Evolutionary and Functional Insights into Rice Universal Stress Proteins in Response to Abiotic Stresses
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of OsUSP Genes in Rice
2.2. Multiple Comparisons and Phylogenetic Analysis
2.3. Collinearity and Synteny Analysis of the OsUSP Genes
2.4. Gene Structure, Conserved Domains, and Protein Motif Analysis
2.5. Cis-Acting Element Analysis
2.6. Expression Patterns Analysis of OsUSP Genes
2.7. Plant Materials and Treatments
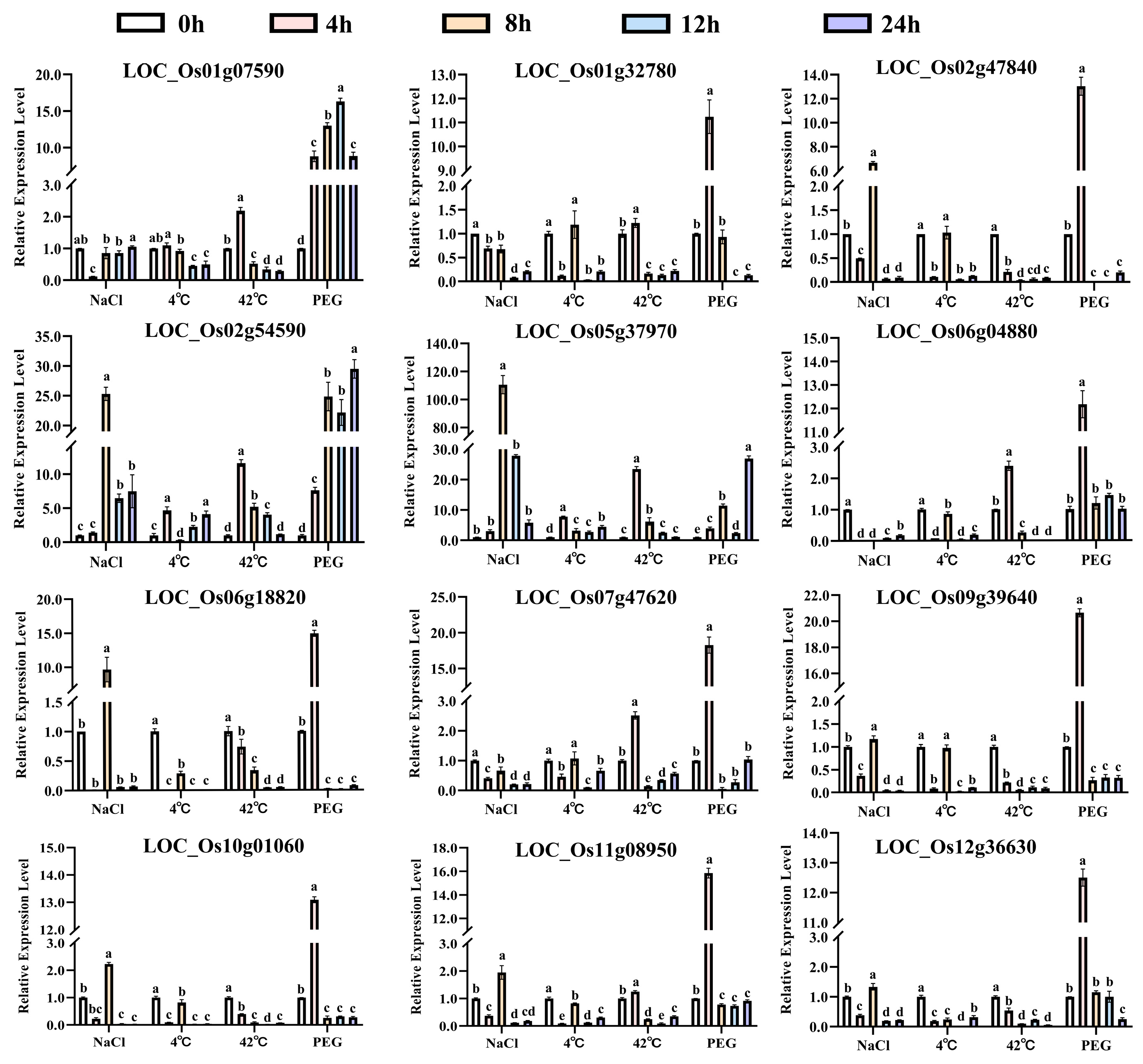
2.8. Quantitative PCR Validation
2.9. PPI Network Prediction of OsUSP Proteins
2.10. Statistical Analysis
3. Results
3.1. Identification and Molecular Characterization of USP Proteins in Rice
3.2. Protein Conserved Motif, Domain, and Gene Structure Analysis of OsUSPs
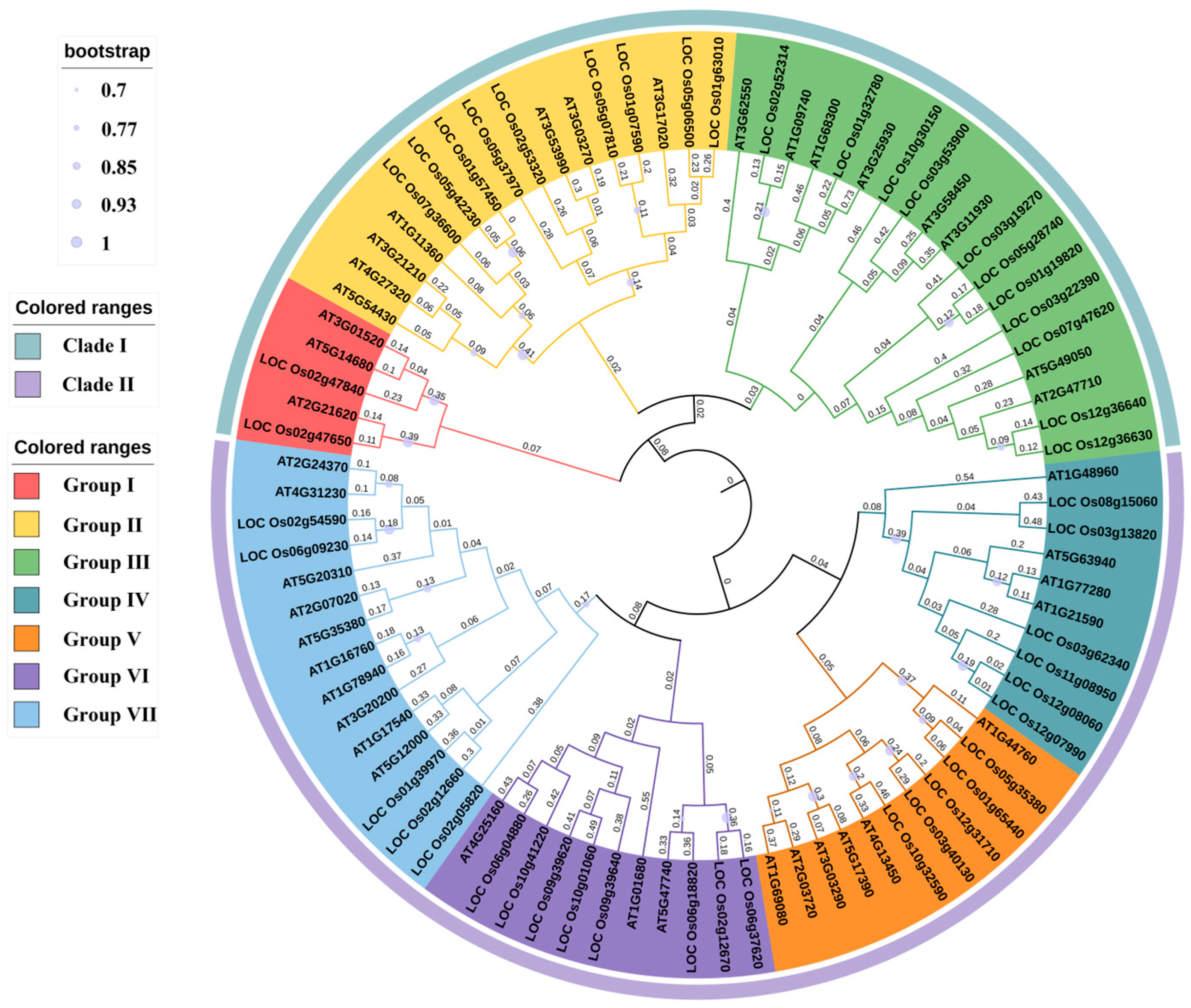
3.3. Phylogenetic Analysis of OsUSPs in Rice
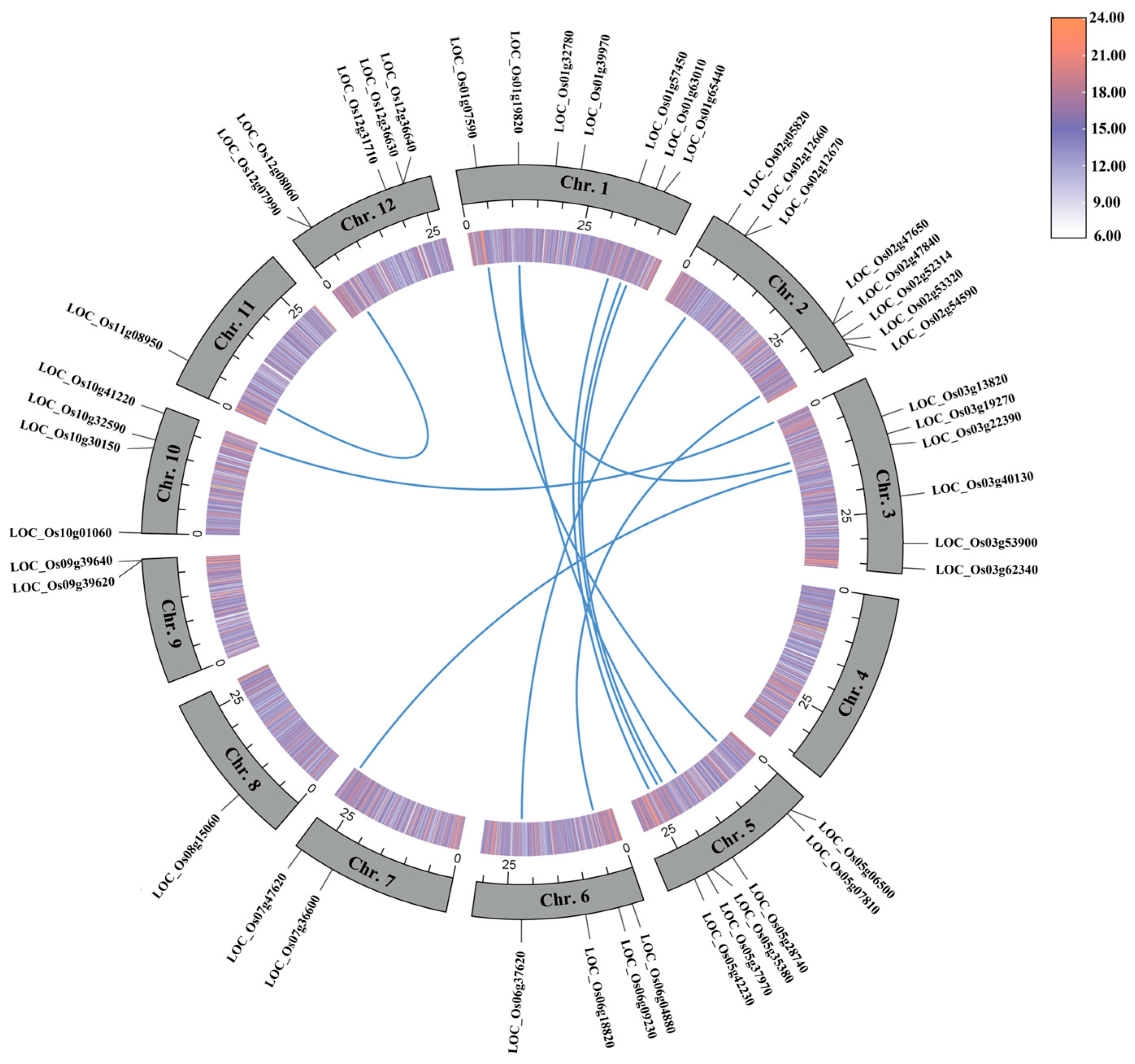
3.4. Collinearity Analysis of OsUSPs in Rice
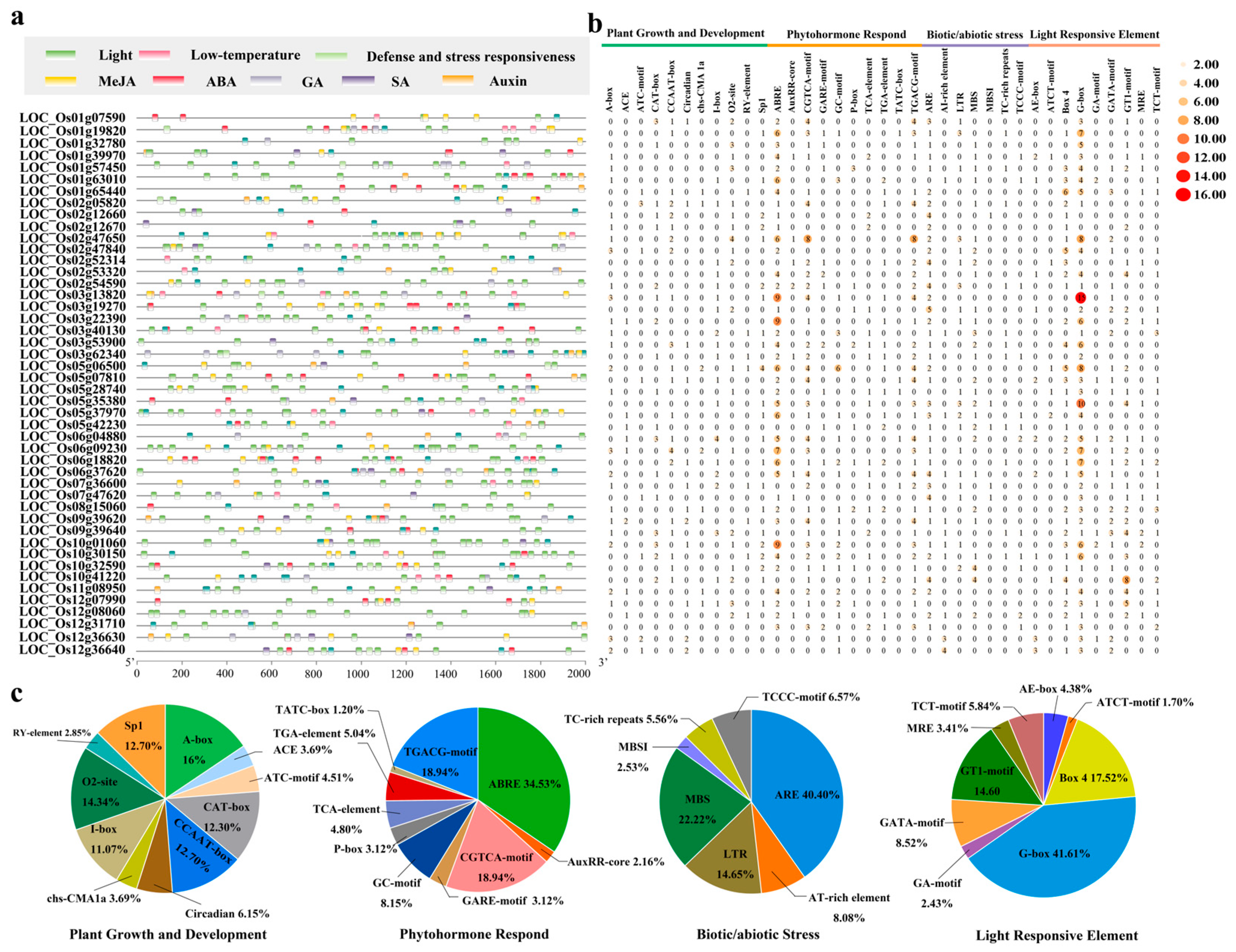
3.5. Characterization of Cis-Acting Elements in OsUSP Promoters Regions
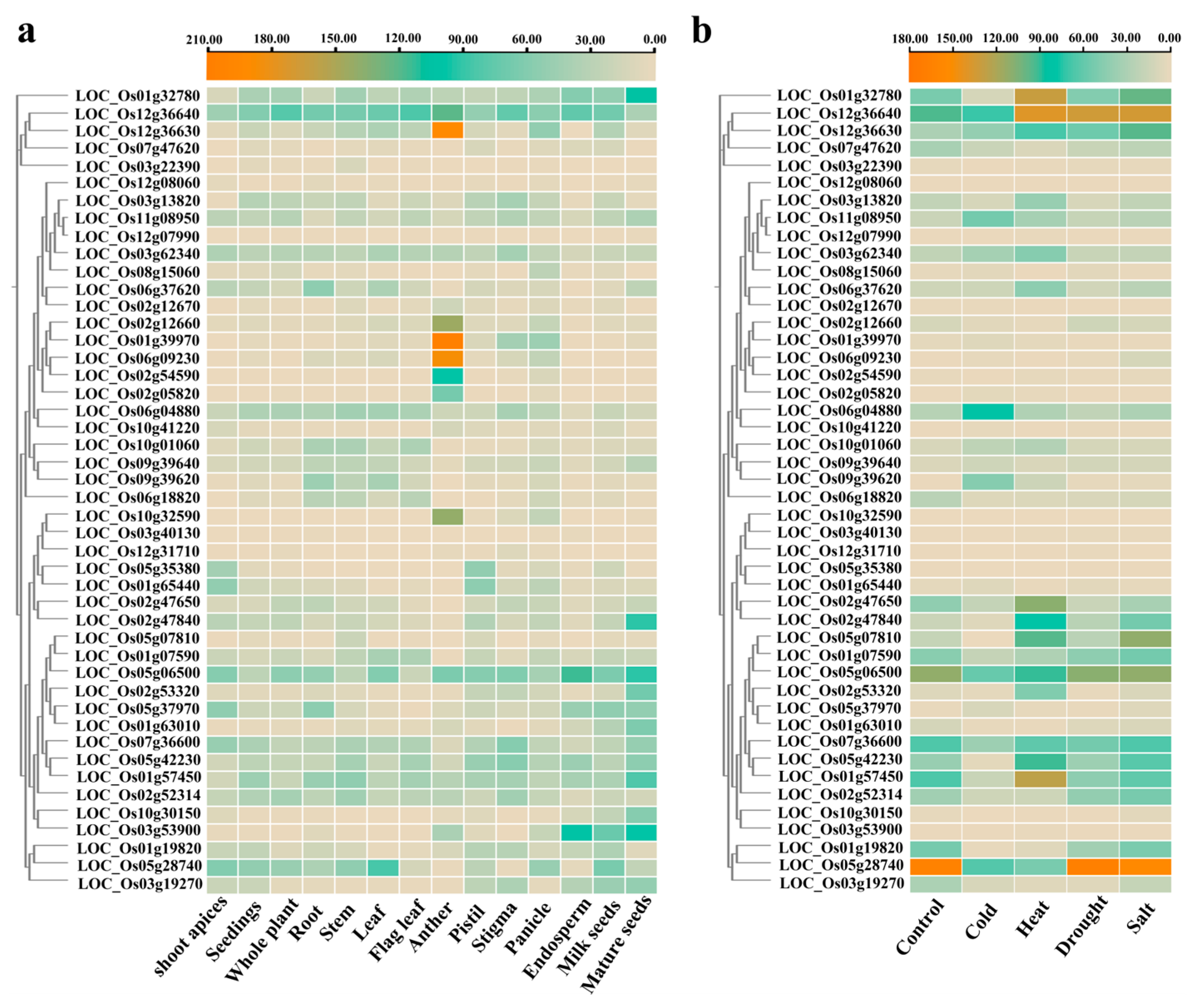
3.6. Expression Patterns of OsUSPs in Different Tissues and Under Abiotic Stress Conditions
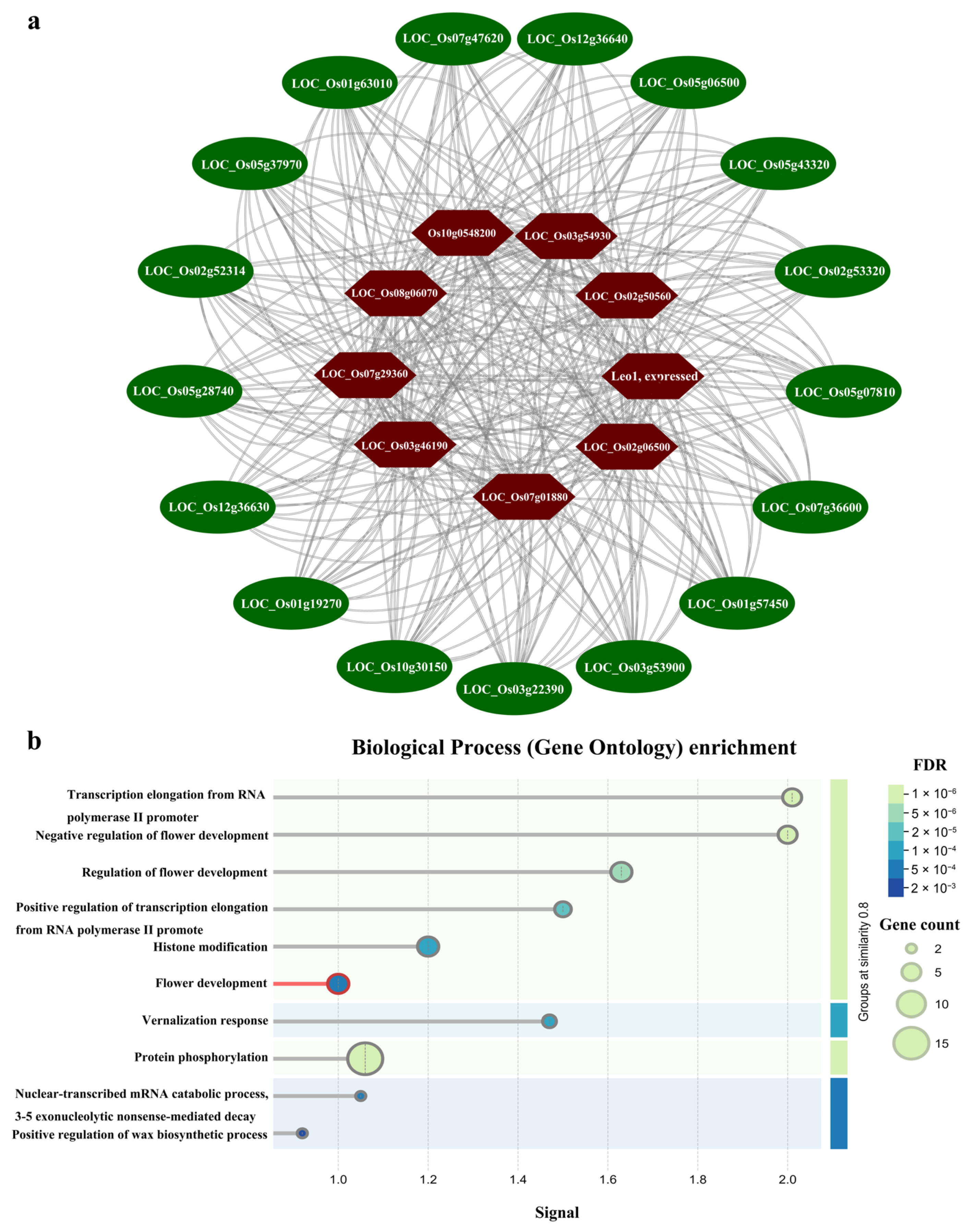
3.7. Protein–Protein Interaction (PPI) Network of OsUSP in Rice
4. Discussion
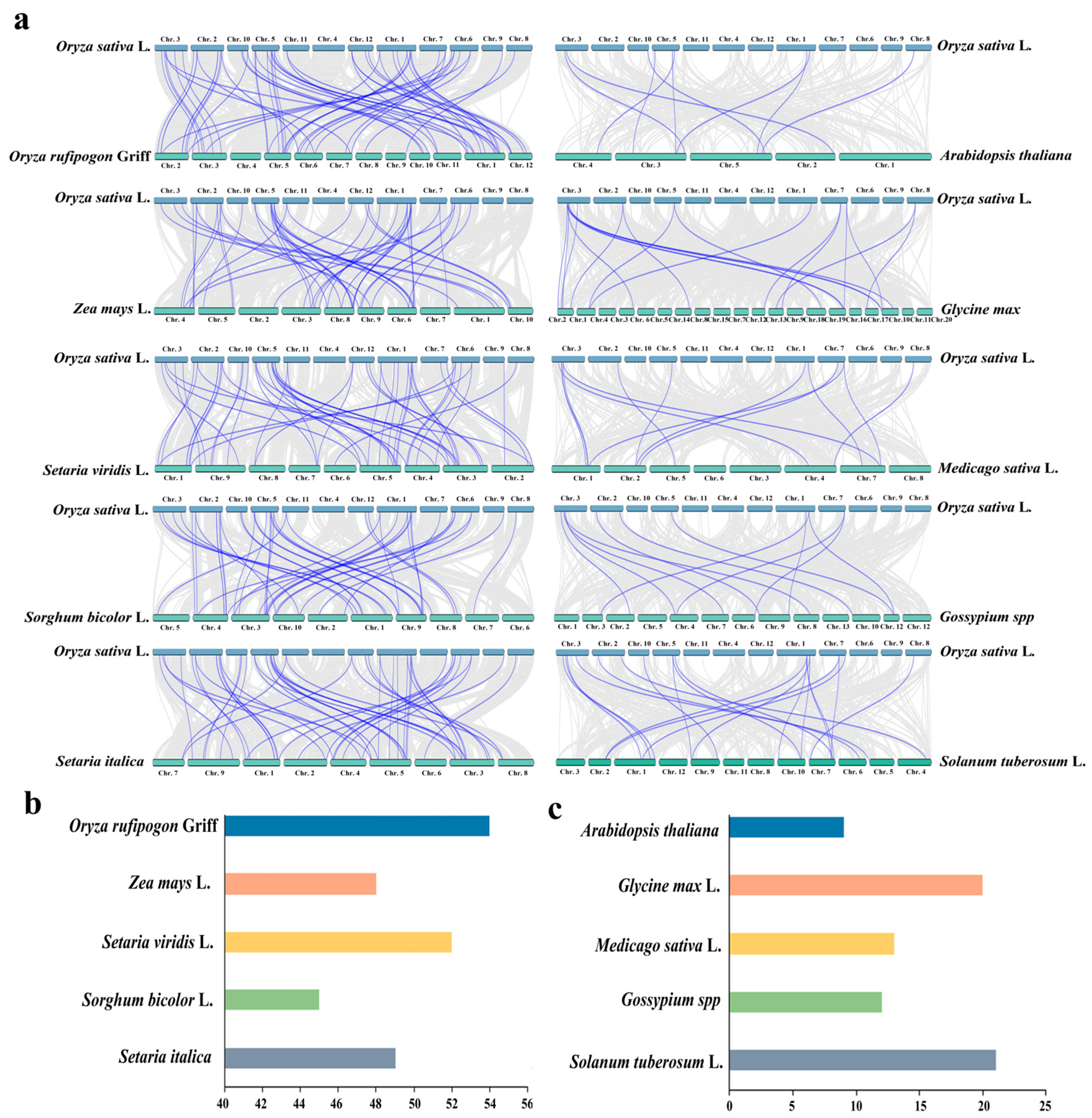
4.1. Distribution of USPs in Plants
4.2. Structural and Functional Diversity of USPs
4.2.1. Phylogenetic Classification and Domain Architecture Underpin Functional Diversification
4.2.2. Cis-Regulatory Elements Reveal Hormonal and Stress-Responsive Regulation
4.2.3. Integrated Structural and Regulatory Features Support Roles in Stress Adaptation
4.3. Potential Molecular Mechanism Underlying Abiotic Stress Responses of USPs
4.4. Future Research Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CDD | Conserved Domain Database |
| Co-IP | Co-immunoprecipitation |
| Ka | Non-synonymous substitution rate |
| Ks | Synonymous substitution rate |
| MW | Molecular weight |
| pI | Theoretical isoelectric point |
| PPI | Protein–protein interaction |
| qPCR | Quantitative PCR |
| SRA | Sequence Read Archive |
| TPR | Tetratricopeptide repeat |
| USP | Universal Stress Protein |
| UTRs | Untranslated regions |
| Y2H | Yeast two-hybrid |
References
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.; Li, J.; Wang, P.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Kopecká, R.; Kameniarová, M.; Černý, M.; Brzobohatý, B.; Novák, J. Abiotic stress in crop production. Int. J. Mol. Sci. 2023, 24, 6603. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Z.; Chong, K.; Xu, Y. Chilling tolerance in rice: Past and present. J. Plant Physiol. 2022, 268, 153576. [Google Scholar] [CrossRef]
- Heredia, M.C.; Kant, J.; Prodhan, M.A.; Dixit, S.; Wissuwa, M. Breeding rice for a changing climate by improving adaptations to water saving technologies. Theor. Appl. Genet. 2021, 135, 17–33. [Google Scholar] [CrossRef]
- Khush, G.S. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 2005, 59, 1–6. [Google Scholar] [CrossRef]
- Fan, M.; Gao, S.; Yang, Y.; Yang, S.; Wang, H.; Shi, L. Genome-wide identification and expression analysis of the universal stress protein (USP) gene family in Arabidopsis thaliana, Zea mays, and Oryza sativa. Genetica 2024, 152, 119–132. [Google Scholar] [CrossRef]
- Vollmer, A.C.; Bark, S.J. Twenty-five years of investigating the universal stress protein: Function, structure, and applications. Adv. Appl. Microbiol. 2018, 102, 1–36. [Google Scholar]
- Nyström, T.; Neidhardt, F.C. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 1992, 6, 3178–3198. [Google Scholar] [CrossRef]
- Kvint, K.; Nachin, L.; Diez, A.; Nyström, T. The bacterial universal stress protein: Function and regulation. Curr. Opin. Microbiol. 2003, 6, 140–145. [Google Scholar] [CrossRef]
- Sauter, M.; Rzewuski, G.; Marwedel, T.; Lorbiecke, R. The novel ethylene-regulated gene OsUsp1 from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 2002, 53, 2325–2331. [Google Scholar]
- Park, S.C.; Jung, Y.J.; Lee, Y.; Kim, I.R.; Seol, M.A.; Kim, E.J.; Jang, M.K.; Lee, J.R. Functional characterization of the Arabidopsis universal stress protein AtUSP with an antifungal activity. Biochem. Biophys. Res. Commun. 2017, 486, 923–929. [Google Scholar] [CrossRef]
- Qi, T.; He, F.; Zhang, X.; Wang, J.; Zhang, Z.; Jiang, H.; Zhao, B.; Du, C.; Che, Y.; Feng, X.; et al. Genome-wide identification and expression profiling of potato (Solanum tuberosum L.) universal stress proteins reveal essential roles in mechanical damage and deoxynivalenol stress. Int. J. Mol. Sci. 2024, 25, 1341. [Google Scholar] [CrossRef]
- Gutiérrez-Beltrán, E.; Personat, J.M.; de la Torre, F.; del Pozo, O. A universal stress protein involved in oxidative stress is a phosphorylation target for protein kinase CIPK6. Plant Physiol. 2017, 173, 836–852. [Google Scholar] [PubMed]
- Feng, Y.J.; Zhao, W.; Li, Y.L.; Shen, Y.J.; Sun, Y.C.; Meng, X.Y.; Li, J.; Wu, W.; Zhang, G.X.; Liu, M.Y.; et al. Genome-wide identification of wheat USP gene family and functional dissection of TaUSP85 involved in heat tolerance. Plant Physiol. Biochem. 2025, 219, 109359. [Google Scholar]
- Chi, Y.H.; Koo, S.S.; Oh, H.T.; Lee, E.S.; Park, J.H.; Phan, K.A.T.; Wi, S.D.; Bae, S.B.; Paeng, S.K.; Chae, H.B.; et al. The physiological functions of universal stress proteins and their molecular mechanism to protect plants from environmental stresses. Front. Plant Sci. 2019, 10, 750. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Wu, Z.; Bai, Q.; Zhang, Y.; Huang, M.; Huang, Y.; Li, X. Universal Stress Proteins: From Gene to Function. Int. J. Mol. Sci. 2023, 24, 4725. [Google Scholar] [CrossRef]
- Gorshkova, D.S.; Getman, I.A.; Voronkov, A.S.; Chizhova, S.I.; Kuznetsov, V.V.; Pojidaeva, E.S. The gene encoding the universal stress protein AtUSP is regulated by phytohormones and involved in seed germination of Arabidopsis thaliana. Dokl. Biochem. Biophys. 2018, 479, 105–107. [Google Scholar] [CrossRef]
- Gorshkova, D.S.; Getman, I.A.; Sergeeva, L.I.; Kuznetsov, V.V.; Pojidaeva, E.S. GRUSP, an universal stress protein, is involved in gibberellin-dependent induction of flowering in Arabidopsis thaliana. Dokl. Biochem. Biophys. 2021, 499, 233–237. [Google Scholar] [CrossRef]
- Gonzali, S.; Loreti, E.; Cardarelli, F.; Novi, G.; Parlanti, S.; Pucciariello, C.; Bassolino, L.; Banti, V.; Licausi, F.; Perata, P. Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nat. Plants 2015, 1, 15151. [Google Scholar] [CrossRef]
- Isokpehi, R.D.; Mahmud, O.; Mbah, A.N.; Simmons, S.S.; Avelar, L.; Rajnarayanan, R.V.; Udensi, U.K.; Ayensu, W.K.; Cohly, H.H.; Brown, S.D.; et al. Developmental regulation of genes encoding universal stress proteins in Schistosoma mansoni. Gene Regul. Syst. Bio. 2011, 5, 61–74. [Google Scholar]
- Loukehaich, R.; Wang, T.; Ouyang, B.; Ziaf, K.; Li, H.; Zhang, J.; Lu, Y.; Ye, Z. SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J. Exp. Bot. 2012, 63, 5593–5606. [Google Scholar]
- Maqbool, A.; Zahur, M.; Husnain, T.; Riazuddin, S. GUSP1 and GUSP2, two drought-responsive genes in Gossypium arboreum have homology to universal stress proteins. Plant Mol. Biol. Report. 2008, 27, 109–114. [Google Scholar] [CrossRef]
- Hassan, S.; Ahmad, A.; Batool, F.; Rashid, B.; Husnain, T. Genetic modification of Gossypium arboreum universal stress protein (GUSP1) improves drought tolerance in transgenic cotton (Gossypium hirsutum). Physiol. Mol. Biol. Plants 2021, 27, 1779–1794. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhao, F.a.; Fang, W.; Xie, D.; Hou, J.; Yang, X.; Zhao, Y.; Tang, Z.; Nie, L.; Lv, S. Identification of early salt stress responsive proteins in seedling roots of upland cotton (Gossypium hirsutum L.) employing iTRAQ-based proteomic technique. Front. Plant Sci. 2015, 6, 1179–1794. [Google Scholar]
- Isokpehi, R.D.; Simmons, S.S.; Cohly, H.H.P.; Ekunwe, S.I.N.; Begonia, G.B.; Ayensu, W.K. Identification of drought-responsive universal stress proteins in viridiplantae. Bioinform. Biol. Insights 2011, 5, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, U.; Richard, O.; Zamboni, A.; Gapper, C.; Geisler, M.; Pogson, B.; Karpinski, S.; Mullineaux, P.M. Impact of chloroplastic- and extracellular-sourced ROS on high light-responsive gene expression in Arabidopsis. J. Exp. Bot. 2008, 59, 121–133. [Google Scholar]
- Diao, J.; Gu, W.; Jiang, Z.; Wang, J.; Zou, H.; Zong, C.; Ma, L. Comprehensive analysis of universal stress protein family genes and their expression in Fusarium oxysporum response of Populus davidiana × P. alba var. pyramidalis louche based on the transcriptome. Int. J. Mol. Sci. 2023, 24, 5405. [Google Scholar]
- Espinola, S.M.; Cancela, M.P.; Brisolara Corrêa, L.; Zaha, A. Evolutionary fates of universal stress protein paralogs in Platyhelminthes. BMC Evol. Biol. 2018, 18, 10. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Lozano, R.; Hamblin, M.T.; Prochnik, S.; Jannink, J.L. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genom. 2015, 16, 360. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A "one for all, all for one" bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinf. 2021, 8, 77–80. [Google Scholar]
- Lang, H.; He, Y.; Zeng, F.; Xu, F.; Zhao, M.; Ma, D. Comparative transcriptomic and physiological analyses of weedy rice and cultivated rice to identify vital differentially expressed genes and pathways regulating the ABA response. Sci. Rep. 2021, 11, 12881. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, H.; Chern, M.; Yin, J.; Chen, Y.; Wang, J.; Zhu, X.; Chen, Z.; Yuan, C.; Zhao, W.; et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 3174–3179. [Google Scholar]
- Fukagawa, N.K.; Ziska, L.H. Rice: Importance for global nutrition. J. Nutr. Sci. Vitaminol. 2019, 65, S2–S3. [Google Scholar] [CrossRef]
- Sharif, M.K.; Butt, M.S.; Anjum, F.M.; Khan, S.H. Rice bran: A novel functional ingredient. Crit. Rev. Food Sci. Nutr. 2014, 54, 807–816. [Google Scholar]
- Arabia, S.; Sami, A.A.; Akhter, S.; Sarker, R.H.; Islam, T. Comprehensive in silico characterization of universal stress proteins in rice (Oryza sativa L.) with insight into their stress-specific transcriptional modulation. Front. Plant Sci. 2021, 12, 712607. [Google Scholar] [CrossRef]
- Li, W.T.; Wei, Y.M.; Wang, J.R.; Liu, C.J.; Lan, X.J.; Jiang, Q.T.; Pu, Z.E.; Zheng, Y.L. Identification, localization, and characterization of putative USP genes in barley. Theor. Appl. Genet. 2010, 121, 907–917. [Google Scholar] [CrossRef]
- Sinha, P.; Pazhamala, L.T.; Singh, V.K.; Saxena, R.K.; Krishnamurthy, L.; Azam, S.; Khan, A.W.; Varshney, R.K. Identification and validation of selected universal stress protein domain containing drought-responsive genes in pigeonpea (Cajanus cajan L.). Front. Plant Sci. 2016, 6, 1065. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, A.; Li, Z.; Wang, H.; Wang, J.; Dong, Z.; Yao, L.; Han, X.; Wei, F. Characterization and gene expression analysis reveal universal stress proteins respond to abiotic stress in Gossypium hirsutum. BMC Genom. 2024, 25, 98. [Google Scholar] [CrossRef] [PubMed]
- Oudelaar, A.M.; Higgs, D.R. The relationship between genome structure and function. Nat. Rev. Genet. 2021, 22, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.J.; Melencion, S.M.B.; Lee, E.S.; Park, J.H.; Alinapon, C.V.; Oh, H.T.; Yun, D.J.; Chi, Y.H.; Lee, S.Y. Universal stress protein exhibits a redox-dependent chaperone function in Arabidopsis and enhances plant tolerance to heat shock and oxidative stress. Front. Plant Sci. 2015, 6, 1141. [Google Scholar] [CrossRef] [PubMed]
- Melencion, S.M.B.; Chi, Y.H.; Pham, T.T.; Paeng, S.K.; Wi, S.D.; Lee, C.; Ryu, S.W.; Koo, S.S.; Lee, S.Y. RNA Chaperone function of a universal stress protein in Arabidopsis confers enhanced cold stress tolerance in plants. Int. J. Mol. Sci. 2017, 18, 2546. [Google Scholar] [CrossRef]
- Merkouropoulos, G.; Andreasson, E.; Hess, D.; Boller, T.; Peck, S.C. An Arabidopsis protein phosphorylated in response to microbial elicitation, AtPHOS32, is a substrate of MAP Kinases 3 and 6. J. Biol. Chem. 2008, 283, 10493–10499. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2019, 46, 197–212. [Google Scholar] [CrossRef]
- Bhuria, M.; Goel, P.; Kumar, S.; Singh, A.K. AtUSP17 negatively regulates salt stress tolerance through modulation of multiple signaling pathways in Arabidopsis. Physiol. Plant 2022, 174, 13635. [Google Scholar]
- Zahur, M.; Maqbool, A.; Irfan, M.; Khan Barozai, M.Y.; Rashid, B.; Riazuddin, S.; Husnain, T. Isolation and functional analysis of cotton universal stress protein promoter in response to phytohormones and abiotic stresses. Mol. Biol. 2009, 43, 578–585. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lang, H.; Jiang, Y.; Xie, Y.; Wu, J.; Wang, Y.; Jiang, M. Evolutionary and Functional Insights into Rice Universal Stress Proteins in Response to Abiotic Stresses. Biology 2025, 14, 1359. https://doi.org/10.3390/biology14101359
Lang H, Jiang Y, Xie Y, Wu J, Wang Y, Jiang M. Evolutionary and Functional Insights into Rice Universal Stress Proteins in Response to Abiotic Stresses. Biology. 2025; 14(10):1359. https://doi.org/10.3390/biology14101359
Chicago/Turabian StyleLang, Hong, Yuxi Jiang, Yan Xie, Jiayin Wu, Yubo Wang, and Mingliang Jiang. 2025. "Evolutionary and Functional Insights into Rice Universal Stress Proteins in Response to Abiotic Stresses" Biology 14, no. 10: 1359. https://doi.org/10.3390/biology14101359
APA StyleLang, H., Jiang, Y., Xie, Y., Wu, J., Wang, Y., & Jiang, M. (2025). Evolutionary and Functional Insights into Rice Universal Stress Proteins in Response to Abiotic Stresses. Biology, 14(10), 1359. https://doi.org/10.3390/biology14101359





