Unique and Under Pressure: Conservation Genetics of an Isolated Alpine Salamander Population
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
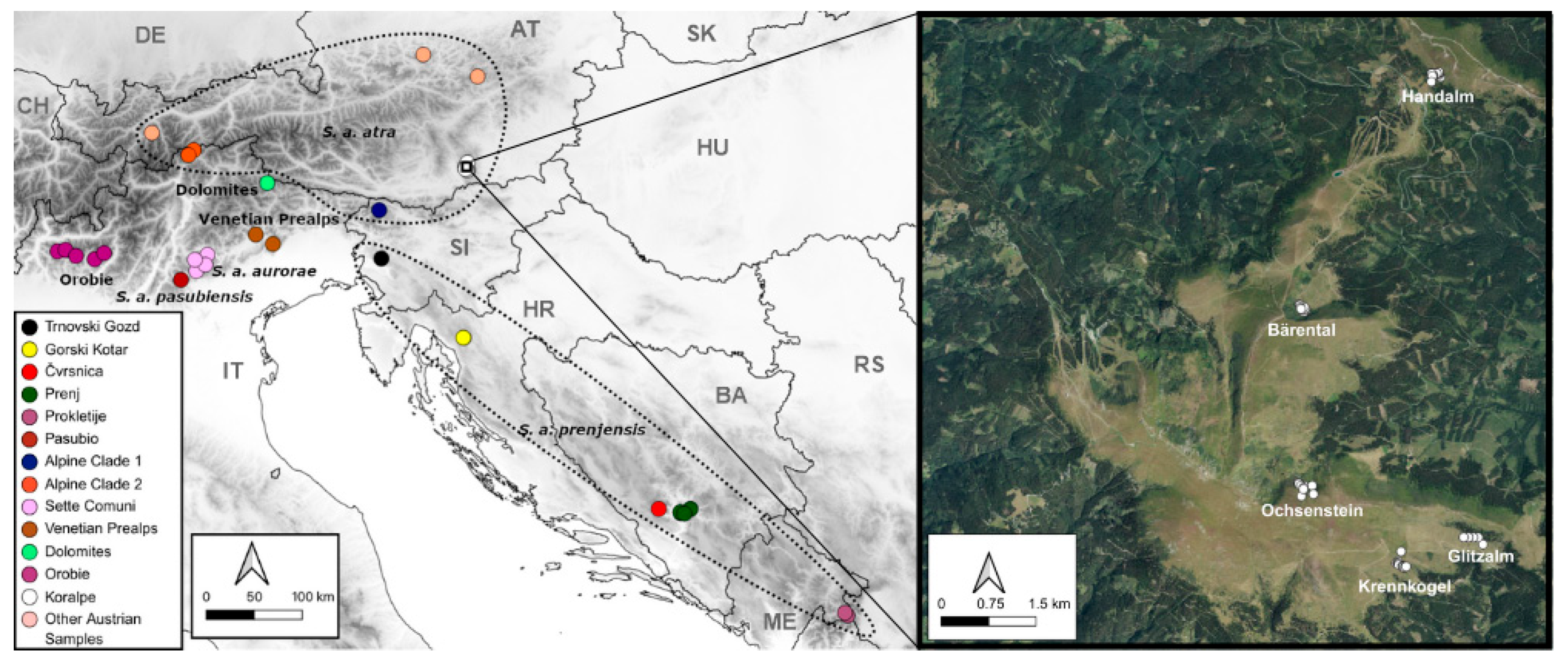
2.1. Sampling and DNA Extraction
2.2. Mitochondrial DNA
2.3. Microsatellite Markers
3. Results
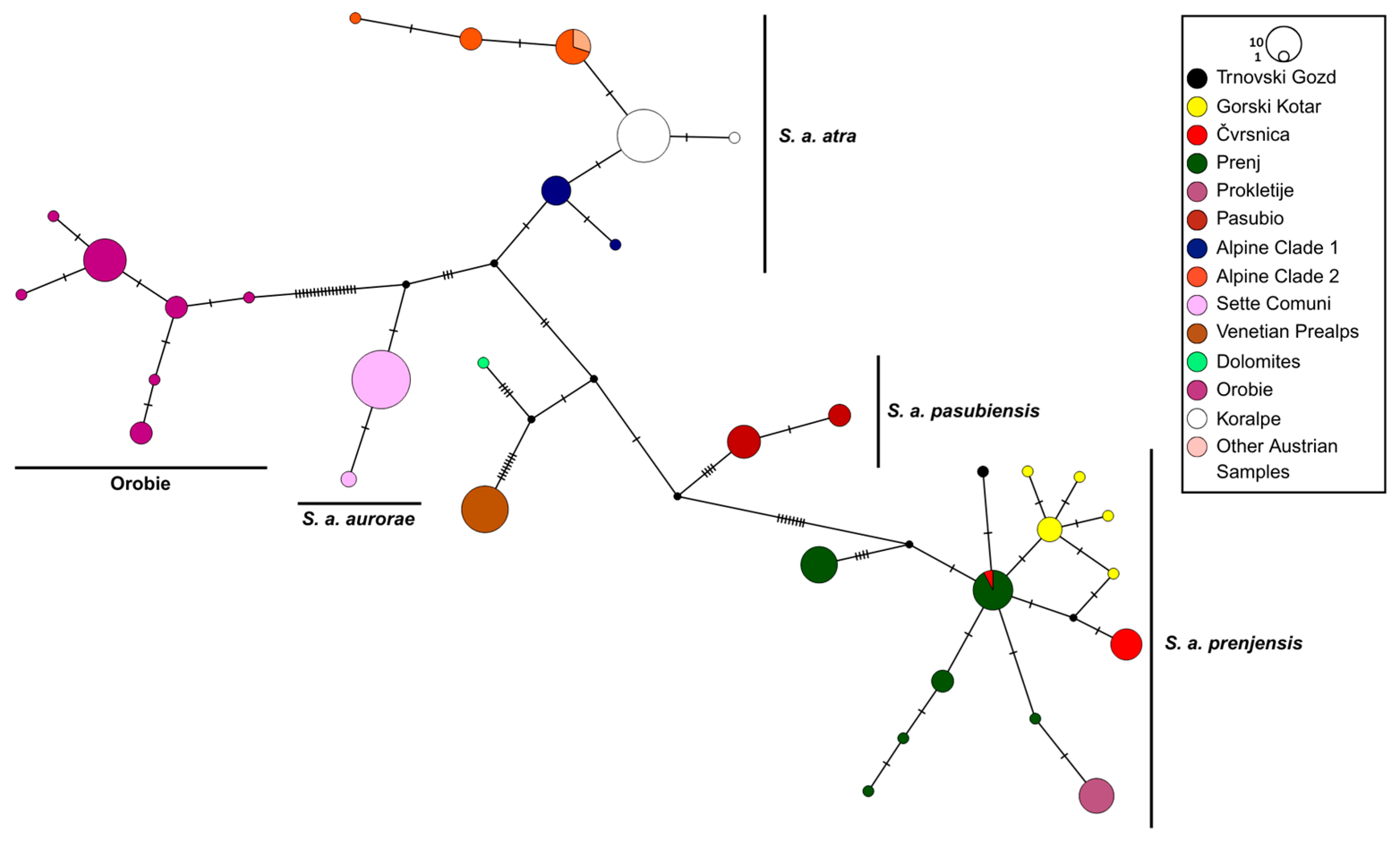
3.1. Mitochondrial DNA–Phylogenetic Placement of Salamandra atra from the Koralpe Mountain Range
3.2. Microsatellites–Population Structure of Salamandra atra in the Koralpe Mountain Range
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cardinale, B.J.; Duffy, J.E.; Gonzalez, A.; Hooper, D.U.; Perrings, C.; Venail, P.; Narwani, A.; Mace, G.M.; Tilman, D.; Wardle, D.A.; et al. Biodiversity loss and its impact on humanity. Nature 2012, 486, 59–67. [Google Scholar] [CrossRef]
- Kurtz, T.; Wübbels, G.; Portafaix, A.; Meyer zum Felde, A.; Zielcke, S. The Biodiversity Crisis is a Business Crisis; Boston Consulting Group: Boston, MA, USA, 2021. [Google Scholar]
- Hermoso, V.; Carvalho, S.B.; Giakoumi, S.; Goldsborough, D.; Katsanevakis, S.; Leontiou, S.; Markantonatou, V.; Rumes, B.; Vogiatzakis, I.N.; Yates, K.L. The EU Biodiversity Strategy for 2030: Opportunities and challenges on the path towards biodiversity recovery. Environ. Sci. Policy 2022, 127, 263–271. [Google Scholar] [CrossRef]
- Hallmann, C.A.; Sorg, M.; Jongejans, E.; Siepel, H.; Hofland, N.; Schwan, H.; Stenmans, W.; Müller, A.; Sumser, H.; Hörren, T.; et al. More than 75 percent decline over 27 years in total flying insect biomass in protected areas. PLoS ONE 2017, 12, e0185809. [Google Scholar] [CrossRef]
- Habel, J.C.; Schmitt, T. Vanishing of the common species: Empty habitats and the role of genetic diversity. Biol. Conserv. 2018, 218, 211–216. [Google Scholar] [CrossRef]
- Noss, R.F. Indicators for monitoring biodiversity: A hierarchical approach. Conserv. Biol. 1990, 4, 355–364. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Sgrò, C.M.; Kristensen, T.N. Revisiting adaptive potential, population size, and conservation. Trends Ecol. Evol. 2017, 32, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Allendorf, F.W.; Luikart, G.; Aitken, S.N. Conservation and the Genetics of Populations, 2nd ed.; Wiley-Blackwell: Oxford, UK, 2013. [Google Scholar]
- EU Commission. Communication from the Commission for the European Parliament, the Council, the European Economic and Social Committee and the Committee of Regions—EU Biodiversity Strategy for 2030: Bringing Nature Back into Our Lives; COM(2020) 380 final; EU Commission: Brussels, Belgium, 2020. [Google Scholar]
- Mimura, M.; Yahara, T.; Faith, D.P.; Vázquez-Domínguez, E.; Colautti, R.I.; Araki, H.; Javadi, F.; Núñez-Farfán, J.; Mori, A.S.; Zhou, S.; et al. Understanding and monitoring the consequences of human impacts on intraspecific variation. Evol. Appl. 2017, 10, 121–139. [Google Scholar] [CrossRef]
- Paz-Vinas, I.; Loot, G.; Hermoso, V.; Veyssière, C.; Poulet, N.; Grenouillet, G.; Blanchet, S. Systematic conservation planning for intraspecific genetic diversity. Proc. R. Soc. B 2018, 285, 20172746. [Google Scholar] [CrossRef]
- Gasc, J.-P.; Cabela, A.; Crnobrnja-Isailovic, J.; Dolmen, D.; Grossenbacher, K.; Haffner, P.; Lescure, J.; Martens, H.; Martinez Rica, J.P.; Maurin, H.; et al. Atlas of Amphibians and Reptiles in Europe; Societas Europaea Herpetologica & Museum National d’Histoire Naturelle: Paris, France, 1997. [Google Scholar]
- Meikl, M.; Reinthaler-Lottermoser, U.; Weinke, E.; Schwarzenbacher, R. Collection of fire salamander (Salamandra salamandra) and Alpine salamander (Salamandra atra) distribution data in Austria using a new, community-based approach. J. Prot. Mt. Areas Res. Manag. 2010, 2, 59–65. [Google Scholar]
- Šunje, E.; Zuazu Bermejo, A.; Van Damme, E.; Bakeljau, T.; Pojskić, N.; Lukić Bilela, L.; Kalamujić Stroil, B. Genetic diversity and differentiation of alpine salamanders from the Dinarides—An evolutionary perspective with insights for species conservation. Salamandra 2021, 57, 75–88. [Google Scholar]
- Council of the European Communities. In Council Directive 92/43/EEC of 21 May 1992 on the Conservation of Natural Habitats and of Wild Fauna and Flora; European Union: Brussels, Belgium, 1992; Volume 206, pp. 7–50.
- Gollmann, G. Rote Liste gefährdeter Lurche (Amphibia) und Kriechtiere (Reptilia) Österreichs. In Grüne Reihe Band 14/2: Rote Liste gefährdeter Tiere Österreichs: Kriechtiere, Lurche, Fische, Nachtfalter, Weichtiere. Checklisten, Gefährdungsanalysen, Handlungsbedarf; Zulka, K.P., Wallner, R.M., Eds.; Böhlau: Vienna, Austria, 2007; pp. 37–60. [Google Scholar]
- Kammel, W. Lurche (Amphibia). In Rote Listen der Tiere der Steiermark, Teil 2A.; Ökoteam, Ed.; unpublished project report on behalf of the Austrian Youth Nature Conservation Association for the State of Styria; Nature Conservation: Graz, Austria, 2021; pp. 35–64. [Google Scholar]
- Lamprecht, J.; Komposch, C.; Gutleb, B.; Petutschnig, W. Amphibien (Amphibia). In Rote Liste gefährdeter Tiere Kärntens; Komposch, C., Ed.; Verlag des Naturwissenschaftlichen Vereins für Kärnten: Klagenfurt am Wörthersee, Austria, 2023; pp. 281–300. [Google Scholar]
- Riberon, A.; Miaud, C.; Grossenbacher, K.; Taberlet, P. Phylogeography of the Alpine salamander, Salamandra atra (Salamandridae) and the influence of the Pleistocene climatic oscillations on population divergence. Mol. Ecol. 2001, 10, 2555–2560. [Google Scholar] [CrossRef] [PubMed]
- Razpet, A.; Šunje, E.; Kalamujić, B.; Tulić, U.; Pojskić, N.; Stanković, D.; Krizmanić, I.; Marić, S. Genetic differentiation and population dynamics of Alpine salamanders (Salamandra atra, Laurenti 1768) in southeastern Alps and Dinarides. Herpetetol. J. 2016, 26, 109–119. [Google Scholar]
- Bonato, L.; Corbetta, A.; Giovine, G.; Romanazzi, E.; Šunje, E.; Vernesi, E.; Crestanello, B. Diversity among peripheral population: Genetic and evolutionary differentiation of Salamandra atra at the southern edge of the Alps. J. Zool. Syst. Evol. Res. 2018, 56, 533–548. [Google Scholar] [CrossRef]
- Sedlmayr, I.; Bernhart, E.; Fachbach, G.; Holzinger, W.E.; Kammel, W.; Lipovnik, C. Erster Nachweis des Alpensalamanders (Salamandra atra Laurenti, 1768) und aktuelle Bestandsaufnahmen im steirischen Koralmgebiet. Joannea Zool. 2020, 18, 25–32. [Google Scholar]
- Komposch, C.; Paill, W.; Aurenhammer, S.; Graf, W.; Degasperi, G.; Dejaco, T.; Friess, T.; Holzinger, W.; Leitner, A.; Rabitsch, W.; et al. Endemitenberg Koralpe—Erste Zusammenfassende Darstellung (Literaturauswertung) der Zoologischen und Botanischen Endemiten Dieses Einzigartigen Gebirgsstocks; unpublished project report commissioned; The Environmental Ombudswomen of the State of Styria: Graz, Austria, 2016. [Google Scholar]
- Berg, C.; Drescher, A. Hochmontane Lebensräume auf der endemitenreichen Koralpe. Beiheft Nr. 2018, 11, 149–169. [Google Scholar]
- Tribsch, A. Areas of endemism of vascular plants in the Eastern Alps in relation to Pleistocene glaciation. J. Biogeogr. 2004, 31, 747–760. [Google Scholar] [CrossRef]
- Schönswetter, P.; Stehlik, I.; Holderegger, R.; Tribsch, A. Molecular evidence for glacial refugia of mountain plants in European Alps. Mol. Ecol. 2005, 14, 3547–3555. [Google Scholar] [CrossRef]
- Haubrich, K.; Schmitt, T. Cryptic differentiation in alpine-endemic, high-altitude butterflies reveals down-slope glacial refugia. Mol. Ecol. 2007, 16, 3643–3658. [Google Scholar] [CrossRef]
- Zangl, L.; Daill, D.; Schweiger, S.; Gassner, G.; Koblmüller, S. A reference DNA barcode library for Austrian amphibians and reptiles. PLoS ONE 2020, 15, e0229454. [Google Scholar] [CrossRef]
- Steinfartz, S.; Veith, M.; Tautz, D. Mitochondrial sequence analysis of Salamandra taxa suggests old splits of major lineages and postglacial recolonizations of Central Europe from distinct source populations of Salamandra salamandra. Mol. Ecol. 2000, 9, 397–410. [Google Scholar] [CrossRef]
- Vences, M.; Sanchez, E.; Hauswaldt, J.S.; Eikelmann, D.; Rodríguez, A.; Carranza, S.; Donaire, D.; Gehara, M.; Helfer, V.; Lötters, S.; et al. Nuclear and mitochondrial multilocus phylogeny and survey of alkaloid content in true salamanders of the genus Salamandra (Salamandridae). Mol. Phylogenet. Evol. 2014, 73, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Koblmüller, S.; Resl, P.; Klar, N.; Bauer, H.; Zangl, L.; Hahn, C. DNA barcoding for species identification of moss-dwelling invertebrates: Performance of nanopore sequencing and coverage in reference database. Diversity 2024, 16, 196. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Templeton, A.R.; Crandall, K.A.; Sing, C.F. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data III. Cladogram estimation. Genetics 1992, 132, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Hasesler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Rodríguez, A.; Burgon, J.D.; Lyra, M.; Irisarri, I.; Baurain, D.; Blaustein, L.; Göçmen, B.; Künzel, S.; Mable, B.K.; Nolte, A.W.; et al. Inferring the shallow phylogeny of true salamanders (Salamandra) by multiple phylogenomic approaches. Mol. Phylogenet. Evol. 2017, 115, 16–26. [Google Scholar] [CrossRef]
- Zhang, P.; Papenfuss, T.J.; Wake, M.H.; Qu, L.; Wake, D.B. Phylogeny and biogeography of the family Salamandridae (Amphibia: Caudata) inferred from complete mitochondrial genomes. Mol. Phylogenet. Evol. 2008, 49, 586–597. [Google Scholar] [CrossRef]
- Hendrix, R.; Hauswaldt, J.S.; Veith, M.; Steinfartz, S. Strong correlation between cross-amplification success and genetic distance across all members of ‘True Salamanders’ (Amphibia: Salamandridae) revealed by Salamandra salamandra-specific microsatellite loci. Mol. Ecol. Res. 2010, 10, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Steinfartz, S.; Küsters, D.; Tautz, D. Isolation and characterization of polymorphic tetranucleotide microsatellite loci in the Fire salamander Salamandra salamandra (Amphibia: Caudata). Mol. Ecol. Notes 2004, 4, 626–628. [Google Scholar] [CrossRef]
- Helfer, V.; Gimeno, A.; Balzarini, L.; Schwarzenbacher, R.; Ferri, V. Genetic characterization of alpine salamander populations in Valtelline (Lombardia, Italy): Preliminary results. Panura 2011, 27, 91–94. [Google Scholar]
- Helfer, V.; Broquet, T.; Fumagalli, L. Sex-specific estimates of dispersal show female philopatry and male dispersal in a promiscuous amphibian, the alpine salamander (Salamandra atra). Mol. Ecol. 2012, 21, 4706–4720. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2007, 1, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.; Cockerham, C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach for multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, L. CLUMPAK: A program for identifying clustering models and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Wilson, G.A.; Rannala, B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 2003, 163, 1177–1191. [Google Scholar] [CrossRef] [PubMed]
- Lischer, H.E.L.; Excoffier, L. PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 2012, 28, 298–299. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Kuhner, M.K. Coalescent genealogy samplers: Windows into population history. Trends Ecol. Evol. 2009, 24, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Faubet, P.; Waples, R.S.; Gaggiotti, O.E. Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol. Ecol. 2007, 16, 1149–1166. [Google Scholar] [CrossRef]
- Spiegelhalter, D.J.; Brest, N.G.; Carlin, B.P.; Van der Linde, A. Bayesian measures of model complexity and fit. J. R. Stat. Soc. Ser. B 2002, 64, 583–639. [Google Scholar] [CrossRef]
- Meirmans, P.G. Nonconvergence in Bayesian estimation of migration rates. Mol. Ecol. Resour. 2014, 14, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize: Implements and enhances circular visualization in R. Bioinformatics 2014, 20, 2811–2812. [Google Scholar] [CrossRef]
- Kivelä, M.; Arnaud-Haond, S.; Saramäki, J. EDENetworks: A use-friendly software to build and analyse networks in biogeography, ecology and population genetics. Mol. Ecol. Resour. 2015, 15, 117–122. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: A largely untapped resource for applied conservation and evolution. Evol. Appl. 2010, 3, 244–262. [Google Scholar] [CrossRef] [PubMed]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Gilbert, K.J.; Whitlock, M.C. Evaluating methods for estimating local effective population size with and without migration. Evolution 2015, 69, 2154–2166. [Google Scholar] [CrossRef]
- Waples, R.S. Practical application of the linkage disequilibrium method for estimating contemporary effective population size: A review. Mol. Ecol. 2024, 24, e13879. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J.-M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Cornuet, J.-M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [CrossRef]
- Luikart, G.; Allendorf, F.W.; Cornuet, J.-M.; Sherwin, W.B. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 1998, 89, 238–247. [Google Scholar] [CrossRef]
- Carlsson, J. Effects of microsatellite ull alleles on assignment testing. J. Hered. 2008, 99, 616–623. [Google Scholar] [CrossRef]
- Wang, J. Estimation of migration rates from marker-based parentage analysis. Mol. Ecol. 2014, 23, 3191–3213. [Google Scholar] [CrossRef]
- Amt der Steiermärkischen Landesregierung. Gutachten Zum Einfluss des Projekts “PSW Koralm” Auf die Lokale Population des Alpensalamanders (GZ: ABT13-205895/2020-44); Abteilung 13 Umwelt und Raumordnung; Official Expert Report: Graz, Austria, 2020. [Google Scholar]
- Klewen, R. Die Landsalamander Europas. In Teil 1: Die Gattungen Salamandra und Mertensiella; Die Neue Brehm-Bücherei: Wittenberg Lutherstadt, Germany, 1988. [Google Scholar]
- Bonato, L.; Fracasso, G. Movements, distribution pattern and density in a population of Salamandra atra aurorae (Caudata: Salamandridae). Amphib.-Reptil. 2003, 24, 251–260. [Google Scholar] [CrossRef]
- Häfli, H.-P. Zur Fortpflanzungsbiologie des Alpensalamanders (Salamandra atra Laur.). Rev. Suisse Zool. 1971, 78, 235–293. [Google Scholar] [CrossRef]
- Greven, H. Survey of the oviduct of salamandrids with special reference to the viviparous species J. Exp. Zool. Part. A Ecol. Genet. Physiol. 1998, 282, 507–525. [Google Scholar] [CrossRef]
- Luiselli, L.; Andreone, F.; Capizzi, D.; Anibaldi, C. Body size, population structure and fecundity traits of a Salamandra atra atra (Amphibia, Urodelia, Salamandridae) population from the northeastern Italian Alps. Ital. J. Zool. 2001, 68, 125–130. [Google Scholar] [CrossRef]
- Fachbach, G. Röhrenknochenentwicklung und Altersbestimmung bei Salamandra atra Laur., 1768 (Urodela, Salamandridae). Zool. Anz. 1988, 221, 188–200. [Google Scholar]
- Waples, R.S.; Antao, T.; Luikart, G. Effects of overlapping generations on linkage disequilibrium estimates of effective population size. Genetics 2014, 197, 769–780. [Google Scholar] [CrossRef]
- Franklin, I.R. Evolutionary change in small populations. In Conservation Biology: An Evolutionary-Ecological Perspective; Soulé, M.E., Wilcox, B.A., Eds.; Sinauer Associates: Sunderland, MA, USA, 1980; pp. 135–149. [Google Scholar]
- Jamieson, I.G.; Allendorf, F.W. How does the 50/500 rule apply to MVPs? Trends Ecol. Evol. 2012, 27, 578–584. [Google Scholar] [CrossRef]
- Frankham, R.; Bradshaw, C.J.A.; Brook, B.W. Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 2014, 170, 56–63. [Google Scholar] [CrossRef]
- Feldmeier, S.; Schmidt, B.R.; Zimmermann, N.E.; Veith, M.; Ficetola, G.F.; Lötters, S. Shifting aspect or elevation? The climate change response of ectotherms in a complex mountain topography. Divers. Distrib. 2020, 26, 1483–1495. [Google Scholar] [CrossRef]
- Dubos, N.; Harvard, A.; Crottini, A.; Seglie, D.; Andreone, F. Predicting future conservation areas while avoiding sympatry in two alpine amphibians severely threatened by climate change. J. Nat. Conserv. 2023, 76, 126490. [Google Scholar] [CrossRef]
- Čengić, M.; Šunje, E.; Bonato, L.; Van Damme, R.; Lenders, R.H.J.; Hujbregts, M.A.; Lukić Bilela, L.; Schipper, A.M. A multi-modelling approach for informing the conservation of a cold-adapted terrestrial amphibian in the face of climate change. J. Biogeogr. 2024, 51, 2469–2483. [Google Scholar] [CrossRef]
- Gobiet, A.; Kotlarski, S. Future climate change in the European Alps. In Oxford Research Encyclopedia of Climate Science; Gobiet, A., Kotlarski, S., Eds.; Oxford University Press: Oxford, UK, 2020. [Google Scholar] [CrossRef]
- Moritz, C. Applications of mitochondrial DNA analysis in conservation: A critical review. Mol. Ecol. 1994, 3, 401–411. [Google Scholar] [CrossRef]
- Lesica, P.; Allendorf, F.W. When are peripheral population valuable for conservation? Conserv. Biol. 1995, 9, 753–760. [Google Scholar] [CrossRef]


| Population | Locus | Average (s.d.) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SalE12 | SalE6 | SalE8 | SST-C3 | SST-E11 | SST-G6 | Sal23 | SalE7 | SalE14 | |||
| HA (N = 21) | NA | 6 | 6 | 2 | 4 | 9 | 2 | 4 | 7 | 6 | 5.11 (2.32) |
| HO | 0.857 | 0.810 | 0.381 | 0.571 | 0.810 | 0.211 | 0.714 | 0.762 | 0.619 | 0.637 (0.218) | |
| HE | 0.775 | 0.722 | 0.372 | 0.553 | 0.830 | 0.341 | 0.612 | 0.800 | 0.763 | 0.640 (0.184) | |
| BA (N = 16) | NA | 5 | 3 | 2 | 4 | 10 | 2 | 4 | 4 | 7 | 4.56 (2.56) |
| HO | 0.875 | 0.563 | 0.250 | 0.500 | 0.875 | 0.286 | 0.538 | 0.917 | 0.625 | 0.603 (0.247) | |
| HE | 0.800 | 0.599 | 0.229 | 0.464 | 0.780 | 0.349 | 0.726 | 0.764 | 0.823 | 0.615 (0.219) | |
| OS (N = 13) | NA | 7 | 7 | 3 | 5 | 8 | 1 | 4 | 6 | 6 | |
| HO | 0.769 | 0.9230 | 0.308 | 0.923 | 0.846 | NA | 0.692 | 0.615 | 0.769 | 0.731 (0.201) | |
| HE | 0.828 | 0.766 | 0.283 | 0.757 | 0.840 | NA | 0.742 | 0.680 | 0.837 | 0.717 (0.184) | |
| KK (N = 20) | NA | 4 | 5 | 2 | 5 | 7 | 2 | 3 | 6 | 4 | 4.22 (1.72) |
| HO | 0.700 | 0.550 | 0.050 | 0.750 | 0.700 | 0.450 | 0.400 | 0.500 | 0.700 | 0.533 (0.221) | |
| HE | 0.673 | 0.709 | 0.050 | 0.745 | 0.629 | 0.512 | 0.650 | 0.685 | 0.763 | 0.602 (0.220) | |
| GA (N = 20) | NA | 5 | 6 | 2 | 4 | 9 | 2 | 4 | 4 | 6 | 4.67 (2.18) |
| HO | 0.750 | 0.500 | 0.300 | 0.700 | 0.850 | 0.300 | 0.650 | 0.350 | 0.750 | 0.572 (0.214) | |
| HE | 0.700 | 0.664 | 0.328 | 0.696 | 0.787 | 0.431 | 0.668 | 0.455 | 0.753 | 0.609 (0.162) | |
| average | NA | 5.4 | 5.4 | 2.2 | 4.4 | 8.6 | 1.8 | 3.8 | 5.4 | 5.8 | |
| total | NA | 7 | 8 | 3 | 6 | 15 | 2 | 5 | 7 | 10 |
| Population | HA | BA | OS | KK |
|---|---|---|---|---|
| BA | 0.0236 ** | |||
| OS | 0.0318 ** | 0.0362 ** | ||
| KK | 0.0947 *** | 0.0770 *** | 0.0467 ** | |
| GA | 0.0618 *** | 0.0618 *** | 0.0185 * | 0.0900 *** |
| Population | TPM | Mode Shift |
|---|---|---|
| HA | 0.213 | No |
| BA | 0.326 | Yes |
| OS | 0.629 | No |
| KK | 0.367 | No |
| GA | 0.590 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koblmüller, S.; Schäffer, S.; Donabaum, R.; Sedlmayr, I.; Kammel, W.; Bernhart, E.; Zangl, L. Unique and Under Pressure: Conservation Genetics of an Isolated Alpine Salamander Population. Biology 2025, 14, 1428. https://doi.org/10.3390/biology14101428
Koblmüller S, Schäffer S, Donabaum R, Sedlmayr I, Kammel W, Bernhart E, Zangl L. Unique and Under Pressure: Conservation Genetics of an Isolated Alpine Salamander Population. Biology. 2025; 14(10):1428. https://doi.org/10.3390/biology14101428
Chicago/Turabian StyleKoblmüller, Stephan, Sylvia Schäffer, Raphael Donabaum, Irmgard Sedlmayr, Werner Kammel, Eva Bernhart, and Lukas Zangl. 2025. "Unique and Under Pressure: Conservation Genetics of an Isolated Alpine Salamander Population" Biology 14, no. 10: 1428. https://doi.org/10.3390/biology14101428
APA StyleKoblmüller, S., Schäffer, S., Donabaum, R., Sedlmayr, I., Kammel, W., Bernhart, E., & Zangl, L. (2025). Unique and Under Pressure: Conservation Genetics of an Isolated Alpine Salamander Population. Biology, 14(10), 1428. https://doi.org/10.3390/biology14101428







