Remodeling of Germ Cell mRNPs for Translational Control
Simple Summary
Abstract
1. Introduction
2. Topical Review
2.1. History of Germ Granules and mRNPs: Microenvironments to Sort, Decorate, and Repress mRNAs
2.2. Translation on the Border: Ribosomes on the Periphery of Germ Cell Condensates
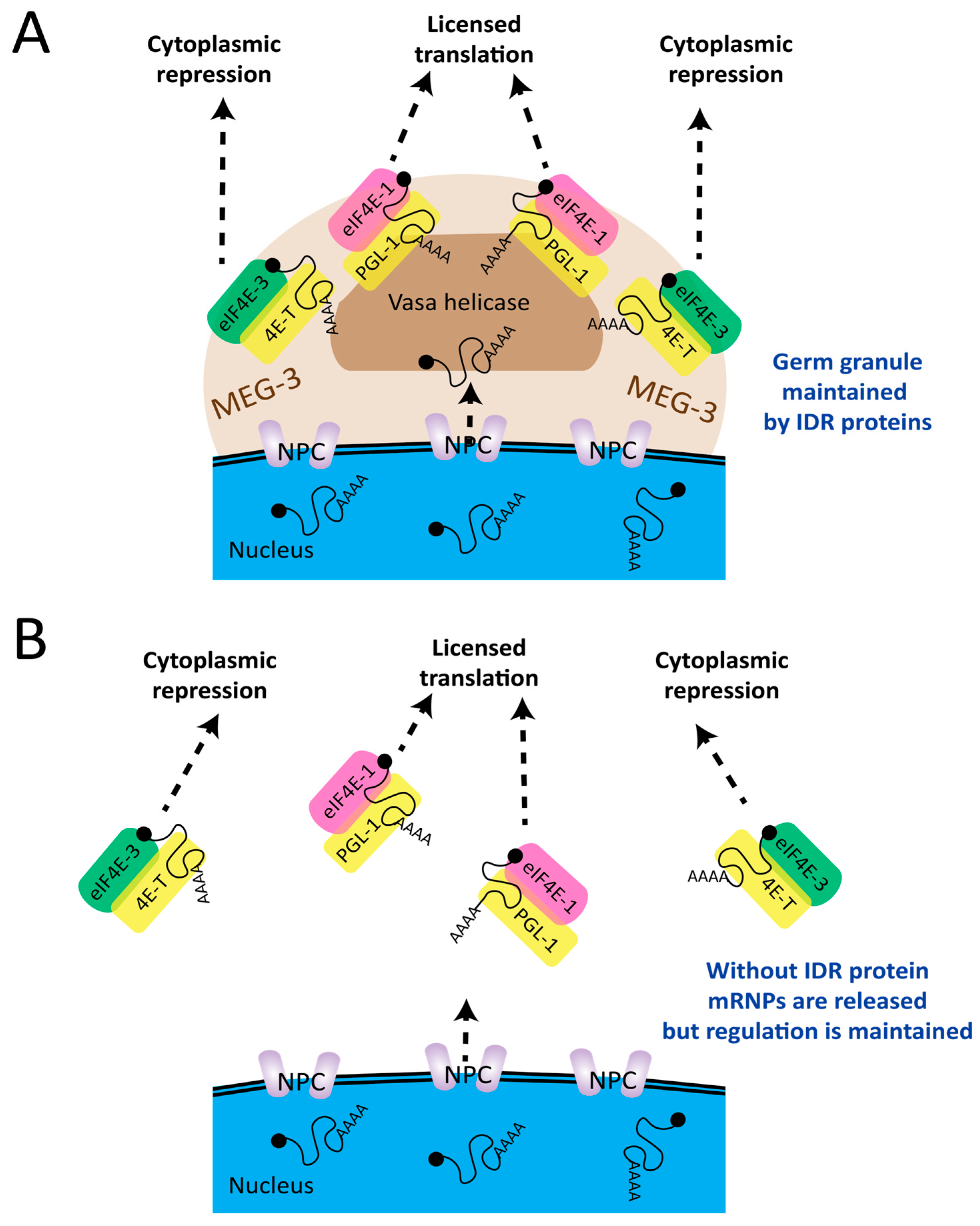
2.3. The Transition to Translational Activation
2.4. mRNP Remodeling During Stress and Aging: Altered Condensate Dynamics
2.5. Proteins with Intrinsically Disordered Regions (IDRs) Promote Condensation
2.6. Stratified Arrangements of Protein and mRNA Within Condensates
2.7. Condensates Collect Regulatory mRNPs but May Not Exert Regulation
3. Conclusions
3.1. A New Way to Envision mRNA–Protein Dynamics in the Cytoplasm
3.2. Emerging Technologies That May Address Dynamics and Causality
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| mRNA | Messenger ribonucleic acid |
| GTP | Guanosine triphosphate |
| LLPS | Liquid–liquid phase separation |
| RBP | RNA-binding protein |
| eIF4E | Eukaryotic initiation factor 4E |
| eIF4G | Eukaryotic initiation factor 4G |
| IDR | Intrinsically disordered region |
| 4EIP | eIF4E-interacting protein |
| 4E-T | eIF4E transport protein |
| Dnd1 | Deadend1 |
| IP-MS | Immunoprecipitation mass spectrometry |
| smFISH | Single-molecule fluorescence in situ hybridization |
References
- Ghosh, S.; Lasko, P. Loss-of-function analysis reveals distinct requirements of the translation initiation factors eIF4E, eIF4E-3, eIF4G and eIF4G2 in Drosophila spermatogenesis. PLoS ONE 2015, 10, e0122519. [Google Scholar] [CrossRef]
- Nousch, M.; Eckmann, C.R. Translational control in the Caenorhabditis elegans germ line. Adv. Exp. Med. Biol. 2013, 757, 205–247. [Google Scholar]
- Keiper, B. Cap-Independent mRNA Translation in Germ Cells. Int. J. Mol. Sci. 2019, 20, 173. [Google Scholar] [CrossRef]
- Keiper, B.D.; Gan, W.; Rhoads, R.E. Protein synthesis initiation factor 4G. Int. J. Biochem. Cell Biol. 1999, 31, 37–41. [Google Scholar] [CrossRef]
- Mendez, R.; Richter, J.D. Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell Biol. 2001, 2, 521–529. [Google Scholar] [CrossRef]
- Delaidelli, A.; Jan, A.; Herms, J.; Sorensen, P.H. Translational control in brain pathologies: Biological significance and therapeutic opportunities. Acta Neuropathol. 2019, 137, 535–555. [Google Scholar] [CrossRef]
- Gkogkas, C.G.; Khoutorsky, A.; Ran, I.; Rampakakis, E.; Nevarko, T.; Weatherill, D.B.; Vasuta, C.; Yee, S.; Truitt, M.; Dallaire, P.; et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 2013, 493, 371–377. [Google Scholar] [CrossRef]
- Gkogkas, C.G.; Sonenberg, N. Translational control and autism-like behaviors. Cell. Logist. 2013, 3, e24551. [Google Scholar] [CrossRef]
- Iacoangeli, A.; Tiedge, H. Translational control at the synapse: Role of RNA regulators. Trends Biochem. Sci. 2013, 38, 47–55. [Google Scholar] [CrossRef]
- Kimelman, D.; Kirschner, M.; Scherson, T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell 1987, 48, 399–407. [Google Scholar] [CrossRef]
- Newport, J.; Kirschner, M. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell 1982, 30, 675–686. [Google Scholar] [CrossRef]
- Chen, R.; Stainier, W.; Dufourt, J.; Lagha, M.; Lehmann, R. Direct observation of translational activation by a ribonucleoprotein granule. Nat. Cell Biol. 2024, 26, 1322–1335. [Google Scholar] [CrossRef]
- Pushpa, K.; Kumar, G.A.; Subramaniam, K. Translational Control of Germ Cell Decisions. In Signaling-Mediated Control of Cell Division: From Oogenesis to Oocyte-to-Embryo Development; Results and Problems in Cell Differentiation; Springer: Cham, Switzerland, 2017; Volume 59, pp. 175–200. [Google Scholar] [CrossRef]
- Saxe, J.P.; Lin, H. Small noncoding RNAs in the germline. Cold Spring Harb. Perspect. Biol. 2011, 3, a002717. [Google Scholar] [CrossRef]
- Huggins, H.P.; Keiper, B.D. Regulation of Germ Cell mRNPs by eIF4E:4EIP Complexes: Multiple Mechanisms, One Goal. Front. Cell Dev. Biol. 2020, 8, 562. [Google Scholar] [CrossRef]
- Friday, A.J.; Keiper, B.D. Positive mRNA Translational Control in Germ Cells by Initiation Factor Selectivity. BioMed Res. Int. 2015, 2015, e327963. [Google Scholar] [CrossRef]
- Lee, M.H.; Mamillapalli, S.S.; Keiper, B.D.; Cha, D.S. A Systematic mRNA Control Mechanism for Germline Stem Cell Homeostasis and Cell Fate Specification. BMB Rep. 2015, 2015, 3259. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Knutson, A.K.; Egelhofer, T.; Rechtsteiner, A.; Strome, S. Germ Granules Prevent Accumulation of Somatic Transcripts in the Adult Caenorhabditis elegans Germline. Genetics 2017, 206, 163–178. [Google Scholar] [CrossRef]
- Updike, D.L.; Knutson, A.K.; Egelhofer, T.A.; Campbell, A.C.; Strome, S. Germ-granule components prevent somatic development in the C. elegans germline. Curr. Biol. 2014, 24, 970–975. [Google Scholar] [CrossRef]
- Scholl, A.; Liu, Y.; Seydoux, G. Caenorhabditis elegans germ granules accumulate hundreds of low translation mRNAs with no systematic preference for germ cell fate regulators. Development 2024, 151, dev202575. [Google Scholar] [CrossRef]
- Liu-Yesucevitz, L.; Bassell, G.J.; Gitler, A.D.; Hart, A.C.; Klann, E.; Richter, J.D.; Warren, S.T.; Wolozin, B. Local RNA translation at the synapse and in disease. J. Neurosci. 2011, 31, 16086–16093. [Google Scholar] [CrossRef]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef]
- Phillips, C.M.; Updike, D.L. Germ granules and gene regulation in the Caenorhabditis elegans germline. Genetics 2022, 220, iyab195. [Google Scholar] [CrossRef]
- Sengupta, M.S.; Boag, P.R. Germ granules and the control of mRNA translation. IUBMB Life 2012, 64, 586–594. [Google Scholar] [CrossRef]
- Puoti, A.; Pugnale, P.; Belfiore, M.; Schlappi, A.C.; Saudan, Z. RNA and sex determination in Caenorhabditis elegans. Post-transcriptional regulation of the sex-determining tra-2 and fem-3 mRNAs in the Caenorhabditis elegans hermaphrodite. EMBO Rep. 2001, 2, 899–904. [Google Scholar] [CrossRef][Green Version]
- Albarqi, M.M.Y.; Ryder, S.P. The role of RNA-binding proteins in orchestrating germline development in Caenorhabditis elegans. Front. Cell Dev. Biol. 2023, 10, 1094295. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef]
- Temme, C.; Simonelig, M.; Wahle, E. Deadenylation of mRNA by the CCR4-NOT complex in Drosophila: Molecular and developmental aspects. Front. Genet. 2014, 5, 143. [Google Scholar] [CrossRef]
- Hird, S.N.; Paulsen, J.E.; Strome, S. Segregation of germ granules in living Caenorhabditis elegans embryos: Cell-type-specific mechanisms for cytoplasmic localisation. Development 1996, 122, 1303–1312. [Google Scholar] [CrossRef]
- Seydoux, G.; Strome, S. Launching the germline in Caenorhabditis elegans: Regulation of gene expression in early germ cells. Development 1999, 126, 3275–3283. [Google Scholar] [CrossRef]
- Schisa, J.A.; Pitt, J.N.; Priess, J.R. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 2001, 128, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Putnam, A.; Lu, T.; He, S.; Ouyang, J.P.T.; Seydoux, G. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. eLife 2020, 9, e52896. [Google Scholar] [CrossRef]
- Rybarska, A.; Harterink, M.; Jedamzik, B.; Kupinski, A.P.; Schmid, M.; Eckmann, C.R. GLS-1, a novel P granule component, modulates a network of conserved RNA regulators to influence germ cell fate decisions. PLoS Genet. 2009, 5, e1000494. [Google Scholar] [CrossRef]
- Sengupta, M.S.; Low, W.Y.; Patterson, J.R.; Kim, H.M.; Traven, A.; Beilharz, T.H.; Colaiacovo, M.P.; Schisa, J.A.; Boag, P.R. ifet-1 is a broad-scale translational repressor required for normal P granule formation in C. elegans. J. Cell Sci. 2013, 126, 850–859. [Google Scholar] [CrossRef]
- Voronina, E.; Paix, A.; Seydoux, G. The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development 2012, 139, 3732–3740. [Google Scholar] [CrossRef]
- Amiri, A.; Keiper, B.D.; Kawasaki, I.; Fan, Y.; Kohara, Y.; Rhoads, R.E.; Strome, S. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development 2001, 128, 3899–3912. [Google Scholar] [CrossRef]
- Kawasaki, I.; Amiri, A.; Fan, Y.; Meyer, N.; Dunkelbarger, S.; Motohashi, T.; Karashima, T.; Bossinger, O.; Strome, S. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics 2004, 167, 645–661. [Google Scholar] [CrossRef]
- Kawasaki, I.; Shim, Y.H.; Kirchner, J.; Kaminker, J.; Wood, W.B.; Strome, S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 1998, 94, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Shimada, M.; Yokosawa, H.; Kawahara, H. OMA-1 is a P granules-associated protein that is required for germline specification in Caenorhabditis elegans embryos. Genes Cells 2006, 11, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Julicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef]
- Wan, G.; Fields, B.D.; Spracklin, G.; Shukla, A.; Phillips, C.M.; Kennedy, S. Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 2018, 557, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Marnik, E.A.; Fuqua, J.H.; Sharp, C.S.; Rochester, J.D.; Xu, E.L.; Holbrook, S.E.; Updike, D.L. Germline Maintenance Through the Multifaceted Activities of GLH/Vasa in Caenorhabditis elegans P Granules. Genetics 2019, 213, 923–939. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Arkov, A.L. Next generation organelles: Structure and role of germ granules in the germline. Mol. Reprod. Dev. 2013, 80, 610–623. [Google Scholar] [CrossRef]
- Zheng, H.; Peng, K.; Gou, X.; Ju, C.; Zhang, H. RNA recruitment switches the fate of protein condensates from autophagic degradation to accumulation. J. Cell Biol. 2023, 222, e202210104. [Google Scholar] [CrossRef]
- Sheth, U.; Pitt, J.; Dennis, S.; Priess, J.R. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 2010, 137, 1305–1314. [Google Scholar] [CrossRef]
- Reed, K.J.; Svendsen, J.M.; Brown, K.C.; Montgomery, B.E.; Marks, T.N.; Vijayasarathy, T.; Parker, D.M.; Nishimura, E.O.; Updike, D.L.; Montgomery, T.A. Widespread roles for piRNAs and WAGO-class siRNAs in shaping the germline transcriptome of Caenorhabditis elegans. Nucleic Acids Res. 2020, 48, 1811–1827. [Google Scholar] [CrossRef] [PubMed]
- Campbell, A.C.; Updike, D.L. CSR-1 and P granules suppress sperm-specific transcription in the C. elegans germline. Development 2015, 142, 1745–1755. [Google Scholar] [CrossRef]
- Du, Z.; Shi, K.; Brown, J.S.; He, T.; Wu, W.-S.; Zhang, Y.; Lee, H.-C.; Zhang, D. Condensate cooperativity underlies transgenerational gene silencing. Cell Rep. 2023, 42, 112859. [Google Scholar] [CrossRef]
- Thomson, T.; Liu, N.; Arkov, A.; Lehmann, R.; Lasko, P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech. Dev. 2008, 125, 865–873. [Google Scholar] [CrossRef]
- Navarro, R.E.; Shim, E.Y.; Kohara, Y.; Singson, A.; Blackwell, T.K. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development 2001, 128, 3221–3232. [Google Scholar] [CrossRef]
- Gajjar, G.; Huggins, H.P.; Kim, E.S.; Huang, W.; Bonnet, F.X.; Updike, D.L.; Keiper, B.D. Two eIF4E paralogs occupy separate germ granule messenger ribonucleoproteins that mediate mRNA repression and translational activation. Genetics 2025, 230, iyaf053. [Google Scholar] [CrossRef]
- Nakamura, A.; Sato, K.; Hanyu-Nakamura, K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell 2004, 6, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Huggins, H.P.; Subash, J.S.; Stoffel, H.; Henderson, M.A.; Hoffman, J.L.; Buckner, D.S.; Sengupta, M.S.; Boag, P.R.; Lee, M.H.; Keiper, B.D. Distinct roles of two eIF4E isoforms in the germline of Caenorhabditis elegans. J. Cell Sci. 2020, 133, jcs.237990. [Google Scholar] [CrossRef]
- Curtis, D.; Lehmann, R.; Zamore, P.D. Translational regulation in development. Cell 1995, 81, 171–178. [Google Scholar] [CrossRef]
- Gavis, E.R.; Lehmann, R. Localization of nanos RNA controls embryonic polarity. Cell 1992, 71, 301–313. [Google Scholar] [CrossRef]
- Jadhav, S.; Rana, M.; Subramaniam, K. Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development 2008, 135, 1803–1812. [Google Scholar] [CrossRef] [PubMed]
- Konwerski, J.; Senchuk, M.; Petty, E.; Lahaie, D.; Schisa, J.A. Cloning and expression analysis of pos-1 in the nematodes Caenorhabditis briggsae and Caenorhabditis remanei. Dev. Dyn. 2005, 233, 1006–1012. [Google Scholar] [CrossRef]
- Tabara, H.; Hill, R.J.; Mello, C.C.; Priess, J.R.; Kohara, Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 1999, 126, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Barrios, F.; Filipponi, D.; Pellegrini, M.; Paronetto, M.P.; Di Siena, S.; Geremia, R.; Rossi, P.; De Felici, M.; Jannini, E.A.; Dolci, S. Opposing effects of retinoic acid and FGF9 on Nanos2 expression and meiotic entry of mouse germ cells. J. Cell Sci. 2010, 123, 871–880. [Google Scholar] [CrossRef]
- Westerich, K.J.; Tarbashevich, K.; Schick, J.; Gupta, A.; Zhu, M.; Hull, K.; Romo, D.; Zeuschner, D.; Goudarzi, M.; Gross-Thebing, T.; et al. Spatial organization and function of RNA molecules within phase-separated condensates in zebrafish are controlled by Dnd1. Dev. Cell 2023, 58, 1578–1592.e5. [Google Scholar] [CrossRef]
- Ogura, K.; Kishimoto, N.; Mitani, S.; Gengyo-Ando, K.; Kohara, Y. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development 2003, 130, 2495–2503. [Google Scholar] [CrossRef]
- Albarqi, M.M.Y.; Ryder, S.P. The endogenous mex-3 3’UTR is required for germline repression and contributes to optimal fecundity in C. elegans. PLoS Genet. 2021, 17, e1009775. [Google Scholar] [CrossRef]
- Mercer, M.; Jang, S.; Ni, C.; Buszczak, M. The Dynamic Regulation of mRNA Translation and Ribosome Biogenesis During Germ Cell Development and Reproductive Aging. Front. Cell Dev. Biol. 2021, 9, 710186. [Google Scholar] [CrossRef]
- Lai, F.; King, M.L. Repressive translational control in germ cells. Mol. Reprod. Dev. 2013, 80, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Cassani, M.; Seydoux, G. Specialized germline P-bodies are required to specify germ cell fate in Caenorhabditis elegans embryos. Development 2022, 149, dev200920. [Google Scholar] [CrossRef] [PubMed]
- Hoege, C.; Hyman, A.A. Principles of PAR polarity in Caenorhabditis elegans embryos. Nat. Rev. Mol. Cell Biol. 2013, 14, 315–322. [Google Scholar] [CrossRef]
- Cho, P.F.; Gamberi, C.; Cho-Park, Y.A.; Cho-Park, I.B.; Lasko, P.; Sonenberg, N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr. Biol. 2006, 16, 2035–2041. [Google Scholar] [CrossRef]
- Thio, G.L.; Ray, R.P.; Barcelo, G.; Schupbach, T. Localization of gurken RNA in drosophila oogenesis requires elements in the 5′ and 3′ regions of the transcript. Dev. Biol. 2000, 221, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Mahowald, A.P.; Hennen, S. Ultrastructure of the “germ plasm” in eggs and embryos of Rana pipiens. Dev. Biol. 1971, 24, 37–53. [Google Scholar] [CrossRef]
- Davidson, E.H. Gene Activity in Early Development, 3rd ed.; Academic Press, Inc.: Orlando, FL, USA, 1986; pp. 193–303. [Google Scholar]
- Lasko, P. mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a012294. [Google Scholar] [CrossRef]
- Macdonald, P.M.; Smibert, C.A. Translational regulation of maternal mRNAs. Curr. Opin. Genet. Dev. 1996, 6, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Lasko, P. Translational control in oocyte development. Cold Spring Harb. Perspect. Biol. 2011, 3, a002758. [Google Scholar] [CrossRef]
- Frydryskova, K.; Masek, T.; Borcin, K.; Mrvova, S.; Venturi, V.; Pospisek, M. Distinct recruitment of human eIF4E isoforms to processing bodies and stress granules. BMC Mol. Biol. 2016, 17, 21. [Google Scholar] [CrossRef]
- Hanazawa, M.; Kawasaki, I.; Kunitomo, H.; Gengyo-Ando, K.; Bennett, K.L.; Mitani, S.; Iino, Y. The Caenorhabditis elegans eukaryotic initiation factor 5A homologue, IFF-1, is required for germ cell proliferation, gametogenesis and localization of the P-granule component PGL-1. Mech. Dev. 2004, 121, 213–224. [Google Scholar] [CrossRef]
- Minshall, N.; Reiter, M.H.; Weil, D.; Standart, N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biol. Chem. 2007, 282, 37389–37401. [Google Scholar] [CrossRef]
- Standart, N.; Minshall, N. Translational control in early development: CPEB, P-bodies and germinal granules. Biochem. Soc. Trans. 2008, 36, 671–676. [Google Scholar] [CrossRef]
- Mair, G.R.; Lasonder, E.; Garver, L.S.; Franke-Fayard, B.M.; Carret, C.K.; Wiegant, J.C.; Dirks, R.W.; Dimopoulos, G.; Janse, C.J.; Waters, A.P. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 2010, 6, e1000767. [Google Scholar] [CrossRef] [PubMed]
- Schisa, J.A.; Elaswad, M.T. An Emerging Role for Post-translational Modifications in Regulating RNP Condensates in the Germ Line. Front. Mol. Biosci. 2021, 8, 658020. [Google Scholar] [CrossRef]
- Schisa, J.A. Germ Cell Responses to Stress: The Role of RNP Granules. Front. Cell Dev. Biol. 2019, 7, 220. [Google Scholar] [CrossRef] [PubMed]
- Henderson, M.A.; Cronland, E.; Dunkelbarger, S.; Contreras, V.; Strome, S.; Keiper, B.D. A germ line-specific isoform of eIF4E (IFE-1) is required for efficient translation of stored mRNAs and maturation of both oocytes and sperm. J. Cell Sci. 2009, 122, 1529–1539. [Google Scholar] [CrossRef]
- Friday, A.J.; Henderson, M.A.; Morrison, J.K.; Hoffman, J.L.; Keiper, B.D. Spatial and temporal translational control of germ cell mRNAs mediated by the eIF4E isoform IFE-1. J. Cell Sci. 2015, 128, 4487–4498. [Google Scholar] [CrossRef]
- Kawasaki, I.; Jeong, M.H.; Shim, Y.H. Regulation of sperm-specific proteins by IFE-1, a germline-specific homolog of eIF4E, in C. elegans. Mol. Cells 2011, 31, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Keiper, B.D.; Lamphear, B.J.; Deshpande, A.M.; Jankowska-Anyszka, M.; Aamodt, E.J.; Blumenthal, T.; Rhoads, R.E. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 2000, 275, 10590–10596. [Google Scholar] [CrossRef]
- Lorenzo-Orts, L.; Strobl, M.; Steinmetz, B.; Leesch, F.; Pribitzer, C.; Roehsner, J.; Schutzbier, M.; Dürnberger, G.; Pauli, A. eIF4E1b is a non-canonical eIF4E protecting maternal dormant mRNAs. EMBO Rep. 2024, 25, 404–427. [Google Scholar] [CrossRef] [PubMed]
- Browning, K.S.; Bailey-Serres, J. Mechanism of cytoplasmic mRNA translation. Arab. Book 2015, 13, e0176. [Google Scholar] [CrossRef] [PubMed]
- Gray, N.K.; Wickens, M. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 1998, 14, 399–458. [Google Scholar] [CrossRef]
- Merrick, W.C.; Hershey, J.W.B. (Eds.) The Pathway and Mechanism of Eukaryotic Protein Synthesis; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 1996; pp. 31–69. [Google Scholar]
- Rhoads, R.E. Cap recognition and the entry of mRNA into the protein synthesis initiation cycle. Trends Biochem. Sci. 1988, 13, 52–56. [Google Scholar] [CrossRef]
- Romasko, E.J.; Amarnath, D.; Midic, U.; Latham, K.E. Association of maternal mRNA and phosphorylated EIF4EBP1 variants with the spindle in mouse oocytes: Localized translational control supporting female meiosis in mammals. Genetics 2013, 195, 349–358. [Google Scholar] [CrossRef]
- Truitt, M.L.; Conn, C.S.; Shi, Z.; Pang, X.; Tokuyasu, T.; Coady, A.M.; Seo, Y.; Barna, M.; Ruggero, D. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell 2015, 162, 59–71. [Google Scholar] [CrossRef]
- Hernandez, G.; Proud, C.G.; Preiss, T.; Parsyan, A. On the Diversification of the Translation Apparatus across Eukaryotes. Comp. Funct. Genom. 2012, 2012, 256848. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Hyman, A.A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 196–213. [Google Scholar] [CrossRef]
- Aulas, A.; Fay, M.M.; Lyons, S.M.; Achorn, C.A.; Kedersha, N.; Anderson, P.; Ivanov, P. Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 2017, 130, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Nunes, C.; Mestre, I.; Marcelo, A.; Koppenol, R.; Matos, C.A.; Nóbrega, C. MSGP: The first database of the protein components of the mammalian stress granules. Database 2019, 2019, baz031. [Google Scholar] [CrossRef]
- Voronina, E.; Seydoux, G.; Sassone-Corsi, P.; Nagamori, I. RNA granules in germ cells. Cold Spring Harb. Perspect. Biol. 2011, 3, a002774. [Google Scholar] [CrossRef] [PubMed]
- Riggs, C.L.; Kedersha, N.; Ivanov, P.; Anderson, P. Mammalian stress granules and P bodies at a glance. J. Cell Sci. 2020, 133, jcs242487. [Google Scholar] [CrossRef]
- Sanders, D.W.; Kedersha, N.; Lee, D.S.W.; Strom, A.R.; Drake, V.; Riback, J.A.; Bracha, D.; Eeftens, J.M.; Iwanicki, A.; Wang, A.; et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 2020, 181, 306–324.e28. [Google Scholar] [CrossRef]
- Putnam, A.; Thomas, L.; Seydoux, G. RNA granules: Functional compartments or incidental condensates? Genes Dev. 2023, 37, 354–376. [Google Scholar] [CrossRef]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef]
- Decker, C.J.; Teixeira, D.; Parker, R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007, 179, 437–449. [Google Scholar] [CrossRef]
- Brengues, M.; Teixeira, D.; Parker, R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 2005, 310, 486–489. [Google Scholar] [CrossRef]
- Markmiller, S.; Soltanieh, S.; Server, K.L.; Mak, R.; Jin, W.; Fang, M.Y.; Luo, E.-C.; Krach, F.; Yang, D.; Sen, A.; et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172, 590–604.e13. [Google Scholar] [CrossRef]
- Youn, J.-Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.R.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M.; et al. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol. Cell 2018, 69, 517–532.e11. [Google Scholar] [CrossRef] [PubMed]
- Rao, B.S.; Parker, R. Numerous interactions act redundantly to assemble a tunable size of P bodies in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2017, 114, E9569–E9578. [Google Scholar] [CrossRef]
- Thomas, L.; Putnam, A.; Folkmann, A. Germ granules in development. Development 2023, 150, dev201037. [Google Scholar] [CrossRef]
- Jud, M.C.; Czerwinski, M.J.; Wood, M.P.; Young, R.A.; Gallo, C.M.; Bickel, J.S.; Petty, E.L.; Mason, J.M.; Little, B.A.; Padilla, P.A.; et al. Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev. Biol. 2008, 318, 38–51. [Google Scholar] [CrossRef]
- Elaswad, M.T.; Watkins, B.M.; Sharp, K.G.; Munderloh, C.; Schisa, J.A. Large RNP granules in Caenorhabditis elegans oocytes have distinct phases of RNA-binding proteins. G3 (Bethesda) 2022, 12, jkac173. [Google Scholar] [CrossRef]
- Helton, N.S.; Dodd, B.; Moon, S.L. Ribosome association inhibits stress-induced gene mRNA localization to stress granules. Genes Dev. 2025, 39, 826–848. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Putnam, A.; Rasoloson, D.; Seydoux, G. Protein-based condensation mechanisms drive the assembly of RNA-rich P granules. eLife 2021, 10, e63698. [Google Scholar] [CrossRef] [PubMed]
- Kaur, T.; Raju, M.; Alshareedah, I.; Davis, R.B.; Potoyan, D.A.; Banerjee, P.R. Sequence-encoded and composition-dependent protein-RNA interactions control multiphasic condensate morphologies. Nat. Commun. 2021, 12, 872. [Google Scholar] [CrossRef]
- Marnik, E.A.; Updike, D.L. Membraneless organelles: P granules in Caenorhabditis elegans. Traffic 2019, 20, 373–379. [Google Scholar] [CrossRef]
- Folkmann, A.W.; Putnam, A.; Lee, C.F.; Seydoux, G. Regulation of biomolecular condensates by interfacial protein clusters. Science 2021, 373, 1218–1224. [Google Scholar] [CrossRef]
- Bontems, F.; Stein, A.; Marlow, F.; Lyautey, J.; Gupta, T.; Mullins, M.C.; Dosch, R. Bucky Ball Organizes Germ Plasm Assembly in Zebrafish. Curr. Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Fang, H.; Hu, S.; Yang, B.; Zhou, J.; Grifone, R.; Li, P.; Lu, T.; Wang, Z.; et al. Rbm24a dictates mRNA recruitment for germ granule assembly in zebrafish. EMBO J. 2025, 44, 3121–3149. [Google Scholar] [CrossRef]
- Feric, M.; Misteli, T. Function moves biomolecular condensates in phase space. Bioessays 2022, 44, e2200001. [Google Scholar] [CrossRef] [PubMed]
- Yeong, V.; Werth, E.G.; Brown, L.M.; Obermeyer, A.C. Formation of biomolecular condensates in bacteria by tuning protein electrostatics. ACS Cent. Sci. 2020, 6, 2301–2310. [Google Scholar] [CrossRef] [PubMed]
- Boeynaems, S.; Holehouse, A.S.; Weinhardt, V.; Kovacs, D.; Van Lindt, J.; Larabell, C.; Van Den Bosch, L.; Das, R.; Tompa, P.S.; Pappu, R.V. Spontaneous driving forces give rise to protein− RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. USA 2019, 116, 7889–7898. [Google Scholar] [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Boeynaems, S.; De Decker, M.; Tompa, P.; Van Den Bosch, L. Arginine-rich Peptides Can Actively Mediate Liquid-liquid Phase Separation. Bio-Protocol 2017, 7, e2525. [Google Scholar] [CrossRef]
- Rippe, K. Liquid-Liquid Phase Separation in Chromatin. Cold Spring Harb. Perspect. Biol. 2022, 14, a040683. [Google Scholar] [CrossRef]
- Alberti, S.; Arosio, P.; Best, R.B.; Boeynaems, S.; Cai, D.; Collepardo-Guevara, R.; Dignon, G.L.; Dimova, R.; Elbaum-Garfinkle, S.; Fawzi, N.L.; et al. Current practices in the study of biomolecular condensates: A community comment. Nat. Commun. 2025, 16, 7730. [Google Scholar] [CrossRef] [PubMed]
- Trcek, T.; Douglas, T.E.; Grosch, M.; Yin, Y.; Eagle, W.V.I.; Gavis, E.R.; Shroff, H.; Rothenberg, E.; Lehmann, R. Sequence-Independent Self-Assembly of Germ Granule mRNAs into Homotypic Clusters. Mol. Cell 2020, 78, 941–950.E12. [Google Scholar] [CrossRef] [PubMed]
- Cochard, A.; Navarro, M.G.-J.; Piroska, L.; Kashida, S.; Kress, M.; Weil, D.; Gueroui, Z. RNA at the surface of phase-separated condensates impacts their size and number. Biophys. J. 2022, 121, 1675–1690. [Google Scholar] [CrossRef] [PubMed]
- Contreras, V.; Richardson, M.A.; Hao, E.; Keiper, B.D. Depletion of the cap-associated isoform of translation factor eIF4G induces germline apoptosis in C. elegans. Cell Death Differ. 2008, 15, 1232–1242. [Google Scholar] [CrossRef]
- Raesch, F.; Weber, R.; Izaurralde, E.; Igreja, C. 4E-T-bound mRNAs are stored in a silenced and deadenylated form. Genes Dev. 2020, 34, 847–860. [Google Scholar] [CrossRef]
- Dostie, J.; Ferraiuolo, M.; Pause, A.; Adam, S.A.; Sonenberg, N. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 2000, 19, 3142–3156. [Google Scholar] [CrossRef]
- Uebel, C.J.; Rajeev, S.; Phillips, C.M. Caenorhabditis elegans germ granules are present in distinct configurations and assemble in a hierarchical manner. Development 2023, 150, dev202284. [Google Scholar] [CrossRef]
- Marnik, E.A.; Almeida, M.V.; Cipriani, P.G.; Chung, G.; Caspani, E.; Karaulanov, E.; Gan, H.H.; Zinno, J.; Isolehto, I.J.; Kielisch, F.; et al. The Caenorhabditis elegans TDRD5/7-like protein, LOTR-1, interacts with the helicase ZNFX-1 to balance epigenetic signals in the germline. PLoS Genet. 2022, 18, e1010245. [Google Scholar] [CrossRef]
- Keiper, B.D. Translation of mRNAs in Xenopus oocytes. In Encyclopedia of Life Sciences; Nature Publishing Group: London, UK, 2003; Available online: www.els.net (accessed on 14 December 2012).
- Messina, V.; Di Sauro, A.; Pedrotti, S.; Adesso, L.; Latina, A.; Geremia, R.; Rossi, P.; Sette, C. Differential contribution of the MTOR and MNK pathways to the regulation of mRNA translation in meiotic and postmeiotic mouse male germ cells. Biol. Reprod. 2010, 83, 607–615. [Google Scholar] [CrossRef]
- Joshi, B.; Lee, K.; Maeder, D.L.; Jagus, R. Phylogenetic analysis of eIF4E-family members. BMC Evol. Biol. 2005, 5, 48. [Google Scholar] [CrossRef]
- Gruner, S.; Peter, D.; Weber, R.; Wohlbold, L.; Chung, M.Y.; Weichenrieder, O.; Valkov, E.; Igreja, C.; Izaurralde, E. The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation. Mol. Cell 2016, 2765, 020. [Google Scholar] [CrossRef]
- Gruner, S.; Weber, R.; Peter, D.; Chung, M.Y.; Igreja, C.; Valkov, E.; Izaurralde, E. Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast. Nucleic Acids Res. 2018, 46, 6893–6908. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; He, Z.; Sha, Y.; Kee, K.; Li, L. Eif4enif1 haploinsufficiency disrupts oocyte mitochondrial dynamics and leads to subfertility. Development 2023, 150, dev202151. [Google Scholar] [CrossRef]
- Heim, A.; Cheng, S.; Orth, J.; Stengel, F.; Schuh, M.; Mayer, T.U. Translational repression by 4E-T is crucial to maintain the prophase-I arrest in vertebrate oocytes. Nat. Commun. 2025, 16, 8051. [Google Scholar] [CrossRef]
- Shang, L.; Ren, S.; Yang, X.; Zhang, F.; Jin, L.; Zhang, X.; Wu, Y. EIF4ENIF1 variants in two patients with non-syndromic premature ovarian insufficiency. Eur. J. Med. Genet. 2022, 65, 104597. [Google Scholar] [CrossRef]
- Waghray, S.; Williams, C.; Coon, J.J.; Wickens, M. Xenopus CAF1 requires NOT1-mediated interaction with 4E-T to repress translation in vivo. RNA 2015, 21, 1335–1345. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Dai, S.; Tang, X.; Li, L.; Ishidate, T.; Ozturk, A.R.; Chen, H.; Dube, A.L.; Yan, Y.-H.; Dong, M.-Q.; Shen, E.-Z. A family of C. elegans VASA homologs control Argonaute pathway specificity and promote transgenerational silencing. Cell Rep. 2022, 40, 111265. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xu, J.; Gao, E.; Fan, X.; Wei, J.; Ye, B.; Xu, S.; Ma, W. Enhanced single RNA imaging reveals dynamic gene expression in live animals. eLife 2023, 12, e82178. [Google Scholar] [CrossRef]
- Li, W.; Maekiniemi, A.; Sato, H.; Osman, C.; Singer, R.H. An improved imaging system that corrects MS2-induced RNA destabilization. Nat. Methods 2022, 19, 1558–1562. [Google Scholar] [CrossRef]
- Boersma, S.; Khuperkar, D.; Verhagen, B.M.P.; Sonneveld, S.; Grimm, J.B.; Lavis, L.D.; Tanenbaum, M.E. Multi-Color Single-Molecule Imaging Uncovers Extensive Heterogeneity in mRNA Decoding. Cell 2019, 178, 458–472.e19. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keiper, B.D.; Huggins, H.P. Remodeling of Germ Cell mRNPs for Translational Control. Biology 2025, 14, 1430. https://doi.org/10.3390/biology14101430
Keiper BD, Huggins HP. Remodeling of Germ Cell mRNPs for Translational Control. Biology. 2025; 14(10):1430. https://doi.org/10.3390/biology14101430
Chicago/Turabian StyleKeiper, Brett D., and Hayden P. Huggins. 2025. "Remodeling of Germ Cell mRNPs for Translational Control" Biology 14, no. 10: 1430. https://doi.org/10.3390/biology14101430
APA StyleKeiper, B. D., & Huggins, H. P. (2025). Remodeling of Germ Cell mRNPs for Translational Control. Biology, 14(10), 1430. https://doi.org/10.3390/biology14101430






