Consensus-Guided Construction of H5N1-Specific and Universal Influenza a Multiepitope Vaccines
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval
2.2. Frequency Analysis of Residues in the HA RBD for Vaccine Targeting
2.3. Epitope Prediction
2.3.1. B-Cell Epitope Prediction
2.3.2. CTL Epitope Prediction
2.3.3. HTL Epitope Prediction
2.4. Identification of IFN-γ–Inducing Epitopes
2.5. Toxicity Prediction
2.6. Vaccine Design
2.6.1. Adjuvant Development
2.6.2. Epitope-Based Vaccine Construct Targeting the RBD of H5N1 Hemagglutinin
2.6.3. Structural Segment-Based Vaccine Derived from the H5N1 HA RBD
2.6.4. Universal Influenza a Vaccine
2.6.5. Full-Length HA Fragment Control
2.7. Structural Modeling, Refinement, and Structural Quality Assessment
2.8. Molecular Docking Analysis of Vaccine–TLR Interactions
2.9. In Silico Simulation of Host Immune Response
2.10. Population Coverage Estimation
2.11. Codon Adaptation and In Silico Cloning
3. Results
3.1. Protein Sequence Retrieval
3.2. Comparative Analysis of Residue Conservation in Influenza a Subtypes
3.2.1. Consensus HA Sequence Among H5N1 Isolates
3.2.2. Conservation of the HA Sequence Across Influenza a Subtypes
3.3. Epitopes Prediction
3.4. Vaccine Construction
3.5. Toxicity Prediction and Allerginicity
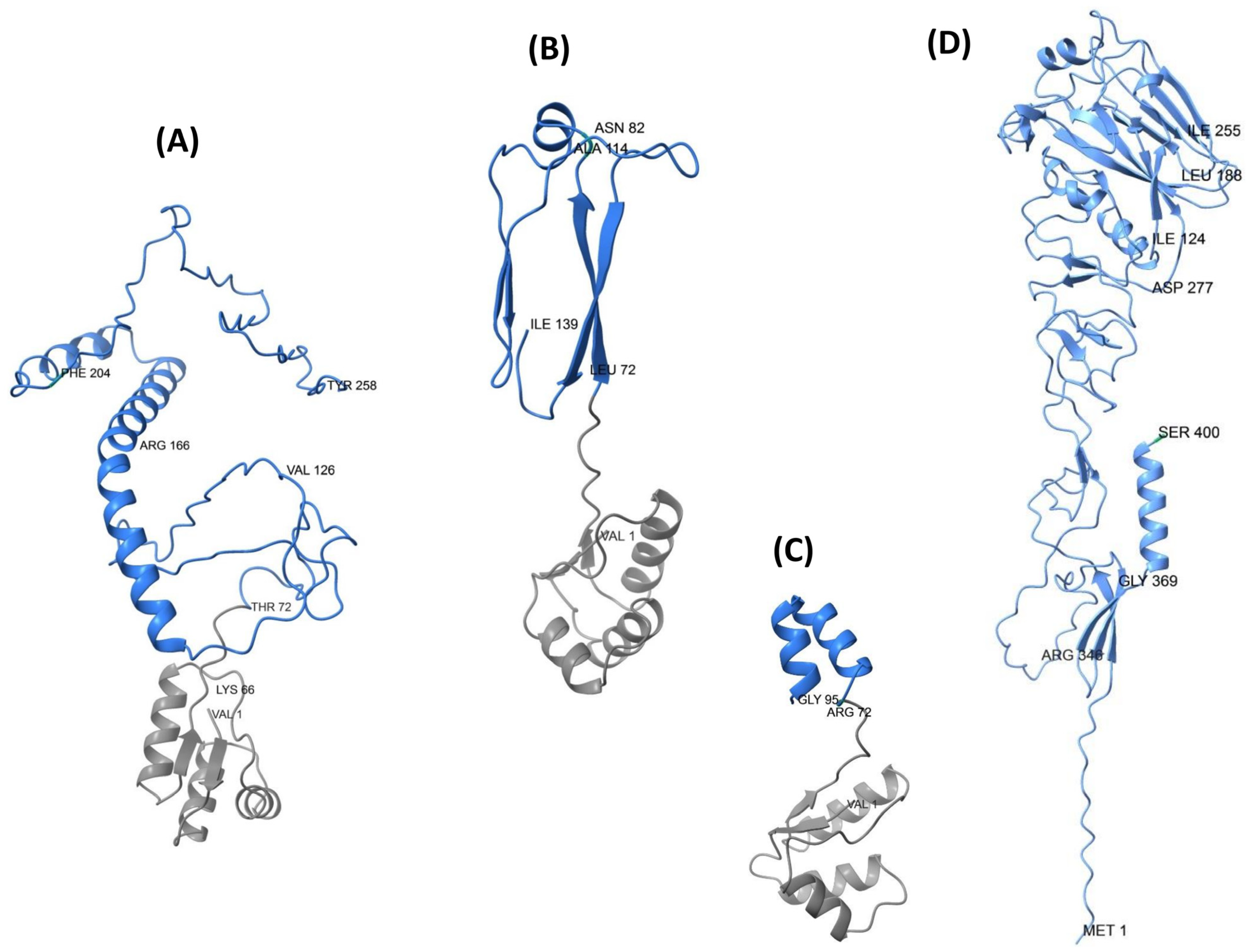
3.6. Structural Prediction
3.7. Structural Refinement and Structural Quality Assessment
3.8. Molecular Docking Analysis
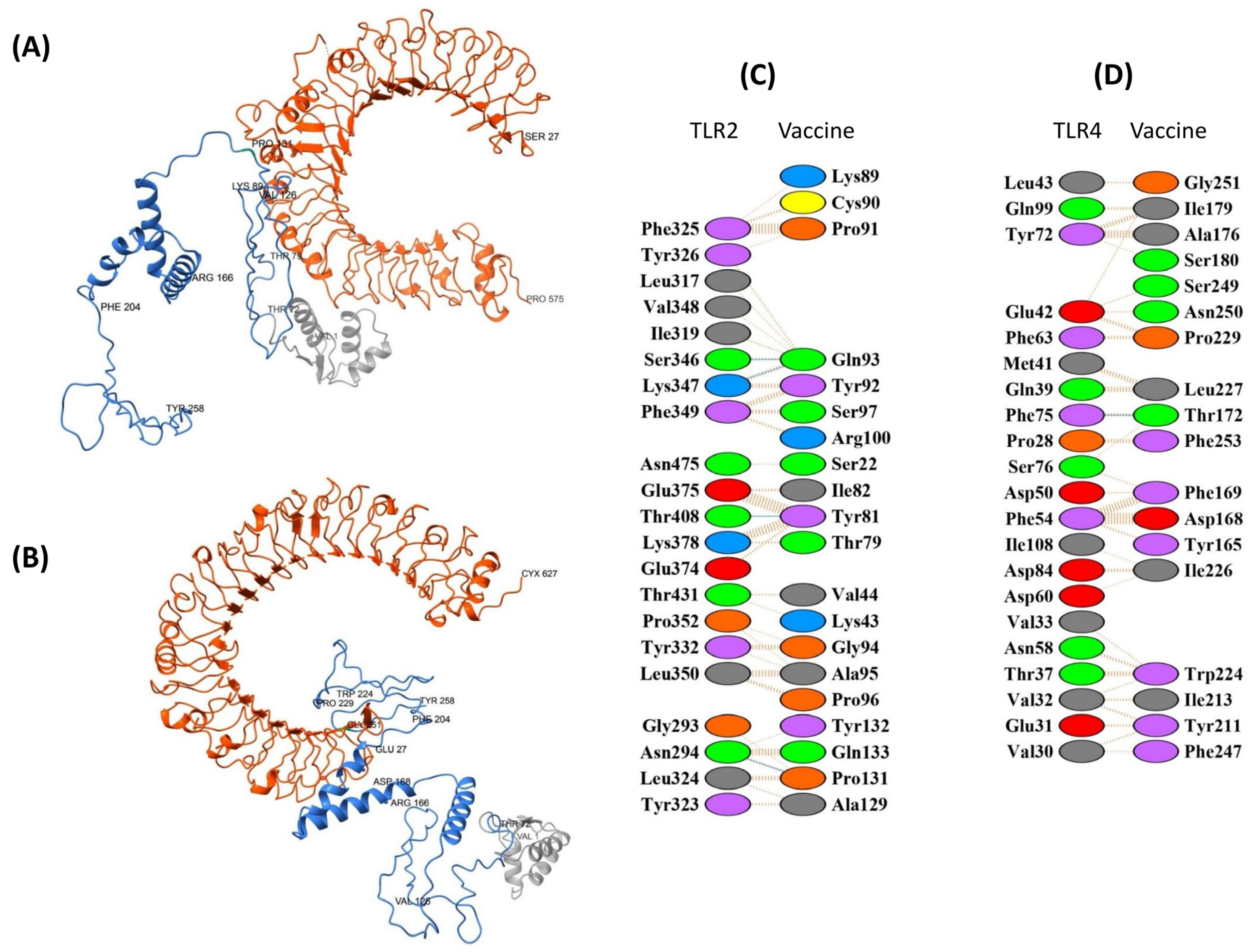
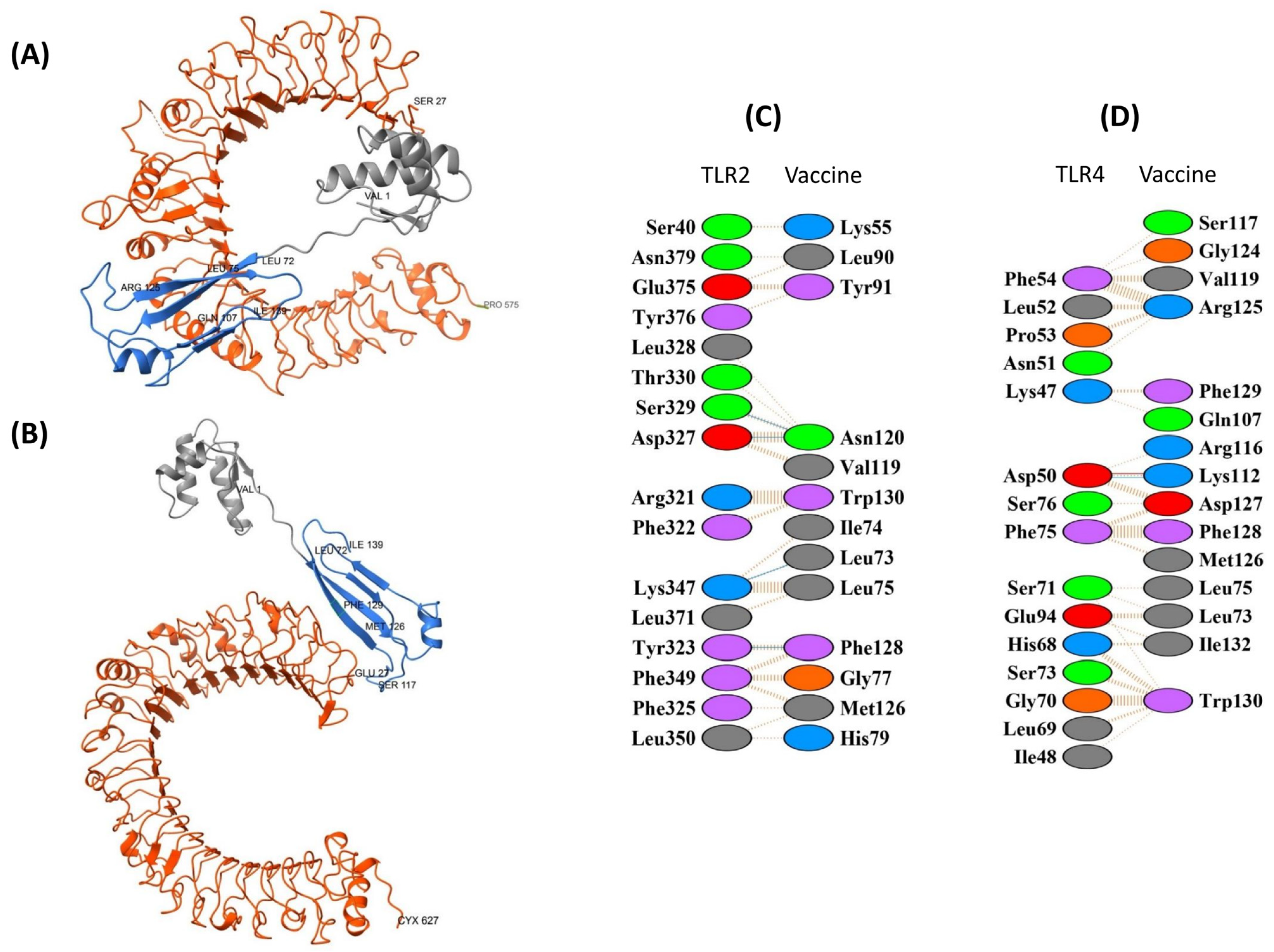
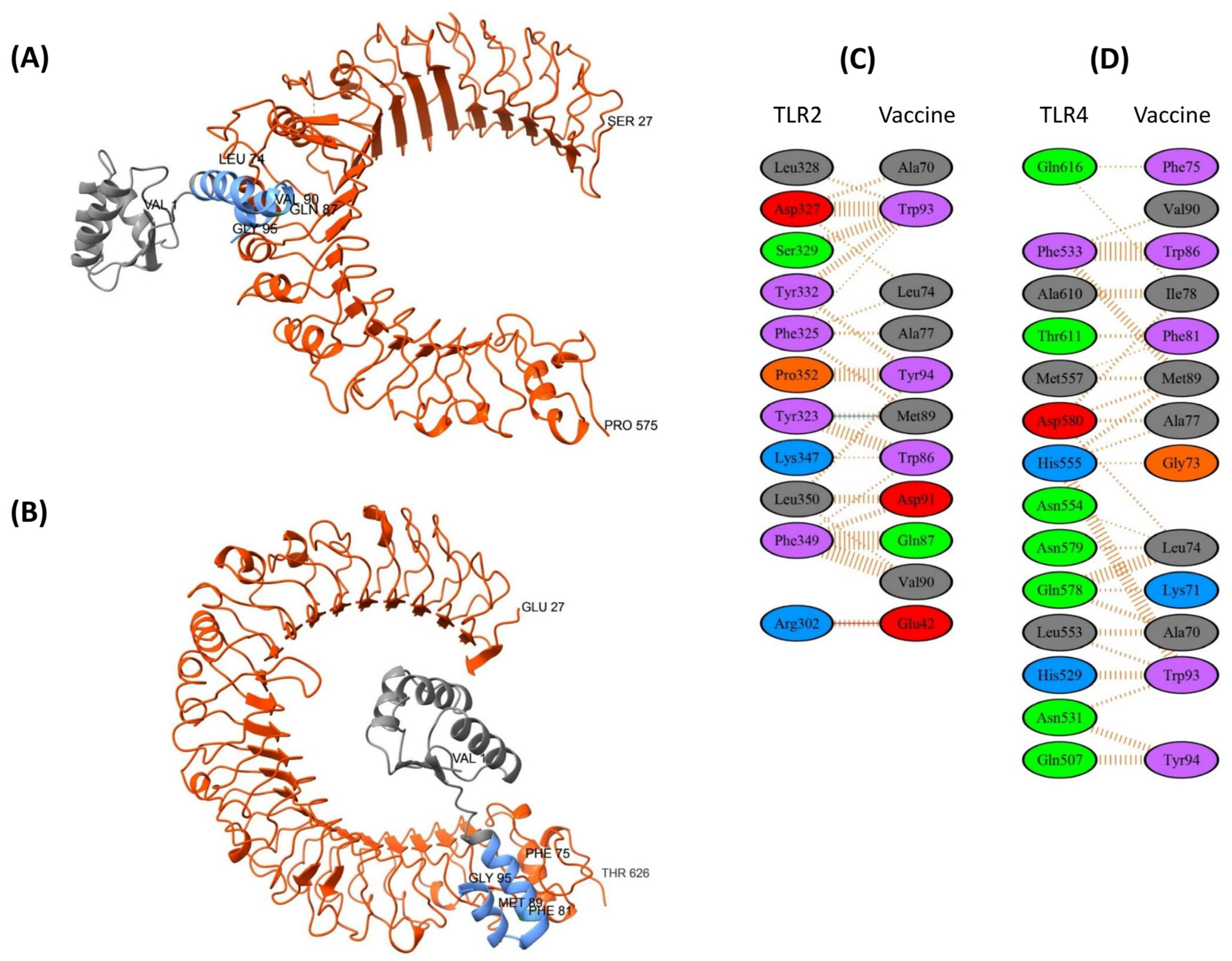
3.8.1. TLR2 and TLR4 Complexes with the EpitoCore-HA-VX Vaccine
3.8.2. TLR2 and TLR4 Complexes with the StructiRBD-HA-VX Vaccine
3.8.3. TLR2 and TLR4 Complexes with the FusiCon-HA-VX Vaccine
3.8.4. TLR2 and TLR4 Complexes with the 400aa Ha Fragment
3.9. Immune Simulation
3.10. Population Coverage Analysis
3.11. Codon Optimization and In Silico Cloning of Vaccine Constructs
3.12. Comparative Evaluation of Vaccine Constructs and HA Fragment
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mostafa, A.; Abdelwhab, E.M.; Mettenleiter, T.C.; Pleschka, S. Zoonotic Potential of Influenza A Viruses: A Comprehensive Overview. Viruses 2018, 10, 497. [Google Scholar] [CrossRef]
- Charostad, J.; Rezaei Zadeh Rukerd, M.; Mahmoudvand, S.; Bashash, D.; Hashemi, S.M.A.; Nakhaie, M.; Zandi, K. A Comprehensive Review of Highly Pathogenic Avian Influenza (HPAI) H5N1: An Imminent Threat at Doorstep. Travel Med. Infect. Dis. 2023, 55, 102638. [Google Scholar] [CrossRef]
- Imai, M.; Kawaoka, Y. The Role of Receptor Binding Specificity in Interspecies Transmission of Influenza Viruses. Curr. Opin. Virol. 2012, 2, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.C.; Wilson, I.A. Influenza Hemagglutinin Structures and Antibody Recognition. Cold Spring Harb. Perspect. Med. 2020, 10, a038778. [Google Scholar] [CrossRef] [PubMed]
- Bullard, B.L.; Weaver, E.A. Strategies Targeting Hemagglutinin as a Universal Influenza Vaccine. Vaccines 2021, 9, 257. [Google Scholar] [CrossRef]
- Padilla-Quirarte, H.O.; Lopez-Guerrero, D.V.; Gutierrez-Xicotencatl, L.; Esquivel-Guadarrama, F. Protective Antibodies Against Influenza Proteins. Front. Immunol. 2019, 10, 1677. [Google Scholar] [CrossRef] [PubMed]
- Sunita; Sajid, A.; Singh, Y.; Shukla, P. Computational Tools for Modern Vaccine Development. Hum. Vaccines Immunother. 2020, 16, 723–735. [Google Scholar] [CrossRef]
- Alexander, J.; Fikes, J.; Hoffman, S.; Franke, E.; Sacci, J.; Appella, E.; Chisari, F.V.; Guidotti, L.G.; Chesnut, R.W.; Livingston, B.; et al. The Optimization of Helper T Lymphocyte (HTL) Function in Vaccine Development. Immunol. Res. 1998, 18, 79–92. [Google Scholar] [CrossRef]
- Shcherbinin, D.N.; Alekseeva, S.V.; Shmarov, M.M.; Smirnov, Y.A.; Naroditskiy, B.S.; Gintsburg, A.L. The Analysis of B-Cell Epitopes of Influenza Virus Hemagglutinin. Acta Naturae 2016, 8, 13–20. [Google Scholar] [CrossRef]
- Feng, Q.; Huang, X.-Y.; Feng, Y.-M.; Sun, L.; Sun, J.-Y.; Li, Y.; Xie, X.; Hu, J.; Guo, C.-Y. Identification and Analysis of B Cell Epitopes of Hemagglutinin of H1N1 Influenza Virus. Arch. Microbiol. 2022, 204, 594. [Google Scholar] [CrossRef]
- Lucchese, G.; Stufano, A.; Kanduc, D. Proteome-Guided Search for Influenza A B-Cell Epitopes. FEMS Immunol. Med. Microbiol. 2009, 57, 88–92. [Google Scholar] [CrossRef] [PubMed][Green Version]
- NCBI Virus; National Library of Medicine (US): Bethesda, MD, USA. Available online: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/ (accessed on 20 September 2025).[Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. ALeaves Facilitates On-Demand Exploration of Metazoan Gene Family Trees on MAFFT Sequence Alignment Server with Enhanced Interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P.S. Prediction of Continuous B-cell Epitopes in an Antigen Using Recurrent Neural Network. Proteins Struct. Funct. Bioinform. 2006, 65, 40–48. [Google Scholar] [CrossRef]
- Høie, M.H.; Gade, F.S.; Johansen, J.M.; Würtzen, C.; Winther, O.; Nielsen, M.; Marcatili, P. DiscoTope-3.0: Improved B-Cell Epitope Prediction Using Inverse Folding Latent Representations. Front. Immunol. 2024, 15, 1322712. [Google Scholar] [CrossRef]
- Larsen, M.V.; Lundegaard, C.; Lamberth, K.; Buus, S.; Lund, O.; Nielsen, M. Large-Scale Validation of Methods for Cytotoxic T-Lymphocyte Epitope Prediction. BMC Bioinform. 2007, 8, 424. [Google Scholar] [CrossRef]
- Nilsson, J.B.; Kaabinejadian, S.; Yari, H.; Kester, M.G.D.; van Balen, P.; Hildebrand, W.H.; Nielsen, M. Accurate Prediction of HLA Class II Antigen Presentation across All Loci Using Tailored Data Acquisition and Refined Machine Learning. Sci. Adv. 2023, 9, eadj6367. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Vir, P.; Raghava, G.P. Designing of Interferon-Gamma Inducing MHC Class-II Binders. Biol. Direct 2013, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.N.; Krutz, N.L.; Limviphuvadh, V.; Lopata, A.L.; Gerberick, G.F.; Maurer-Stroh, S. AllerCatPro 2.0: A Web Server for Predicting Protein Allergenicity Potential. Nucleic Acids Res. 2022, 50, W36–W43. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Shin, S.J.; Lee, M.H.; Lee, M.-G.; Kang, T.H.; Park, W.S.; Soh, B.Y.; Park, J.H.; Shin, Y.K.; Kim, H.W.; et al. A Potential Protein Adjuvant Derived from Mycobacterium tuberculosis Rv0652 Enhances Dendritic Cells-Based Tumor Immunotherapy. PLoS ONE 2014, 9, e104351. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein Structure Refinement Driven by Side-Chain Repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Jiménez-García, B.; Pons, C.; Fernández-Recio, J. PyDockWEB: A Web Server for Rigid-Body Protein–Protein Docking Using Electrostatics and Desolvation Scoring. Bioinformatics 2013, 29, 1698–1699. [Google Scholar] [CrossRef]
- Rapin, N.; Lund, O.; Bernaschi, M.; Castiglione, F. Computational Immunology Meets Bioinformatics: The Use of Prediction Tools for Molecular Binding in the Simulation of the Immune System. PLoS ONE 2010, 5, e9862. [Google Scholar] [CrossRef]
- Bui, H.-H.; Sidney, J.; Dinh, K.; Southwood, S.; Newman, M.J.; Sette, A. Predicting Population Coverage of T-Cell Epitope-Based Diagnostics and Vaccines. BMC Bioinform. 2006, 7, 153. [Google Scholar] [CrossRef]
- Grote, A.; Hiller, K.; Scheer, M.; Munch, R.; Nortemann, B.; Hempel, D.C.; Jahn, D. JCat: A Novel Tool to Adapt Codon Usage of a Target Gene to Its Potential Expression Host. Nucleic Acids Res. 2005, 33, W526–W531. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.-H. The Codon Adaptation Index-a Measure of Directional Synonymous Codon Usage Bias, and Its Potential Applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- Du, L.; Zhao, G.; Sun, S.; Zhang, X.; Zhou, X.; Guo, Y.; Li, Y.; Zhou, Y.; Jiang, S. A Critical HA1 Neutralizing Domain of H5N1 Influenza in an Optimal Conformation Induces Strong Cross-Protection. PLoS ONE 2013, 8, e53568. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Wang, B.; Chen, P.; Jiang, Y.; Liu, J. Analysis of the Conserved Protective Epitopes of Hemagglutinin on Influenza A Viruses. Front. Immunol. 2023, 14, 1086297. [Google Scholar] [CrossRef] [PubMed]
- Staneková, Z.; Varečková, E. Conserved Epitopes of Influenza A Virus Inducing Protective Immunity and Their Prospects for Universal Vaccine Development. Virol. J. 2010, 7, 351. [Google Scholar] [CrossRef] [PubMed]
- Whittle, J.R.R.; Zhang, R.; Khurana, S.; King, L.R.; Manischewitz, J.; Golding, H.; Dormitzer, P.R.; Haynes, B.F.; Walter, E.B.; Moody, M.A.; et al. Broadly Neutralizing Human Antibody That Recognizes the Receptor-Binding Pocket of Influenza Virus Hemagglutinin. Proc. Natl. Acad. Sci. USA 2011, 108, 14216–14221. [Google Scholar] [CrossRef]
- Carter, D.M.; Darby, C.A.; Lefoley, B.C.; Crevar, C.J.; Alefantis, T.; Oomen, R.; Anderson, S.F.; Strugnell, T.; Cortés-Garcia, G.; Vogel, T.U.; et al. Design and Characterization of a Computationally Optimized Broadly Reactive Hemagglutinin Vaccine for H1N1 Influenza Viruses. J. Virol. 2016, 90, 4720–4734. [Google Scholar] [CrossRef]
- Steel, J.; Lowen, A.C.; Wang, T.T.; Yondola, M.; Gao, Q.; Haye, K.; García-Sastre, A.; Palese, P. Influenza Virus Vaccine Based on the Conserved Hemagglutinin Stalk Domain. mBio 2010, 1, e00018-10. [Google Scholar] [CrossRef]
- Jang, Y.H.; Seong, B.L. The Quest for a Truly Universal Influenza Vaccine. Front. Cell. Infect. Microbiol. 2019, 9, 344. [Google Scholar] [CrossRef]
- Staneková, Z.; Király, J.; Stropkovská, A.; Mikušková, T.; Mucha, V.; Kostolanský, F.; Varečková, E. Heterosubtypic Protective Immunity against Influenza A Virus Induced by Fusion Peptide of the Hemagglutinin in Comparison to Ectodomain of M2 Protein. Acta Virol. 2011, 55, 61–67. [Google Scholar] [CrossRef]
- Janulíková, J.; Staneková, Z.; Mucha, V.; Kostolanský, F.; Varečková, E. Two Distinct Regions of HA2 Glycopolypeptide of Influenza Virus Hemagglutinin Elicit Cross-Protective Immunity against Influenza. Acta Virol. 2012, 56, 169–176. [Google Scholar] [CrossRef]
- Maleki, A.; Russo, G.; Parasiliti Palumbo, G.A.; Pappalardo, F. In Silico Design of Recombinant Multi-Epitope Vaccine against Influenza A Virus. BMC Bioinform. 2022, 22, 617. [Google Scholar] [CrossRef] [PubMed]
- Momajadi, L.; Khanahmad, H.; Mahnam, K. Designing a Multi-Epitope Influenza Vaccine: An Immunoinformatics Approach. Sci. Rep. 2024, 14, 25382. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Salinas, G.L.; García-Machorro, J.; Rojas-Hernández, S.; Campos-Rodríguez, R.; de Oca, A.C.-M.; Gomez, M.M.; Luciano, R.; Zimic, M.; Correa-Basurto, J. Bioinformatics Design and Experimental Validation of Influenza A Virus Multi-Epitopes That Induce Neutralizing Antibodies. Arch. Virol. 2020, 165, 891–911. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan, A.; Gravel, C.; Harris, G.; Hashem, A.M.; Zhang, W.; Safronetz, D.; Van Domselaar, G.; Krammer, F.; Sauve, S.; Rosu-Myles, M.; et al. Universal Antibody Targeting the Highly Conserved Fusion Peptide Provides Cross-Protection in Mice. Hum. Vaccin. Immunother. 2022, 18, 2083428. [Google Scholar] [CrossRef]
- Hasan, M.; Ghosh, P.P.; Azim, K.F.; Mukta, S.; Abir, R.A.; Nahar, J.; Hasan Khan, M.M. Reverse Vaccinology Approach to Design a Novel Multi-Epitope Subunit Vaccine against Avian Influenza A (H7N9) Virus. Microb. Pathog. 2019, 130, 19–37. [Google Scholar] [CrossRef]
- Behbahani, M.; Moradi, M.; Mohabatkar, H. In Silico Design of a Multi-Epitope Peptide Construct as a Potential Vaccine Candidate for Influenza A Based on Neuraminidase Protein. Silico Pharmacol. 2021, 9, 36. [Google Scholar] [CrossRef]
- Yuan, L.; Li, X.; Li, M.; Bi, R.; Li, Y.; Song, J.; Li, W.; Yan, M.; Luo, H.; Sun, C.; et al. In Silico Design of a Broad-Spectrum Multiepitope Vaccine against Influenza Virus. Int. J. Biol. Macromol. 2024, 254, 128071. [Google Scholar] [CrossRef]
- Samantaray, M.; Pushan, S.S.; Rajagopalan, M.; Abrol, K.; Basumatari, J.; Murthy, T.P.K.; Ramaswamy, A. Designing a Multi-Epitope Vaccine Candidate against Pandemic Influenza a Virus: An Immunoinformatics and Structural Vaccinology Approach. Mol. Divers. 2025. [Google Scholar] [CrossRef]
- Chuck, A.W.; Jacobs, P.; Tyrrell, G.; Kellner, J.D. Pharmacoeconomic Evaluation of 10- and 13-Valent Pneumococcal Conjugate Vaccines. Vaccine 2010, 28, 5485–5490. [Google Scholar] [CrossRef]
- Cox, M.M.J. Recombinant Protein Vaccines Produced in Insect Cells. Vaccine 2012, 30, 1759–1766. [Google Scholar] [CrossRef]
- Margine, I.; Krammer, F. Animal Models for Influenza Viruses: Implications for Universal Vaccine Development. Pathogens 2014, 3, 845–874. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-R.; Yu, Y.-H.; Tseng, Y.-C.; Chiang, W.-L.; Chiang, M.-F.; Ko, Y.-A.; Chiu, Y.-K.; Ma, H.-H.; Wu, C.-Y.; Jan, J.-T.; et al. Vaccination of Monoglycosylated Hemagglutinin Induces Cross-Strain Protection against Influenza Virus Infections. Proc. Natl. Acad. Sci. USA 2014, 111, 2476–2481. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Du, Y.; Chen, L.; Su, W.; Wei, H.; Liu, A.; Jiang, X.; Guo, J.; Dai, C.; Xu, Y.; et al. A Quadrivalent Recombinant Influenza Hemagglutinin Vaccine Induced Strong Protective Immune Responses in Animal Models. Vaccine 2024, 42, 126008. [Google Scholar] [CrossRef] [PubMed]
- Dashti, F.; Raisi, A.; Pourali, G.; Razavi, Z.S.; Ravaei, F.; Sadri Nahand, J.; Kourkinejad-Gharaei, F.; Mirazimi, S.M.A.; Zamani, J.; Tarrahimofrad, H.; et al. A Computational Approach to Design a Multiepitope Vaccine against H5N1 Virus. Virol. J. 2024, 21, 67. [Google Scholar] [CrossRef]
- DuBois, R.M.; Aguilar-Yañez, J.M.; Mendoza-Ochoa, G.I.; Oropeza-Almazán, Y.; Schultz-Cherry, S.; Alvarez, M.M.; White, S.W.; Russell, C.J. The Receptor-Binding Domain of Influenza Virus Hemagglutinin Produced in Escherichia Coli Folds into Its Native, Immunogenic Structure. J. Virol. 2011, 85, 865–872. [Google Scholar] [CrossRef]




| Influenza A Subtypes | Number of HA Sequences |
|---|---|
| H1N1 | 49,050 |
| H1N2 | 7030 |
| H3N2 | 56,119 |
| H5N1 | 20,039 |
| H5N2 | 1835 |
| H5N3 | 278 |
| H5N6 | 926 |
| H5N8 | 1229 |
| H6N1 | 798 |
| H6N2 | 1045 |
| H7N2 | 448 |
| H7N3 | 1068 |
| H7N4 | 50 |
| H7N7 | 511 |
| H7N9 | 1502 |
| H9N2 | 13,270 |
| H10N3 | 184 |
| H10N4 | 105 |
| H10N5 | 158 |
| H10N7 | 826 |
| H10N8 | 141 |
| Rank | Sequence | Start Position * | Score |
|---|---|---|---|
| 1 | TNLYKNPTTYISVGTS | 81 | 0.94 |
| 2 | CPYQGAPSFFRNVVWL | 28 | 0.94 |
| 3 | SQVNGQRGRMDFFWTI | 110 | 0.89 |
| 4 | DLLILWGIHHSNNAEE | 64 | 0.86 |
| 5 | TIKISYNNTNREDLLI | 52 | 0.86 |
| 6 | IQIIPKSSWPNHETSL | 7 | 0.84 |
| Sequence Number | Sequence | Score | Supertype |
|---|---|---|---|
| 1 | RMDFFWTIL | 0.9613 | A2 |
| 2 | YISVGTSTL | 0.9443 | A2 |
| 3 | RLAPKIATR | 1.3918 | A3 |
| 4 | TLNQRLAPK | 1.3917 | A3 |
| 5 | MDFFWTILK | 1.3062 | A3 |
| 6 | NLYKNPTTY | 1.1584 | A3 |
| 7 | FIAPEYAYK | 1.1506 | A3 |
| 8 | APSFFRNVV | 1.6389 | B7 |
| 9 | CPYQGAPSF | 1.4493 | B7 |
| 10 | APKIATRSQ | 1.1018 | B7 |
| 11 | APEYAYKIV | 1.0909 | B7 |
| 12 | YISVGTSTL | 0.9547 | B7 |
| Rank | Allele | Epitope | Method | Percentile Rank | Score | IF-g Inducer | |
|---|---|---|---|---|---|---|---|
| Result | Score | ||||||
| 1 | HLA-DRB1 08:02 | FIAPEYAYKIVKKGD | netmhciipan_el 4.1 | 0.11 | 0.8720 | Positive | 1 |
| 2 | HLA-DRB3 01:01 | WTILKPDDAIHFESN | netmhciipan_el 4.1 | 0.12 | 0.8699 | Positive | 0.45 |
| 3 | HLA-DQA1 04:01/DQB1 04:02 | FIAPEYAYKIVKKGD | netmhciipan_el 4.1 | 0.15 | 0.3705 | Positive | 1 |
| 4 | HLA-DRB3 01:01 | FWTILKPDDAIHFES | netmhciipan_el 4.1 | 0.15 | 0.8370 | Positive | 0.47 |
| 5 | HLA-DRB3 01:01 | TILKPDDAIHFESNG | netmhciipan_el 4.1 | 0.18 | 0.7780 | Negative | 1 |
| 6 | HLA-DRB3 02:02 | AIHFESNGNFIAPEY | netmhciipan_el 4.1 | 0.18 | 0.8016 | Positive | 0.5 |
| 7 | HLA-DRB1 07:01 | PTTYISVGTSTLNQR | netmhciipan_el 4.1 | 0.21 | 0.8852 | Positive | 0.54 |
| 8 | HLA-DRB3 02:02 | DAIHFESNGNFIAPE | netmhciipan_el 4.1 | 0.22 | 0.7782 | Positive | 0.54 |
| 9 | HLA-DRB3 01:01 | FFWTILKPDDAIHFE | netmhciipan_el 4.1 | 0.26 | 0.7497 | Positive | 0.52 |
| 10 | HLA-DRB3 02:02 | QTNLYKNPTTYISVG | netmhciipan_el 4.1 | 0.31 | 0.7183 | Positive | 0.55 |
| Feature | EpitoCore-HA-VX | StructiRBD-HA-VX | FusiCon-HA-VX | HA | HA-13–263-Fd-His |
|---|---|---|---|---|---|
| Epitope Source | Multi-epitope collation (HA-RBD) | Structure-preserved RBD fragment (188–255) | Fusion peptide (HA2, 24 aa) | HA fragment (400 aa) | RBD (HA1, H5N1; 13–263) |
| Conservation | Consensus from >20 k H5N1 | Same | Ultra-conserved across influenza A subtypes | Consensus from >20 k H5N1 | Strain-specific (A/Anhui/1/2005) |
| Antigenicity & Safety | Antigenic; non-allergen; non-toxic | Antigenic; non-allergen | Antigenic; non-allergen | Antigenic; non-allergen | Antigenic; non-allergen |
| Structural Quality Assessment | Stable after refinement (97.7% favored residues, G-factor 0.21) | High stereochemical quality (96.7% favored, no clashes) | Excellent stereochemistry (97.4% favored, G-factor 0.29) | Model 2 selected; 92.6% favored residues, 0.3% disallowed; minimal deviations (5.8), 1 clash, G-factor 0.11 | Good stereochemistry (91.7% favored; 0.8% disallowed; no clashes; G-factor 0.09) |
| Docking—TLR2 | Strong (–43.9); 1062 Å2; engages B-cell epitopes | Strong (–41.1); 952 Å2; engages RBD motifs | Strong (–46.5); 772 Å2; engages FP residues | Two modes: FP site (–34,338; 1920 Å2; 34 residues; 4 H-bonds, 206 contacts); RBD site (–25,085; 1966 Å2; 36 residues; 9 H-bonds, 194 contacts) | Strong (–29.33); 1635 Å2; three binding residues in RBD domain |
| Docking—TLR4 | Very strong (–76.7); 1088 Å2; engages CTL + HTL epitopes | Balanced (–44.5); 684 Å2; engages RBD residues | Good (702–778 Å2; 138 contacts, hydrophobic) | Two modes: FP site (–40,770; 1635 Å2; 29 residues; 2 H-bonds, 174 contacts); RBD site (–30,679; 1457 Å2; 29 residues; 3 H-bonds, 154 contacts) | Strong (–30.1); 1874 Å2; predominantly RBD-mediated |
| Immune Simulation | IgG1 durable >300 d; CTL + Th1 memory strong; IFN-γ/IL-2 high | Similar profile; strong CTL + Th1; durable memory | IgG1 durable >300 d; strong Th1; no CTL induction | Similar to EpitoCore/StructiRBD; in some antibody responses slightly higher | Similar to EpitoCore/StructiRBD; in some antibody responses slightly higher |
| Population Coverage | 100% combined (class I ~64%, class II 100%) | Same | Same | Similar to EpitoCore/StructiRBD (broad global coverage) | Same |
| Codon Optimization | CAI 0.997; GC 49.7%; cloned in pET28a(+) | CAI 1.0; GC 49.4% | CAI 1.0; GC 48.4% | CAI 1.0; GC 49.1% | CAI 1.0; GC 50.3% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palma, M. Consensus-Guided Construction of H5N1-Specific and Universal Influenza a Multiepitope Vaccines. Biology 2025, 14, 1327. https://doi.org/10.3390/biology14101327
Palma M. Consensus-Guided Construction of H5N1-Specific and Universal Influenza a Multiepitope Vaccines. Biology. 2025; 14(10):1327. https://doi.org/10.3390/biology14101327
Chicago/Turabian StylePalma, Marco. 2025. "Consensus-Guided Construction of H5N1-Specific and Universal Influenza a Multiepitope Vaccines" Biology 14, no. 10: 1327. https://doi.org/10.3390/biology14101327
APA StylePalma, M. (2025). Consensus-Guided Construction of H5N1-Specific and Universal Influenza a Multiepitope Vaccines. Biology, 14(10), 1327. https://doi.org/10.3390/biology14101327






