Mitogenomic Characterization of Microhyla fissipes and Its Implications for Phylogenetic Analysis in Microhylidae
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Sample Collection and DNA Extraction
2.3. PCR Amplification and Sequencing
2.4. Mitogenome Assembly, Annotation and Sequence Analysis
2.5. Phylogenetic Analysis
2.6. Divergence Time Estimates Focused on Microhylidae
2.7. Ka and Ks Analysis
3. Results
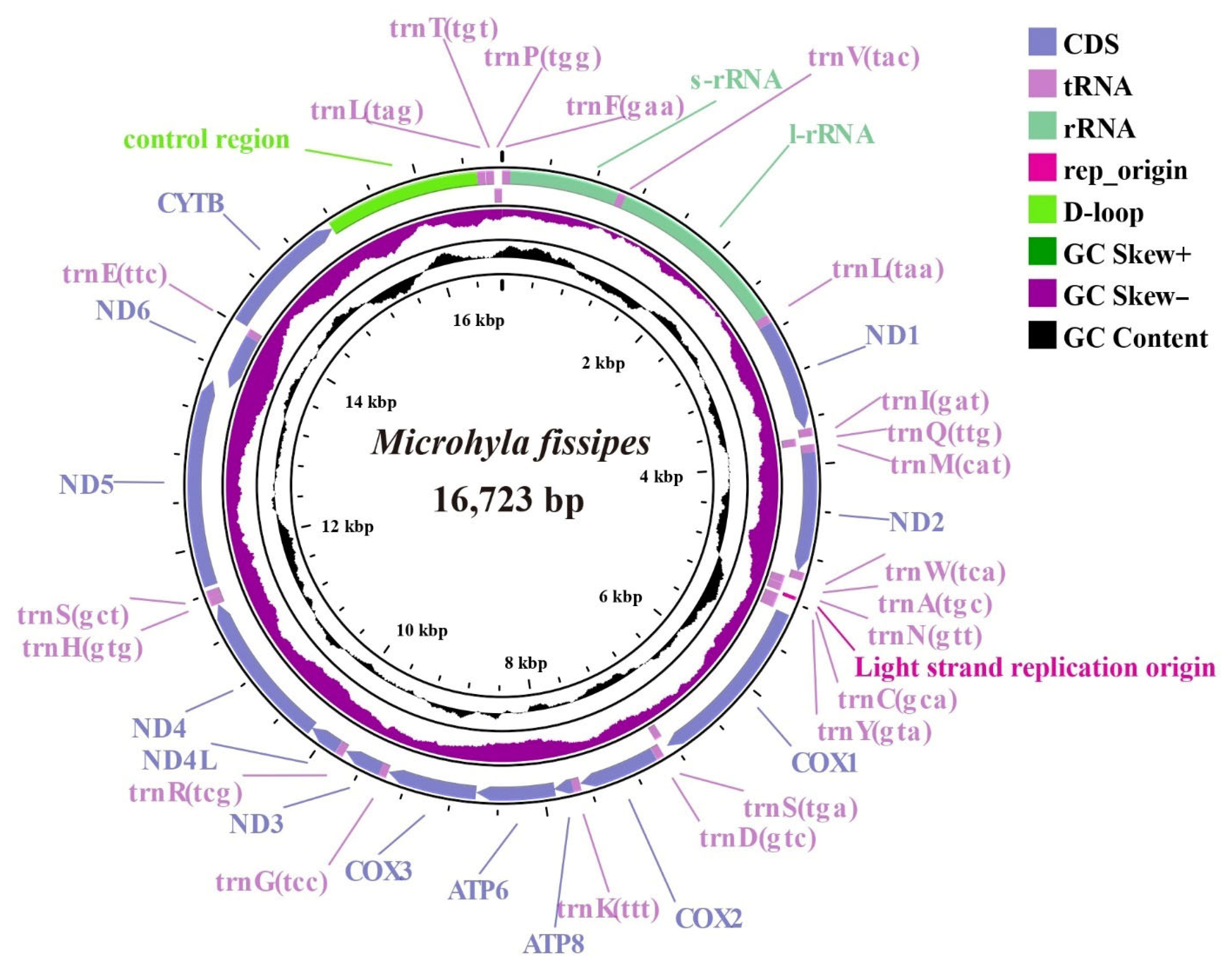
3.1. Composition of the M. fissipes Mitochondrial Genome
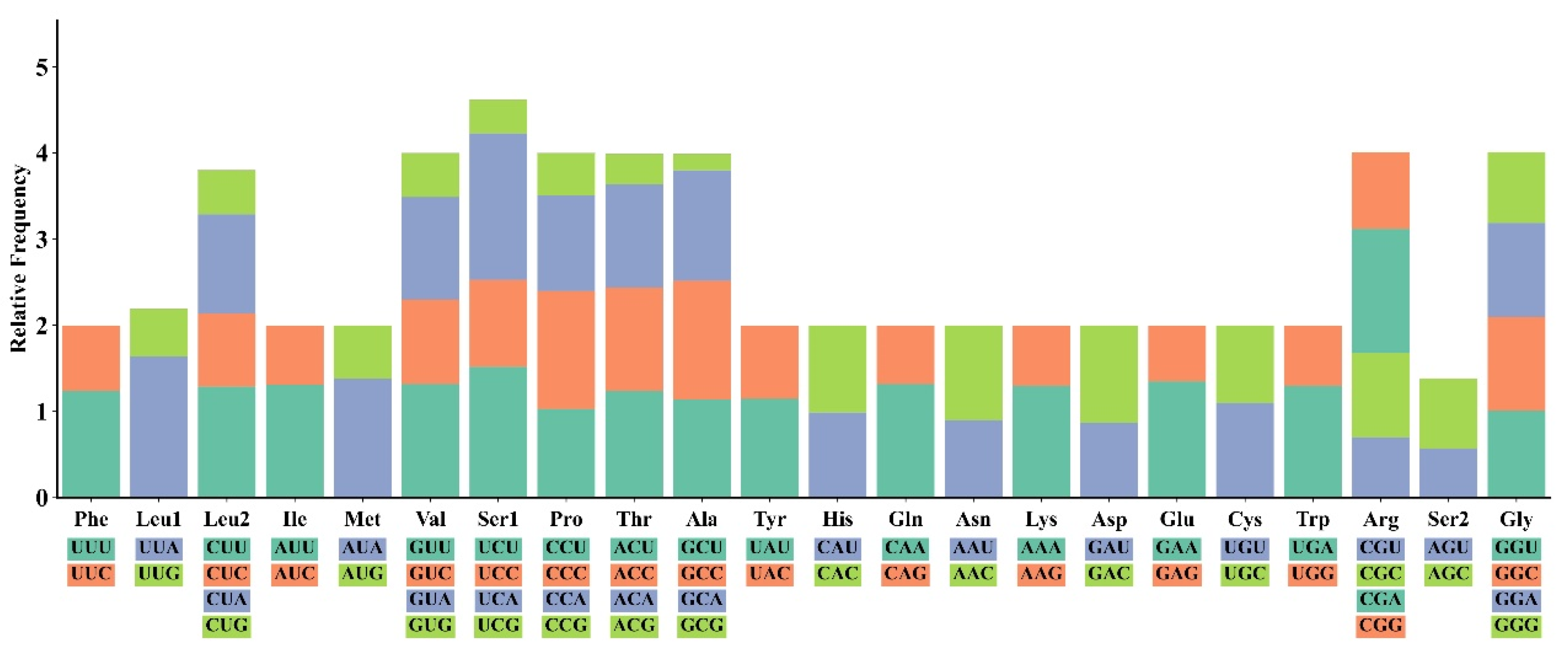
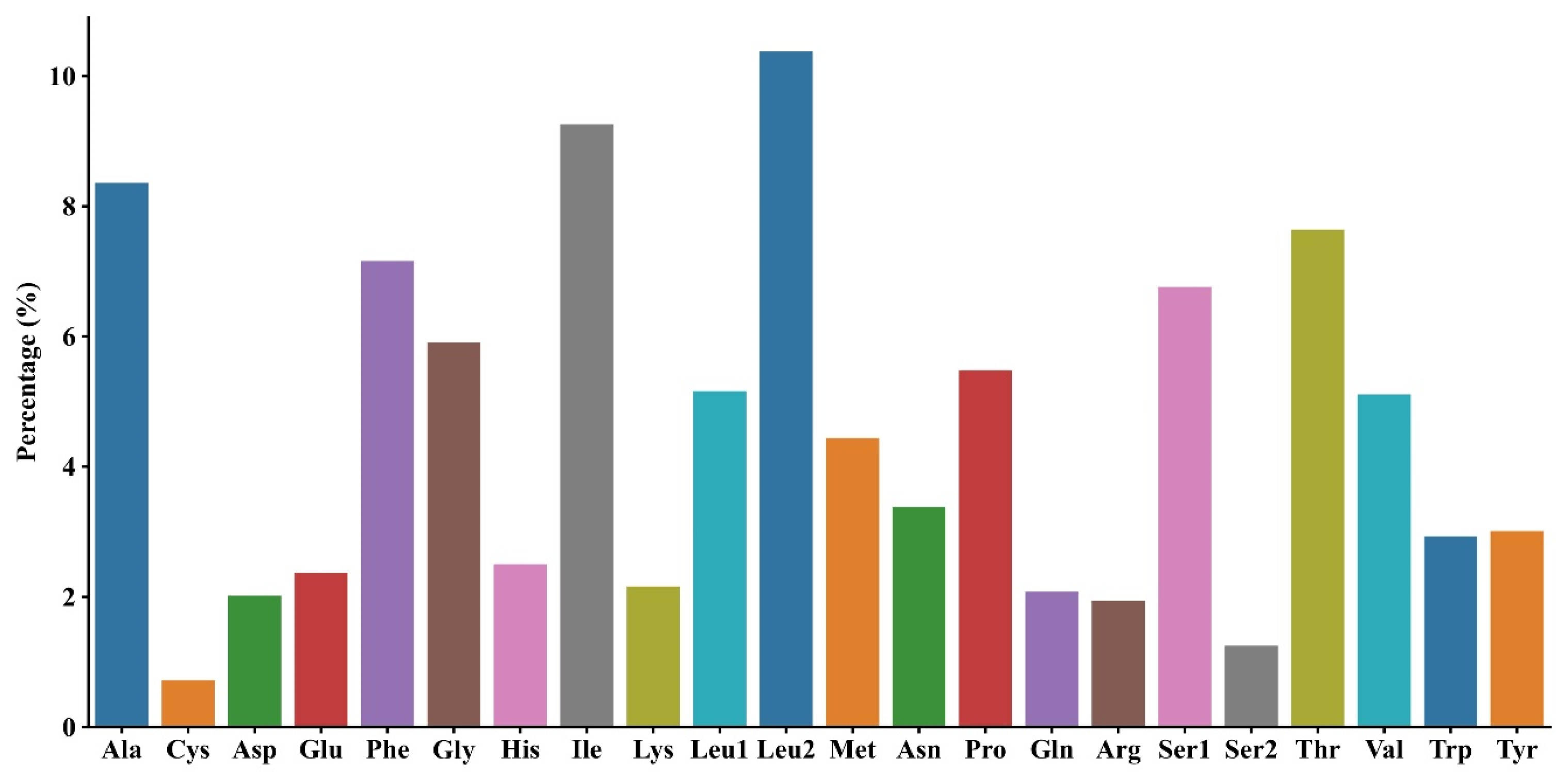
3.2. PCGs and Codon Usage Patterns of M. fissipes
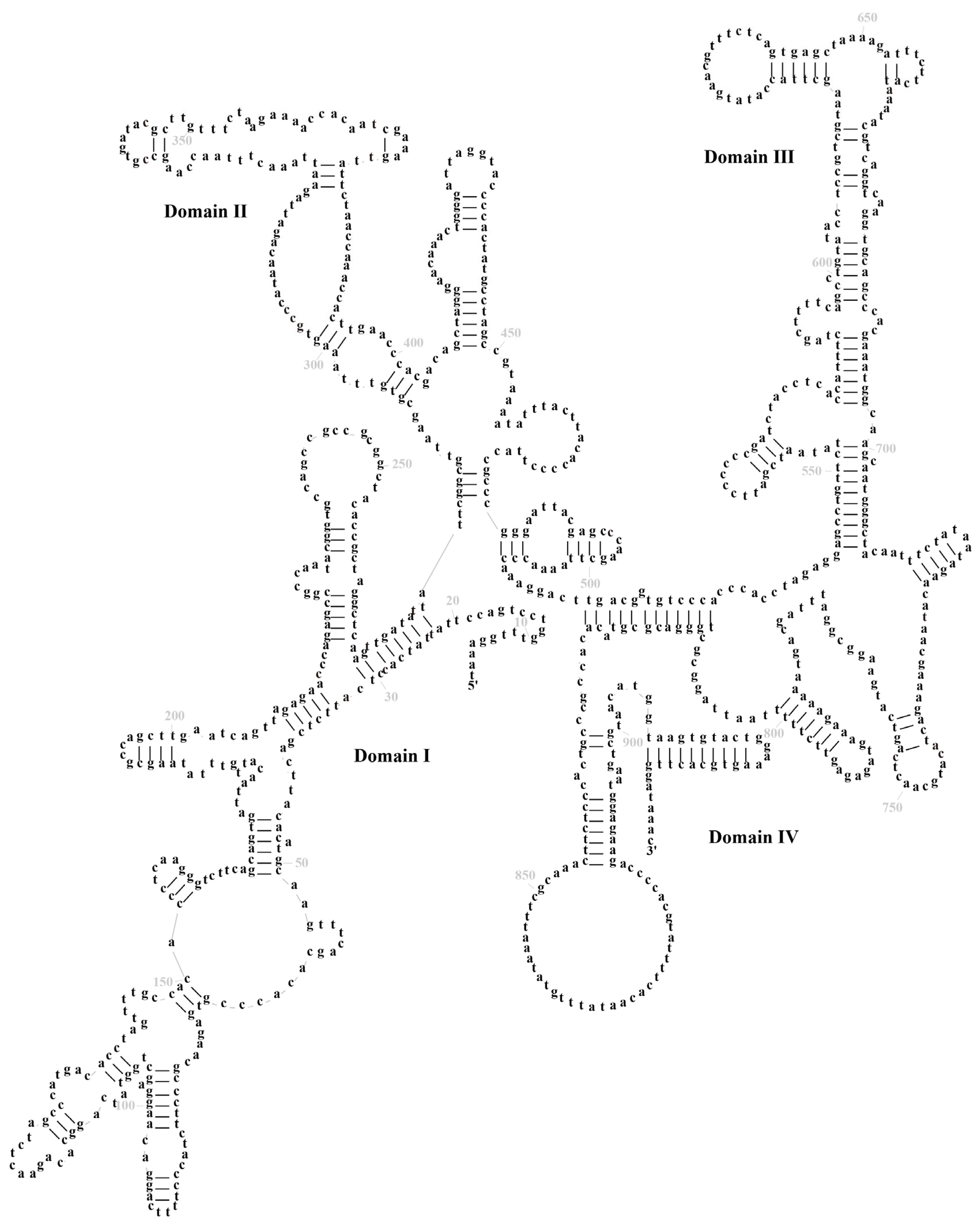
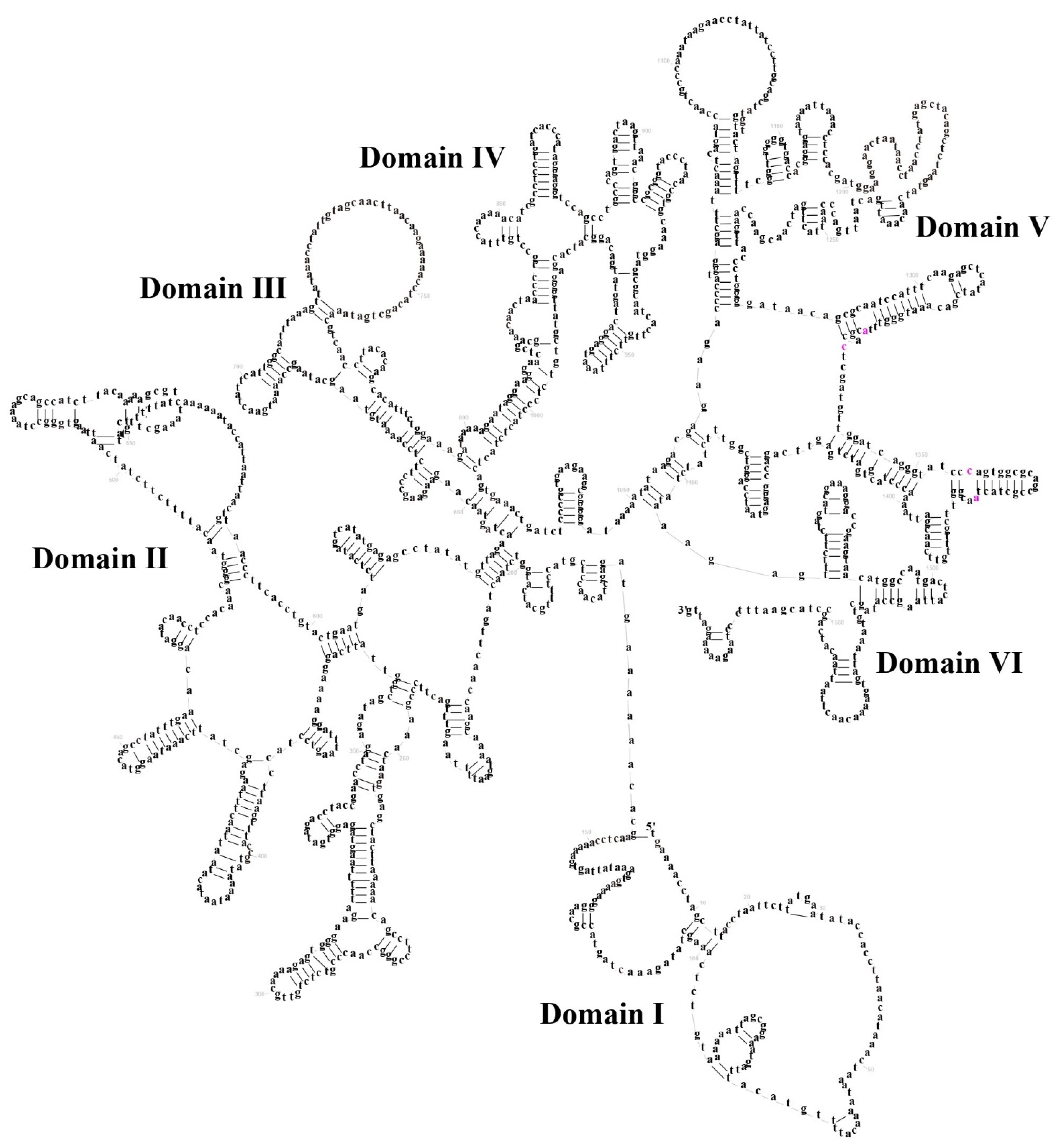
3.3. Transfer RNA and Ribosomal RNA Genes
3.4. The Control Region
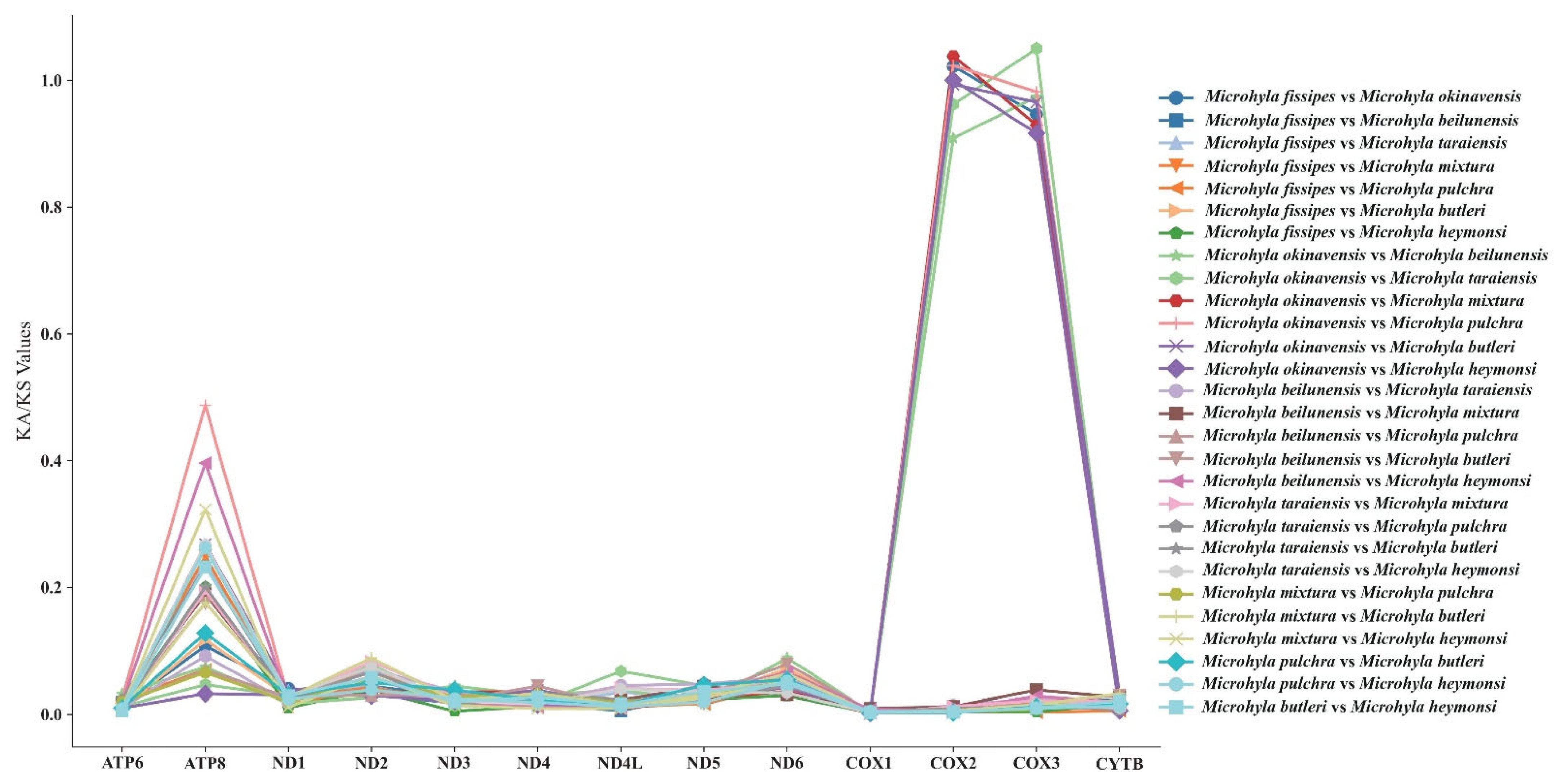
3.5. Synonymous and Nonsynonymous Substitution Rate
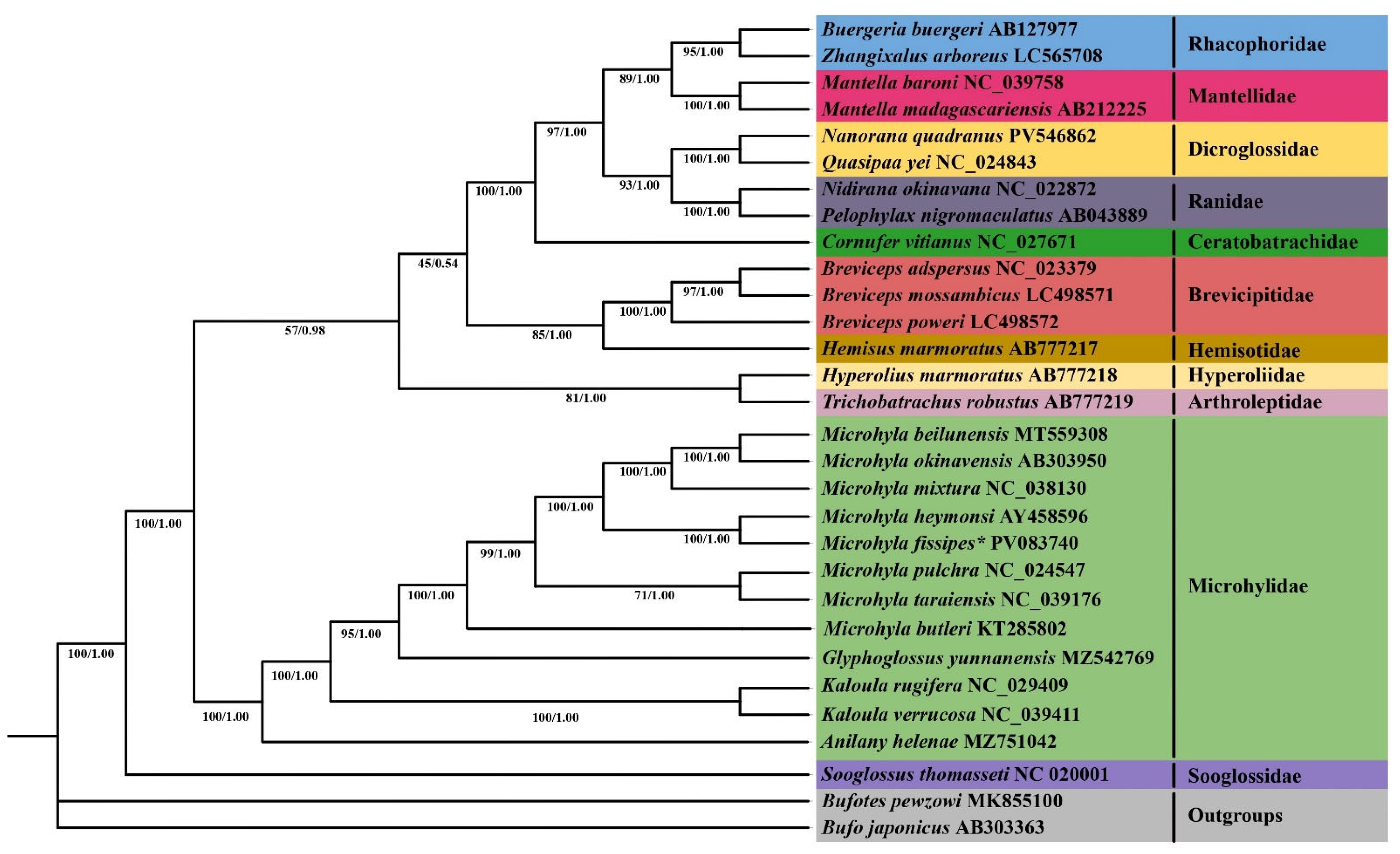
3.6. Phylogenetic Analyses
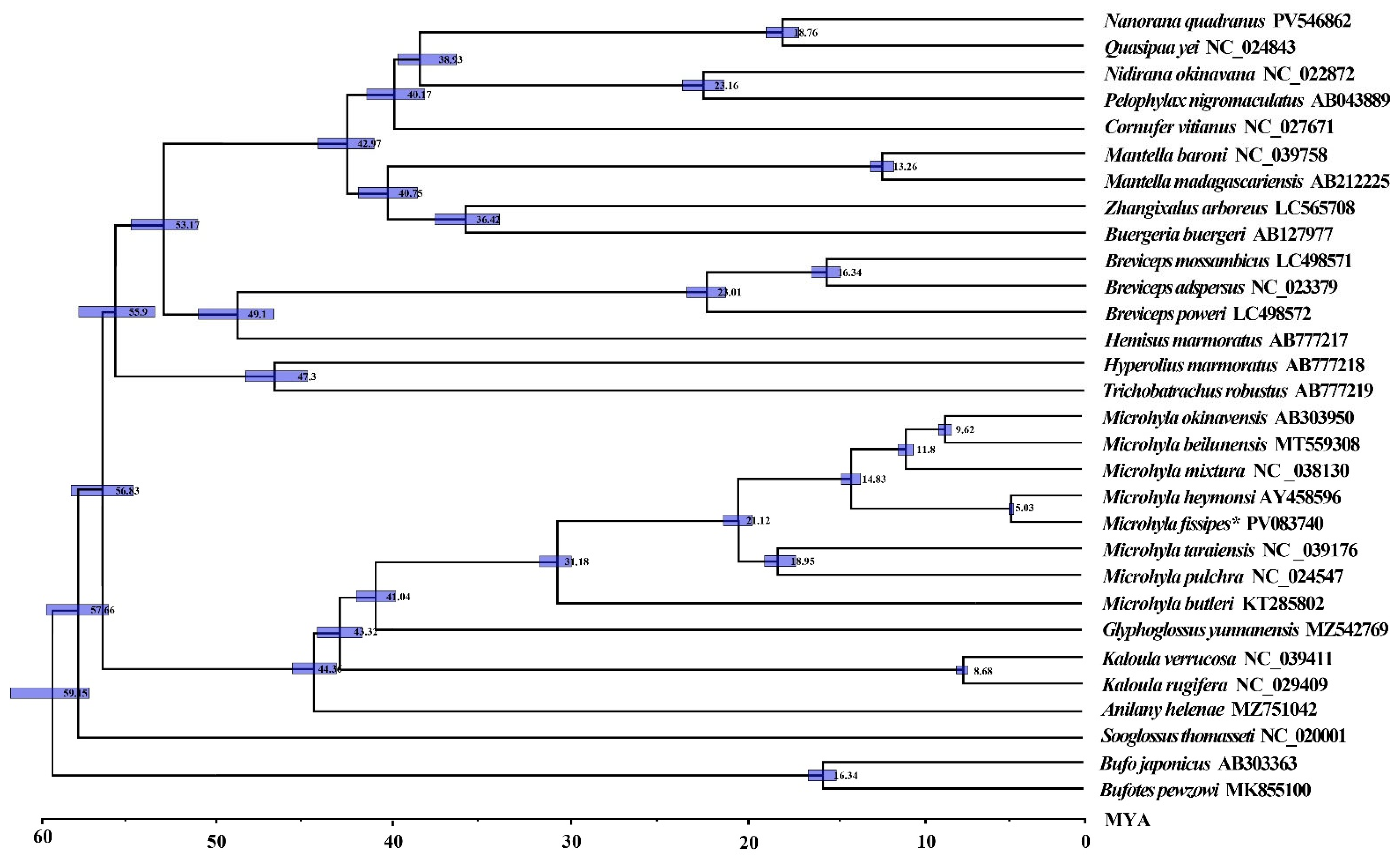
3.7. Divergence Time Estimation of Microhylidae Species
4. Discussion
4.1. Structural and Compositional Features of the M. fissipes Mitochondrial Genome
4.2. Phylogenetic Analysis of Microhylidae
4.3. Divergence Time and Selective Pressure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Han, N.; Wu, Z.; Zhang, L.; Wei, X. The complete mitogenome of Microhyla fissipes (Anura: Microhylidae) and phylogenetic analysis using GenBank data mining. Mitochondrial DNA Part B 2019, 4, 3049–3050. [Google Scholar] [CrossRef]
- Lin, J.S.; Liu, F.R. The complete mitochondrial genome of Microhyla fissipes (Amphidia, Anura, Microhylidae) from Taiwan. Mitochondrial DNA Part B 2017, 2, 930–931. [Google Scholar] [CrossRef][Green Version]
- Keeling, P.J.; Archibald, J.M. Organelle evolution: What’s in a name? Curr. Biol. 2008, 18, R345–R347. [Google Scholar] [CrossRef]
- Murat, F.; Mbengue, N.; Winge, S.B.; Trefzer, T.; Leushkin, E.; Sepp, M.; Cardoso-Moreira, M.; Schmidt, J.; Schneider, C.; Mößinger, K.; et al. The molecular evolution of spermatogenesis across mammals. Nature 2023, 613, 308–316. [Google Scholar] [CrossRef]
- Wang, R.; Tursun, M.; Shan, W. Complete Mitogenomes of Xinjiang Hares and Their Selective Pressure Considerations. Int. J. Mol. Sci. 2024, 25, 11925. [Google Scholar] [CrossRef] [PubMed]
- Kurabayashi, A.; Sumida, M. Afrobatrachian mitochondrial genomes: Genome reorganization, gene rearrangement mechanisms, and evolutionary trends of duplicated and rearranged genes. BMC Genom. 2013, 14, 633. [Google Scholar] [CrossRef]
- Gorin, V.A.; Solovyeva, E.N.; Hasan, M.; Okamiya, H.; Karunarathna, D.M.S.S.; Pawangkhanant, P.; de Silva, A.; Juthong, W.; Milto, K.D.; Nguyen, L.T.; et al. A little frog leaps a long way: Compounded colonizations of the Indian Subcontinent discovered in the tiny Oriental frog genus Microhyla (Amphibia: Microhylidae). PeerJ 2020, 8, e9411. [Google Scholar] [CrossRef]
- Jiang, L.; Zhao, L.; Cheng, D.; Zhu, L.; Zhang, M.; Ruan, Q.; Chen, W. The complete mitochondrial genome sequence of the Sichuan Digging Frog, Kaloula rugifera (Anura: Microhylidae) and its phylogenetic implications. Gene 2017, 626, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Meng, H.; Su, L. The complete mitochondrial genome of the mixtured pygmy frog Microhyla mixtura (Anura, Microhylidae). Conserv. Genet. Resour. 2018, 10, 427–430. [Google Scholar] [CrossRef]
- Khatiwada, J.R.; Wang, S.; Shu, G.; Xie, F.; Jiang, J. The mitochondrial genome of the Microhyla taraiensis (Anura: Microhylidae) and related phylogenetic analyses. Conserv. Genet. Resour. 2018, 10, 441–444. [Google Scholar] [CrossRef]
- Fedorin, N.D.; Chuykova, O.V.; Eprintsev, T.A. Modification of the Method for Isolating MicroRNA from Plants by Phenol–Chloroform Extraction Using Polyethylene Glycol 1500. Biochem. Suppl. Ser. B Biomed. Chem. 2023, 17, 26–30. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Sweeney, B.A.; Hoksza, D.; Nawrocki, E.P.; Ribas, C.E.; Madeira, F.; Cannone, J.J.; Gutell, R.; Maddala, A.; Meade, C.D.; Williams, L.D.; et al. R2DT is a framework for predicting and visualising RNA secondary structure using templates. Nat. Commun. 2021, 12, 3494. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- Tamura, K.; Glen, S.; Sudhir, K. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Li, W.X.; Jakovli’c, I.; Zhou, H.; Zhang, J.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Baele, G.; Carvalho, L.M.; Brusselmans, M.; Dudas, G.; Ji, X.; McCrone, J.T.; Lemey, P.; Suchard, M.A.; Rambaut, A. HIPSTR: Highest independent posterior subtree reconstruction in TreeAnnotator X. bioRxiv 2024, 12, 627395. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.-Q.; Wang, J.; Wong, G.K.-S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through Model Selection and Model Averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating Selective Pressure on Coding and Non-Coding Sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal Mitochondrial Genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Hassanin, A.; Léger, N.; Deutsch, J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef]
- Liu, Z.Q.; Wang, Y.Q.; Bing, S. The mitochondrial genome organization of the rice frog, Fejervarya limnocharis, (Amphibia: Anura): A new gene order in the vertebrate mtDNA. Gene 2005, 346, 145–151. [Google Scholar] [CrossRef]
- Wu, X.; Li, Y.; Zhang, H.; Yan, L.; Wu, X.B. The complete mitochondrial genome of Microhyla pulchra (Amphidia, Anura, Microhylidae). Mitochondrial DNA A DNA Mapp. Mitochondrial DNA Part A 2016, 27, 40–41. [Google Scholar] [CrossRef]
- Chen, L.; Lin, Y.; Xiao, Q.; Lin, Y.; Du, Y.; Lin, C.; Ward-Fear, E.; Hu, C.; Qu, Y.; Li, H. Characterization of the complete mitochondrial genome of the many-lined sun skink (Eutropis multifasciata) and comparison with other Scincomorpha species. Genomics 2020, 113, 2526–2536. [Google Scholar] [CrossRef]
- Su, X.; Wu, X.B.; Yan, P.; Cao, S.Y.; Hu, Y.L. Rearrangement of a mitochondrial tRNA gene of the concave-eared torrent frog, Amolops tormotus. Gene 2007, 394, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Hixson, J.E.; Brown, W.M. A comparison of the small ribosomal RNA genes from the mitochondrial DNA of the great apes and humans: Sequence, structure, evolution, and phylogenetic implications. Mol. Biol. Evol. 1986, 3, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Dan, Z.; Zhang, Y.; Chen, Z. Characterization of the Complete Mitochondrial Genome of Three Satyrid Butterfly Species (Satyrinae:Amathusiini) and Reconstructed Phylogeny of Satyrinae. Int. J. Mol. Sci. 2025, 26, 2609. [Google Scholar] [CrossRef]
- Mayfield, J.E.; McKenna, J.F. AT rich sequences in vertebrate DNA. Chromosoma 1978, 67, 157–163. [Google Scholar] [CrossRef]
- Zhang, J.; Luu, B.E.; Yu, D.; Zhang, L.; Al-Attar, R.; Storey, K.B. The complete mitochondrial genome of Dryophytes versicolor: Phylogenetic relationship among Hylidae and mitochondrial protein-coding gene expression in response to freezing and anoxia. Int. J. Biol. Macromol. 2019, 132, 461–469. [Google Scholar] [CrossRef]
- Cai, Y.; Li, Q.; Zhang, J.; Storey, K.B.; Yu, D. Characterization of the mitochondrial genomes of two toads, Anaxyrus americanus (Anura: Bufonidae) and Bufotes pewzowi (Anura: Bufonidae), with phylogenetic and selection pressure analyses. PeerJ 2020, 8, e8901. [Google Scholar] [CrossRef]
- Kurabayashi, A.; Ueshima, R. Complete sequence of the mitochondrial DNA of the primitive opisthobranch gastropod Pupa strigosa: Systematic implication of the genome organization. Mol. Biol. Evol. 2000, 17, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999, 14, 394–398. [Google Scholar] [CrossRef]
- Ohtsuki, T.; Kawai, G.; Watanabe, K. The Minimal tRNA: Unique Structure of Ascaris suum Mitochondrial tRNASerUCU Having a Short T Arm and Lacking the Entire D Arm. FEBS Lett. 2002, 514, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. USA 2000, 97, 13738–13742. [Google Scholar] [CrossRef]
- Meegaskumbura, M.; Bossuyt, F.; Pethiyagoda, R.; Manamendra-Arachchi, K.; Bahir, M.; Milinkovitch, M.C.; Schneider, C.J. Sri Lanka: An amphibian hot spot. Science 2002, 298, 379. [Google Scholar] [CrossRef][Green Version]
- Oliver, P.M.; Iannella, A.; Richards, S.J.; Lee, M.S.Y. Mountain colonisation, miniaturisation and ecological evolution in a radiation of direct-developing New Guinea Frogs (Choerophryne, Microhylidae). PeerJ 2017, 5, e3077. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Garg, S.; Hamidy, A.; Smith, E.N.; Biju, S.D. A new species of Micryletta frog (Microhylidae) from Northeast India. PeerJ 2019, 7, e7012. [Google Scholar] [CrossRef]
- Poyarkov, N.A.; Nguyen, T.V.; Duong, T.V.; Gorin, V.A.; Yang, J. Anew limestone-dwelling species of Micryletta (Amphibia: Anura: Microhylidae) from northern Vietnam. PeerJ 2018, 6, e5771. [Google Scholar] [CrossRef] [PubMed]
- Alhadi, F.; Hamidy, A.; Farajallah, A.; Munir, M.; Atmaja, V.Y.; Garg, S.; Biju, S.D.; Smith, E.N. Rediscovery of Micryletta inornata (Boulenger, 1890) from Sumatra: Redescription, molecular identity and taxonomic implications. Zootaxa 2019, 4613, 111–126. [Google Scholar] [CrossRef]
- Kurabayashi, A.; Matsui, M.; Belabut, D.M.; Yong, H.S.; Ahmad, N.; Sudin, A.; Kuramoto, M.; Hamidy, A.; Sumida, M. From Antarctica or Asia? New colonization scenario for Australian-New Guinean narrow mouth toads suggested from the findings on a mysterious genus Gastrophrynoides. BMC Evol. Biol. 2011, 11, 175. [Google Scholar] [CrossRef]
- Yong, H.; Song, S.; Lim, P.; Eamsobhana, P.; Tan, J. Complete mitochondrial genome and phylogeny of Microhyla butleri (Amphibia: Anura: Microhylidae). Biochem. Syst. Ecol. 2016, 66, 243–253. [Google Scholar] [CrossRef]
- Matsui, M.; Hamidy, A.; Belabut, D.M.; Ahmad, N.; Panha, S.; Sudin, A.; Khonsue, W.; Oh, H.; Yong, H.; Jiang, J.; et al. Systematic relationships of Oriental tiny frogs of the family Microhylidae (Amphibia, Anura) as revealed by mtDNA genealogy. Mol. Phylogenet. Evol. 2011, 61, 167–176. [Google Scholar] [CrossRef]
- deSá, R.O.; Streicher, J.W.; Sekonyela, R.; Forlani, M.C.; Loader, S.P.; Greenbaum, E.; Haddad, C.F. Molecular phylogeny of microhylid frogs (Anura: Microhylidae) with emphasis on relationships among New World genera. BMC Evol. Biol. 2012, 12, 241. [Google Scholar] [CrossRef]
- Pyron, R.A.; Wiens, J.J. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 2011, 61, 543–583. [Google Scholar] [CrossRef]
- Howlader, M.S.A.; Nair, A.; Gopalan, S.V.; Merilä, J. A new species of Microhyla (Anura: Microhylidae) from Nilphamari, Bangladesh. PLoS ONE 2015, 10, e0119825. [Google Scholar] [CrossRef][Green Version]
- Meijden, A.V.; Vences, M.; Hoegg, S.; Boistel, R.; Channing, A.; Meyer, A. Nuclear gene phylogeny of narrow-mouthed toads (Family: Microhylidae) and a discussion of competing hypotheses concerning their biogeographical origins. Mol. Phylogenet. Evol. 2007, 44, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Blackburn, D.C.; Liang, D.; Hillis, D.M.; Wake, D.B.; Cannatella, D.C.; Zhang, P. Phylogenomics reveals rapid, simultaneous diversification of three major clades of Gondwanan frogs at the Cretaceous-Paleogene boundary. Proc. Natl. Acad. Sci. USA 2017, 114, E5864–E5870. [Google Scholar] [CrossRef]
- Garg, S.; Biju, S.D. New microhylid frog genus from Peninsular India with Southeast Asian affinity suggests multiple Cenozoic biotic exchanges between India and Eurasia. Sci. Rep. 2019, 9, 1906. [Google Scholar] [CrossRef] [PubMed]
- Peloso, P.L.V.; Frost, D.R.; Richards, S.J.; Rodrigues, M.T.; Donnellan, S.; Matsui, M.; Raxworthy, C.J.; Biju, S.D.; Lemmon, E.M.; Lemmon, A.R.; et al. The impact of anchored phylogenomics and taxon sampling on phylogenetic inference in narrow–mouthed frogs (Anura, Microhylidae). Cladistics 2016, 32, 113–140. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Y.; Zhu, C.; Flögel, S.; Fang, X.; Sun, J. Modeling the effects of global cooling and the Tethyan Seaway closure on North African and South Asian climates during the Middle Miocene Climate Transition. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2023, 619, 111541. [Google Scholar] [CrossRef]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, Rhythms, and Aberrations in Global Climate 65 Ma to Present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef]
- Gissi, C.; Iannelli, F.; Pesole, G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 2008, 101, 301–320. [Google Scholar] [CrossRef]
- Wang, J.; Tai, J.; Zhang, W.; He, K.; Lan, H.; Liu, H. Comparison of Seven Complete Mitochondrial Genomes from Lamprologus and Neolamprologus (Chordata, Teleostei, Perciformes) and the Phylogenetic Implications for Cichlidae. ZooKeys 2023, 1184, 115–132. [Google Scholar] [CrossRef]
- Ballard, J.W.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef]
- Basu, U.; Bostwick, A.M.; Das, K.; Dittenhafer-Reed, K.E.; Patel, S.S. Structure, mechanism, and regulation of mitochondrial DNA transcription initiation. J. Biol. Chem. 2020, 295, 18406–18425. [Google Scholar] [CrossRef] [PubMed]
- Havird, J.C.; Noe, G.R.; Link, L.; Torres, A.; Logan, D.C.; Sloan, D.B.; Chicco, A.J. Do angiosperms with highly divergent mitochondrial genomes have altered mitochondrial function? Mitochondrion 2019, 49, 1–11. [Google Scholar] [CrossRef]
- Shen, Y.; Shi, P.; Sun, Y.; Zhang, Y. Relaxation of selective constraints on avian mitochondrial DNA following the degeneration of flight ability. Genome Res. 2009, 19, 1760–1765. [Google Scholar] [CrossRef]
- Garvin, M.R.; Bielawski, J.P.; Gharrett, A.J. Positive Darwinian selection in the piston that powers proton pumps in complex I of the mitochondria of Pacific salmon. PLoS ONE 2011, 6, e24127. [Google Scholar] [CrossRef]
- da-Fonseca, R.R.; Johnson, W.E.; O’Brien, S.J.; Ramos, M.J.; Antunes, A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genom. 2008, 9, 119. [Google Scholar] [CrossRef] [PubMed]
| Region | Size (bp) | A (%) | T (%) | G (%) | C (%) | A + T (%) | G + C (%) | AT Skew | GC Skew |
|---|---|---|---|---|---|---|---|---|---|
| Mitogenome | 16,723 | 28.9 | 31.01 | 14.58 | 25.51 | 59.91 | 40.09 | −0.035 | −0.272 |
| tRNAs | 1534 | 28.68 | 28.29 | 22.82 | 20.21 | 56.98 | 43.02 | 0.007 | 0.061 |
| rRNAs | 2516 | 33.47 | 25.16 | 18.36 | 23.01 | 58.62 | 41.38 | 0.142 | −0.112 |
| PCGs | 11,301 | 26.83 | 33.63 | 14.34 | 25.21 | 60.45 | 39.55 | −0.112 | −0.275 |
| PCGs (1st) | 3765 | 28.15 | 25.79 | 23.72 | 22.34 | 53.94 | 46.06 | 0.044 | 0.030 |
| PCGs (2nd) | 3765 | 17.58 | 41.43 | 12.8 | 28.18 | 59.02 | 40.98 | −0.404 | −0.375 |
| PCGs (3rd) | 3765 | 34.79 | 33.55 | 6.51 | 25.15 | 68.34 | 31.66 | 0.018 | −0.589 |
| CR | 1339 | 28.01 | 33.08 | 14.04 | 24.87 | 61.09 | 38.91 | −0.083 | −0.278 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shan, S.; Chen, S.; Li, C.; Huang, H.; Liao, Y.; Jiang, L. Mitogenomic Characterization of Microhyla fissipes and Its Implications for Phylogenetic Analysis in Microhylidae. Biology 2025, 14, 1342. https://doi.org/10.3390/biology14101342
Shan S, Chen S, Li C, Huang H, Liao Y, Jiang L. Mitogenomic Characterization of Microhyla fissipes and Its Implications for Phylogenetic Analysis in Microhylidae. Biology. 2025; 14(10):1342. https://doi.org/10.3390/biology14101342
Chicago/Turabian StyleShan, Siqi, Simin Chen, Chengmin Li, Huiling Huang, Yaqing Liao, and Lichun Jiang. 2025. "Mitogenomic Characterization of Microhyla fissipes and Its Implications for Phylogenetic Analysis in Microhylidae" Biology 14, no. 10: 1342. https://doi.org/10.3390/biology14101342
APA StyleShan, S., Chen, S., Li, C., Huang, H., Liao, Y., & Jiang, L. (2025). Mitogenomic Characterization of Microhyla fissipes and Its Implications for Phylogenetic Analysis in Microhylidae. Biology, 14(10), 1342. https://doi.org/10.3390/biology14101342







