Targeting Hypoxia and HIF1α in Triple-Negative Breast Cancer: New Insights from Gene Expression Profiling and Implications for Therapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. ROS Detection
2.3. Cell Viability Assay
2.4. In Vitro Migration Assay
2.5. Western Blot Analysis
2.6. Quantitative Real-Time PCR Analysis
2.7. RNA-Seq and Data Analysis
3. Results
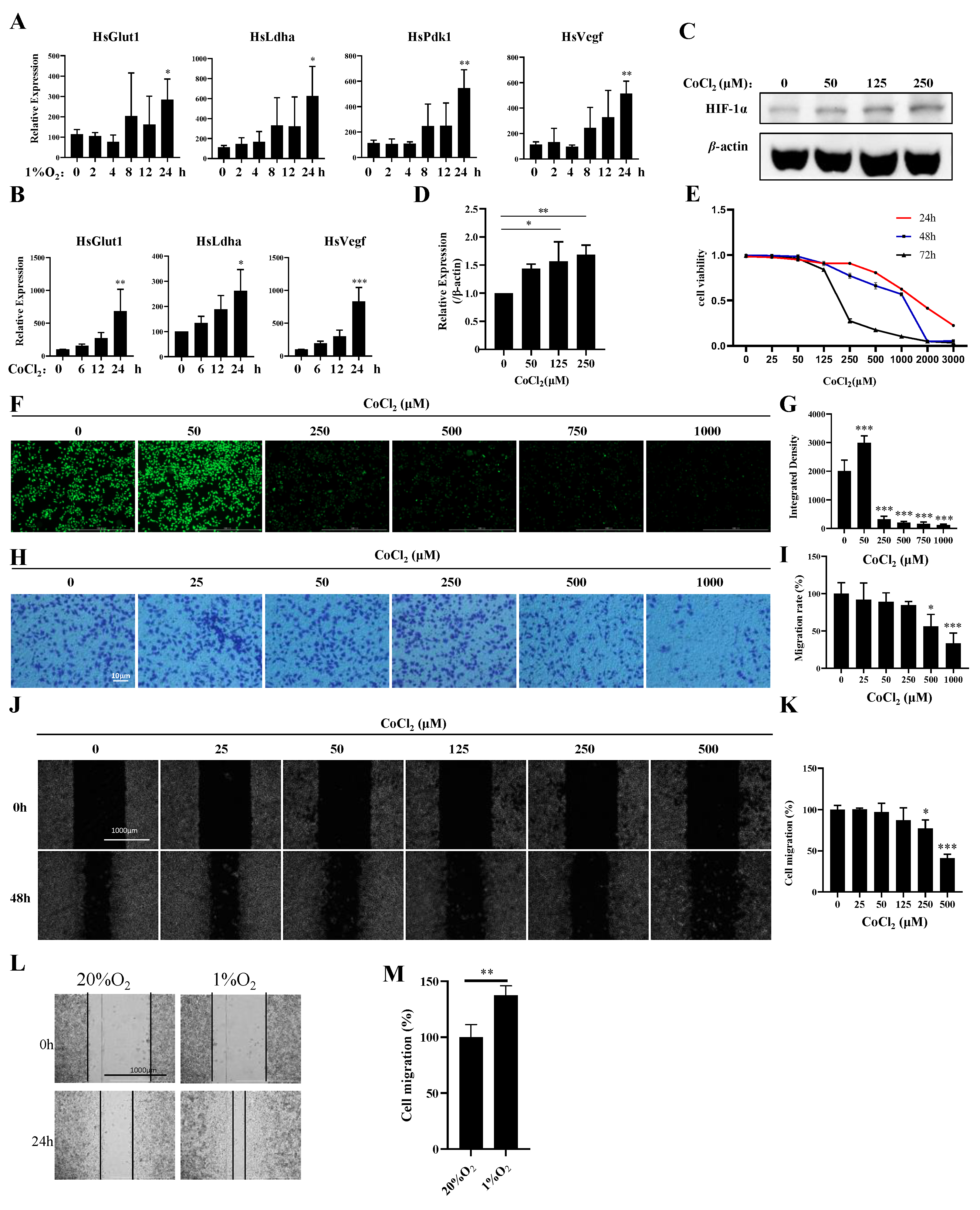
3.1. Establishment of a Hypoxia Model in MDA-MB-231 Cells
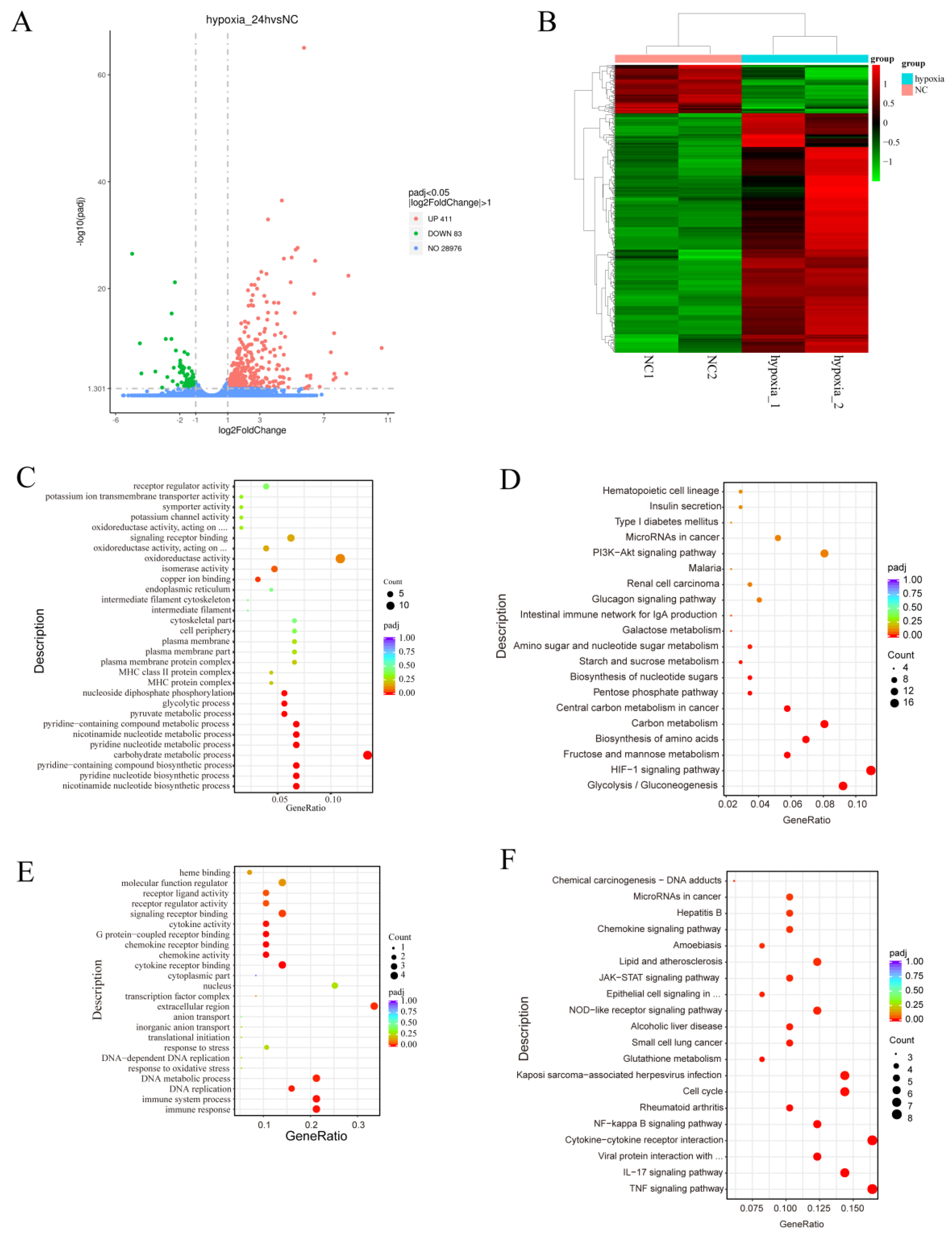
3.2. Transcriptomic Profiles of Normoxic and Hypoxic MDA-MB-231 Cells Reveal Common and Divergent Transcriptomic Patterns
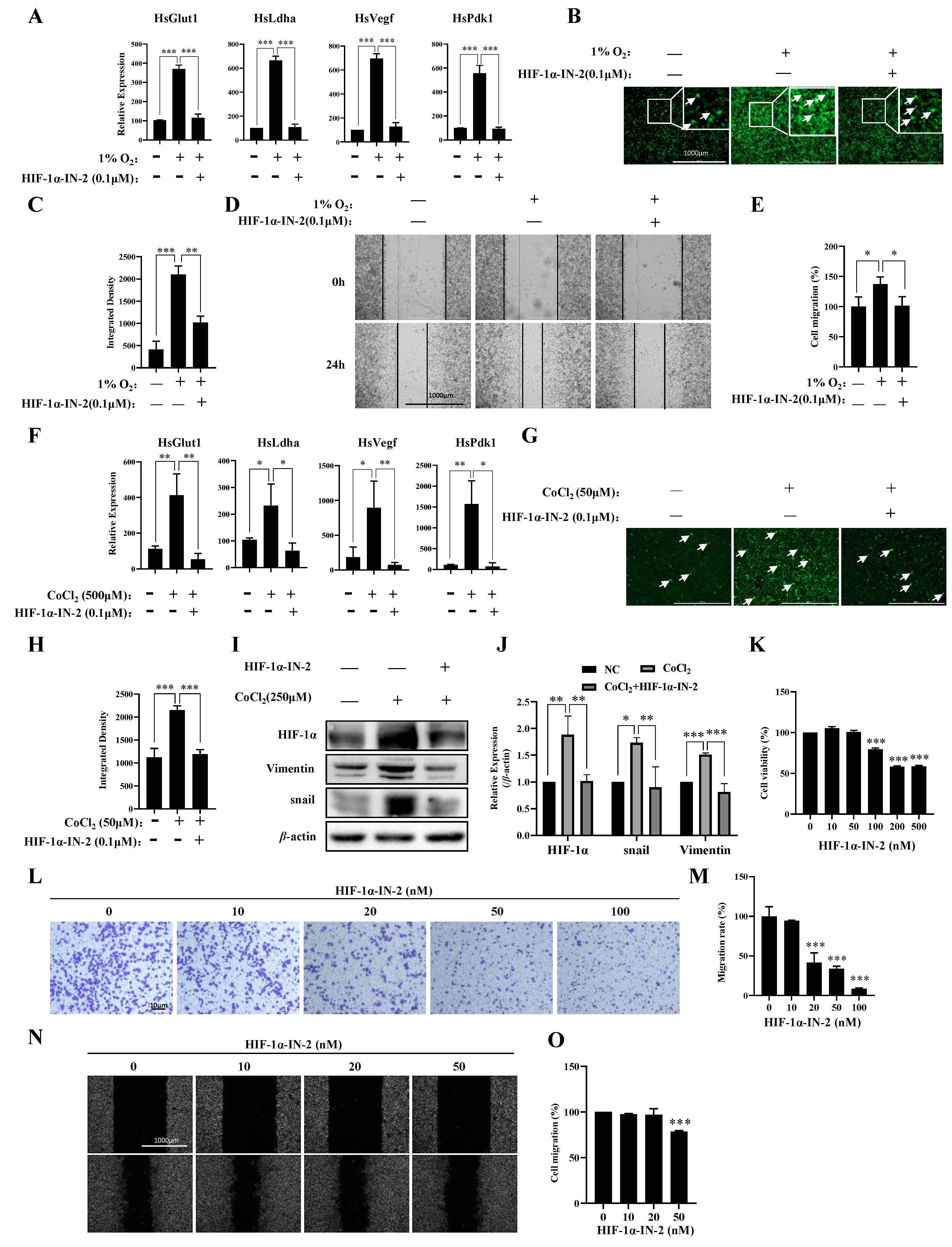
3.3. HIF1α Regulated the Responses of Most Genes to Hypoxia
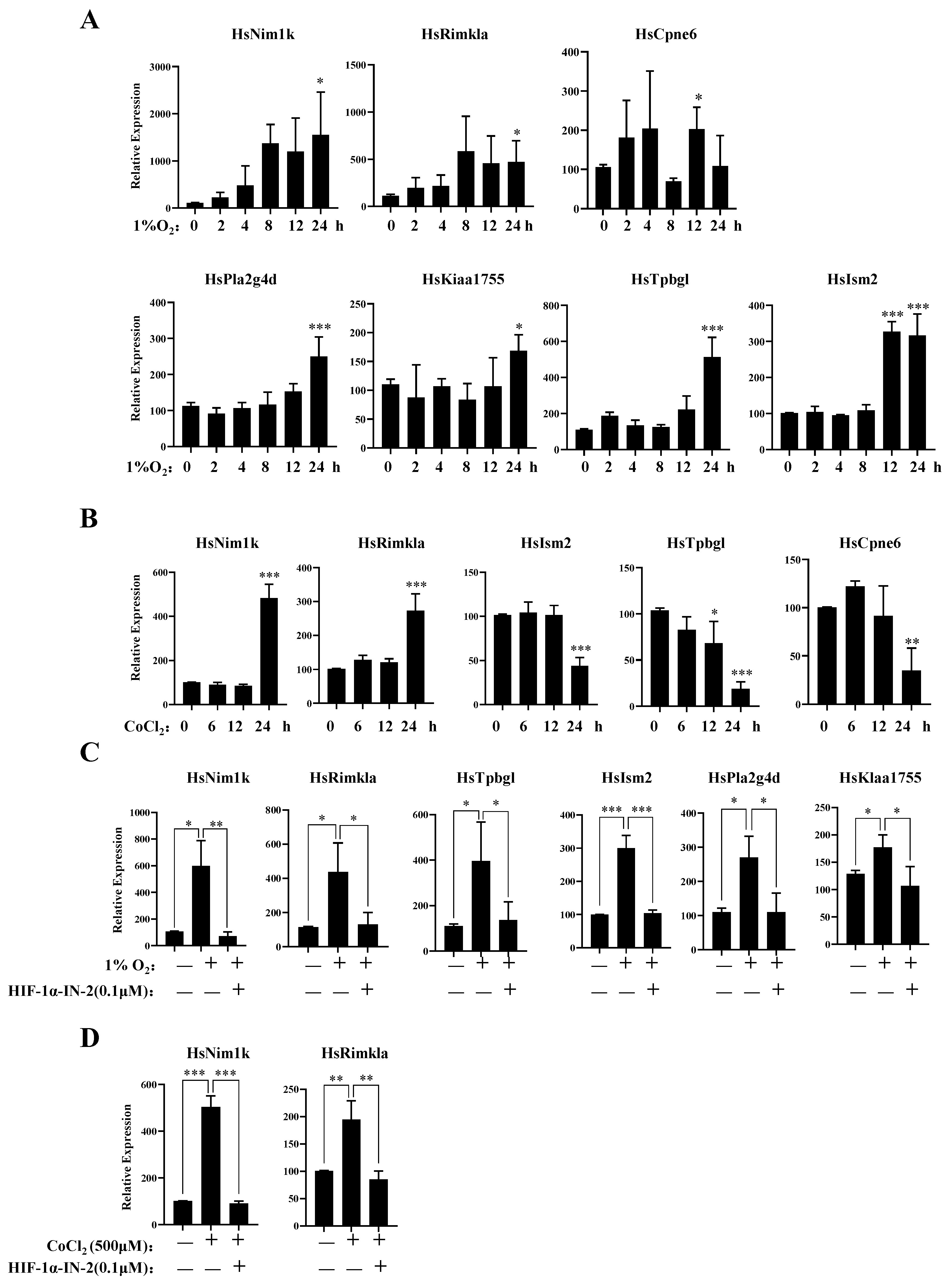
3.4. New Genes Related to the Hypoxia Response of MDA-MB-231 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, M.H.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 2020, 84, 106535. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Derakhshan, F.; Reis-Filho, J.S. Pathogenesis of Triple-Negative Breast Cancer. Annu. Rev. Pathol. 2022, 17, 181–204. [Google Scholar] [CrossRef] [PubMed]
- Rossi, L.; Mazzara, C.; Pagani, O. Diagnosis and Treatment of Breast Cancer in Young Women. Curr. Treat. Options Oncol. 2019, 20, 86. [Google Scholar] [CrossRef]
- Peart, O. Breast intervention and breast cancer treatment options. Radiol. Technol. 2015, 86, 535M–558M, quiz 559–562. [Google Scholar] [PubMed]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Francis, P.A.; Pagani, O.; Fleming, G.F.; Walley, B.A.; Colleoni, M.; Lang, I.; Gomez, H.L.; Tondini, C.; Ciruelos, E.; Burstein, H.J.; et al. Tailoring Adjuvant Endocrine Therapy for Premenopausal Breast Cancer. N. Engl. J. Med. 2018, 379, 122–137. [Google Scholar] [CrossRef]
- Masuda, N.; Lee, S.J.; Ohtani, S.; Im, Y.H.; Lee, E.S.; Yokota, I.; Kuroi, K.; Im, S.A.; Park, B.W.; Kim, S.B.; et al. Adjuvant Capecitabine for Breast Cancer after Preoperative Chemotherapy. N. Engl. J. Med. 2017, 376, 2147–2159. [Google Scholar] [CrossRef]
- Fisher, B.; Anderson, S.; Bryant, J.; Margolese, R.G.; Deutsch, M.; Fisher, E.R.; Jeong, J.H.; Wolmark, N. Twenty-year follow-up of a randomized trial comparing total mastectomy, lumpectomy, and lumpectomy plus irradiation for the treatment of invasive breast cancer. N. Engl. J. Med. 2002, 347, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Hanker, A.B.; Sudhan, D.R.; Arteaga, C.L. Overcoming Endocrine Resistance in Breast Cancer. Cancer Cell 2020, 37, 496–513. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Singh, R. Nanotherapy: Targeting the tumour microenvironment. Nat. Rev. Cancer 2022, 22, 258. [Google Scholar] [CrossRef] [PubMed]
- Kinnel, B.; Singh, S.K.; Oprea-Ilies, G.; Singh, R. Targeted Therapy and Mechanisms of Drug Resistance in Breast Cancer. Cancers 2023, 15, 1320. [Google Scholar] [CrossRef] [PubMed]
- Soysal, S.D.; Tzankov, A.; Muenst, S.E. Role of the Tumor Microenvironment in Breast Cancer. Pathobiology 2015, 82, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Brown, N.J.; Holen, I. The breast tumor microenvironment: Role in cancer development, progression and response to therapy. Expert Rev. Mol. Diagn. 2018, 18, 227–243. [Google Scholar] [CrossRef]
- Kudelova, E.; Smolar, M.; Holubekova, V.; Hornakova, A.; Dvorska, D.; Lucansky, V.; Koklesova, L.; Kudela, E.; Kubatka, P. Genetic Heterogeneity, Tumor Microenvironment and Immunotherapy in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2022, 23, 14937. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, J.; Huang, Y.; Shangguan, D.; Zhang, P. Single-Cell Profiling to Explore Immunological Heterogeneity of Tumor Microenvironment in Breast Cancer. Front. Immunol. 2021, 12, 643692. [Google Scholar] [CrossRef] [PubMed]
- Deepak, K.G.K.; Vempati, R.; Nagaraju, G.P.; Dasari, V.R.; Nagini, S.; Rao, D.N.; Malla, R.R. Tumor microenvironment: Challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol. Res. 2020, 153, 104683. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, H.; Song, X.; Yang, Q. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin. Cancer Biol. 2020, 60, 14–27. [Google Scholar] [CrossRef]
- Zou, Y.; Ye, F.; Kong, Y.; Hu, X.; Deng, X.; Xie, J.; Song, C.; Ou, X.; Wu, S.; Wu, L.; et al. The Single-Cell Landscape of Intratumoral Heterogeneity and The Immunosuppressive Microenvironment in Liver and Brain Metastases of Breast Cancer. Adv. Sci. 2023, 10, e2203699. [Google Scholar] [CrossRef] [PubMed]
- Knowles, H.J.; Harris, A.L. Hypoxia and oxidative stress in breast cancer. Hypoxia and tumourigenesis. Breast Cancer Res. 2001, 3, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Schwab, L.P.; Peacock, D.L.; Majumdar, D.; Ingels, J.F.; Jensen, L.C.; Smith, K.D.; Cushing, R.C.; Seagroves, T.N. Hypoxia-inducible factor 1alpha promotes primary tumor growth and tumor-initiating cell activity in breast cancer. Breast Cancer Res. 2012, 14, R6. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, N.; Myers, C. Breast cancer-induced angiogenesis: Multiple mechanisms and the role of the microenvironment. Breast Cancer Res. 2003, 5, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Semenza, G.L.; Zhang, H.F. Hypoxia-inducible factor 1 and breast cancer metastasis. J. Zhejiang Univ. Sci. B 2015, 16, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Gilkes, D.M.; Semenza, G.L. Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncol. 2013, 9, 1623–1636. [Google Scholar] [CrossRef]
- Sforna, L.; Cenciarini, M.; Belia, S.; D’Adamo, M.C.; Pessia, M.; Franciolini, F.; Catacuzzeno, L. The role of ion channels in the hypoxia-induced aggressiveness of glioblastoma. Front. Cell Neurosci. 2014, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Michelucci, A.; Sforna, L.; Franciolini, F.; Catacuzzeno, L. Hypoxia, Ion Channels and Glioblastoma Malignancy. Biomolecules 2023, 13, 1742. [Google Scholar] [CrossRef]
- Sforna, L.; Cenciarini, M.; Belia, S.; Michelucci, A.; Pessia, M.; Franciolini, F.; Catacuzzeno, L. Hypoxia Modulates the Swelling-Activated Cl Current in Human Glioblastoma Cells: Role in Volume Regulation and Cell Survival. J. Cell Physiol. 2017, 232, 91–100. [Google Scholar] [CrossRef]
- Sceneay, J.; Parker, B.S.; Smyth, M.J.; Moller, A. Hypoxia-driven immunosuppression contributes to the pre-metastatic niche. Oncoimmunology 2013, 2, e22355. [Google Scholar] [CrossRef]
- Chen, Y.; Luo, L.; Deng, L.; Tian, X.; Chen, S.; Xu, A.; Yuan, S. New Insights Into the Lineage-Specific Expansion and Functional Diversification of Lamprey AID/APOBEC Family. Front. Immunol. 2022, 13, 822616. [Google Scholar] [CrossRef]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef]
- Essemine, J.; Li, J.; Chen, G.; Qu, M. Analytical dataset of short-term heat stress induced reshuffling of metabolism and transcriptomes in maize grown under elevated CO2. Data Brief. 2020, 28, 105004. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, D.C.; Gorham, J.M.; Herman, D.S.; Seidman, J.G. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Curr. Protoc. Mol. Biol. 2011, 94, 4.12.1–4.12.11. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, M.C.; McLellan, A.S. Whole-Exome Sequencing (WES) for Illumina Short Read Sequencers Using Solution-Based Capture. Methods Mol. Biol. 2020, 2076, 85–108. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xue, R.; Qin, W.; Yang, X.; Ye, Q.; Wu, Q. Performance and transcriptome analysis of Salmonella enterica serovar Enteritidis PT 30 under persistent desiccation stress: Cultured by lawn and broth methods. Food Microbiol. 2023, 115, 104323. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Yuan, X.; Song, Y.; Liu, Y.; Wang, X.F. First report of maize yellow mosaic virus (MaYMV) naturally infecting wheat in China. Plant Dis. 2022, 106, 2763. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Shi, X.; Sheng, K.; Han, G.; Li, W.; Zhao, Q.; Jiang, B.; Feng, J.; Li, J.; Gu, Y. PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (Review). Mol. Med. Rep. 2019, 19, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Chun, Y.; Kim, J. AMPK-mTOR Signaling and Cellular Adaptations in Hypoxia. Int. J. Mol. Sci. 2021, 22, 9765. [Google Scholar] [CrossRef] [PubMed]
- Yourick, D.L.; Koenig, M.L.; Durden, A.V.; Long, J.B. N-acetylaspartylglutamate and beta-NAAG protect against injury induced by NMDA and hypoxia in primary spinal cord cultures. Brain Res. 2003, 991, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Bratek, E.; Ziembowicz, A.; Salinska, E. N-Acetylaspartylglutamate (NAAG) Pretreatment Reduces Hypoxic-Ischemic Brain Damage and Oxidative Stress in Neonatal Rats. Antioxidants 2020, 9, 877. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, J.R.; Kriz, A.; Galic, M.; Angliker, N.; Rajalu, M.; Vogt, K.E.; Ruegg, M.A. The calcium sensor Copine-6 regulates spine structural plasticity and learning and memory. Nat. Commun. 2016, 7, 11613. [Google Scholar] [CrossRef] [PubMed]
- Cross, B.M.; Breitwieser, G.E.; Reinhardt, T.A.; Rao, R. Cellular calcium dynamics in lactation and breast cancer: From physiology to pathology. Am. J. Physiol. Cell Physiol. 2014, 306, C515–C526. [Google Scholar] [CrossRef] [PubMed]
- Jardin, I.; Lopez, J.J.; Salido, G.M.; Rosado, J.A. Store-Operated Ca2+ Entry in Breast Cancer Cells: Remodeling and Functional Role. Int. J. Mol. Sci. 2018, 19, 4053. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, C.; Liu, S.; Zhou, W.; Du, J.; Jiang, Y.; Dai, J.; Jin, G.; Ma, H.; Hu, Z.; et al. Potential functional variants of KIAA genes are associated with breast cancer risk in a case control study. Ann. Transl. Med. 2021, 9, 549. [Google Scholar] [CrossRef]
- Khan, S.A.; Ilies, M.A. The Phospholipase A2 Superfamily: Structure, Isozymes, Catalysis, Physiologic and Pathologic Roles. Int. J. Mol. Sci. 2023, 24, 1353. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Shao, P.; Gao, Z.; Zhang, S.; Ma, J.; Bai, J.; Wei, Y. Homocysteine and Lp-PLA2 levels: Diagnostic value in coronary heart disease. Medicine 2023, 102, e35982. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Shu, D.; Wu, S.; Cai, P.; Liang, T. Higher serum Lp-PLA2 is associated with cognitive impairment in Parkinson’s disease patients. Front. Neurosci. 2024, 18, 1374567. [Google Scholar] [CrossRef] [PubMed]
- Bazan, N.G.; Colangelo, V.; Lukiw, W.J. Prostaglandins and other lipid mediators in Alzheimer’s disease. Prostaglandins Other Lipid Mediat. 2002, 68–69, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Yamashita, J.; Sakamoto, K.; Inada, K.; Nakashima, Y.; Murata, K.; Saishoji, T.; Nomura, K.; Ogawa, M. Increased expression of membrane-associated phospholipase A2 shows malignant potential of human breast cancer cells. Cancer 1993, 71, 3058–3064. [Google Scholar] [CrossRef] [PubMed]
- Gimenes, S.N.C.; Lopes, D.S.; Alves, P.T.; Azevedo, F.; Vecchi, L.; Goulart, L.R.; Rodrigues, T.C.S.; Santos, A.L.Q.; Brites, V.L.C.; Teixeira, T.L.; et al. Antitumoral effects of gammaCdcPLI, a PLA(2) inhibitor from Crotalus durissus collilineatus via PI3K/Akt pathway on MDA-MB-231 breast cancer cell. Sci. Rep. 2017, 7, 7077. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Bai, J.; Hu, L.; Jiang, D. Hypoxia-mediated activation of hypoxia-inducible factor-1alpha in triple-negative breast cancer: A review. Medicine 2023, 102, e35493. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Xu, J.; She, Y.; Jiang, T.; Zhou, S.; Shi, H.; Li, C. Necrostatin-1 protects C2C12 myotubes from CoCl2-induced hypoxia. Int. J. Mol. Med. 2018, 41, 2565–2572. [Google Scholar] [CrossRef]
- Nguyen, T.M.H.; Lai, Y.S.; Chen, Y.C.; Lin, T.C.; Nguyen, N.T.; Chiu, W.T. Hypoxia-induced YAP activation and focal adhesion turnover to promote cell migration in mesenchymal TNBC cells. Cancer Med. 2023, 12, 9723–9737. [Google Scholar] [CrossRef]
- Rong, Z.; Li, L.; Fei, F.; Luo, L.; Qu, Y. Combined treatment of glibenclamide and CoCl2 decreases MMP9 expression and inhibits growth in highly metastatic breast cancer. J. Exp. Clin. Cancer Res. 2013, 32, 32. [Google Scholar] [CrossRef]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef]
- Vishnubalaji, R.; Alajez, N.M. Transcriptional landscape associated with TNBC resistance to neoadjuvant chemotherapy revealed by single-cell RNA-seq. Mol. Ther. Oncolytics 2021, 23, 151–162. [Google Scholar] [CrossRef]
- Bergin, A.R.T.; Loi, S. Triple-negative breast cancer: Recent treatment advances. F1000Research 2019, 8, 1342. [Google Scholar] [CrossRef]
- Emens, L.A. Immunotherapy in Triple-Negative Breast Cancer. Cancer J. 2021, 27, 59–66. [Google Scholar] [CrossRef]
- Ferrari, P.; Scatena, C.; Ghilli, M.; Bargagna, I.; Lorenzini, G.; Nicolini, A. Molecular Mechanisms, Biomarkers and Emerging Therapies for Chemotherapy Resistant TNBC. Int. J. Mol. Sci. 2022, 23, 1665. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Guan, C.; Liu, C.; Li, H.; Wu, J.; Sun, C. Targeting hypoxia-inducible factor-1alpha: A new strategy for triple-negative breast cancer therapy. Biomed. Pharmacother. 2022, 156, 113861. [Google Scholar] [CrossRef]
- Semenza, G.L. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 2001, 13, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.H.; Rue, E.; Wang, G.L.; Roe, R.; Semenza, G.L. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 17771–17778. [Google Scholar] [CrossRef]
- Semenza, G.L.; Agani, F.; Booth, G.; Forsythe, J.; Iyer, N.; Jiang, B.H.; Leung, S.; Roe, R.; Wiener, C.; Yu, A. Structural and functional analysis of hypoxia-inducible factor 1. Kidney Int. 1997, 51, 553–555. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Potluri, N.; Lu, J.; Kim, Y.; Rastinejad, F. Structural integration in hypoxia-inducible factors. Nature 2015, 524, 303–308. [Google Scholar] [CrossRef]
- Semenza, G.L.; Jiang, B.H.; Leung, S.W.; Passantino, R.; Concordet, J.P.; Maire, P.; Giallongo, A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef] [PubMed]
- Tsuzuki, Y.; Fukumura, D.; Oosthuyse, B.; Koike, C.; Carmeliet, P.; Jain, R.K. Vascular endothelial growth factor (VEGF) modulation by targeting hypoxia-inducible factor-1alpha--> hypoxia response element--> VEGF cascade differentially regulates vascular response and growth rate in tumors. Cancer Res. 2000, 60, 6248–6252. [Google Scholar] [PubMed]
- Zhao, X.Y.; Chen, T.T.; Xia, L.; Guo, M.; Xu, Y.; Yue, F.; Jiang, Y.; Chen, G.Q.; Zhao, K.W. Hypoxia inducible factor-1 mediates expression of galectin-1: The potential role in migration/invasion of colorectal cancer cells. Carcinogenesis 2010, 31, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Kang, Y. Hypoxia and hypoxia-inducible factors: Master regulators of metastasis. Clin. Cancer Res. 2010, 16, 5928–5935. [Google Scholar] [CrossRef] [PubMed]
- Terry, S.; Engelsen, A.S.T.; Buart, S.; Elsayed, W.S.; Venkatesh, G.H.; Chouaib, S. Hypoxia-driven intratumor heterogeneity and immune evasion. Cancer Lett. 2020, 492, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.S.; Lee, H.; Kim, K.W. HIF-1alpha: A valid therapeutic target for tumor therapy. Cancer Res. Treat. 2004, 36, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.J.; Wang, L.Y.; Chodosh, L.A.; Keith, B.; Simon, M.C. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol. Cell Biol. 2003, 23, 9361–9374. [Google Scholar] [CrossRef]
| Key TF | # of Overlapped Genes | p Value | Q Value | List of Overlapped Genes |
|---|---|---|---|---|
| HIF1A | 19 | 3.06 × 10−16 | 2.69 × 10−14 | LOX, CA9, VEGFA, FAM162A, PFKFB3, RORA, BNIP3, PFKFB4, PGK1, ITGB2, MUC1, EGLN3, GAPDH, ENO1, ALDOA, LDHA, SLC2A1, EGLN1, VEGFB |
| SP1 | 19 | 0.000841 | 0.0185 | ITGB2, RORA, PPL, ITGA5, CDC42BPG, EPOR, NOS3, CA9, VEGFA, TGFB1, ITGB3, KRT18, LDHA, CDKN1A, TIMP3, KCTD11, NKX2-1, P4HA1, CAV1 |
| JUN | 7 | 0.0174 | 0.0383 | SOX7, CDKN1A, PDK1, NOS3, NEFL, VEGFA, LDHA |
| MYC | 6 | 0.00895 | 0.0272 | VEGFA, NDRG1, LDHA, SFRP1, CDKN1A, PGK1 |
| SP3 | 6 | 0.0157 | 0.0363 | CA9, CDKN1A, RORA, NKX2-1, VEGFA, NOS3 |
| STAT3 | 6 | 0.0417 | 0.068 | VEGFA, VEGFB, TGFB1, CDKN1A, HSPB1, MUC1 |
| TP53 | 6 | 0.0733 | 0.104 | CAV1, VEGFA, CDKN1A, SLC2A1, DUSP1, PLAGL1 |
| PPARG | 5 | 0.00639 | 0.0238 | CDKN1A, CAV1, ANGPTL4, PLIN2, VLDLR |
| HDAC1 | 5 | 0.00866 | 0.0272 | NOS3, RUNX3, POU5F1, SFRP1, CDKN1A |
| ETS1 | 5 | 0.0134 | 0.0327 | CDKN1A, ITGB3, NDRG1, TMEM158, CA9 |
| AR | 5 | 0.0253 | 0.0473 | NDRG1, CDKN1A, MUC1, KISS1R, VEGFA |
| ATM | 4 | 0.00056 | 0.0164 | CDKN1A, DUSP1, SLC2A1, VEGFA |
| DNMT1 | 4 | 0.00212 | 0.0191 | TIMP3, RUNX3, SFRP1, VEGFA |
| SMAD3 | 4 | 0.00212 | 0.0191 | VEGFA, ANGPTL4, TGFB1, CDKN1A |
| EZH2 | 4 | 0.00544 | 0.0238 | CDKN1A, RUNX3, ADAMTS1, SFRP1 |
| VDR | 4 | 0.00648 | 0.0238 | BHLHE40, DDIT4, CDKN1A, HLA-DRB1 |
| MYCN | 4 | 0.00828 | 0.0272 | DKK3, NDRG1, CDKN1A, MXI1 |
| E2F1 | 4 | 0.217 | 0.244 | CDKN1A, DUSP1, VEGFA, ISYNA1 |
| NFKB1 | 4 | 0.791 | 0.8 | CDKN1A, VEGFA, TGFB1, NOS3 |
| Key TF | # of Overlapped Genes | p-Value | Q Value | List of Overlapped Genes |
|---|---|---|---|---|
| SP1 | 7 | 0.00413 | 0.00744 | CYP1B1, GDA, AREG, IL6, PTGS2, CXCL1, ODC1 |
| E2F1 | 6 | 0.000024 | 0.000216 | RRM2, CDC6, CDCA7, MEFV, UHRF1, PCNA |
| RELA | 6 | 0.00186 | 0.00408 | IL6, PTGS2, CCL20, CXCL2, OLR1, CXCL1 |
| NFKB1 | 6 | 0.00193 | 0.00408 | CCL20, PTGS2, IL6, CXCL1, CXCL2, OLR1 |
| JUND | 4 | 1.32 × 10−5 | 0.000159 | IL6, NQO1, GADD45A, PTGS2 |
| EP300 | 4 | 9.76 × 10−5 | 0.000442 | PTGS2, CYP1B1, PCNA, IL6 |
| BRCA1 | 4 | 0.000105 | 0.000442 | CYP1B1, GADD45A, AREG, CXCL1 |
| CREB1 | 4 | 0.000608 | 0.00199 | SLC20A1, ODC1, IL6, PTGS2 |
| MYC | 4 | 0.000903 | 0.0027 | CDCA7, IL6, PCNA, ODC1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, D.; Li, Z.; Luo, L.; Jiang, H. Targeting Hypoxia and HIF1α in Triple-Negative Breast Cancer: New Insights from Gene Expression Profiling and Implications for Therapy. Biology 2024, 13, 577. https://doi.org/10.3390/biology13080577
Han D, Li Z, Luo L, Jiang H. Targeting Hypoxia and HIF1α in Triple-Negative Breast Cancer: New Insights from Gene Expression Profiling and Implications for Therapy. Biology. 2024; 13(8):577. https://doi.org/10.3390/biology13080577
Chicago/Turabian StyleHan, Delong, Zeyu Li, Lingjie Luo, and Hezhong Jiang. 2024. "Targeting Hypoxia and HIF1α in Triple-Negative Breast Cancer: New Insights from Gene Expression Profiling and Implications for Therapy" Biology 13, no. 8: 577. https://doi.org/10.3390/biology13080577
APA StyleHan, D., Li, Z., Luo, L., & Jiang, H. (2024). Targeting Hypoxia and HIF1α in Triple-Negative Breast Cancer: New Insights from Gene Expression Profiling and Implications for Therapy. Biology, 13(8), 577. https://doi.org/10.3390/biology13080577





