AI and Machine Learning in Biology: From Genes to Proteins
Abstract
Simple Summary
Abstract
1. Introduction
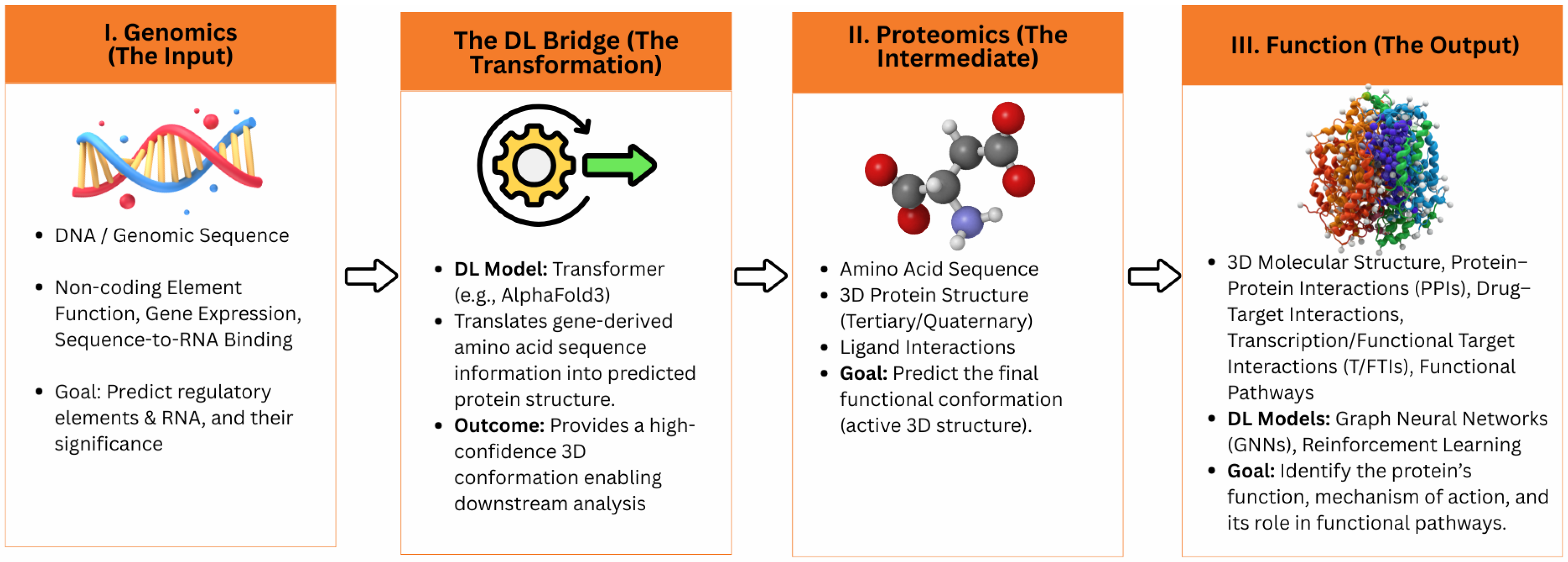
1.1. Genomics and Protein Structure Prediction: A Unified Frontier Enabled by Deep Learning
1.2. Brief History and Evolution of Deep Learning
2. Advantages and Challenges of Using Deep Learning in Computational Biology
2.1. Advantages of Using Deep Learning
2.2. Challenges of Using Deep Learning
3. Interconnecting Genomics and Protein Structure Prediction Through Deep Learning
3.1. Role of Deep Learning in Genomic Variant Detection and Precision Medicine
3.2. Advancements in Deep Learning for Epigenetic Data Analysis
3.3. Applications of Deep Learning in Protein Structure Prediction
4. Deep Learning Models for Prediction of Protein Structure from Sequence Data
4.1. Applications of Deep Learning in Protein–Protein Interaction Prediction and Drug Discovery
4.2. Recent Developments in Deep Learning-Based Techniques for Analyzing Protein Function and Evolution
4.3. Challenges and Future Directions
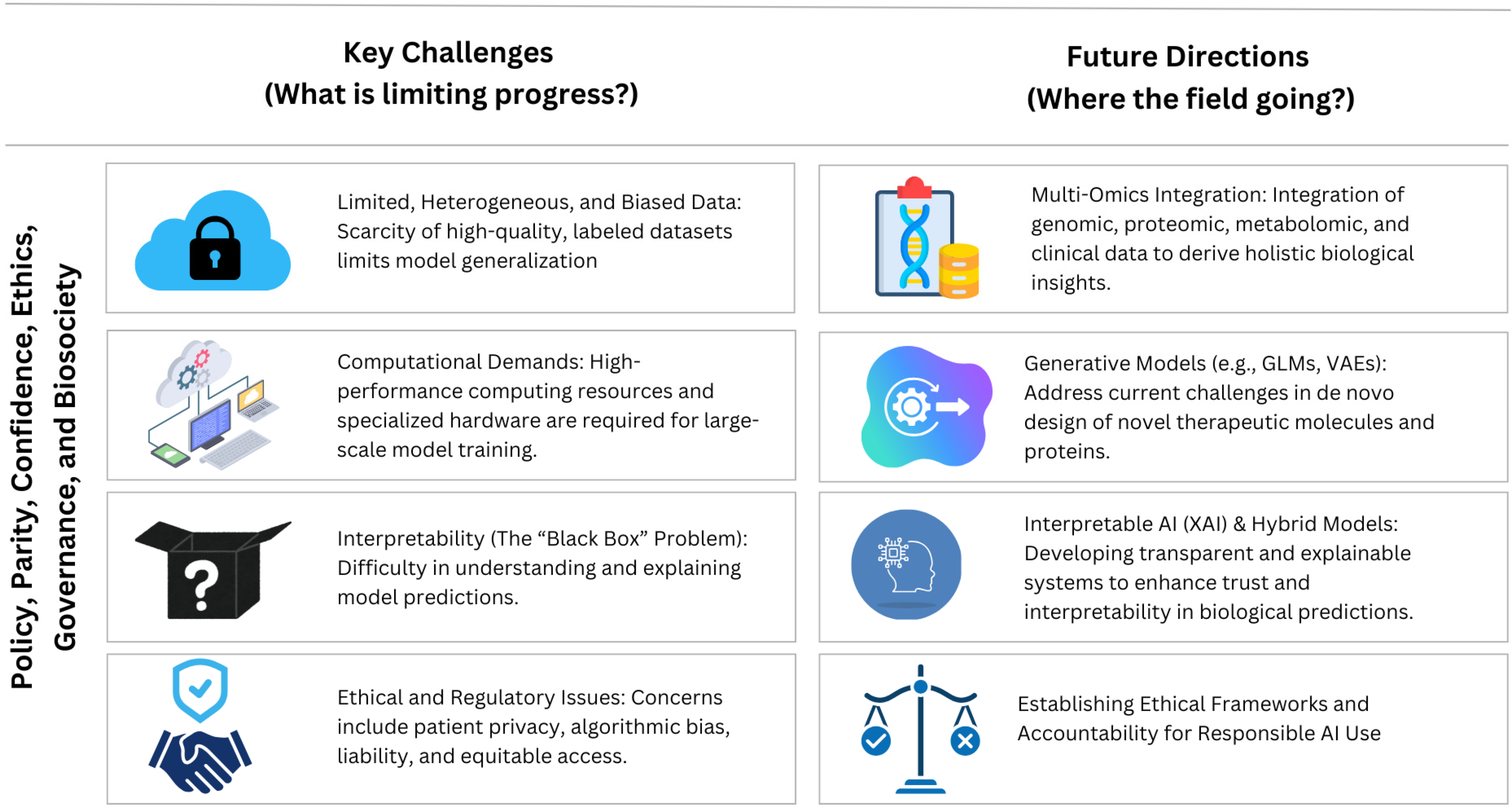
5. Key Challenges and Future Directions
5.1. Emerging Areas of Research and Potential Applications
5.2. Ethical and Social Implications
6. Future Prospects and Potential Impact of Deep Learning on Biological Research and Clinical Practice
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ANNs | Artificial neural networks |
| BLSTM | Bidirectional long short-term memory |
| CASP | Critical Assessment of Structure Prediction |
| CNNs | Convolutional neural networks |
| CRNN | Convolutional Recurrent Neural Network |
| DBN | Deep Belief Network |
| DBM | Deep Boltzmann Machine |
| DGMs | Deep generative models |
| DNA | Deoxyribonucleic Acid |
| DNN | Deep Neural Network |
| DQN | Deep Q-Network |
| GAN | Generative adversarial network |
| GCN | Graph Convolutional Network |
| gLMs | genomic language models |
| GNNs | Graph neural networks |
| LLMs | Large Language Models |
| LSTMs | Long short-term memory networks |
| PLMs | Protein language models |
| PPI | Protein–Protein Interaction |
| PSSM | Protein-positioning specific scoring matrix |
| QA | Quality assessment |
| RL | Reinforcement Learning |
| RNA | Ribonucleic Acid |
| RNNs | Recurrent neural networks |
| scGNN | single-cell analysis using graph neural networks |
| SNPs | Single-nucleotide polymorphisms |
| VAE | Variational Autoencoder |
| VGAE | Variational Graph Autoencoder |
References
- Way, G.P.; Greene, C.S.; Carninci, P.; Carvalho, B.S.; de Hoon, M.; Finley, S.D.; Gosline, S.J.C.; Lê Cao, K.-A.; Lee, J.S.H.; Marchionni, L.; et al. A field guide to cultivating computational biology. PLoS Biol. 2021, 19, e3001419. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Alipanahi, B.; Delong, A.; Weirauch, M.T.; Frey, B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015, 33, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Machine Learning Industry Trends Report Data Book, 2022–2030. Available online: https://www.grandviewresearch.com/sector-report/machine-learning-industry-data-book (accessed on 9 May 2023).
- Wang, H.; Raj, B. On the origin of deep learning. arXiv 2017, arXiv:1702.07800. [Google Scholar]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Jumper, J.M. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Feng, C.; Wang, W.; Han, R.; Wang, Z.; Ye, L.; Du, Z.; Wei, H.; Zhang, F.; Peng, Z.; Yang, J. Accurate de novo prediction of RNA 3D structure with transformer network. bioRxiv 2022. [Google Scholar] [CrossRef]
- Li, C.; Yan, Y.; Lin, W.; Zhang, Y. Enhancing cancer subtype classification through convolutional neural networks: A deepinsight analysis of TCGA gene expression data. Health Inf. Sci. Syst. 2025, 13, 33. [Google Scholar] [CrossRef]
- Jukič, M.; Bren, U. Machine learning in antibacterial drug design. Front. Pharmacol. 2022, 13, 1284. [Google Scholar] [CrossRef]
- Ieremie, I.; Ewing, R.M.; Niranjan, M. TransformerGO: Predicting protein–protein interactions by modelling the attention between sets of gene ontology terms. Bioinformatics 2022, 38, 2269–2277. [Google Scholar] [CrossRef]
- Mao, G.; Pang, Z.; Zuo, K.; Liu, J. Gene regulatory network inference using convolutional neural networks from scRNA-seq data. J. Comput. Biol. 2023, 30, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, U.A.; Tang, H.; Wu, G.; Marjan, S.; Hussain, A. Deep learning with graph convolutional networks: An overview and latest applications in computational intelligence. Int. J. Intell. Syst. 2023, 2023, 8342104. [Google Scholar] [CrossRef]
- Mreyoud, Y.; Song, M.; Lim, J.; Ahn, T.H. MegaD: Deep learning for rapid and accurate disease status prediction of metagenomic samples. Life 2022, 12, 669. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Sun, S.; Hu, C.; Yao, Y.; Yan, Y.; Zhang, Y. DeepPPI: Boosting Prediction of Protein-Protein Interactions with Deep Neural Networks. J. Chem. Inf. Model. 2017, 57, 1499–1510. [Google Scholar] [CrossRef]
- Yang, K.; Huang, H.; Vandans, O.; Murali, A.; Tian, F.; Yap, R.H.; Dai, L. Applying deep reinforcement learning to the HP model for protein structure prediction. Phys. A Stat. Mech. Its Appl. 2023, 609, 128395. [Google Scholar] [CrossRef]
- Angermueller, C.; Lee, H.J.; Reik, W.; Stegle, O. DeepCpG: Accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol. 2017, 18, 67. [Google Scholar]
- Xu, J.; Wang, S. Analysis of distance-based protein structure prediction by deep learning in CASP13. Proteins Struct. Funct. Bioinform. 2019, 87, 1069–1081. [Google Scholar] [CrossRef]
- Yassi, M.; Chatterjee, A.; Parry, M. Application of deep learning in cancer epigenetics through DNA methylation analysis. Brief. Bioinform. 2023, 24, bbad411. [Google Scholar] [CrossRef]
- Lin, E.; Lin, C.H.; Lane, H.Y. De novo peptide and protein design using generative adversarial networks: An update. J. Chem. Inf. Model. 2022, 62, 761–774. [Google Scholar] [CrossRef]
- Miller, R.F.; Singh, A.; Otaki, Y.; Tamarappoo, B.; Kavanagh, P.; Parekh, T.; Hu, L.; Gransar, H.; Sharir, T.; Einstein, A.J.; et al. Mitigating bias in deep learning for diagnosis of coronary artery disease from myocardial perfusion SPECT images. Eur. J. Nucl. Med. Mol. Imaging 2022, 50, 387–397. [Google Scholar] [CrossRef]
- Svensson, V.; Gayoso, A.; Yosef, N.; Pachter, L. Interpretable factor models of single-cell RNA- seq via variational autoencoders. Bioinformatics 2020, 36, 3418–3421. [Google Scholar] [CrossRef]
- Li, M.; Cai, X.; Xu, S.; Ji, H. Metapath-aggregated heterogeneous graph neural network for drug-target interaction prediction. Brief. Bioinform. 2023, 24, bbac578. [Google Scholar] [CrossRef] [PubMed]
- Veturi, Y.A.; Woof, W.; Lazebnik, T.; Moghul, I.; Woodward-Court, P.; Wagner, S.K.; de Guimarães, T.A.; Varela, M.D.; Liefers, B.; Patel, P.J.; et al. SynthEye: Investigating the Impact of Synthetic Data on Artificial Intelligence-assisted Gene Diagnosis of Inherited Retinal Disease. Ophthalmol. Sci. 2023, 3, 100258. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.K.; Azizan, F.L.; Wong, Y.S.; Omar, N. DeepFinder: An integration of feature-based and deep learning approach for DNA motif discovery. Biotechnol. Biotechnol. Equip. 2018, 32, 759–768. [Google Scholar] [CrossRef]
- Baek, M.; Baker, D. Deep learning and protein structure modeling. Nat. Methods 2022, 19, 13–14. [Google Scholar] [CrossRef]
- Gao, M.; Zhou, H.; Skolnick, J. DESTINI: A deep-learning approach to contact- driven protein structure prediction. Sci. Rep. 2019, 9, 3514. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- de Lima, L.M.; de Assis, M.C.; Soares, J.P.; Grão-Velloso, T.R.; de Barros, L.A.; Camisasca, D.R.; Krohling, R.A. On the importance of complementary data to histopathological image analysis of oral leukoplakia and carcinoma using deep neural networks. Intell. Med. 2023, 3, 258–266. [Google Scholar] [CrossRef]
- Popova, M.; Isayev, O.; Tropsha, A. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018, 4, eaap7885. [Google Scholar] [CrossRef]
- Li, Z.; Gao, E.; Zhou, J.; Han, W.; Xu, X.; Gao, X. Applications of deep learning in understanding gene regulation. Cell Rep. Methods 2023, 3, 100384. [Google Scholar] [CrossRef]
- Hou, Z.; Yang, Y.; Ma, Z.; Wong, K.C.; Li, X. Learning the protein language of proteome-wide protein-protein binding sites via explainable ensemble deep learning. Commun. Biol. 2023, 6, 73. [Google Scholar] [CrossRef]
- Lin, P.; Yan, Y.; Huang, S.Y. DeepHomo2.0: Improved protein–protein contact prediction of homodimers by transformer-enhanced deep learning. Brief. Bioinform. 2023, 24, bbac499. [Google Scholar] [CrossRef]
- Torrisi, M.; Pollastri, G.; Le, Q. Deep learning methods in protein structure prediction. Comput. Struct. Biotechnol. J. 2020, 18, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Jha, K.; Saha, S.; Singh, H. Prediction of protein-protein interaction using graph neural networks. Sci. Rep. 2022, 12, 8360. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zheng, H. Drug-target interaction prediction based on spatial consistency constraint and graph convolutional autoencoder. BMC Bioinform. 2023, 24, 151. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. Prediction of protein–ligand binding affinity via deep learning models. Brief. Bioinform. 2024, 25, bbae081. [Google Scholar] [CrossRef]
- Suhartono, D.; Majiid, M.R.; Handoyo, A.T.; Wicaksono, P.; Lucky, H. Towards a more general drug target interaction prediction model using transfer learning. Procedia Comput. Sci. 2023, 216, 370–376. [Google Scholar] [CrossRef]
- Jo, T.; Nho, K.; Bice, P.; Saykin, A.J. Deep learning-based identification of genetic variants: Application to Alzheimer’s disease classification. Brief. Bioinform. 2022, 23, bbac022. [Google Scholar] [CrossRef]
- Sharma, P.; Dahiya, S.; Kaur, P.; Kapil, A. Computational biology: Role and scope in taming antimicrobial resistance. Indian J. Med. Microbiol. 2023, 41, 33–38. [Google Scholar] [CrossRef]
- Hinton, G.E.; Sabour, S.; Frosst, N. Matrix capsules with EM routing. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 6 May 2018. [Google Scholar]
- Giri, S.J.; Dutta, P.; Halani, P.; Saha, S. MultiPredGO: Deep multi-modal protein function prediction by amalgamating protein structure, sequence, and interaction information. IEEE J. Biomed. Health Inform. 2020, 25, 1832–1838. [Google Scholar] [CrossRef]
- Mohamed, T.; Sayed, S.; Salah, A.; Houssein, E.H. Long short-term memory neural networks for RNA viruses mutations prediction. Math. Probl. Eng. 2021, 2021, 9980347. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Xia, Y.; Liang, Z.; Du, X.; Cao, D.; Li, J.; Sun, L.; Guo, S. Design of function-regulating RNA via deep learning and AlphaFold 3. Brief. Bioinform. 2025, 26, bbaf419. [Google Scholar] [CrossRef] [PubMed]
- Stegle, O.; Pärnamaa, T.; Parts, L. Deep learning for computational biology. Mol. Syst. Biol. 2016, 12, 878. [Google Scholar]
- Ferrucci, D.A.; Brown, E.D.; Chu-Carroll, J.; Fan, J.Z.; Gondek, D.C.; Kalyanpur, A.; Lally, A.; Murdock, J.W.; Nyberg, E.; Prager, J.M.; et al. Building Watson: An Overview of the DeepQA Project. AI Mag. 2010, 31, 59. [Google Scholar] [CrossRef]
- Larrañaga, P.; Calvo, B.; Santana, R.; Bielza, C.; Galdiano, J.; Inza, I.; Lozano, J.A.; Armañanzas, R.; Santafé, G.; Pérez, A.; et al. Machine learning in bioinformatics. Brief. Bioinform. 2006, 7, 86–112. [Google Scholar] [CrossRef]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.D.; Kovacs, I.E.; De Kaa Ca, H.; Bult, P.; Van Ginneken, B.; Van Der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef]
- Mamoshina, P.; Vieira, A.; Putin, E.; Zhavoronkov, A. Applications of Deep Learning in Biomedicine. Mol. Pharm. 2016, 13, 1445–1454. [Google Scholar] [CrossRef]
- Dinov, I.D.; Heavner, B.; Tang, M.; Glusman, G.; Chard, K.; D’Arcy, M.; Madduri, R.; Pa, J.; Spino, C.; Kesselman, C.; et al. Predictive Big Data Analytics: A Study of Parkinson’s Disease Using Large, Complex, Heterogeneous, Incongruent, Multi-Source and Incomplete Observations. PLoS ONE 2016, 11, e0157077. [Google Scholar] [CrossRef]
- Carracedo-Reboredo, P.; Liñares-Blanco, J.; Rodriguez-Fernandez, N.; Cedrón, F.; Novoa, F.J.; Carballal, A.; Maojo, V.; Pazos, A.; Fernandez-Lozano, C. A review on machine learning approaches and trends in drug discovery. Comput. Struct. Biotechnol. J. 2021, 19, 4538–4558. [Google Scholar] [CrossRef]
- Zhao, L.; Ciallella, H.L.; Aleksunes, L.M.; Zhu, H. Advancing computer-aided drug discovery (CADD) by big data and data-driven machine learning modeling. Drug Discov. Today 2020, 25, 1624–1638. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; Wang, H.; Yan, J. Application of deep learning in genomics. Sci. China Life Sci. 2020, 63, 1860–1878. [Google Scholar] [CrossRef]
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Brief. Bioinform. 2016, 18, 851–869. [Google Scholar] [CrossRef]
- Ma, Q.; Xu, D. Deep learning shapes single-cell data analysis. Nat. Rev. Mol. Cell Biol. 2022, 23, 303–304. [Google Scholar] [CrossRef]
- Romão, M.C.; Castro, N.F.; Pedro, R.; Vale, T. Transferability of deep learning models in searches for new physics at colliders. Phys. Rev. D 2020, 101, 035042. [Google Scholar] [CrossRef]
- Prabhu, S.P. Ethical challenges of machine learning and deep learning algorithms. Lancet Oncol. 2019, 20, 621–622. [Google Scholar] [CrossRef] [PubMed]
- Data sharing in the age of deep learning. Nat. Biotechnol. 2023, 41, 433. [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.Q.; Duan, Y.; Al-Shamma, O.; Santamaría, J.V.G.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Wang, X.; Li, F.; Zhang, Y.; Imoto, S.; Shen, H.-H.; Li, S.; Guo, Y.; Yang, J.; Song, J. Deep learning approaches for non-coding genetic variant effect prediction: Current progress and future prospects. Brief. Bioinform. 2024, 25, bbae446. [Google Scholar] [CrossRef]
- Terranova, N.; Venkatakrishnan, K.; Benincosa, L.J. Application of Machine Learning in Translational Medicine: Current Status and Future Opportunities. AAPS J. 2021, 23, 74. [Google Scholar] [CrossRef]
- Yelmen, B.; Jay, F. An Overview of Deep Generative Models in Functional and Evolutionary Genomics. Annu. Rev. Biomed. Data Sci. 2023, 6, 173–189. [Google Scholar] [CrossRef]
- Liu, Y.; Qu, H.-Q.; Mentch, F.D.; Qu, J.; Chang, X.; Nguyen, K.; Tian, L.; Glessner, J.; Sleiman, P.M.A.; Hakonarson, H. Application of deep learning algorithm on whole genome sequencing data uncovers structural variants associated with multiple mental disorders in African American patients. Mol. Psychiatry 2022, 27, 1469–1478. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, S.J.; Tellam, R.L.; Morrison, J.L.; Muhlhausler, B.S.; Molloy, P.L. Recent developments on the role of epigenetics in obesity and metabolic disease. Clin. Epigenetics 2015, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Wu, M.; Liu, Q.; Lv, H.; Jiang, R. DeepHistone: A deep learning approach to predicting histone modifications. BMC Genom. 2019, 20 (Suppl. S2), 11–23. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-López, O.A.; Montesinos-López, A.; Pérez-Rodríguez, P.; Barrón-López, J.A.; Martini, J.W.R.; Fajardo-Flores, S.B.; Gaytan-Lugo, L.S.; Santana-Mancilla, P.C.; Crossa, J. A review of deep learning applications for genomic selection. BMC Genom. 2021, 22, 19. [Google Scholar] [CrossRef]
- Avsec, Ž.; Agarwal, V.; Visentin, D.; Ledsam, J.R.; Grabska-Barwinska, A.; Taylor, K.R.; Assael, Y.; Jumper, J.; Kohli, P.; Kelley, D.R. Effective gene expression prediction from sequence by integrating long-range interactions. Nat. Methods 2021, 18, 1196–1203. [Google Scholar] [CrossRef]
- Treppner, M.; Binder, H.; Hess, M. Interpretable generative deep learning: An illustration with single cell gene expression data. Hum. Genet. 2022, 141, 1481–1498. [Google Scholar] [CrossRef]
- Galkin, F.; Mamoshina, P.; Kochetov, K.; Sidorenko, D.; Zhavoronkov, A. DeepMAge: A Methylation Aging Clock Developed with Deep Learning. Aging Dis. 2021, 12, 1252. [Google Scholar] [CrossRef]
- Zhang, B.; Li, J.; Lü, Q. Prediction of 8-state protein secondary structures by a novel deep learning architecture. BMC Bioinform. 2018, 19, 293. [Google Scholar] [CrossRef]
- AlQuraishi, M. AlphaFold at CASP13. Bioinformatics 2019, 35, 4862–4865. [Google Scholar] [CrossRef]
- Feng, X.; Zhang, H.; Lin, H.; Long, H. Single-cell RNA-seq data analysis based on directed graph neural network. Methods 2023, 211, 48–60. [Google Scholar] [CrossRef]
- Wang, S.; Mo, B.; Zhao, J.; Zhao, J.; Zhao, J. Theory-based residual neural networks: A synergy of discrete choice models and deep neural networks. Transp. Res. Part B 2021, 146, 333–358. [Google Scholar] [CrossRef]
- Cramer, P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ma, A.; Chang, Y.; Gong, J.; Jiang, Y.; Qi, R.; Wang, C.; Fu, H.; Ma, Q.; Xu, D. scGNN is a novel graph neural network framework for single-cell RNA-Seq analyses. Nat. Commun. 2021, 12, 1882. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Edwards, M.D.; Liu, G.; Gifford, D.K. Convolutional neural network architectures for predicting DNA–protein binding. Bioinformatics 2016, 32, i121–i127. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.P.; Hou, T.S.; Su, Y.; Zheng, C.H. scSSA: A clustering method for single cell RNA-seq data based on semi-supervised autoencoder. Methods 2022, 208, 66–74. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, J.; Hu, H.; Gong, H.; Chen, L.; Cheng, C.; Zeng, J. A deep learning framework for modeling structural features of RNA-binding protein targets. Nucleic Acids Res. 2016, 44, e32. [Google Scholar] [CrossRef]
- Hashemifar, S.; Neyshabur, B.; Khan, A.A.; Xu, J. Predicting protein-protein interactions through sequence-based deep learning. Bioinformatics 2018, 34, i802–i810. [Google Scholar] [CrossRef]
- Yao, Y.; Du, X.; Diao, Y.; Zhu, H. An integration of deep learning with feature embedding for protein–protein interaction prediction. PeerJ 2019, 2019, e7126. [Google Scholar] [CrossRef]
- Yang, F.; Fan, K.; Song, D.; Lin, H. Graph-based prediction of Protein-protein interactions with attributed signed graph embedding. BMC Bioinform. 2020, 21, 323. [Google Scholar] [CrossRef]
- Dara, S.; Dhamercherla, S.; Jadav, S.S.; Babu, C.M.; Ahsan, M.J. Machine Learning in Drug Discovery: A Review. Artif. Intell. Rev. 2022, 55, 1947–1999. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Zhang, Z.; Niu, S.; Sha, H.; Yang, R.; Yun, Y.; Lu, H. Deep-Learning- Based Drug-Target Interaction Prediction. J. Proteome Res. 2017, 16, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- Suh, D.; Lee, J.W.; Choi, S.; Lee, Y. Recent applications of deep learning methods on evolution-and contact-based protein structure prediction. Int. J. Mol. Sci. 2021, 22, 6032. [Google Scholar] [CrossRef] [PubMed]
- Moussad, B.; Roche, R.; Bhattacharya, D. The transformative power of transformers in protein structure prediction. Proc. Natl. Acad. Sci. USA 2023, 120, e2303499120. [Google Scholar] [CrossRef]
- Pan, X.; Yang, Y.; Xia, C.Q.; Mirza, A.H.; Shen, H.B. Recent methodology progress of deep learning for RNA–protein interaction prediction. Wiley Interdiscip. Rev. RNA 2019, 10, e1544. [Google Scholar] [CrossRef]
- Sapoval, N.; Aghazadeh, A.; Nute, M.G.; Antunes, D.A.; Balaji, A.; Baraniuk, R.; Barberan, C.J.; Dannenfelser, R.; Dun, C.; Edrisi, M.; et al. Current progress and open challenges for applying deep learning across the biosciences. Nat. Commun. 2022, 13, 1728. [Google Scholar] [CrossRef]
- Akay, A.; Hess, H. Deep Learning: Current and Emerging Applications in Medicine and Technology. IEEE J. Biomed. Health Inform. 2019, 23, 906–920. [Google Scholar] [CrossRef]
- Choudhary, K.; DeCost, B.; Chen, C.; Jain, A.; Tavazza, F.; Cohn, R.; Park, C.W.; Choudhary, A.; Agrawal, A.; Billinge, S.J.L.; et al. Recent advances and applications of deep learning methods in materials science. NPJ Comput. Mater. 2022, 8, 59. [Google Scholar] [CrossRef]
- O’Connor, O.M.; Alnahhas, R.N.; Lugagne, J.; Dunlop, M.J. DeLTA 2.0: A deep learning pipeline for quantifying single-cell spatial and temporal dynamics. PLOS Comput. Biol. 2022, 18, e1009797. [Google Scholar] [CrossRef]
- Fernandez-Quilez, A. Deep learning in radiology: Ethics of data and on the value of algorithm transparency, interpretability and explainability. AI Ethics 2023, 3, 257–265. [Google Scholar] [CrossRef]
- Zhang, Y.; Ye, T.; Xi, H.; Juhas, M.; Li, J.J. Deep Learning Driven Drug Discovery: Tackling Severe Acute Respiratory Syndrome Coronavirus 2. Front. Microbiol. 2021, 12, 739684. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Feng, C.; Ling, T.; Chen, M. Deep learning frameworks for protein–protein interaction prediction. Comput. Struct. Biotechnol. J. 2022, 20, 3223–3233. [Google Scholar] [CrossRef] [PubMed]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020, 18, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Bai, Q.; Liu, S.; Tian, Y.; Xu, T.; Banegas-Luna, A.J.; Pérez-Sánchez, H.; Huang, J.; Liu, H.; Yao, X. Application advances of deep learning methods for de novo drug design and molecular dynamics simulation. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2021, 12, e1581. [Google Scholar] [CrossRef]
- Pan, X.; Lin, X.; Cao, D.; Zeng, X.; Yu, P.S.; He, L.; Nussinov, R.; Cheng, F. Deep learning for drug repurposing: Methods, databases, and applications. arXiv 2022. [Google Scholar] [CrossRef]
- Jobin, A.; Vayena, E. The global landscape of AI ethics guidelines. Nat. Mach. Intell. 2019, 1, 389–399. [Google Scholar] [CrossRef]
- Jing, Y.; Bian, Y.; Hu, Z.; Wang, L.; Xie, X.Q.S. Deep learning for drug design: An artificial intelligence paradigm for drug discovery in the big data era. AAPS J. 2018, 20, 58. [Google Scholar] [CrossRef]
- Lo Piano, S. Ethical principles in machine learning and artificial intelligence: Cases from the field and possible ways forward. Humanit. Soc. Sci. Commun. 2020, 7, 1–7. [Google Scholar] [CrossRef]
- Kuang, M.; Liu, Y.; Gao, L. DLPAlign: A deep learning based progressive alignment method for multiple protein sequences. In Proceedings of the CSBio’20, Eleventh International Conference on Computational Systems-Biology and Bioinformatics, New York, NY, USA, 19–21 November 2020; pp. 83–92. [Google Scholar]
- Sharma, P.B.; Ramteke, P. Recommendation for Selecting Smart Village in India through Opinion Mining Using Big Data Analytics. Indian Sci. J. Res. Eng. Manag. 2023, 07, 105–112. [Google Scholar] [CrossRef]
- Chen, H.; Engkvist, O.; Wang, Y.; Olivecrona, M.; Blaschke, T. The rise of deep learning in drug discovery. Drug Discov. Today 2018, 23, 1241–1250. [Google Scholar] [CrossRef]
- Braun, M.; Hummel, P.; Beck, S.C.; Dabrock, P. Primer on an ethics of AI-based decision support systems in the clinic. J. Med. Ethics 2021, 47, e3. [Google Scholar] [CrossRef] [PubMed]
- Jarrahi, M.H.; Askay, D.A.; Eshraghi, A.; Smith, P. Artificial intelligence and knowledge management: A partnership between human and AI. Bus. Horiz. 2022, 66, 87–99. [Google Scholar] [CrossRef]
- Hartl, D.; De Luca, V.; Kostikova, A.; Laramie, J.M.; Kennedy, S.D.; Ferrero, E.; Siegel, R.M.; Fink, M.; Ahmed, S.; Millholland, J.; et al. Translational precision medicine: An industry perspective. J. Transl. Med. 2021, 19, 245. [Google Scholar] [CrossRef] [PubMed]
- Zitnik, M.; Nguyen, F.; Wang, B.; Leskovec, J.; Goldenberg, A.; Hoffman, M.M. Machine learning for integrating data in biology and medicine: Principles, practice, and opportunities. Inf. Fusion 2018, 50, 71–91. [Google Scholar] [CrossRef]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Comput. Sci. 2021, 2, 1–20. [Google Scholar] [CrossRef]
- Li, S.; Hua, H.; Chen, S. Graph neural networks for single-cell omics data: A review of approaches and applications. Brief. Bioinform. 2025, 26, bbaf109. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Tan, R.K.; Liu, Y.; Xie, L. Reinforcement learning for systems pharmacology-oriented and personalized drug design. Expert Opin. Drug Discov. 2022, 17, 849–863. [Google Scholar] [CrossRef]
- Wang, J.; Xie, X.; Shi, J.; He, W.; Chen, Q.; Chen, L.; Gu, W.; Zhou, T. Denoising autoencoder, a deep learning algorithm, aids the identification of a novel molecular signature of lung adenocarcinoma. Genom. Proteom. Bioinform. 2020, 18, 468–480. [Google Scholar] [CrossRef]
- Zou, J.; Huss, M.; Abid, A.; Mohammadi, P.; Torkamani, A.; Telenti, A. A primer on deep learning in genomics. Nat. Genet. 2019, 51, 12–18. [Google Scholar] [CrossRef]
- .Sarkar, C.; Das, B.; Rawat, V.S.; Wahlang, J.B.; Nongpiur, A.; Tiewsoh, I.; Lyngdoh, N.M.; Das, D.; Bidarolli, M.; Sony, H.T. Artificial Intelligence and Machine Learning Technology Driven Modern Drug Discovery and Development. Int. J. Mol. Sci. 2023, 24, 2026. [Google Scholar] [CrossRef]
- Das, D.; Chakrabarty, B.; Srinivasan, R.; Roy, A. Gex2SGen: Designing Drug-like Molecules from Desired Gene Expression Signatures. J. Chem. Inf. Model. 2023, 63, 1882–1893. [Google Scholar] [CrossRef]
- Bansal, H.; Luthra, H.; Chaurasia, A. Impact of Machine Learning Practices on Biomedical Informatics, Its Challenges and Future Benefits. In Artificial Intelligence Technologies for Computational Biology; CRC Press: Boca Raton, FL, USA, 2023; pp. 273–294. [Google Scholar]
- Dou, H.; Tan, J.; Wei, H.; Wang, F.; Yang, J.; Ma, X.G.; Wang, J.; Zhou, T. Transfer inhibitory potency prediction to binary classification: A model only needs a small training set. Comput. Methods Programs Biomed. 2022, 215, 106633. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sinha, D. Synthetic attack data generation model applying generative adversarial network for intrusion detection. Comput. Secur. 2023, 125, 103054. [Google Scholar] [CrossRef]
- Gupta, A.; Zou, J. Feedback GAN for DNA optimizes protein functions. Nat. Mach. Intell. 2019, 1, 105–111. [Google Scholar] [CrossRef]
- Hamdy, R.; Maghraby, F.A.; Omar, Y.M. ConvChrome: Predicting gene expression based on histone modifications using deep learning techniques. Curr. Bioinform. 2022, 17, 273–283. [Google Scholar] [CrossRef]
- Cai, J.; Wang, S.; Xu, C.; Guo, W. Unsupervised deep clustering via contractive feature representation and focal loss. Pattern Recognit. 2022, 123, 108386. [Google Scholar] [CrossRef]
- Stepniewska-Dziubinska, M.M.; Zielenkiewicz, P.; Siedlecki, P. Development and evaluation of a deep learning model for protein–ligand binding affinity prediction. Bioinformatics 2018, 34, 3666–3674. [Google Scholar] [CrossRef]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: Review, opportunities and challenges. Brief. Bioinform. 2018, 19, 1236–1246. [Google Scholar] [CrossRef]

| Date | Developed by | Evolution |
|---|---|---|
| 1873 | A. Bain | Introduced earliest models of neural networks, called “Neural Groupings,” for associative memory, influencing later Hebbian learning concepts. |
| 1943 | Warren McCulloch & Walter Pitts | Introduced the McCulloch-Pitts (MCP) model, considered the precursor to artificial neural networks. |
| 1949 | Donald Hebb | Introduced Hebbian Learning Rule, fundamental to neural network learning; considered the “father” of neural learning concepts. |
| 1958 | Frank Rosenblatt | Introduced the perceptron, the first learning algorithm for neural networks resembling modern perceptrons. |
| 1969 | Marvin Minsky & Seymour Papert | Published “Perceptrons,” critically analyzing limitations of single-layer neural networks. |
| 1974 | Paul Werbos | First proposed backpropagation algorithm for training multilayer networks (widely popularized later). |
| 1980 | Teuvo Kohonen; Kunihiko Fukushima | Kohonen introduced Self-Organizing Maps (SOM); Fukushima introduced Neocognitron, a basis for modern CNNs. |
| 1982 | John Hopfield | Introduced Hopfield Network, a recurrent associative memory model. |
| 1985 | Geoffrey Hinton & Terry Sejnowski | Introduced the Boltzmann machine for probabilistic learning with undirected networks. |
| 1986 | Paul Smolensky; Michael I. Jordan | Smolensky introduced the Harmonium (precursor to Restricted Boltzmann Machines); Jordan conceptualized RNNs. |
| 1990 | Yann LeCun | Introduced LeNet, a convolutional neural network for handwriting recognition demonstrating deep learning’s potential. |
| 1997 | Mike Schuster & Kuldip K. Paliwal; Sepp Hochreiter & Jürgen Schmidhuber | Schuster & Paliwal introduced Bidirectional RNNs; Hochreiter & Schmidhuber introduced LSTM networks solving vanishing gradient issues. |
| 2006 | Geoffrey Hinton | Introduced Deep Belief Networks with layer-wise pretraining, marking a leap in modern deep learning. |
| 2009 | Ruslan Salakhutdinov & Geoffrey Hinton | Introduced Deep Boltzmann Machines for multilayer generative modeling. |
| 2012 | Geoffrey Hinton | Dropout technique introduced for efficient training and to reduce overfitting. |
| 2012 | Alex Krizhevsky, Ilya Sutskever, Geoffrey Hinton | Introduced AlexNet, a CNN architecture that won ImageNet competition and sparked the deep learning revolution. |
| 2014 | Ian Goodfellow, Yoshua Bengio, Aaron Courville | Introduced Generative Adversarial Networks (GANs) for image generation and data synthesis. |
| 2020 | Various researchers | Deep learning expanded to tasks like self-driving cars, medical imaging, financial trading using advanced CNNs and RNNs. |
| Group | Deep Learning Algorithm | Applications in Computational Biology | References |
|---|---|---|---|
| Supervised DL | Convolutional Neural Networks (CNNs) | Gene expression analysis Histopathology image analysis | [8,9,10] |
| Recurrent Neural Networks (RNNs) | DNA sequence analysis Protein secondary structure prediction and gene co-regulation (with gLMs) | [11,12] | |
| Long Short-Term Memory (LSTM) | RNA splicing prediction | [13] | |
| Convolutional Recurrent Neural Networks (CRNN) | Chromatin state prediction | [14] | |
| Deep Neural Networks (DNNs) | Metagenomic analysis | [15] | |
| Deep Survival Analysis | Cancer survival prediction | [7] | |
| Deep Transfer Learning | Drug response prediction | [16] | |
| Deep Clustering | Cell type identification | [17] | |
| Generative/Unsupervised DL | Generative Adversarial Networks (GANs) | Synthetic biology and protein design Synthetic data generation Synthetic biology and gene synthesis | [9,18,19,20] |
| Variational Autoencoders (VAEs) | Single-cell genomics analysis Metabolomics data analysis Disease gene prioritization (with VGAEs) | [21,22,23] | |
| Autoencoders | Disease diagnosis and prognosis Single-cell epigenomics analysis DNA motif discovery | [18,24,25] | |
| Deep Belief Networks (DBNs) | Protein structure prediction Genetic variant classification Drug side effect prediction | [16,26,27] | |
| Adversarial Autoencoders | Gene expression imputation Image-based phenotypic screening | [28,29] | |
| Deep Boltzmann Machines (DBMs) | Epigenetic data analysis | [30] | |
| Deep Generative Models | DNA sequence generation | [31] | |
| Advanced Architectures | Transformer Networks | RNA structure prediction Transcriptomics analysis Protein–protein interaction network analysis Protein contact prediction | [8,19,32,33] |
| Graph Neural Networks (GNNs) | Protein–protein interaction prediction Drug repurposing Cell type classification in single-cell transcriptomics | [22,34,35] | |
| Graph Convolutional Networks (GCNs) | Drug–target interaction prediction Drug response prediction Drug–target interaction network analysis | [36,37,38] | |
| Reinforcement Learning (RL)/Deep RL | Drug discovery and optimization Drug target identification Protein folding Protein–ligand binding affinity prediction Antibiotic resistance prediction Gene regulatory network inference Genome sequence assembly | [25,37,39,40] | |
| Capsule Networks | Protein structure classification Protein–protein interaction prediction Protein function prediction Cancer subtype classification | [8,9,41,42] | |
| Attention Mechanisms | Single-cell RNA sequencing analysis | [11] | |
| Deep Q-Networks (DQNs) | Drug toxicity prediction | [43] |
| Model Type | Primary Application | Key Advancement (2020–2025) | Performance Metrics | Computational Requirements | Limitations | References |
|---|---|---|---|---|---|---|
| CNNs | DNA methylation analysis | DeepCpG predicts methylation states with high accuracy (2017–2024) | AUC: 0.92 | Moderate (GPU required) | Requires large datasets, risk of overfitting | [33,61] |
| RNNs | Gene expression prediction | Improved sequential modeling, but limited by long-range dependencies | Accuracy: 85% | Moderate | Struggles with long sequences | [69,120] |
| Transformers | Protein structure prediction | AlphaFold 3 predicts protein complexes and ligand interactions (2024) | RMSD: <1 Å | High (TPU/GPU clusters) | Computationally intensive | [13,27] |
| GNNs | Single-cell omics | scGNN advances cell type interaction modeling (2024) | F1 Score: 0.89 | High | Interpretability issues | [77] |
| gLMs | Gene co-regulation prediction | Transformer-based gLMs predict single-cell co-regulation (2024) | AUC: 0.90 | High | Limited to specific datasets | [43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hein, Z.M.; Guruparan, D.; Okunsai, B.; Che Mohd Nassir, C.M.N.; Ramli, M.D.C.; Kumar, S. AI and Machine Learning in Biology: From Genes to Proteins. Biology 2025, 14, 1453. https://doi.org/10.3390/biology14101453
Hein ZM, Guruparan D, Okunsai B, Che Mohd Nassir CMN, Ramli MDC, Kumar S. AI and Machine Learning in Biology: From Genes to Proteins. Biology. 2025; 14(10):1453. https://doi.org/10.3390/biology14101453
Chicago/Turabian StyleHein, Zaw Myo, Dhanyashri Guruparan, Blaire Okunsai, Che Mohd Nasril Che Mohd Nassir, Muhammad Danial Che Ramli, and Suresh Kumar. 2025. "AI and Machine Learning in Biology: From Genes to Proteins" Biology 14, no. 10: 1453. https://doi.org/10.3390/biology14101453
APA StyleHein, Z. M., Guruparan, D., Okunsai, B., Che Mohd Nassir, C. M. N., Ramli, M. D. C., & Kumar, S. (2025). AI and Machine Learning in Biology: From Genes to Proteins. Biology, 14(10), 1453. https://doi.org/10.3390/biology14101453











