host interactions under salinity stress. Microbiol. Res. 2016, 183, 8–18. [Google Scholar] [CrossRef]
- Kouadria, R.; Bouzouina, M.; Lotmani, B. Endophytic Fungi from Salt Adapted Plants Confer Salt Tolerance in Barley. Tunis. J. Plant Prot. 2023, 18, 63–70. [Google Scholar] [CrossRef]
- Li, X.; Han, S.; Wang, G.; Liu, X.; Amombo, E.; Xie, Y.; Fu, J. The fungus Aspergillus aculeatus enhances salt-stress tolerance, metabolite accumulation, and improves forage quality in perennial ryegrass. Front. Microbiol. 2017, 8, 1664. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhao, L.; Song, Y.; Du, X.; Huo, J.; Mei, W.; Wang, X.; Feng, N.; Zheng, D.; Wu, Z. Changes in Antioxidant and Photosynthetic Capacity in Rice Under Different Substrates. Biology 2025, 14, 34. [Google Scholar] [CrossRef]
- Bao, S. Soil and Agricultural Chemistry Analysis; China Agriculture Publication: Beijing, China, 2000. [Google Scholar]
- Qirom, M.A.; Yuwati, T.W.; Rachmanadi, D.; Halwany, W. The variation of carbon content and bulk density on different time period post fire and peat depth. IOP Conf. Ser. Earth Environ. Sci. 2021, 886, 012096. [Google Scholar] [CrossRef]
- Zhou, P.; Liu, G.; Wen, A. Study on characters of soil water content and water conservation of arbor and shrub lands with different forest ages in the loess hilly-gullied region. Res. Soil Water Conserv. 2010, 17, 188–193. [Google Scholar]
- Zhang, G.; Bai, J.; Jia, J.; Wang, W.; Wang, D.; Zhao, Q.; Wang, C.; Chen, G. Soil microbial communities regulate the threshold effect of salinity stress on SOM decomposition in coastal salt marshes. Fundam. Res. 2023, 3, 868–879. [Google Scholar] [CrossRef]
- Challacombe, J.F.; Hesse, C.N.; Bramer, L.M.; McCue, L.A.; Lipton, M.; Purvine, S.; Nicora, C.; Gallegos-Graves, L.V.; Porras-Alfaro, A.; Kuske, C.R. Genomes and secretomes of Ascomycota fungi reveal diverse functions in plant biomass decomposition and pathogenesis. BMC Genom. 2019, 20, 976. [Google Scholar] [CrossRef]
- Yuan, M.; Zhang, D.; Wang, Z.; Zhu, Z.; Sun, H.; Wang, W.; Han, D.; Qu, Z.; Ma, B.; Wang, J. Salt altered rhizosphere fungal community and induced soybean recruit specific species to ameliorate salt stress. Front. Microbiol. 2023, 14, 1142780. [Google Scholar] [CrossRef]
- Gupta, V.K. New and Future Developments in Microbial Biotechnology and Bioengineering: Aspergillus System Properties and Applications; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Berg, G.; Zachow, C.; Lottmann, J.; Götz, M.; Costa, R.; Smalla, K. Impact of plant species and site on rhizosphere-associated fungi antagonistic to Verticillium dahliae Kleb. Appl. Environ. Microbiol. 2005, 71, 4203–4213. [Google Scholar] [CrossRef]
- Elias, F.; Woyessa, D.; Muleta, D. Phosphate solubilization potential of rhizosphere fungi isolated from plants in Jimma Zone, Southwest Ethiopia. Int. J. Microbiol. 2016, 2016, 5472601. [Google Scholar] [CrossRef]
- Park, M.S.; Lee, J.W.; Kim, S.H.; Park, J.-H.; You, Y.-H.; Lim, Y.W. Penicillium from rhizosphere soil in terrestrial and coastal environments in South Korea. Mycobiology 2020, 48, 431–442. [Google Scholar] [CrossRef]
- Galeano, R.M.S.; Silva, S.M.; Yonekawa, M.K.A.; de Alencar Guimarães, N.C.; Giannesi, G.C.; Masui, D.C.; Corrêa, B.O.; da Silva Brasil, M.; Zanoelo, F.F. Penicillium chrysogenum strain 34-P promotes plant growth and improves initial development of maize under saline conditions. Rhizosphere 2023, 26, 100710. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Hee Lee, S.; Jeong Jeon, S.; Burm Lee, H. First records of rare Ascomycete fungi, Acrostalagmus luteoalbus, Bartalinia robillardoides, and Collariella carteri from freshwater samples in Korea. Mycobiology 2019, 47, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Guo, H.; Zhang, K.; Zhao, M.; Ruan, J.; Chen, J. Trichoderma and its role in biological control of plant fungal and nematode disease. Front. Microbiol. 2023, 14, 1160551. [Google Scholar] [CrossRef]
- Bruyant, P.; Moënne-Loccoz, Y.; Almario, J. Root-associated Helotiales fungi: Overlooked players in plant nutrition. Soil Biol. Biochem. 2024, 191, 109363. [Google Scholar] [CrossRef]
- Newsham, K.K. A meta-analysis of plant responses to dark septate root endophytes. New Phytol. 2011, 190, 783–793. [Google Scholar] [CrossRef]
- Rissi, D.V.; Ijaz, M.; Baschien, C. Comparative Genomics of Different Lifestyle Fungi in Helotiales (Leotiomycetes) Reveals Temperature and Ecosystem Adaptations. J. Fungi 2024, 10, 869. [Google Scholar] [CrossRef]
- Pem, D.; Jeewon, R.; Chethana, K.W.T.; Hongsanan, S.; Doilom, M.; Suwannarach, N.; Hyde, K.D. Species concepts of Dothideomycetes: Classification, phylogenetic inconsistencies and taxonomic standardization. Fungal Divers. 2021, 109, 283–319. [Google Scholar] [CrossRef]
- Sun, H.; Liu, X.; Wang, T.; Liu, S.; Zhang, R.; Guo, X.; Yu, Z.; Zhao, Y.; Shen, P.; Zhang, Y. Rhizosphere microbiomes are closely linked to seagrass species: A comparative study of three coastal seagrasses. Appl. Environ. Microbiol. 2024, 90, e01754-24. [Google Scholar] [CrossRef]
- Hanrahan-Tan, D.G.; Henderson, L.; Kertesz, M.A.; Lilje, O. The Effects of Nitrogen and Phosphorus on Colony Growth and Zoospore Characteristics of Soil Chytridiomycota. J. Fungi 2022, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- Hibbett, D.S.; Donoghue, M.J. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Syst. Biol. 2001, 50, 215–242. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.; Jiang, J.; Post, W.; Classen, A.T. Decomposition by ectomycorrhizal fungi alters soil carbon storage in a simulation model. Ecosphere 2015, 6, 1–16. [Google Scholar] [CrossRef]
- Yi, X.; Yi, K.; Fang, K.; Gao, H.; Dai, W.; Cao, L. Microbial community structures and important associations between soil nutrients and the responses of specific taxa to rice-frog cultivation. Front. Microbiol. 2019, 10, 1752. [Google Scholar] [CrossRef]
- Liu, S.; Xiong, C.; Lin, L.; Keyhani, N.O.; Zhu, M.; Zhao, Z.; Zhang, W.; Yang, C.; Su, H.; Liu, P. Assessing the structure and diversity of fungal community in plant soil under different climatic and vegetation conditions. Front. Microbiol. 2023, 14, 1288066. [Google Scholar] [CrossRef]




| Substances | Treatments | Symbols |
|---|---|---|
| S1 | Fresh water | T1 |
| S2 | Fresh water | T2 |
| S3 | Fresh water | T3 |
| S4 | Fresh water | T4 |
| S1 | Salt stress | TY1 |
| S2 | Salt stress | TY2 |
| S3 | Salt stress | TY3 |
| S4 | Salt stress | TY4 |
| Sequencing Region | Primer Name | Primer Sequences (5′-3′) |
|---|---|---|
| ITS1F_ITS2R | ITS1F | CTTGGTCATTTAGAGGAAGTAA |
| ITS2R | GCTGCGTTCTTCATCGATGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Du, X.; Lin, Y.; Zhao, L.; Feng, N.; Zheng, D. ITS Sequencing Reveals the Changing Characteristics of Fungal Communities in Different Rice-Growing Substrates Under Salt Stress. Biology 2025, 14, 1456. https://doi.org/10.3390/biology14101456
Zhou H, Du X, Lin Y, Zhao L, Feng N, Zheng D. ITS Sequencing Reveals the Changing Characteristics of Fungal Communities in Different Rice-Growing Substrates Under Salt Stress. Biology. 2025; 14(10):1456. https://doi.org/10.3390/biology14101456
Chicago/Turabian StyleZhou, Hang, Xiaole Du, Yin Lin, Liming Zhao, Naijie Feng, and Dianfeng Zheng. 2025. "ITS Sequencing Reveals the Changing Characteristics of Fungal Communities in Different Rice-Growing Substrates Under Salt Stress" Biology 14, no. 10: 1456. https://doi.org/10.3390/biology14101456
APA StyleZhou, H., Du, X., Lin, Y., Zhao, L., Feng, N., & Zheng, D. (2025). ITS Sequencing Reveals the Changing Characteristics of Fungal Communities in Different Rice-Growing Substrates Under Salt Stress. Biology, 14(10), 1456. https://doi.org/10.3390/biology14101456




