Genome-Wide Analysis and Functional Characterization of Small Heat Shock Proteins in Allium sativum L. Under Multiple Abiotic Stresses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of HSP20 Genes
2.2. Phylogenetic and Gene Structure Analyses
2.3. Chromosomal Localization, Duplication and Synteny
2.4. Cis-Regulatory Element Analysis, Protein–Protein Interaction Network and Gene Ontology Enrichment
2.5. Plant Material and Stress Treatments
2.6. Quantitative Real-Time PCR
2.7. High-Temperature Stress Assay in Transgenic Yeast
2.8. Subcellular Localization of AsHSP20 Proteins
3. Results
3.1. Identification of Family Members
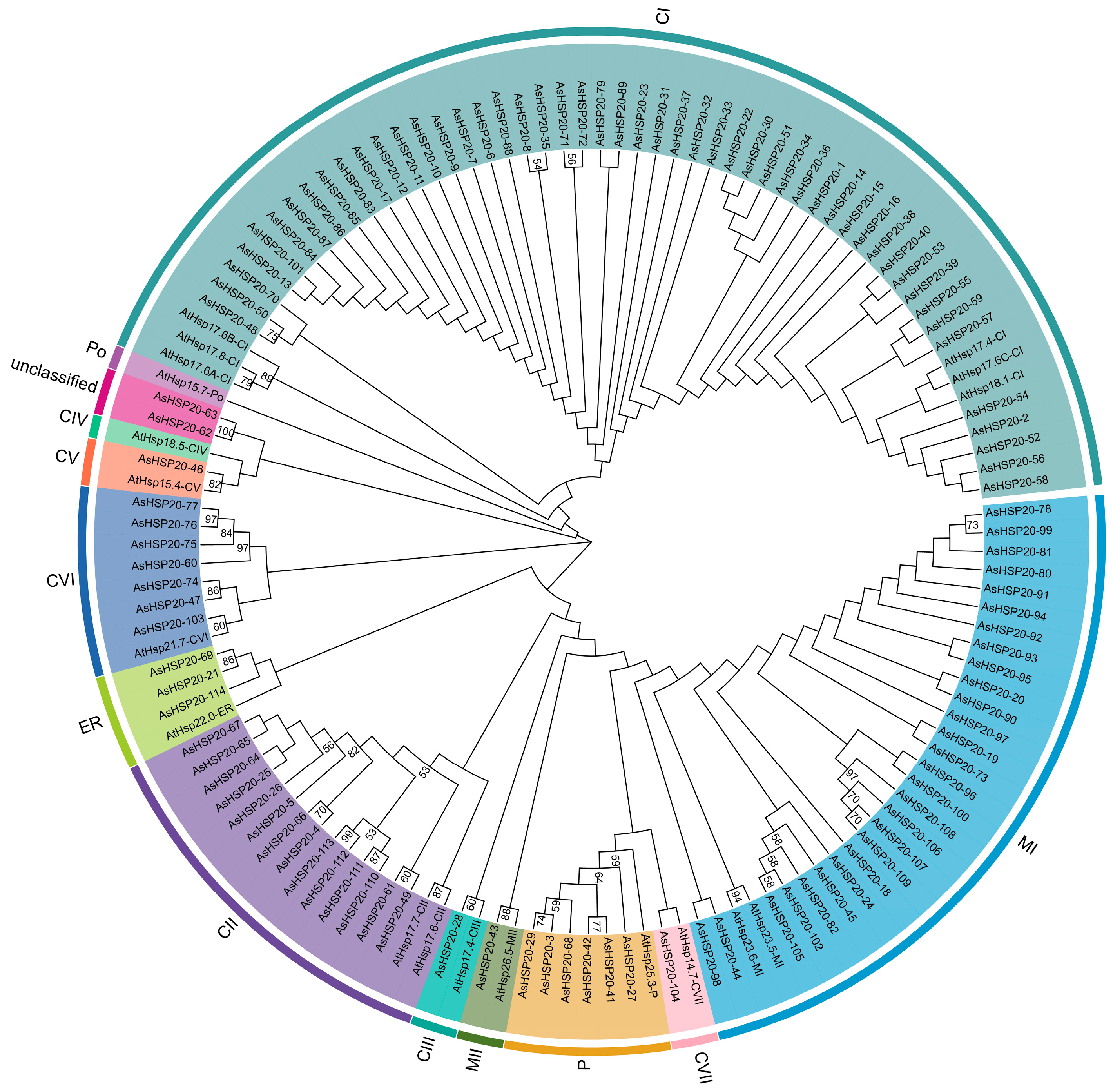
3.2. Phylogenetic Analysis
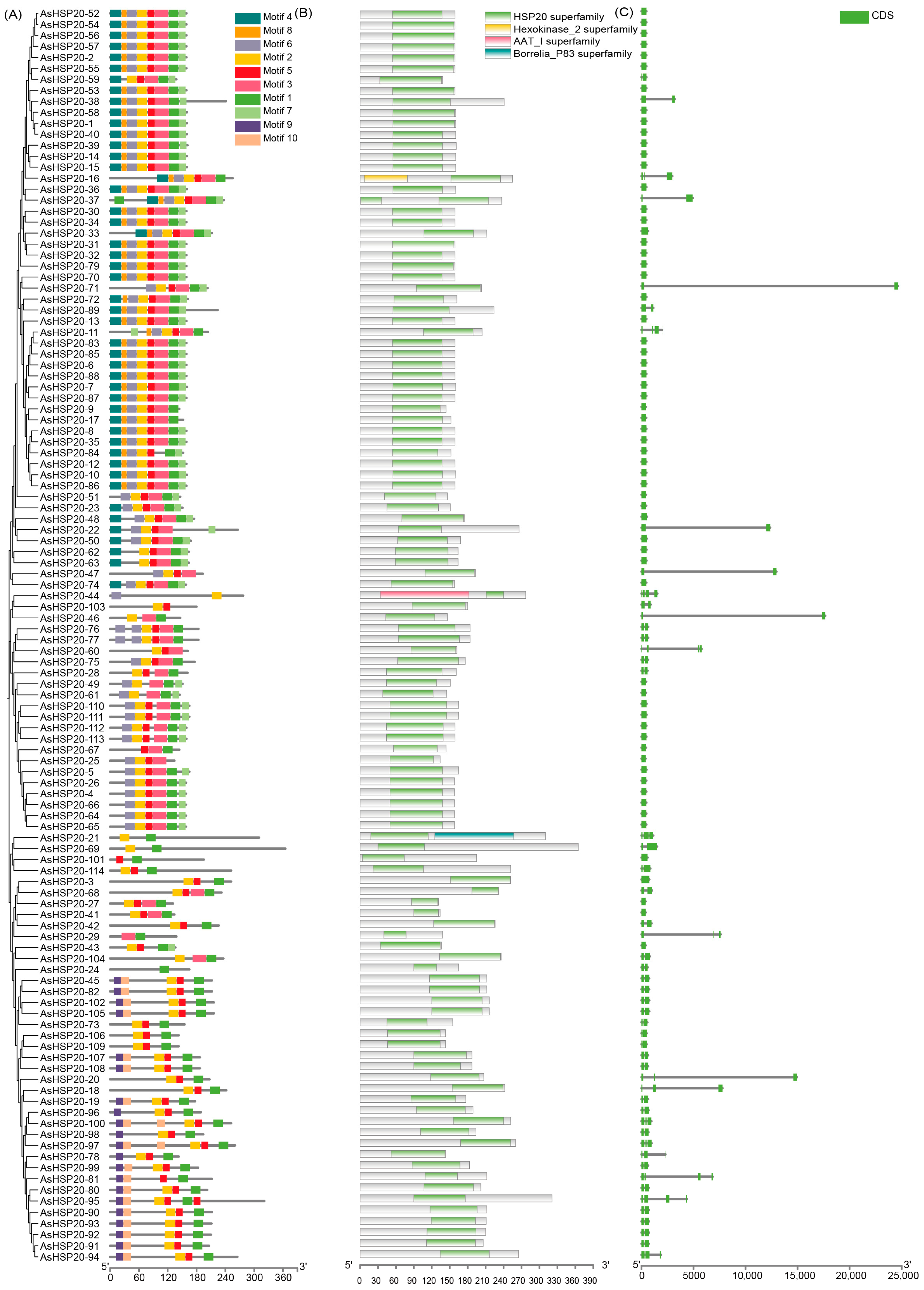
3.3. Conserved Motif and Gene Structure Analysis
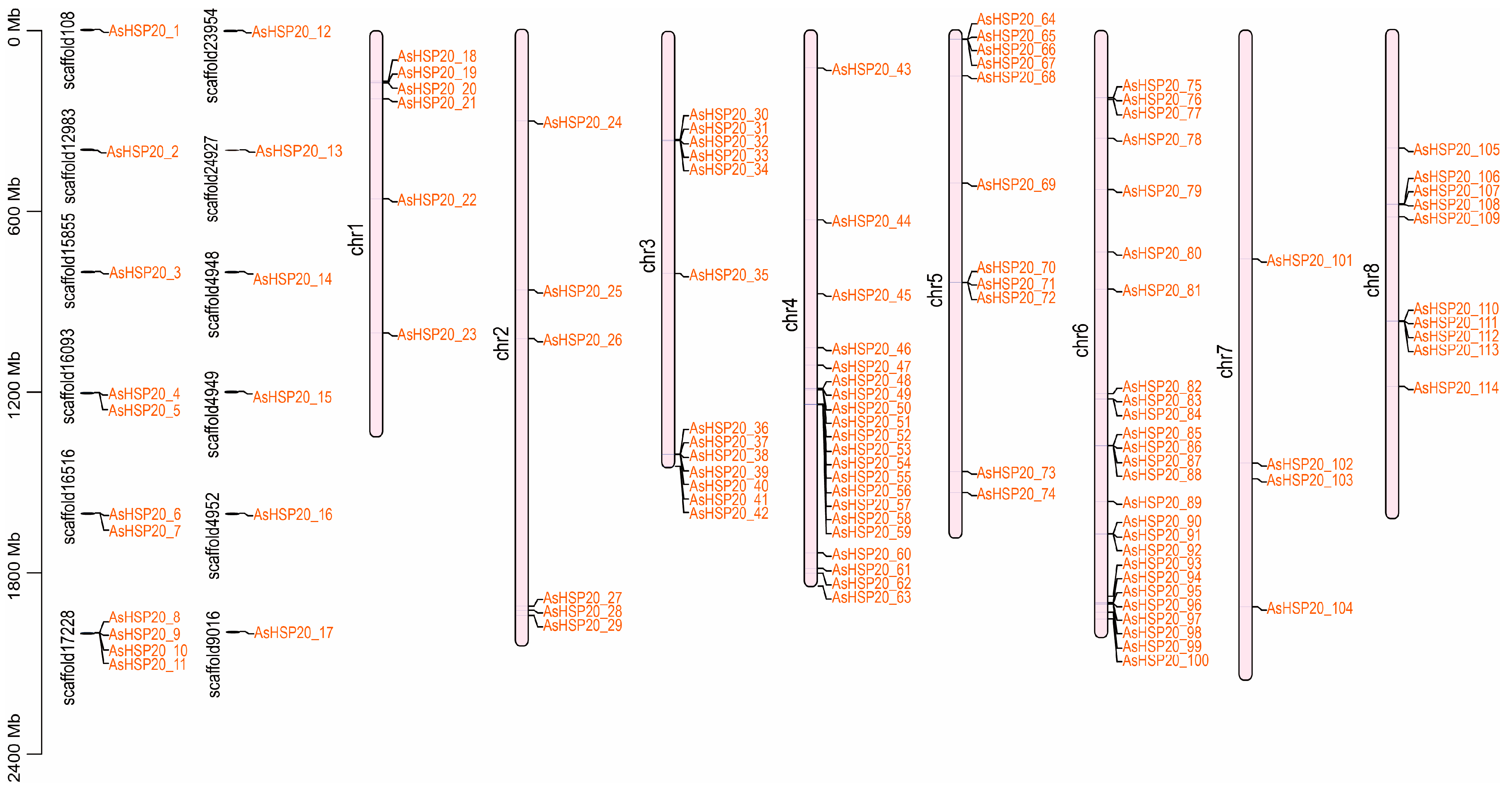
3.4. Chromosomal Localization
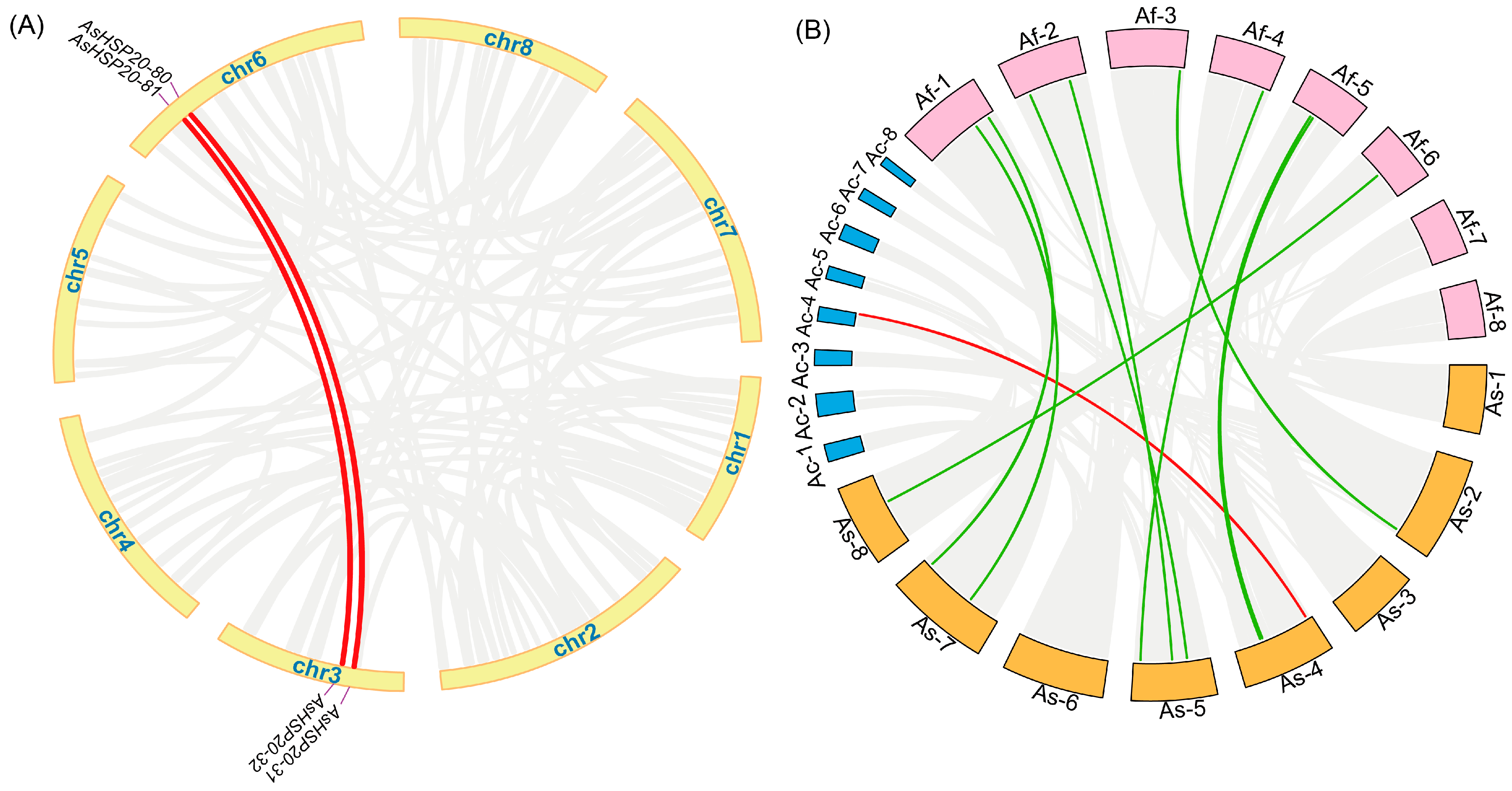
3.5. Inter Species Collinearity Analysis
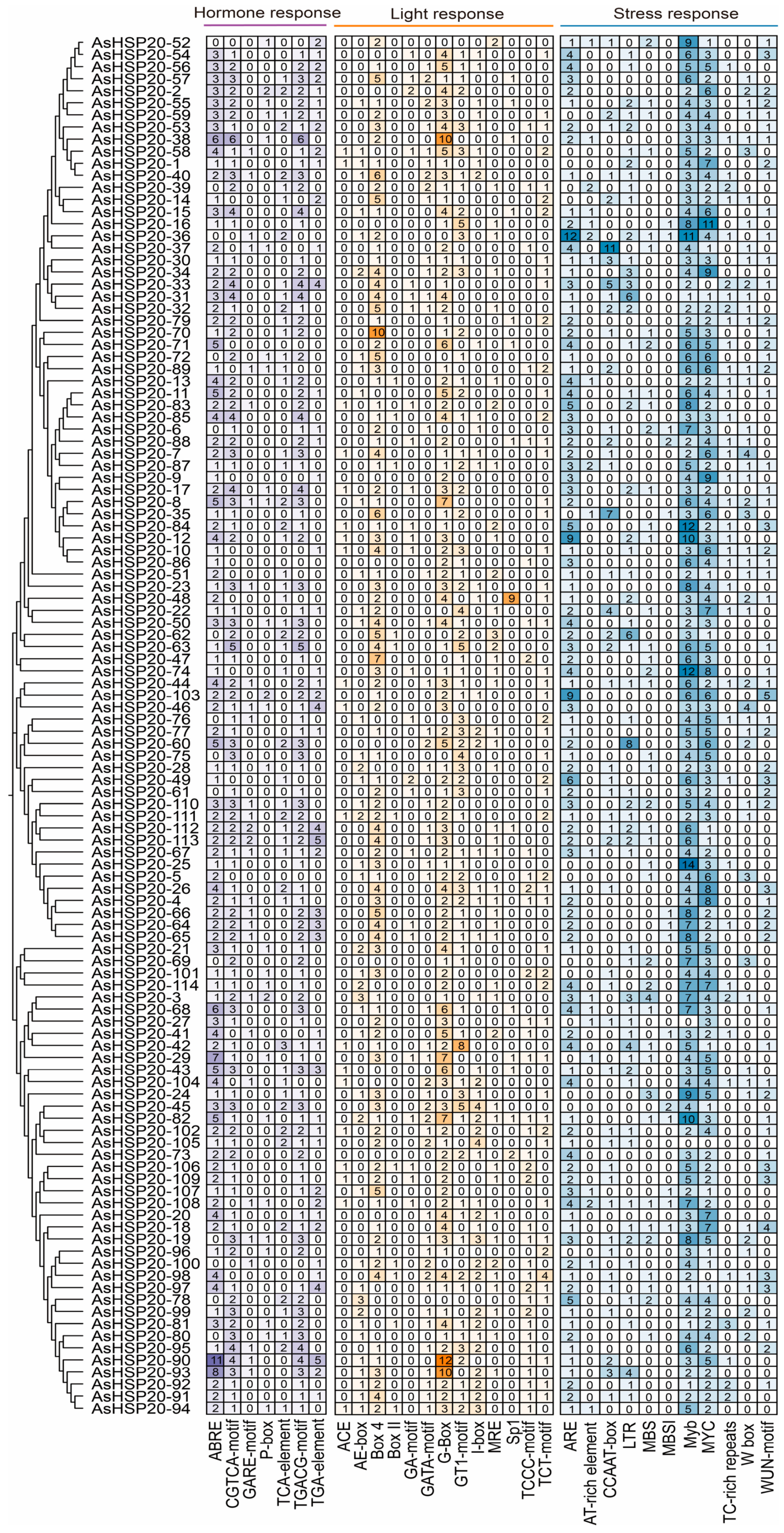
3.6. Analysis of Cis-Acting Elements in Promoter Regions
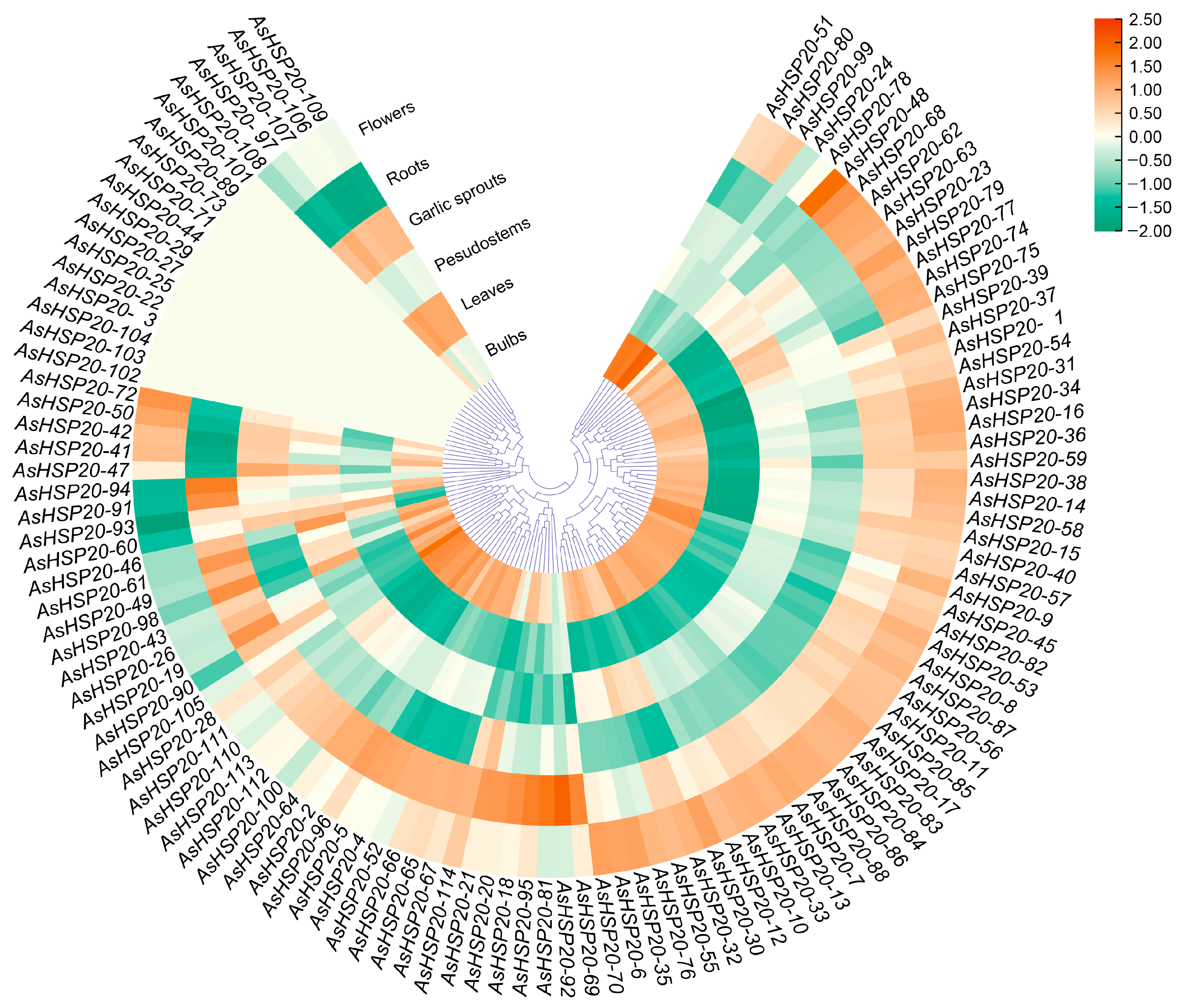
3.7. Expression Pattern Analysis
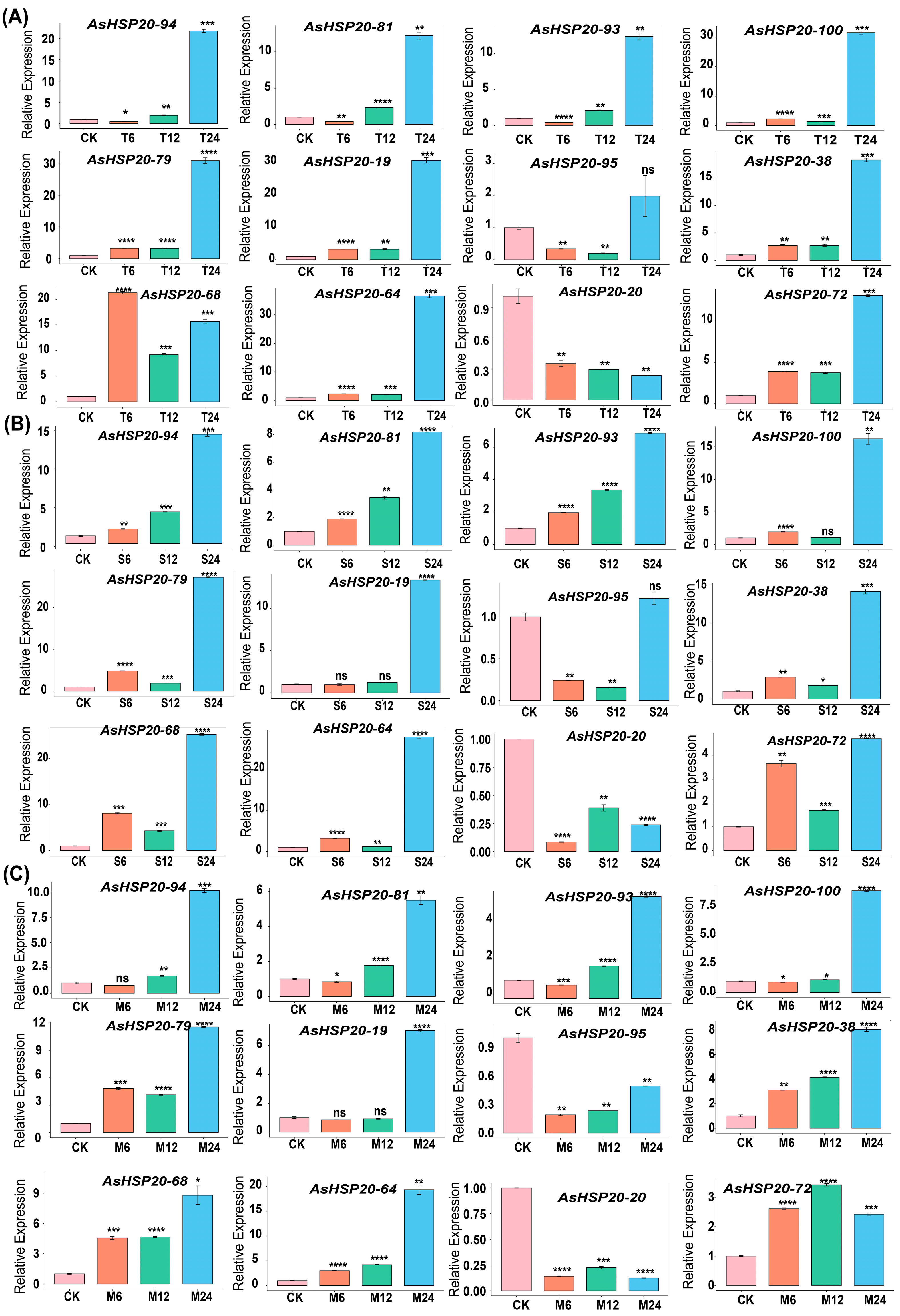
3.8. qRT–PCR Analysis
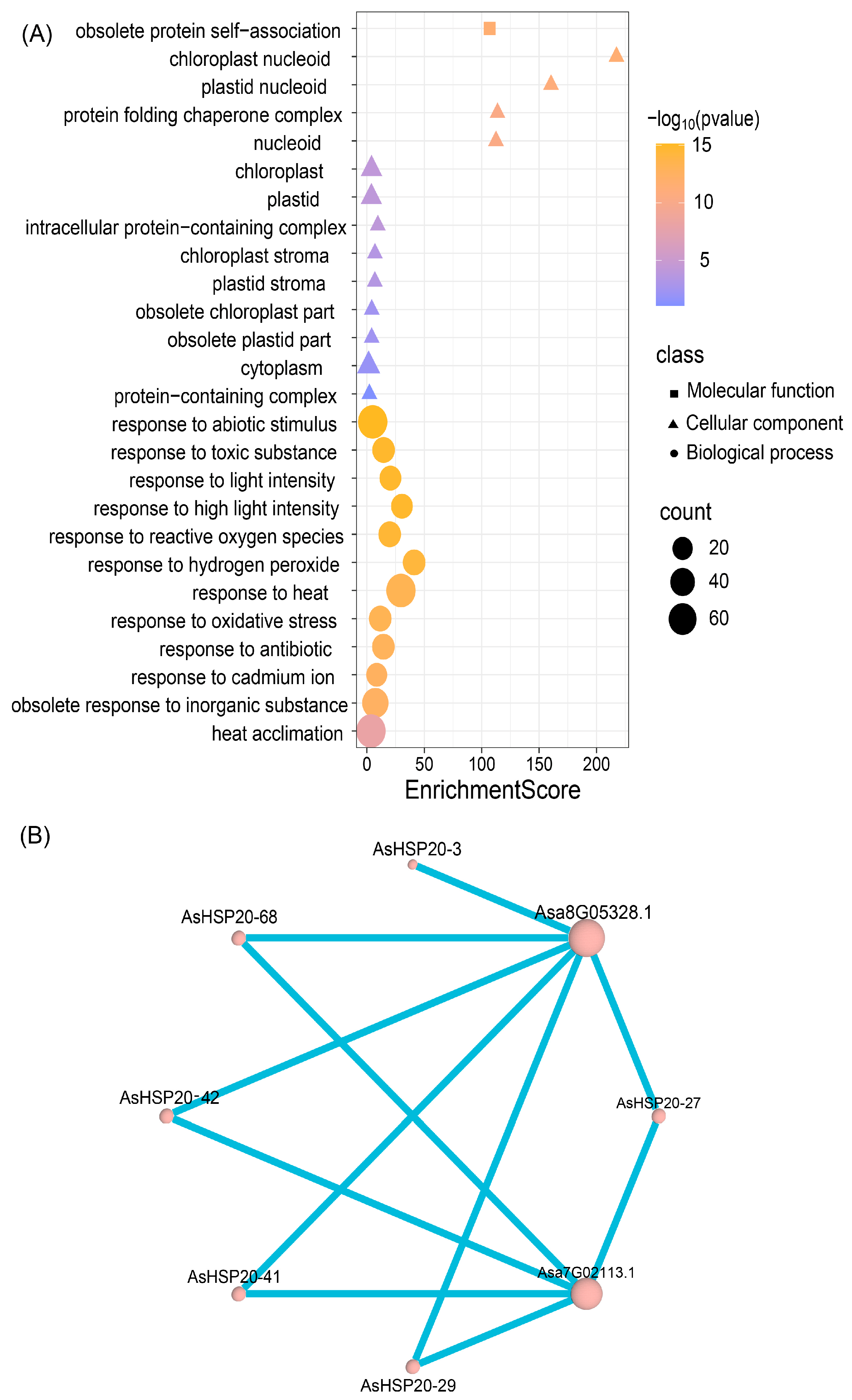
3.9. Protein–Protein Interaction Network and Gene Ontology Enrichment
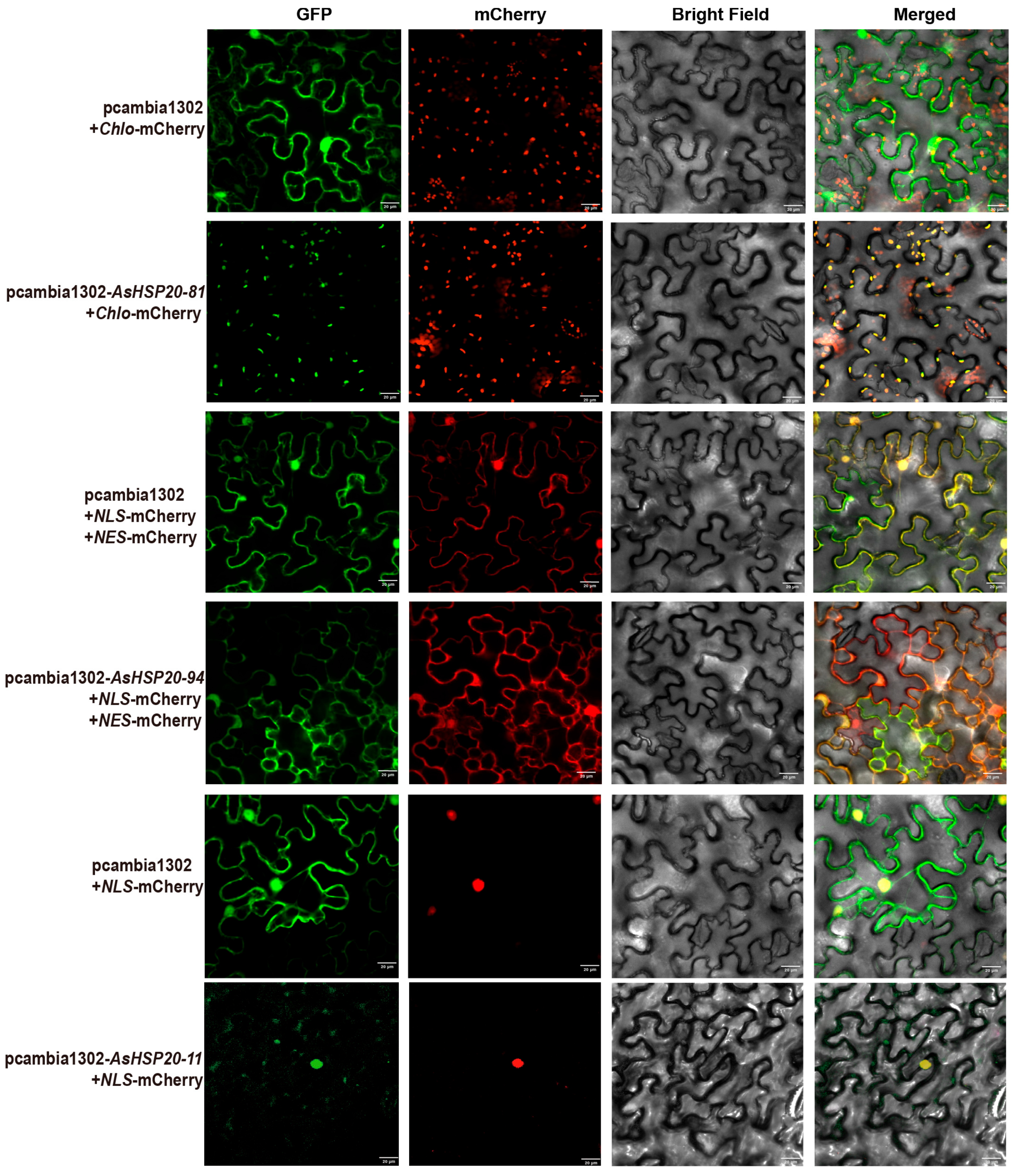
3.10. Subcellular Localization
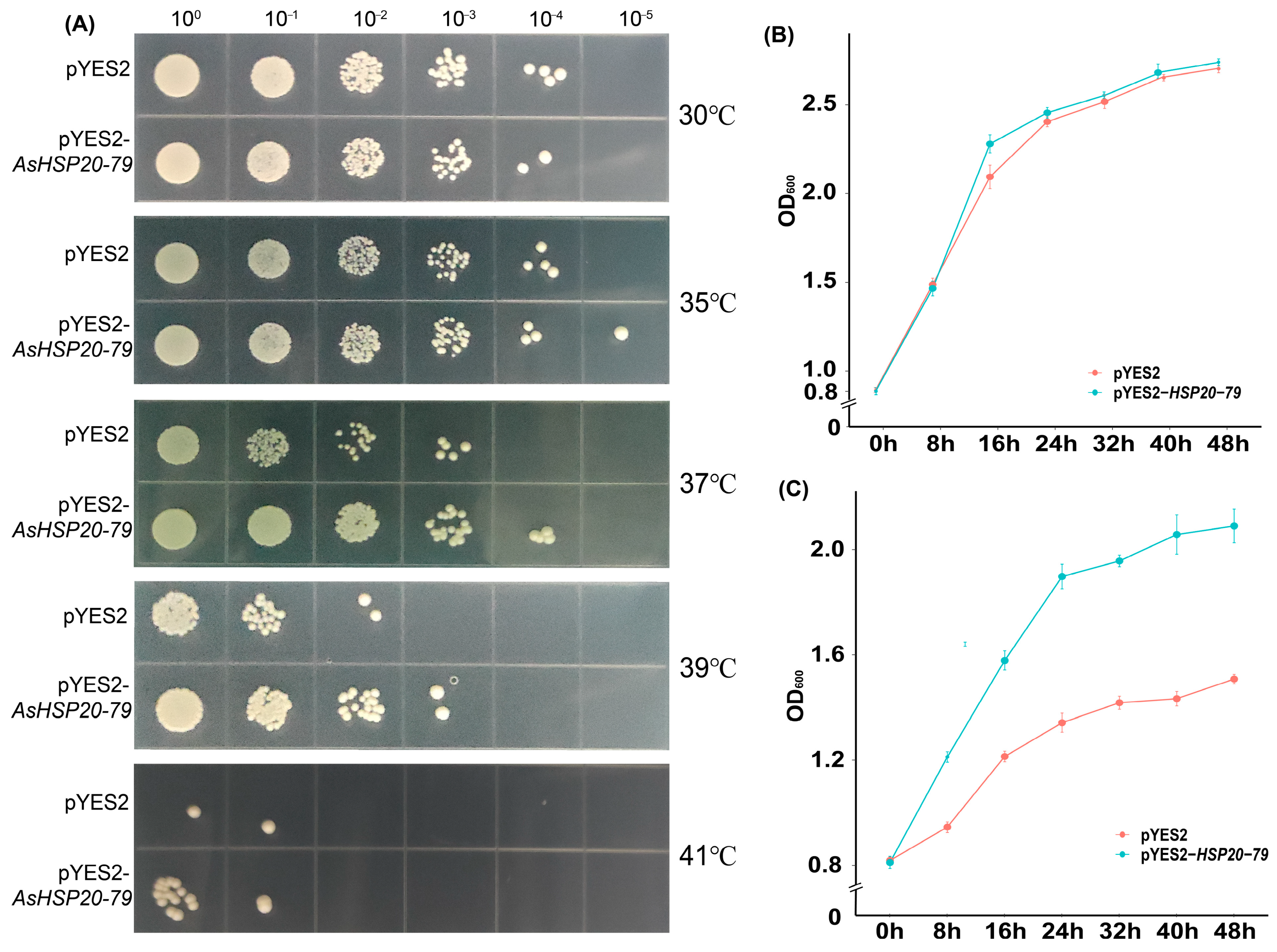
3.11. Yeast Transgenic Verification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.-X.; Xu, J.-H. The heat shock transcription factors regulate response mechanisms to abiotic stresses in plants. Crop Des. 2025, 4, 100109. [Google Scholar] [CrossRef]
- Zahra, N.; Hafeez, M.B.; Ghaffar, A.; Kausar, A.; Zeidi, M.A.; Siddique, K.H.M.; Farooq, M. Plant photosynthesis under heat stress: Effects and management. Environ. Exp. Bot. 2023, 206, 105178. [Google Scholar] [CrossRef]
- Molinier, J.; Ries, G.; Zipfel, C.; Hohn, B. Transgeneration memory of stress in plants. Nature 2006, 442, 1046–1049. [Google Scholar] [CrossRef]
- Tian, F.; Hu, X.-L.; Yao, T.; Yang, X.; Chen, J.-G.; Lu, M.-Z.; Zhang, J. Recent Advances in the Roles of HSFs and HSPs in Heat Stress Response in Woody Plants. Front. Plant Sci. 2021, 12, 704905. [Google Scholar] [CrossRef]
- Naaz, S.; Pande, A.; Laxmi, A. Nitric oxide-mediated thermomemory: A new perspective on plant heat stress resilience. Front. Plant Sci. 2025, 16, 1525336. [Google Scholar] [CrossRef]
- Staacke, T.; Mueller-Roeber, B.; Balazadeh, S. Stress resilience in plants: The complex interplay between heat stress memory and resetting. New Phytol. 2025, 245, 2402–2421. [Google Scholar] [CrossRef] [PubMed]
- Farnsworth, P.; Kamalendra, S. Structure function relationship among α-crystallin related small heat shock proteins. Exp. Eye Res. 2004, 79, 787–794. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of Plant heat-shock Proteins and Molecular Chaperones in the Abiotic Stress Response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Sarkar, N.K.; Kim, Y.-K.; Grover, A. Rice sHsp genes: Genomic organization and expression profiling under stress and development. BMC Genom. 2009, 10, 393. [Google Scholar] [CrossRef]
- Van Montfort, R.; Slingsby, C.; Vierling, E. Structure and function of the small heat shock protein/α-crystallin family of molecular chaperones. Adv. Protein Chem. 2001, 105–156. [Google Scholar] [CrossRef]
- Tan, G.-F.; Wang, Z.; Li, M.; Wang, G.-L.; Jiang, Q.; Xiong, A.-S. De novo assembly and transcriptome characterization: Novel insights into the temperature stress in Cryptotaenia japonica Hassk. Acta Physiol. Plant. 2014, 37, 1–12. [Google Scholar] [CrossRef]
- Zhang, F.J.; Li, Z.Y.; Zhang, D.E.; Ma, N.; Wang, Y.X.; Zhang, T.T.; Zhao, Q.; Zhang, Z.; You, C.X.; Lu, X.Y. Identification of Hsp20 gene family in Malus domestica and functional characterization of Hsp20 class I gene MdHsp18.2b. Physiol. Plant. 2024, 176, e14288. [Google Scholar] [CrossRef]
- Li, N.; Jiang, M.; Li, P.; Li, X. Identification, expression, and functional analysis of Hsf and Hsp20 gene families in Brachypodium distachyon under heat stress. PeerJ 2021, 9, e12267. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Wang, Q.; Li, T.; Gao, H.; Li, H.; Zheng, X.; Wang, X.; Zhang, H.; Cheng, J.; Wang, W.; et al. Phylogenetic and Transcriptional Analyses of the HSP20 Gene Family in Peach Revealed That PpHSP20-32 Is Involved in Plant Height and Heat Tolerance. Int. J. Mol. Sci. 2022, 23, 10849. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhu, S.; Li, N.; Cheng, Y.; Zhao, J.; Qiao, X.; Lu, L.; Liu, S.; Wang, Y.; Liu, C.; et al. A Chromosome-Level Genome Assembly of Garlic (Allium sativum) Provides Insights into Genome Evolution and Allicin Biosynthesis. Mol. Plant 2020, 13, 1328–1339. [Google Scholar] [CrossRef]
- Peška, V.; Mandáková, T.; Ihradská, V.; Fajkus, J. Comparative Dissection of Three Giant Genomes: Allium cepa, Allium sativum, and Allium ursinum. Int. J. Mol. Sci. 2019, 20, 733. [Google Scholar] [CrossRef]
- Zhong, L.; Shi, Y.; Xu, S.; Xie, S.; Huang, X.; Li, Y.; Qu, C.; Liu, J.; Liao, J.; Huang, Y.; et al. Heterologous overexpression of heat shock protein 20 genes of different species of yellow Camellia in Arabidopsis thaliana reveals their roles in high calcium resistance. BMC Plant Biol. 2024, 24, 5. [Google Scholar] [CrossRef]
- Xue, M.; You, Y.; Zhang, L.; Cao, J.; Xu, M.; Chen, S. ZmHsp18 screened from the ZmHsp20 gene family confers thermotolerance in maize. BMC Plant Biol. 2024, 24, 1048. [Google Scholar] [CrossRef]
- Huang, J.; Hai, Z.; Wang, R.; Yu, Y.; Chen, X.; Liang, W.; Wang, H. Genome-wide analysis of HSP20 gene family and expression patterns under heat stress in cucumber (Cucumis sativus L.). Front. Plant Sci. 2022, 13, 968418. [Google Scholar] [CrossRef]
- Hao, X.; He, S. Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Garlic (Allium sativum L.). BMC Plant Biol. 2024, 24, 421. [Google Scholar] [CrossRef]
- Yang, P.; Yuan, Y.; Yan, C.; Jia, Y.; You, Q.; Da, L.; Lou, A.; Lv, B.; Zhang, Z.; Liu, Y. AlliumDB: A central portal for comparative and functional genomics in Allium. Hortic. Res. 2024, 11, uhad285. [Google Scholar] [CrossRef]
- Reiser, L.; Bakker, E.; Subramaniam, S.; Chen, X.; Sawant, S.; Kartik, K.; Trilok, P.; Berardini, T.Z. The Arabidopsis Information Resource in 2024. Genetics 2024, 227, iyae027. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2020, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2022, 51, D384–D388. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; et al. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef]
- Yang, Q.-Q.; Fan, J.-D.; Liu, C.-Y.; Zhao, Y.-Q.; Xu, Z.-S.; Lu, X.-J.; Ge, J.; Zhang, B.-W.; Li, M.-Q.; Yang, Y.; et al. Physiological and transcriptome analysis of changes in endogenous hormone contents and related synthesis and signaling genes during the heat stress in garlic (Allium sativum L.). BMC Plant Biol. 2025, 25, 1–20. [Google Scholar] [CrossRef]
- Wang, G.-L.; Ren, X.-Q.; Liu, J.-X.; Yang, F.; Wang, Y.-P.; Xiong, A.-S. Transcript profiling reveals an important role of cell wall remodeling and hormone signaling under salt stress in garlic. Plant Physiol. Biochem. 2019, 135, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Akan, S.; Tuna Gunes, N.; Yanmaz, R. Methyl jasmonate and low temperature can help for keeping some physicochemical quality parameters in garlic (Allium sativum L.) cloves. Food Chem. 2019, 270, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Tian, C.; Wang, Y.; Wan, F.; Hu, L.; Xiong, A.; Tian, J. Selection of reliable reference genes for quantitative RT-PCR in garlic under salt stress. PeerJ 2019, 7, e7319. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, W.; Zhou, C.; Guo, Y.; Niu, S.; El-Kassaby, Y.A.; Li, W. Genome-wide identification of the Pinus tabuliformis CONSTANS-like gene family and their potential roles in reproductive cone development. Int. J. Biol. Macromol. 2024, 254, 127621. [Google Scholar] [CrossRef]
- Mani, N.; Ramakrishna, K.; Kaza, S. Characterization of rice small heat shock proteins targeted to different cellular organelles. Cell Stress Chaperones 2015, 20, 451–460. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, D.; Wang, R.; Kong, N.; Zhang, C.; Yang, C.; Wu, W.; Ma, H.; Chen, Q. Genome-wide analysis of the potato Hsp20 gene family: Identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 2018, 19, 61. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Widespread Paleopolyploidy in Model Plant Species Inferred from Age Distributions of Duplicate Genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.-H. Importance of Lineage-Specific Expansion of Plant Tandem Duplicates in the Adaptive Response to Environmental Stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-M.; Cao, Y.-Q.; Liu, M.-X.; Liu, B.; Zhou, H.; Xia, Y.-P.; Wang, X.-Y. Phylogenetic and expression analysis of HSP20 gene family in Rhododendron species of different altitudes. Int. J. Biol. Macromol. 2025, 309, 143125. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Long, X.; Kav, N.N.V.; Fang, Y.; Qin, Y.; Yang, J.; Xiao, X. The Hexokinases HbHXK2 and 4 Are Key Enzymes Involved in Glucose Metabolism and Contribute to Rubber Productivity in Hevea Brasiliensis (para Rubber tree). Ind. Crops Prod. 2020, 159, 113025. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, Z.; Liang, C.; Wei, Y.; Li, Y.; Zhang, Y.; Zhang, Y. Genome-Wide Analysis of the Aspartate Aminotransferase Family in Brassica Rapa and the Role of BraASP1 in Response to Nitrogen Starvation. Int. J. Mol. Sci. 2025, 26, 1586. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Lee, G.J.; Vierling, E. A Small Heat Shock Protein Cooperates with Heat Shock Protein 70 Systems to Reactivate a Heat-Denatured Protein. Plant Physiol. 2000, 122, 189–198. [Google Scholar] [CrossRef]
- Neta-Sharir, I.; Isaacson, T.; Lurie, S.; Weiss, D. Dual Role for Tomato Heat Shock Protein 21: Protecting Photosystem II from Oxidative Stress and Promoting Color Changes during Fruit Maturation. Plant Cell 2005, 17, 1829–1838. [Google Scholar] [CrossRef]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Ren, S.-x.; Zou, H.-l.; Cui, J.-w.; Shen, N.; Bao, H.-y.; Gan, Q.; Wang, L.; Lu, Z.-g.; Jin, B. PeHSFA3 is essential for the heat-stress response of Populus × euramericana. Ind. Crops Prod. 2024, 219, 119054. [Google Scholar] [CrossRef]
- Shen, R.-J.; Xing, M.-Y.; Wang, W.-Q.; Su, W.-Y.; Zhu, J.-Z.; Li, K.-F.; Zhang, Y.; Allan, A.C.; Grierson, D.; Yin, X.-R.; et al. Over-expression of heat shock factor AcHsfA2-1 upregulates transcripts of multiple genes and enhances heat tolerance of kiwifruit plants. Environ. Exp. Bot. 2023, 207, 105196. [Google Scholar] [CrossRef]
- Ding, C.-K.; Wang, C.Y.; Gross, K.C.; Smith, D.L. Reduction of chilling injury and transcript accumulation of heat shock proteins in tomato fruit by methyl jasmonate and methyl salicylate. Plant Sci. 2001, 161, 1153–1159. [Google Scholar] [CrossRef]
- Galsurker, O.; Doron-Faigenboim, A.; Teper-Bamnolker, P.; Daus, A.; Lers, A.; Eshel, D. Differential response to heat stress in outer and inner onion bulb scales. J. Exp. Bot. 2018, 69, 4047–4064. [Google Scholar] [CrossRef]
- Dafny-Yelin, M.; Tzfira, T.; Vainstein, A.; Adam, Z. Non-redundant functions of sHSP-CIs in acquired thermotolerance and their role in early seed development in Arabidopsis. Plant Mol. Biol. 2008, 67, 363–373. [Google Scholar] [CrossRef]
- Waters, E.R.; Vierling, E. Plant small heat shock proteins—Evolutionary and functional diversity. New Phytol. 2020, 227, 24–37. [Google Scholar] [CrossRef]
- Wang, A.; Yu, X.; Mao, Y.; Liu, Y.; Liu, G.; Liu, Y.; Niu, X. Overexpression of a small heat-shock-protein gene enhances tolerance to abiotic stresses in rice. Plant Breed. 2015, 134, 384–393. [Google Scholar] [CrossRef]
- Wu, J.; Gao, T.; Hu, J.; Zhao, L.; Yu, C.; Ma, F. Research advances in function and regulation mechanisms of plant small heat shock proteins (sHSPs) under environmental stresses. Sci. Total Environ. 2022, 825, 154054. [Google Scholar] [CrossRef]
- He, Y.; Yao, Y.; Li, L.; Li, Y.; Gao, J.; Fan, M. A heat-shock 20 protein isolated from watermelon (ClHSP22.8) negatively regulates the response of Arabidopsis to salt stress via multiple signaling pathways. PeerJ 2021, 9, e10524. [Google Scholar] [CrossRef]
- Ma, W.; Zhao, T.; Li, J.; Liu, B.; Fang, L.; Hu, Y.; Zhang, T. Identification and characterization of the GhHsp20 gene family in Gossypium hirsutum. Sci. Rep. 2016, 6, 32517. [Google Scholar] [CrossRef]
- Zhai, M.; Sun, Y.; Jia, C.; Peng, S.; Liu, Z.; Yang, G. Over-expression of JrsHSP17.3 gene from Juglans regia confer the tolerance to abnormal temperature and NaCl stresses. J. Plant Biol. 2016, 59, 549–558. [Google Scholar] [CrossRef]
- Sun, J.-T.; Cheng, G.-X.; Huang, L.-J.; Liu, S.; Ali, M.; Khan, A.; Yu, Q.-H.; Yang, S.-B.; Luo, D.-X.; Gong, Z.-H. Modified expression of a heat shock protein gene, CaHSP22.0, results in high sensitivity to heat and salt stress in pepper (Capsicum annuum L.). Sci. Hortic. 2019, 249, 364–373. [Google Scholar] [CrossRef]
- Sun, X.; Sun, C.; Li, Z.; Hu, Q.; Han, L.; Luo, H. AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress. Plant Cell Environ. 2016, 39, 1320–1337. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b Act as Repressors of the Expression of Heat-Inducible Hsfs But Positively Regulate the Acquired Thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Cao, Y.; Dai, J.; Li, G.; Muhammad Aamir, M.; Chen, C.; Deng, H. The Multifaceted Roles of MYC2 in Plants: Toward Transcriptional Reprogramming and Stress Tolerance by Jasmonate Signaling. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Zhong, L.; Zhou, W.; Wang, H.; Ding, S.; Lu, Q.; Wen, X.; Peng, L.; Zhang, L.; Lu, C. Chloroplast Small Heat Shock Protein HSP21 Interacts with Plastid Nucleoid Protein pTAC5 and Is Essential for Chloroplast Development in Arabidopsis under Heat Stress. Plant Cell 2013, 25, 2925–2943. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sharma, S.; Chunduri, V.; Kaur, A.; Kaur, S.; Malhotra, N.; Kumar, A.; Kapoor, P.; Kumari, A.; Kaur, J.; et al. Genome-wide Identification and Characterization of Heat Shock Protein Family Reveals Role in Development and Stress Conditions in Triticum aestivum L. Sci Rep. 2020, 10, 7858. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Schlieker, C.; Friedrich, K.L.; Schönfeld, H.-J.; Vierling, E.; Bukau, B. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J. Biol. Chem. 2003, 278, 31033–31042. [Google Scholar] [CrossRef]
- Kang, X.; Zhao, L.; Liu, X. Calcium signaling and the response to heat shock in crop plants. Int. J. Mol. Sci. 2023, 25, 324. [Google Scholar] [CrossRef]
| No. | Name | Sequence ID | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| 1 | AsHSP20-1 | Asa0G00144.1 | 159 | 18,139.53 | 5.58 | 56.31 | 69.25 | −0.695 | Cytoplasm |
| 2 | AsHSP20-2 | Asa0G00606.1 | 158 | 18,067.53 | 5.84 | 50.66 | 74.62 | −0.687 | Cytoplasm |
| 3 | AsHSP20-3 | Asa0G01177.1 | 251 | 28,755.7 | 9.53 | 50.46 | 83.9 | −0.514 | Chloroplast |
| 4 | AsHSP20-4 | Asa0G01213.1 | 157 | 17,658.25 | 5.79 | 35.18 | 75.16 | −0.597 | Cytoplasm |
| 5 | AsHSP20-5 | Asa0G01214.1 | 164 | 18,416.18 | 6.19 | 33.58 | 70.73 | −0.699 | Cytoplasm |
| 6 | AsHSP20-6 | Asa0G01290.1 | 158 | 18,040.5 | 5.84 | 49.91 | 71.52 | −0.732 | Cytoplasm |
| 7 | AsHSP20-7 | Asa0G01291.1 | 159 | 18,154.52 | 5.56 | 52.42 | 71.07 | −0.751 | Cytoplasm |
| 8 | AsHSP20-8 | Asa0G01440.1 | 158 | 18,056.5 | 5.83 | 52.43 | 70.89 | −0.748 | Cytoplasm |
| 9 | AsHSP20-9 | Asa0G01441.1 | 143 | 16,504.7 | 5.78 | 56.83 | 68.18 | −0.685 | Cytoplasm |
| 10 | AsHSP20-10 | Asa0G01442.1 | 159 | 18,127.54 | 5.82 | 52.95 | 71.07 | −0.736 | Cytoplasm |
| 11 | AsHSP20-11 | Asa0G01443.1 | 203 | 23,184.11 | 5.61 | 38.01 | 64.33 | −0.897 | Nucleus |
| 12 | AsHSP20-12 | Asa0G02746.1 | 158 | 17,999.45 | 5.83 | 49.91 | 71.52 | −0.715 | Cytoplasm |
| 13 | AsHSP20-13 | Asa0G02962.1 | 158 | 18,084.51 | 5.58 | 51.91 | 70.89 | −0.749 | Cytoplasm |
| 14 | AsHSP20-14 | Asa0G04879.1 | 159 | 18,061.43 | 5.43 | 50.45 | 67.99 | −0.713 | Cytoplasm |
| 15 | AsHSP20-15 | Asa0G04881.1 | 159 | 18,121.54 | 5.43 | 50.51 | 67.99 | −0.699 | Cytoplasm |
| 16 | AsHSP20-16 | Asa0G04883.1 | 255 | 28,913.75 | 5.02 | 51.01 | 81.76 | −0.54 | Cytoplasm |
| 17 | AsHSP20-17 | Asa0G05708.1 | 151 | 17,233.36 | 6.33 | 51.53 | 61.99 | −0.845 | Cytoplasm |
| 18 | AsHSP20-18 | Asa1G00674.1 | 241 | 26,723.78 | 4.98 | 50.14 | 102.7 | −0.014 | Mitochondria |
| 19 | AsHSP20-19 | Asa1G00690.1 | 176 | 19,328.19 | 5.8 | 37.6 | 89.83 | −0.241 | Chloroplast |
| 20 | AsHSP20-20 | Asa1G00698.1 | 206 | 23,916.61 | 9.47 | 46.58 | 83.74 | −0.582 | Cytoplasm |
| 21 | AsHSP20-21 | Asa1G00886.1 | 309 | 35,742.26 | 8.83 | 40.37 | 65.05 | −1.144 | Cytoplasm |
| 22 | AsHSP20-22 | Asa1G02085.1 | 265 | 29,746.8 | 9.22 | 47.87 | 71.7 | −0.528 | Chloroplast |
| 23 | AsHSP20-23 | Asa1G03670.1 | 151 | 17,129.22 | 5.06 | 49.2 | 66.49 | −0.784 | Cytoplasm |
| 24 | AsHSP20-24 | Asa2G01049.1 | 164 | 18,497.86 | 5.15 | 49.69 | 73.78 | −0.564 | Peroxisome |
| 25 | AsHSP20-25 | Asa2G03191.1 | 133 | 15,090.12 | 5.74 | 35.3 | 70.45 | −0.614 | Cytoplasm |
| 26 | AsHSP20-26 | Asa2G03812.1 | 157 | 17,651.22 | 5.59 | 32.87 | 74.52 | −0.616 | Cytoplasm |
| 27 | AsHSP20-27 | Asa2G07035.1 | 130 | 15,223.41 | 5.38 | 48.28 | 74.85 | −0.793 | Cytoplasm |
| 28 | AsHSP20-28 | Asa2G07089.1 | 160 | 17,709.24 | 5.76 | 44.12 | 83.31 | −0.458 | Nucleus |
| 29 | AsHSP20-29 | Asa2G07141.1 | 137 | 15,715.82 | 9.17 | 35.29 | 66.13 | −1.131 | Nucleus |
| 30 | AsHSP20-30 | Asa3G01343.1 | 158 | 18,151.61 | 5.99 | 42.95 | 72.78 | −0.749 | Cytoplasm |
| 31 | AsHSP20-31 | Asa3G01354.1 | 158 | 18,062.45 | 5.82 | 43.72 | 73.35 | −0.709 | Cytoplasm |
| 32 | AsHSP20-32 | Asa3G01355.1 | 158 | 18,058.42 | 6.19 | 46.95 | 70.89 | −0.777 | Cytoplasm |
| 33 | AsHSP20-33 | Asa3G01357.1 | 211 | 24,285.48 | 5.77 | 48.47 | 75.78 | −0.626 | Extracellular |
| 34 | AsHSP20-34 | Asa3G01358.1 | 158 | 18,130.54 | 5.82 | 41.93 | 70.32 | −0.779 | Cytoplasm |
| 35 | AsHSP20-35 | Asa3G02930.1 | 158 | 18,198.66 | 5.83 | 57.43 | 71.52 | −0.78 | Cytoplasm |
| 36 | AsHSP20-36 | Asa3G05026.1 | 159 | 17,964.38 | 5.57 | 50.63 | 67.99 | −0.673 | Cytoplasm |
| 37 | AsHSP20-37 | Asa3G05028.1 | 236 | 27,183.15 | 9.54 | 49.51 | 71.4 | −0.795 | Cytoplasm |
| 38 | AsHSP20-38 | Asa3G05030.1 | 240 | 27,130.25 | 5.1 | 45.76 | 65.38 | −0.873 | Cytoplasm |
| 39 | AsHSP20-39 | Asa3G05031.1 | 160 | 18,020.38 | 5.83 | 47.72 | 67.56 | −0.703 | Cytoplasm |
| 40 | AsHSP20-40 | Asa3G05032.1 | 159 | 18,163.59 | 5.59 | 55.11 | 67.99 | −0.734 | Cytoplasm |
| 41 | AsHSP20-41 | Asa3G05113.1 | 133 | 15,672.91 | 5.09 | 39.27 | 68.8 | −0.793 | Cytoplasm |
| 42 | AsHSP20-42 | Asa3G05114.1 | 225 | 25,733.82 | 9.42 | 48.25 | 74.53 | −0.644 | Chloroplast |
| 43 | AsHSP20-43 | Asa4G00429.1 | 135 | 15,283.21 | 5.13 | 43.5 | 84.37 | −0.597 | Chloroplast |
| 44 | AsHSP20-44 | Asa4G02369.1 | 276 | 31,962.66 | 9.07 | 48.86 | 78.01 | −0.519 | Nucleus |
| 45 | AsHSP20-45 | Asa4G03225.1 | 211 | 23,503.47 | 6.66 | 56.59 | 80 | −0.665 | Chloroplast |
| 46 | AsHSP20-46 | Asa4G03797.1 | 145 | 16,458.88 | 9.9 | 55.27 | 84.69 | −0.51 | Cytoplasm |
| 47 | AsHSP20-47 | Asa4G04043.1 | 192 | 21,970.19 | 9.11 | 46.21 | 82.71 | −0.66 | Cytoplasm |
| 48 | AsHSP20-48 | Asa4G04292.1 | 174 | 19,748.48 | 5.6 | 49.47 | 80.57 | −0.444 | Cytoplasm |
| 49 | AsHSP20-49 | Asa4G04294.1 | 150 | 16,984.37 | 5.55 | 36.96 | 79.2 | −0.593 | Cytoplasm |
| 50 | AsHSP20-50 | Asa4G04295.1 | 167 | 18,884.46 | 5.21 | 43.34 | 78.14 | −0.432 | Cytoplasm |
| 51 | AsHSP20-51 | Asa4G04310.1 | 145 | 16,369.66 | 5.82 | 46.6 | 74 | −0.61 | Chloroplast |
| 52 | AsHSP20-52 | Asa4G04540.1 | 158 | 18,022.45 | 5.84 | 53.82 | 73.99 | −0.712 | Cytoplasm |
| 53 | AsHSP20-53 | Asa4G04541.1 | 158 | 18,042.55 | 5.73 | 44.33 | 73.35 | −0.632 | Cytoplasm |
| 54 | AsHSP20-54 | Asa4G04542.1 | 158 | 17,977.45 | 5.58 | 46.32 | 77.66 | −0.637 | Cytoplasm |
| 55 | AsHSP20-55 | Asa4G04543.1 | 158 | 17,868.2 | 5.57 | 55.65 | 74.62 | −0.653 | Cytoplasm |
| 56 | AsHSP20-56 | Asa4G04545.1 | 158 | 17,999.37 | 5.4 | 49.05 | 73.99 | −0.7 | Cytoplasm |
| 57 | AsHSP20-57 | Asa4G04549.1 | 158 | 18,024.42 | 5.84 | 52.17 | 74.62 | −0.694 | Cytoplasm |
| 58 | AsHSP20-58 | Asa4G04550.1 | 159 | 18,168.52 | 5.57 | 55.48 | 68.62 | −0.716 | Cytoplasm |
| 59 | AsHSP20-59 | Asa4G04555.1 | 137 | 15,729.83 | 5.56 | 51.07 | 73.94 | −0.711 | Cytoplasm |
| 60 | AsHSP20-60 | Asa4G06279.1 | 161 | 18,106.77 | 6.84 | 40.72 | 87.14 | −0.313 | Cytoplasm |
| 61 | AsHSP20-61 | Asa4G06421.1 | 144 | 16,262.56 | 5.54 | 45.56 | 77.08 | −0.632 | Cytoplasm |
| 62 | AsHSP20-62 | Asa4G06481.1 | 163 | 18,743.39 | 8.75 | 42.67 | 66.81 | −0.696 | Cytoplasm |
| 63 | AsHSP20-63 | Asa4G06617.1 | 163 | 18,743.39 | 8.75 | 42.67 | 66.81 | −0.696 | Cytoplasm |
| 64 | AsHSP20-64 | Asa5G00107.1 | 157 | 17,684.29 | 5.79 | 34.7 | 74.52 | −0.619 | Cytoplasm |
| 65 | AsHSP20-65 | Asa5G00109.1 | 157 | 17,714.32 | 5.52 | 35.18 | 74.52 | −0.596 | Cytoplasm |
| 66 | AsHSP20-66 | Asa5G00110.1 | 157 | 17,658.25 | 5.79 | 35.18 | 75.16 | −0.597 | Cytoplasm |
| 67 | AsHSP20-67 | Asa5G00111.1 | 144 | 16,457.79 | 9.95 | 72.56 | 59.51 | −1.116 | Chloroplast |
| 68 | AsHSP20-68 | Asa5G00687.1 | 231 | 26,254.38 | 8.7 | 56.53 | 75.06 | −0.615 | Chloroplast |
| 69 | AsHSP20-69 | Asa5G01992.1 | 364 | 41,148.26 | 7.74 | 31.01 | 61.46 | −1.069 | Mitochondria |
| 70 | AsHSP20-70 | Asa5G03263.1 | 158 | 18,139.6 | 6.77 | 44.21 | 70.19 | −0.739 | Cytoplasm |
| 71 | AsHSP20-71 | Asa5G03264.1 | 202 | 23,326.28 | 6.27 | 52.18 | 62.67 | −1.052 | Chloroplast |
| 72 | AsHSP20-72 | Asa5G03265.1 | 161 | 18,468.97 | 6.2 | 48.03 | 68.32 | −0.775 | Cytoplasm |
| 73 | AsHSP20-73 | Asa5G05424.1 | 154 | 18,137.42 | 6.76 | 50.47 | 81.04 | −0.899 | Cytoplasm |
| 74 | AsHSP20-74 | Asa5G05661.1 | 157 | 18,014.73 | 6.21 | 50.18 | 86.24 | −0.657 | Cytoplasm |
| 75 | AsHSP20-75 | Asa6G00868.1 | 175 | 19,933.25 | 6.66 | 34.13 | 94.69 | −0.399 | Extracellular |
| 76 | AsHSP20-76 | Asa6G00869.1 | 183 | 20,814.25 | 6.36 | 35.82 | 92.68 | −0.374 | Chloroplast |
| 77 | AsHSP20-77 | Asa6G00885.1 | 183 | 20,809.21 | 6.36 | 37.57 | 94.26 | −0.38 | Chloroplast |
| 78 | AsHSP20-78 | Asa6G01385.1 | 142 | 15,912.53 | 8.56 | 36.17 | 102.39 | −0.342 | Mitochondria |
| 79 | AsHSP20-79 | Asa6G02013.1 | 158 | 18,175.6 | 5.84 | 51.36 | 66.58 | −0.782 | Cytoplasm |
| 80 | AsHSP20-80 | Asa6G02870.1 | 201 | 22,824.07 | 6.77 | 44.82 | 81.59 | −0.53 | Chloroplast |
| 81 | AsHSP20-81 | Asa6G03303.1 | 211 | 23,694.78 | 9.43 | 44.38 | 75.5 | −0.55 | Chloroplast |
| 82 | AsHSP20-82 | Asa6G04489.1 | 211 | 23,446.46 | 6.67 | 56.22 | 79.53 | −0.631 | Chloroplast |
| 83 | AsHSP20-83 | Asa6G04532.1 | 158 | 18,026.48 | 5.83 | 49.37 | 71.52 | −0.732 | Cytoplasm |
| 84 | AsHSP20-84 | Asa6G04534.1 | 151 | 17,129.42 | 5.56 | 51.47 | 75.5 | −0.756 | Cytoplasm |
| 85 | AsHSP20-85 | Asa6G05099.1 | 158 | 18,070.59 | 5.83 | 48.04 | 72.72 | −0.696 | Cytoplasm |
| 86 | AsHSP20-86 | Asa6G05100.1 | 158 | 17,972.39 | 5.82 | 50.45 | 70.89 | −0.699 | Cytoplasm |
| 87 | AsHSP20-87 | Asa6G05101.1 | 158 | 18,098.58 | 5.83 | 53.27 | 70.89 | −0.775 | Cytoplasm |
| 88 | AsHSP20-88 | Asa6G05102.1 | 158 | 18,022.49 | 5.84 | 52.35 | 70.89 | −0.74 | Cytoplasm |
| 89 | AsHSP20-89 | Asa6G05713.1 | 223 | 25,172.88 | 8.42 | 47.45 | 86.05 | −0.396 | Mitochondria |
| 90 | AsHSP20-90 | Asa6G06028.1 | 211 | 23,594.81 | 9.32 | 51.03 | 74.45 | −0.654 | Chloroplast |
| 91 | AsHSP20-91 | Asa6G06030.1 | 205 | 22,974.09 | 5.85 | 37.56 | 72.93 | −0.497 | Chloroplast |
| 92 | AsHSP20-92 | Asa6G06033.1 | 209 | 23,406.58 | 6.77 | 51.32 | 77.08 | −0.554 | Chloroplast |
| 93 | AsHSP20-93 | Asa6G06784.1 | 210 | 23,730.91 | 6.86 | 44.27 | 78.57 | −0.548 | Chloroplast |
| 94 | AsHSP20-94 | Asa6G06852.1 | 264 | 29,904.88 | 5.74 | 41.81 | 80.61 | −0.528 | Cytoplasm |
| 95 | AsHSP20-95 | Asa6G06861.1 | 320 | 35,578.35 | 3.97 | 51.8 | 54.94 | −1.213 | Chloroplast |
| 96 | AsHSP20-96 | Asa6G06867.1 | 188 | 21,325.27 | 6.78 | 43.29 | 86.06 | −0.621 | Cytoplasm |
| 97 | AsHSP20-97 | Asa6G06873.1 | 259 | 28,998.91 | 8.46 | 48.9 | 81.74 | −0.464 | Cytoplasm |
| 98 | AsHSP20-98 | Asa6G06874.1 | 193 | 22,206.22 | 9.04 | 42.27 | 77.77 | −0.791 | Chloroplast |
| 99 | AsHSP20-99 | Asa6G06974.1 | 182 | 20,251.13 | 7.71 | 44.44 | 92.2 | −0.394 | Chloroplast |
| 100 | AsHSP20-100 | Asa6G07048.1 | 251 | 28,187.72 | 5.66 | 40.82 | 77.69 | −0.563 | Chloroplast |
| 101 | AsHSP20-101 | Asa7G02785.1 | 194 | 21,729.86 | 8.98 | 42.33 | 81.44 | −0.582 | Cytoplasm |
| 102 | AsHSP20-102 | Asa7G05180.1 | 215 | 23,979.15 | 7.77 | 53.94 | 76.23 | −0.601 | Chloroplast |
| 103 | AsHSP20-103 | Asa7G05384.1 | 179 | 20,697.54 | 5.26 | 47.4 | 76.15 | −0.578 | Cytoplasm |
| 104 | AsHSP20-104 | Asa7G06920.1 | 235 | 26,777.34 | 9.11 | 56.92 | 72.17 | −0.769 | Chloroplast |
| 105 | AsHSP20-105 | Asa8G01375.1 | 215 | 23,965.12 | 6.66 | 59.25 | 75.77 | −0.589 | Chloroplast |
| 106 | AsHSP20-106 | Asa8G02037.1 | 142 | 16,716.65 | 4.95 | 47.33 | 63.1 | −1.128 | Cytoplasm |
| 107 | AsHSP20-107 | Asa8G02053.1 | 186 | 21,194.91 | 5.41 | 45.58 | 73.49 | −0.762 | Chloroplast |
| 108 | AsHSP20-108 | Asa8G02054.1 | 186 | 21,078.92 | 5.66 | 44.21 | 78.17 | −0.659 | Chloroplast |
| 109 | AsHSP20-109 | Asa8G02220.1 | 142 | 16,716.65 | 4.95 | 47.33 | 63.1 | −1.128 | Cytoplasm |
| 110 | AsHSP20-110 | Asa8G03628.1 | 164 | 18,250.02 | 6.6 | 33.48 | 78.41 | −0.538 | Cytoplasm |
| 111 | AsHSP20-111 | Asa8G03629.1 | 164 | 18,248.05 | 6.6 | 32.36 | 79.02 | −0.522 | Cytoplasm |
| 112 | AsHSP20-112 | Asa8G03632.1 | 158 | 17,662.33 | 6.53 | 35.32 | 75.13 | −0.63 | Cytoplasm |
| 113 | AsHSP20-113 | Asa8G03635.1 | 158 | 17,648.31 | 6.53 | 34.78 | 75.13 | −0.63 | Cytoplasm |
| 114 | AsHSP20-114 | Asa8G04457.1 | 251 | 28,493.48 | 9.75 | 35.61 | 60.2 | −0.941 | Chloroplast |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, N.; He, B.; Cao, Z. Genome-Wide Analysis and Functional Characterization of Small Heat Shock Proteins in Allium sativum L. Under Multiple Abiotic Stresses. Biology 2025, 14, 1326. https://doi.org/10.3390/biology14101326
Li N, He B, Cao Z. Genome-Wide Analysis and Functional Characterization of Small Heat Shock Proteins in Allium sativum L. Under Multiple Abiotic Stresses. Biology. 2025; 14(10):1326. https://doi.org/10.3390/biology14101326
Chicago/Turabian StyleLi, Na, Bing He, and Zhenyu Cao. 2025. "Genome-Wide Analysis and Functional Characterization of Small Heat Shock Proteins in Allium sativum L. Under Multiple Abiotic Stresses" Biology 14, no. 10: 1326. https://doi.org/10.3390/biology14101326
APA StyleLi, N., He, B., & Cao, Z. (2025). Genome-Wide Analysis and Functional Characterization of Small Heat Shock Proteins in Allium sativum L. Under Multiple Abiotic Stresses. Biology, 14(10), 1326. https://doi.org/10.3390/biology14101326






