Simple Summary
Coronaviruses are serious pathogens for both humans and animals. The name corona was designated because of the crown-like spikes on their surface. Currently, seven coronaviruses have been identified, such as 229E, NL63, OC43, HKU1, MERS-CoV, SARS-CoV, and SARS-CoV-2. Sometimes, animal coronaviruses infect humans and evolve due to genetic mutations, interspecies transmission, host adaptations, and favorable conditions. The main objective of this study was to analyze the genetic diversity and predict the emergence of new variants with novel properties. It has been reported that the spike protein gene plays an important role in host cell attachment and entry into host cells. The S gene has the highest mutation/deletion and is the most utilized target for vaccine/antiviral development. In this work, we discussed the genetic diversity, phylogenetic relationship, and recombination patterns of selected HCoVs with an emphasis on the newly emerged SARS-CoV-2 and MERS-CoV. The findings of this study showed that MERS-CoV and SARS-CoV-2 have significant sequence identities with the selected HCoVs. The phylogenetic and recombination results concluded that new variants may emerge in the future with novel properties that infect both humans and animals. This information will be helpful for global society to design and develop an effective vaccine and disease management strategy.
Abstract
Human coronaviruses (HCoVs) are seriously associated with respiratory diseases in humans and animals. The first human pathogenic SARS-CoV emerged in 2002–2003. The second was MERS-CoV, reported from Jeddah, the Kingdom of Saudi Arabia, in 2012, and the third one was SARS-CoV-2, identified from Wuhan City, China, in late December 2019. The HCoV-Spike (S) gene has the highest mutation/insertion/deletion rate and has been the most utilized target for vaccine/antiviral development. In this manuscript, we discuss the genetic diversity, phylogenetic relationships, and recombination patterns of selected HCoVs with emphasis on the S protein gene of MERS-CoV and SARS-CoV-2 to elucidate the possible emergence of new variants/strains of coronavirus in the near future. The findings showed that MERS-CoV and SARS-CoV-2 have significant sequence identity with the selected HCoVs. The phylogenetic tree analysis formed a separate cluster for each HCoV. The recombination pattern analysis showed that the HCoV-NL63-Japan was a probable recombinant. The HCoV-NL63-USA was identified as a major parent while the HCoV-NL63-Netherland was identified as a minor parent. The recombination breakpoints start in the viral genome at the 142 nucleotide position and end at the 1082 nucleotide position with a 99% CI and Bonferroni-corrected p-value of 0.05. The findings of this study provide insightful information about HCoV-S gene diversity, recombination, and evolutionary patterns. Based on these data, it can be concluded that the possible emergence of new strains/variants of HCoV is imminent.
1. Introduction
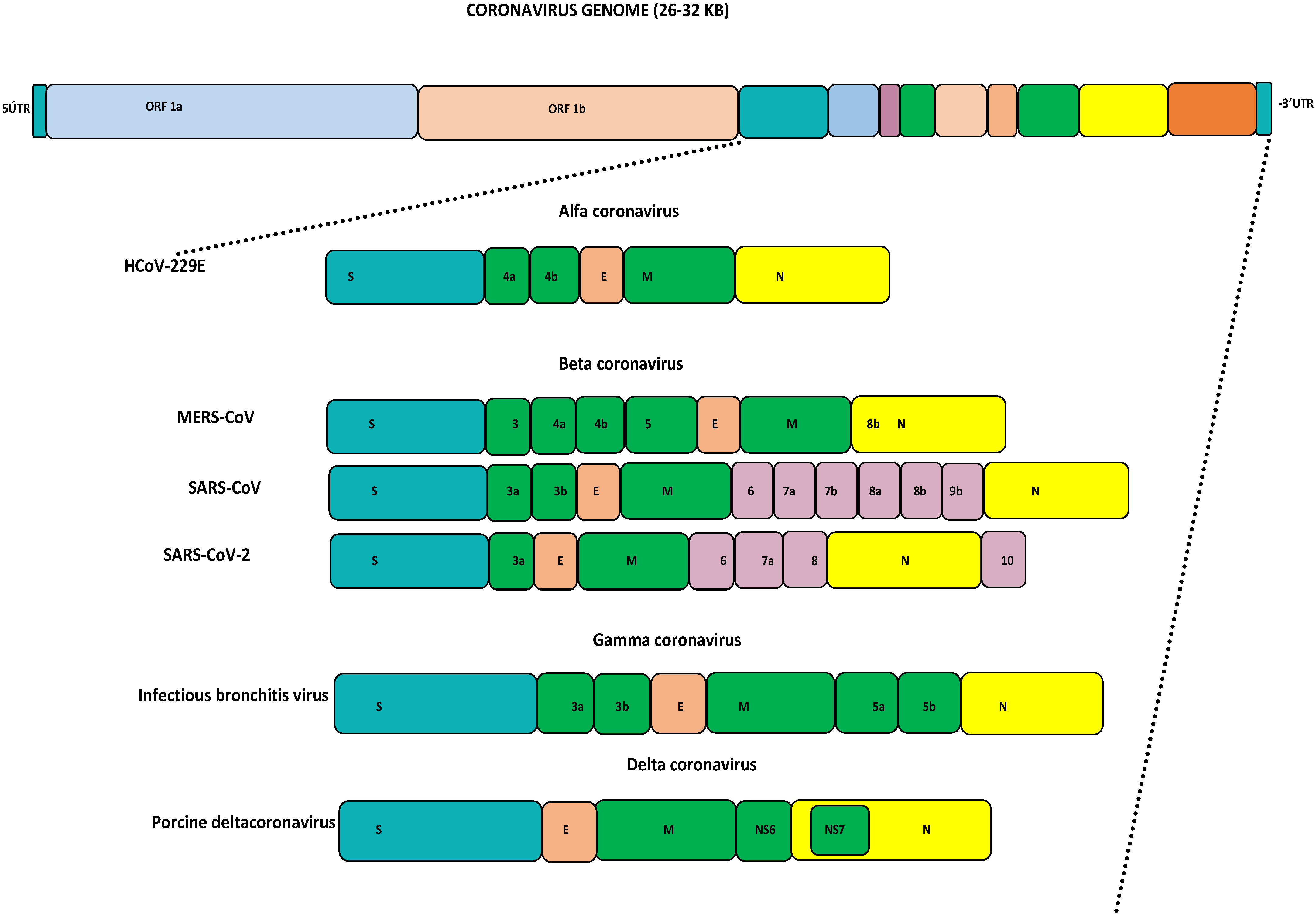
Coronaviruses (CoVs) fall under the Coronviridae family [1]. This family consists of ss +ve sense RNA viruses, which are separated based on their phylogeny into four genera: alpha-CoV, beta-CoV, gamma-CoV, and delta-CoV [2]. Generally, alpha- and beta-CoVs mainly include CoVs of mammalian origin, while the gamma- and delta-CoVs commonly include CoVs of avian origin [3,4,5]. Structurally, all CoVs have a similar organization of their genomes, being approximately 26–32 kb in size with varied G+C contents of 32% to 43%. The major part of the viral genome contains two open reading frames (ORFs) encoding 16 non-structural proteins. The remaining portion contains the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins, which are encoded by other ORFs, as seen in Figure 1 [2,6]. Based on the current reports, human coronaviruses (HCoVs) are well known to transmit easily to other species. Seven HCoVs have emerged so far, causing serious illnesses ranging from mild self-limiting symptoms like the common cold to life-threatening conditions like severe respiratory syndromes [6]. For years, HCoVs such as HKU-1, NL63, 229E, and OC43 were not considered serious human pathogens as they only caused mild illnesses. The first identification of HCoV-HKU1 was completed in 2005 from a patient with pneumonia symptoms in China [7]. HCoV-NL63 was detected for the first time in 2004 in a Dutch child [8]. HCoV-229E was identified in 1966 and finally isolated in 1967 [9]. In 1960, the isolation of HCoV-OC43 was completed from human tracheal explants. SARS-CoV-1 was identified from China in 2002 and designated as the first highly pathogenic HCoV [10,11]. The civet cat was identified as a primary host and due to human-to-human transmission, this virus spread to 26 countries, resulting in a global epidemic that resulted in 8098 cases and 774 deaths by July 2003. This virus disappeared within eighteen months, and no more cases were reported after January 2004 [6,11]. The second pathogenic MERS-CoV was identified from a 60-year-old patient in Jeddah, the Kingdom of Saudi Arabia, in 2012, and to date this virus has been reported in 27 countries [12]. MERS-CoV causes zoonotic respiratory disease and is currently known as a serious pathogen for both humans and camels [13]. Bats and dromedary camels have been identified as primary source for human infection [5,14,15,16,17]. MERS-CoV caused an outbreak in the Arabian Peninsula, African countries, and South Korea, and resulted in more than 2605 cases with 937 deaths [18,19,20,21,22]. The genomic alterations and favorable conditions in a specific location may lead to the re-emergence of pathogenic HCoVs and human infections [4,6]. In late December 2019, the third human pathogenic SARS-CoV-2 emerged because of favorable climatic conditions in Wuhan city, China, that resulted in a global pandemic [23]. As of today, SARS-CoV-2 has spread into 231 countries, with 704,753,890 confirmed cases and 7,010,681 deaths, as well as 675,619,811 recoveries (https://www.worldometers.info/coronavirus/—last accessed on 17 April 2024, 01:00 GMT). All HCoVs are zoonotic viruses that circulate among different animal species before infecting humans. Several pieces of evidence support the theory that most of the HCoVs originated in bats, where they became well adapted [6]. Interspecies transmission of HCoVs and animal coronaviruses continues to increase their genetic diversity and evolutionary rate, resulting in the emergence of novel coronaviruses with diverse characteristics and extended hosts [24,25]. The family Coronaviridae undergoes both homologous and non-homologous recombination, genetic mutation, insertion, and deletion. Among HCoVs, the pattern of recombination was observed for the first time in SARS-CoV-1 in 2004. Additionally in 2006 the recombination was identified in HCoV-HKU1 and HCoV-NL63, followed by the recombination reports in 2011 and 2014 for HCoV-OC43 and MERS-CoV, as well as recently in SARS-CoV-2 in 2020 [26]. The HCoVs-Spike (S) gene has the highest mutation rate site, insertion, and deletion, and has been the most used target for vaccine/antiviral development. The S gene has been identified as being key for host cell attachment and facilitating host cell entry [27]. In MERS-CoV, the S gene attaches DPP4 and CD26 for host cell entry through the receptor-binding domain (RBD), which mediates the interaction, while ACE2 has been identified as the S gene receptor for SARS-CoV-1 and SARS-CoV-2 [28,29,30,31]. In a recent recombination study, co-infections with different MERS-CoV lineages have been reported [22]. Based on recently analyzed samples from Uganda, it was observed that there were many amino acid substitutions in the RBD and recombination in the S1 sub-unit of the S protein gene, which may have facilitated the emergence of MERS-CoV and caused human disease [29,32]. Several significant variations have been identified in the non-structural and structural genes of MERS-CoV among humans and camels, which have significantly impacted virus transmission, disease spread, and the evolution of the virus in various geographical locations, resulting in the emergence of recombinant viruses [6,22,33,34,35,36].
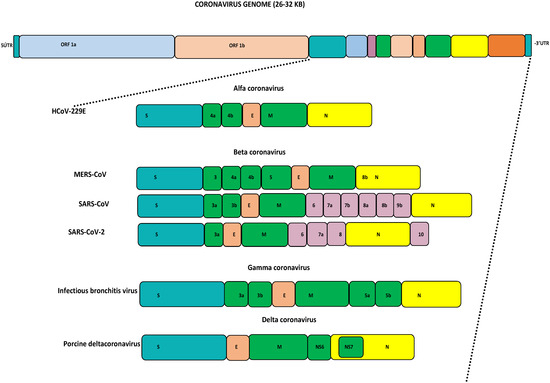
Figure 1.
Genome organization of coronaviruses: HCoV: human coronavirus; MERS-CoV: Middle East respiratory syndrome coronavirus; SARS-CoV: severe acute respiratory syndrome coronavirus; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Currently, many reports have been published about recombination in SARS-CoV-2, which has resulted in the emergence of variants of concern (VOCs) and variants of interest (VOIs) (https://www.who.int/activities/tracking-SARS-CoV-2-variants, accessed on 17 April 2024, https://www.ecdc.europa.eu/en/COVID-19/variants-concern, accessed on 17 April 2024) [2,37,38,39,40,41]. In March 2020, Li et al. reported that the whole RBD of the S gene was introduced through recombination with coronaviruses from pangolins [26], and this was further validated by Zhu et al. in December 2020 [42]. However, based on a recent study using sliding-window bootstrapping, SARS-CoV-2 was defined as a mosaic genome with three bat SCoV2rCs reported from Yunnan, China [4,41,43]. The S gene of SARS-CoV-2 has also been found to have many variants that can affect the virus transmissibility, infectivity, and vaccination efficacy. These variants were classified as variants of interest (VOIs), such as the Lambda and Mu variants, while others were classified as variants of concern (VOCs), such as the Alpha, Beta, Gamma, Delta, and Omicron variants [30,44]. It is essential to consider that the classifications of variants can be changed according to recent updates from global studies [40,44]. Therefore, identifying the genetic diversity of the HCoVs-S gene is essential to understand how evolution can affect the viral pathogenesis and transmission of HCoVs with altered properties to new hosts. Based on the recent status, we performed this work to elucidate the genomic diversity and recombination pattern of the selected HCoVs-S gene. The S gene plays an important role in virus–host cell attachment and entry into infected cells. The S gene has been widely used for vaccine/antiviral development against HCoVs. The main goals of this study were to perform genetic diversity, phylogeny, and recombination pattern analyses of SARS-CoV-2 and MERS-CoV along with other HCoVs. Additionally, we extended our objectives to identify the possible emergence of new variants/strains of HCoV in the near future. This detailed information about the genetic diversity, phylogeny, and recombination pattern of the selected HCoVs-S gene could be extensively used by the scientific community as well as disease control authorities to protect the global human population by designing effective vaccines and antivirals for long-term broad-spectrum protection from coronavirus infections.
2. Materials and Methods
2.1. Retrieval of Viral Genome Sequences
The selected HCoVs-S protein gene sequences from different hosts and locations were retrieved from GenBank, NCBI-PubMed. We included the highest number of S protein gene sequences from SARS-CoV-1, SARS-CoV-2, and MERS-CoV, followed by other HCoVs. A total of 19 sequences of SARS-CoV-2; 19 sequences of SARS-CoV-1; 26 sequences of MERS-CoV; 16 sequences of HCoV-NL63, HCoV-229E, and HCoV-OC43, each; and 8 sequences of HCoV-HKU-1 from different hosts and geographic locations were collected. For the genetic analysis of MERS-CoV with SARS-CoV-2, we selected mostly from the Arabian Peninsula, while for the analysis of SARS-CoV-2 with other HCoVs, we selected and divided the sequences based on their identification and global distribution from multiple hosts and locations. As it has been reported that the MERS-CoV is more prevalent in the Arabian Peninsula than other locations, we focused on the collection and division of these sequences from the Arabian Peninsula region. The selection of viral sequences was made via filtration based on their geographical distributions and spread, as well as their frequency of prevalence globally. Our objective was to collect and analyze the S protein gene sequences of the most prevalent viruses and their number of laboratory-confirmed cases, as well as deaths reported globally. We used SARS-CoV-2 (Accession# MW837148) and MERS-CoV (Accession# NC_019843) as reference sequences to perform all of the analyses because MERS-CoV and SARS-CoV-2 have shown high sequence identity together.
2.2. Genome Analyses of HCoVs
The S protein gene sequences of the selected HCoVs (nucleotide—[NT] and amino acid—[AA]) were aligned by using the CLUSTALW and BioEdit (v.7.2.5) online software tools. The sequence similarity and identity matrices were determined based on the MERS-CoV (Accession# NC_019843) and SARS-CoV-2 (Accession# MW837148) genomes as reference sequences with other HCoVs collected from various parts of the world. To identify the phylogenetic relationships of MERS-CoV and SARS-CoV-2 sequences with other HCoV genomes, the MEGA11 software program was used and a phylogeny dendrogram was generated [45]. Initially, the phylogenetic analysis was performed by using the genome sequences of all HCoVs together. Then, we performed another phylogenetic analysis by using MERS-CoV and HCoVs without the SARS-CoV-2 genome, as well as the nucleotide sequences of SARS-CoV-2 sequences with all selected HCoVs without MERS-CoV. We also performed a phylogenetic analysis using only MESRS-CoV with SARS-CoV-2 and SARS-CoV-1 genome sequences.
2.3. Recombination Pattern Analyses among HCoVs
The selected S protein gene sequences of HCoVs were used to analyze the recombination pattern and elucidate possible recombinants among the minor and major parents by using the recombination detection program (RDP v. 5 program) [46]. The SARS-CoV-2 (Accession# MW837148) S protein gene sequence was used as a reference sequence. The generated FASTA files were exported to the RDP v. 5 program for analysis, and the recombinants with recombination breakpoints and hot spots, including their start and end points in the viral genome, were identified using the software and putative recombination events were identified in the S protein gene sequences of SARS-CoV-2 (MW837148). The putative recombination events were identified with a cut-off p-value (≤0.05).
3. Results
3.1. Genome Analyses of HCoVs
The SARS-CoV-2-S protein gene (Accession# MW837148) sequence was used as a reference sequence to analyze the sequence identity based on nucleotide (NT) and amino acid (AA) sequences with selected HCoV sequences. The highest sequence identities (99.9%—NT and 99.8%—AA) were identified with multiple SARS-CoV-2 isolates, while the lowest identities (32.3%—NT and 19.8%—AA) were identified with an isolate of HCoV-229E-USA (Accession# KY369914). The sequences (NT/AA) from SARS-CoV-2 showed higher sequence identities when compared to MERS-CoV and others such as HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1 collected from various locations during different periods (Table 1).

Table 1.
Sequence identity matrix for the SARS-CoV-2-S protein gene (MW837148) with selected HCoVs.
Additionally, we also analyzed the percent sequence identity matrix based on the NT/AA sequence of the MERS-CoV-S protein gene (Accession# NC_019843) as a reference sequence with only the selected HCoVs. The percent sequence identity matrix ranged from 99.7 to 99.3% for the NTs and from 99.9 to 99.4% for the AAs with the selected MERS-CoV, while the NT sequences of the remaining HCoVs, along with HCoV-OC43, HCoV-NL63, HCoV-HKU1, and HCoV-229E, ranged from 46.2 to 34.6%, and the AA sequences ranged from 28.9 to 18.1% with MERS-CoV (Table 2).

Table 2.
Sequence identity matrix for the MERS-CoV-S protein gene (NC_019843) with selected HCoVs excluding SARS-CoV 1 and SARS-CoV-2.
An analysis was also performed by using the SARS-CoV-2-S protein gene sequence (NT/AA) as a reference sequence with selected HCoVs. The highest identity (99.9%—NT) was observed, while the lowest was 98.5%, and the AA identity varied from 99.9% to 97% with SARS-CoV-2 from various regions. SARS-CoV-1 showed the highest similarity (73.0% NT), the lowest was 72.9%, and the AA identity ranged from 75.8 to 75.4% with SARS-CoV-2. The other HCoVs, such as HCoV-OC43, HCoV-NL63, HCoV-HKU1, and HCoV-229E, showed a variable range of identity for both the NTs and AAs with SARS-CoV-2 (Table 3).

Table 3.
Sequence identity matrix for the SARS-CoV-2-S protein gene (MW837148) with selected HCoVs excluding MERS-CoV.
Another analysis was performed by only using the NT/AA sequence of the MERS-CoV-S protein gene as a reference sequence along with the selected SARS-CoV-1 and SARS-CoV-2-S protein genes. The highest NT identity ranged from 99.7 to 99.2%, and the AA identity ranged from 99.9 to 99.7% for MERS-CoV. The NT sequence identity for SARS-CoV-2 ranged from 44.2 to 44.0%, while the AA identity was 26.7–26.6%. SARS-CoV-1 showed a variable range of sequence identity, which ranged from 44.9 to 44.8% for the NTs and from 26.6 to 26.4% for the MERS-CoV isolates (Table 4).

Table 4.
Sequence identity matrix for the MERS-CoV-S protein gene (NC_019843) with selected SARS-CoV 1 and SARS-CoV-2.
3.2. Phylogenetic Analyses
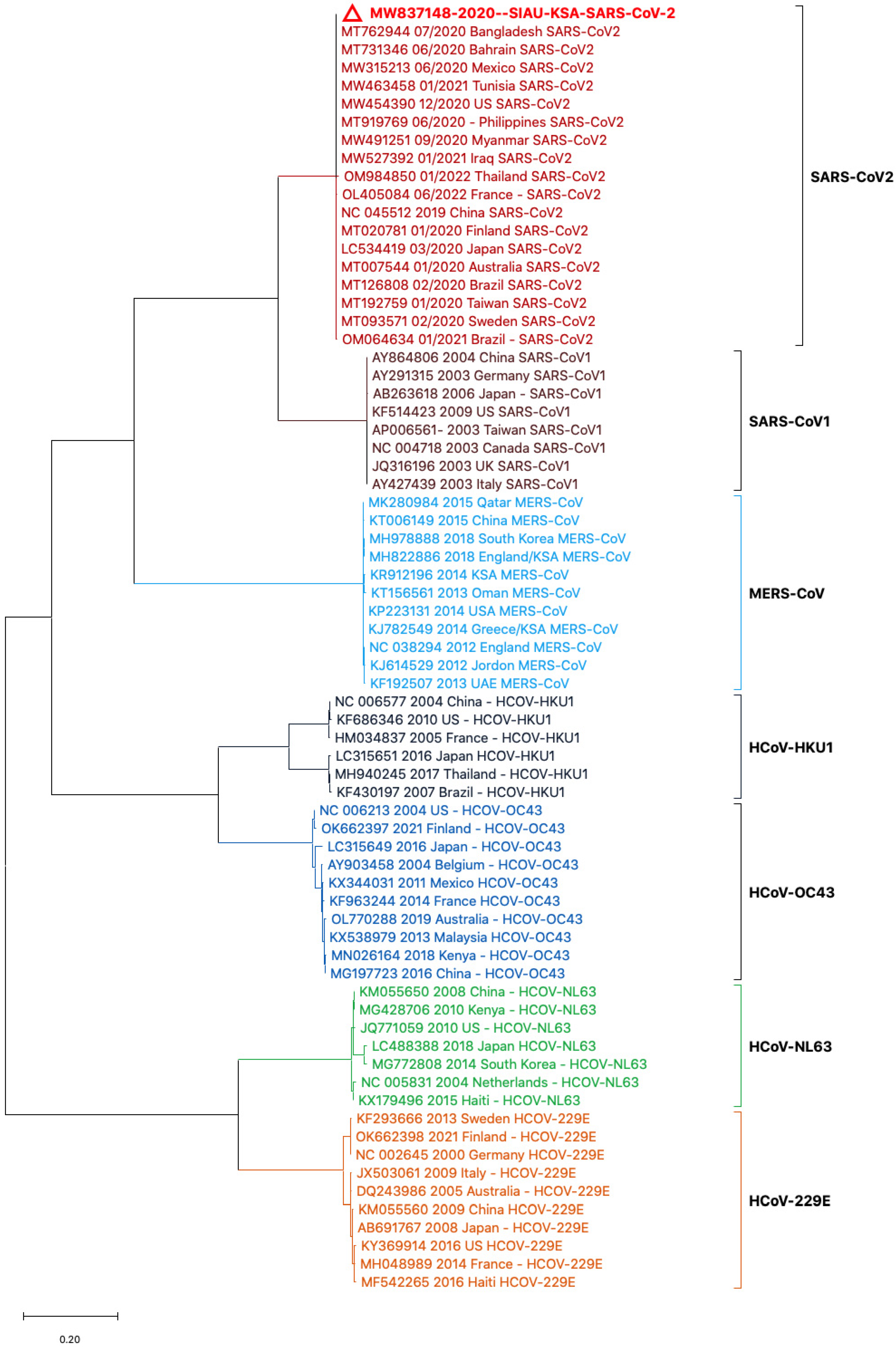
Phylogenetic tree analyses were performed using the nucleotide (NT) sequences of the S protein gene sequences with selected HCoVs. The sequence of SARS-CoV-2 (MW837148) was used as a reference sequence to perform a phylogenetic tree analysis with other viral sequences. The phylogeny was generated by using NT sequences separated into different clusters. All of the HCoVs clustered separately and formed closed clusters with their similar isolates. SARS-CoV-2 (MW837148) only formed a closed cluster with the SARS-CoV-2 isolates selected from different locations (Figure 2).
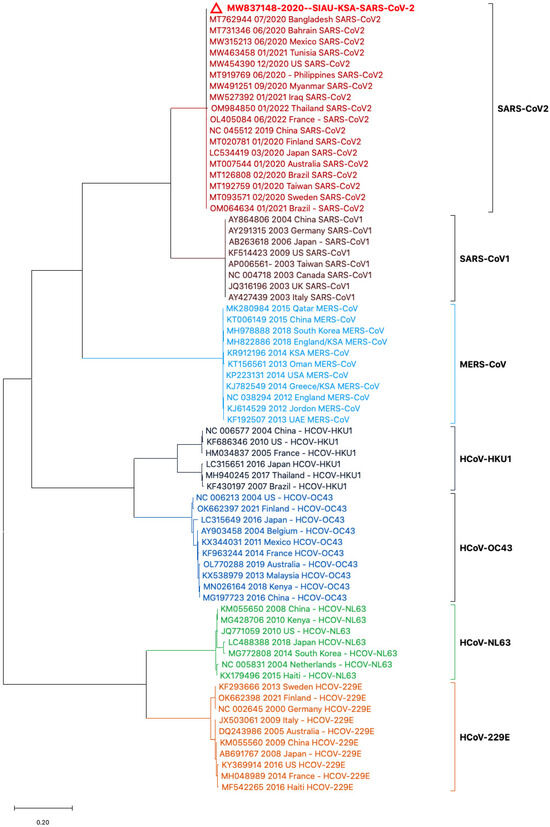
Figure 2.
Phylogeny based on the nucleotide (NT) sequences of the S protein gene of selected HCoVs. The red triangle denotes the SARS-CoV-2 genome sequences from SIAU-KSA.
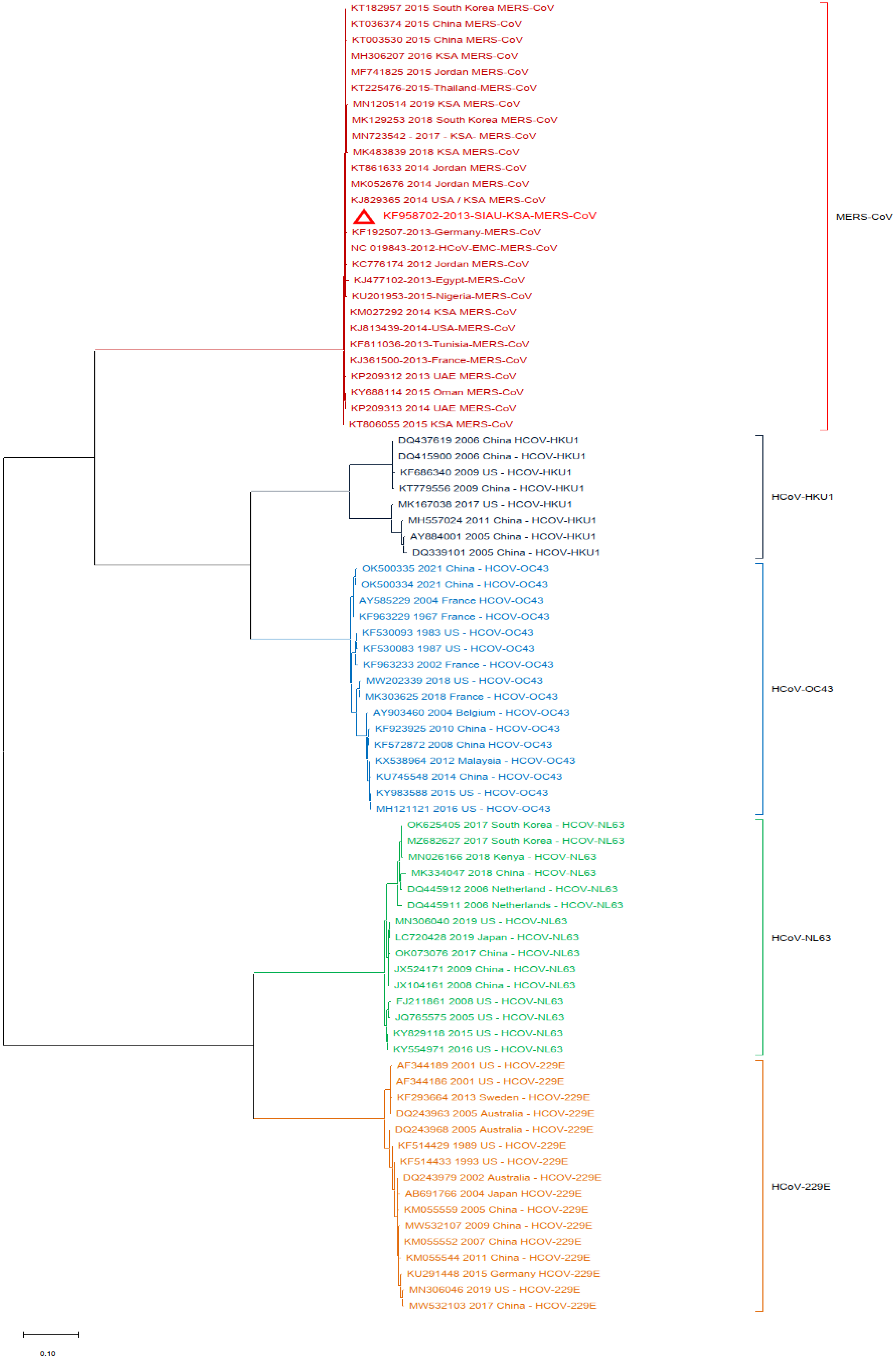
Additionally, a phylogenetic analysis of the MERS-CoV-S protein gene with other HCoVs was performed by using MERS-CoV (KF958702) as the reference sequence. The results showed that multiple clusters were formed with the selected HCoVs. All MERS-CoV samples formed a closed cluster with the selected MERS-CoV isolates. Interestingly, similar clustering was observed with the other HCoVs and their respective virus isolates (Figure 3).
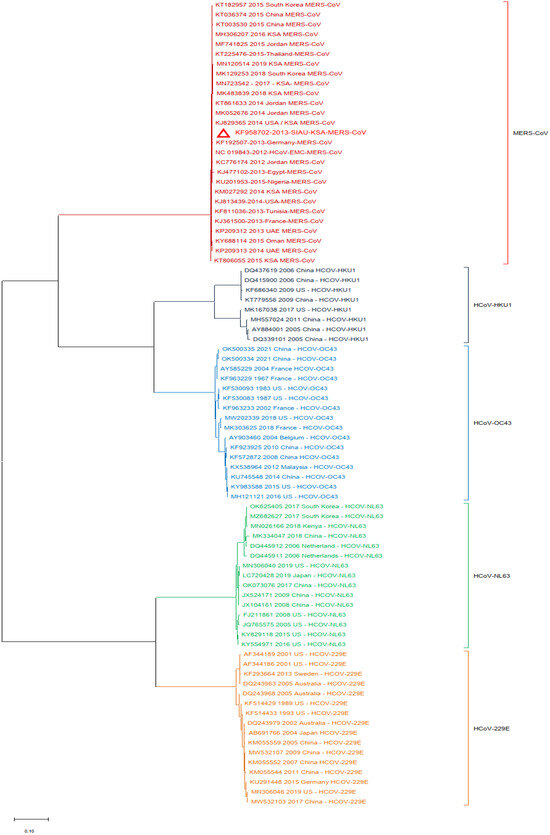
Figure 3.
Phylogeny constructed by using the nucleotide (NT) sequences of the MERS-CoV-S protein gene with selected HCoVs and without SARS-CoV-1 and SARS-CoV-2. The red triangle denotes the MERS-CoV genome sequences from SIAU-KSA.
In another analysis, a similar phylogenetic relationship analysis was performed by excluding the MERS-CoV isolates. SARS-CoV-2 (MW837148) was used as a reference and analyzed with the SARS-CoVs and the selected HCoVs. SARS-CoV-2 (MW837148) clustered with only the selected SARS-CoV-2 isolates; interestingly, none of the SARS-CoV-1 isolates clustered with any SARS-CoV-2. SARS-CoV-1 formed a separate cluster, and similarly, all of the selected HCoVs clustered separately (Figure 4).
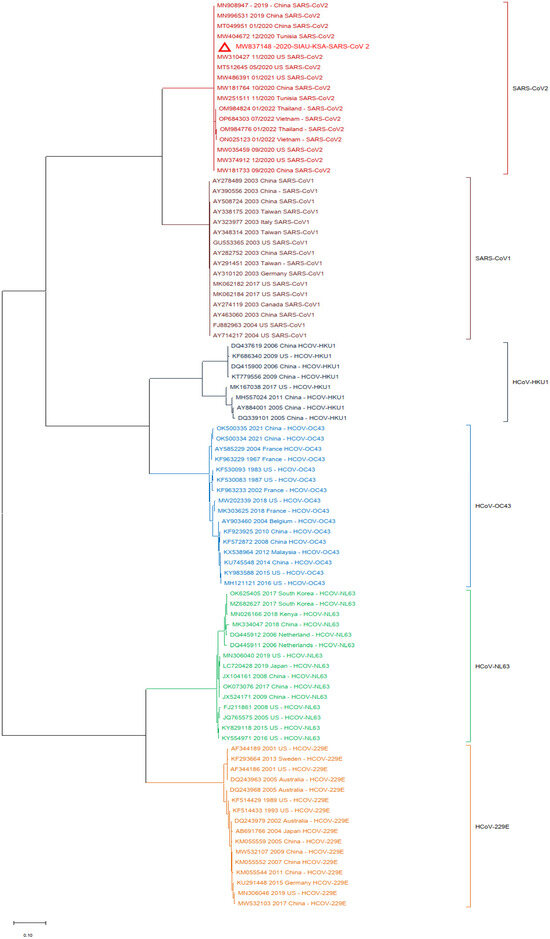
Figure 4.
Phylogeny according to the nucleotide (NT) sequences of the SARS-CoV-2-S protein gene with selected HCoVs without MERS-CoV. The red triangle denotes the SARS-CoV-2 genome sequences from SIAU-KSA.
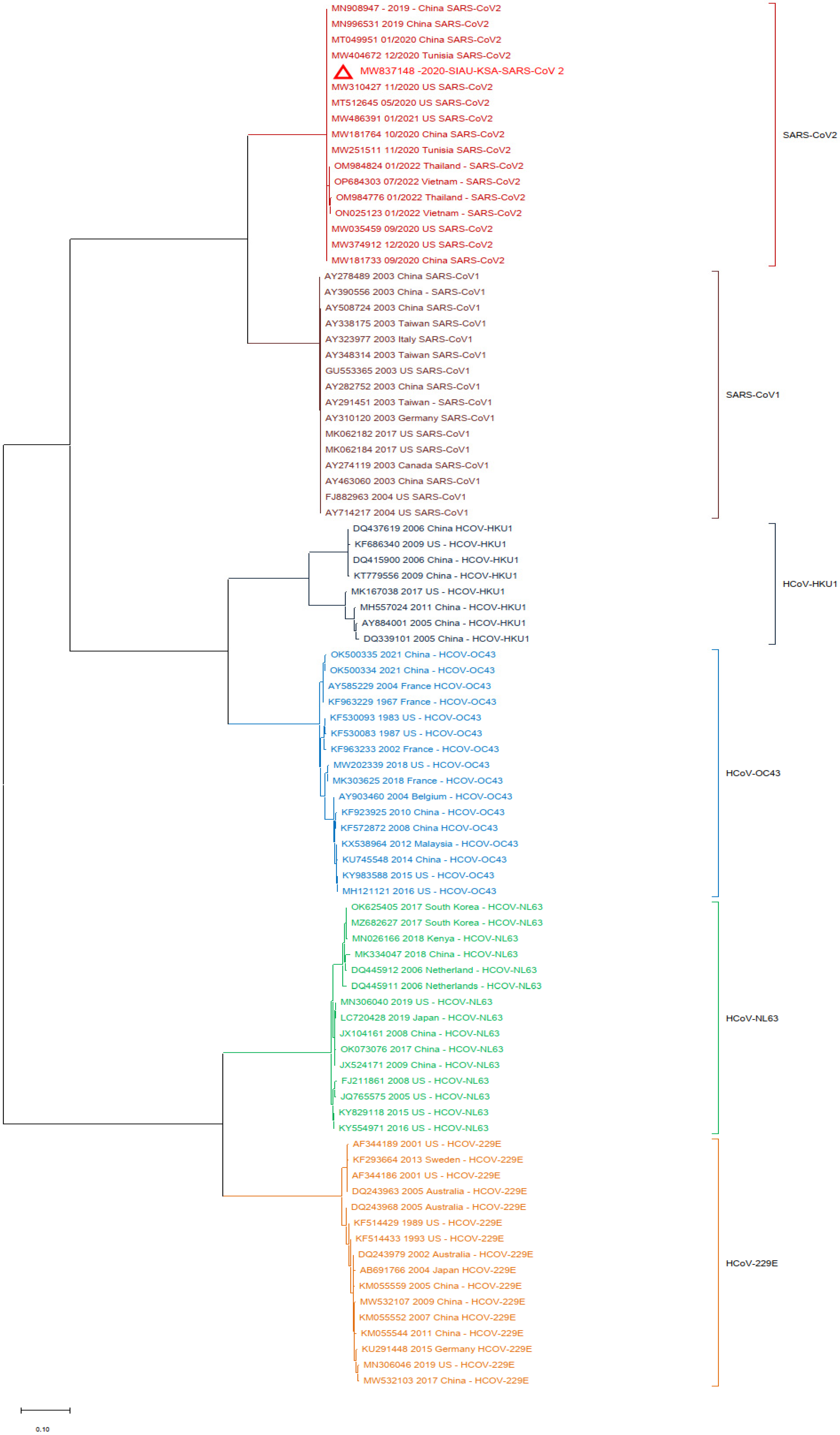
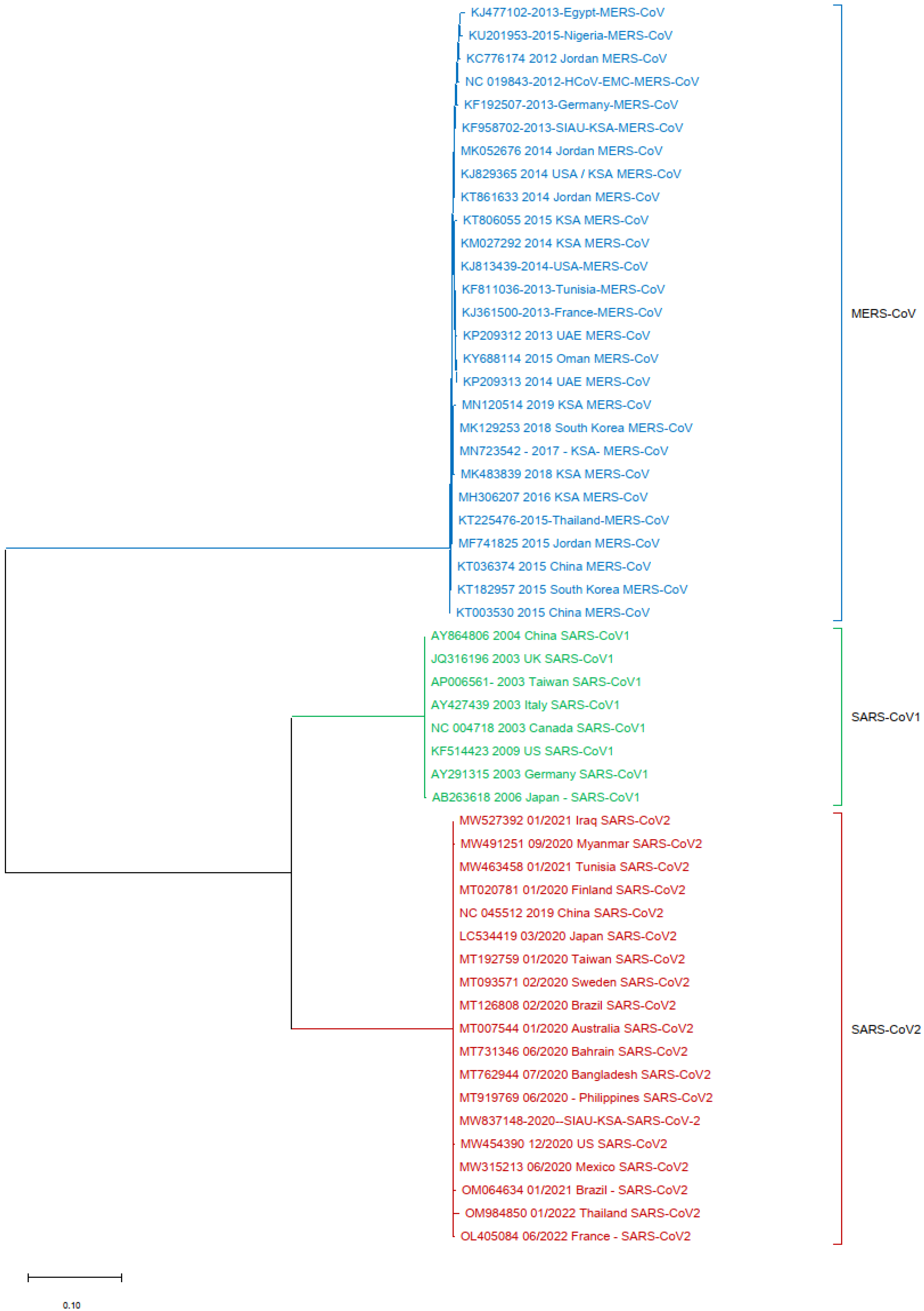
Finally, one more phylogenetic tree analysis was performed by using the MERS-CoV (KF958702) S protein gene sequence as a reference sequence with only SARS-CoV-1 and SARS-CoV-2. It was observed that all of the MERS-CoV samples formed a separate cluster. Interestingly, both SARS-CoV-1 and SARS-CoV-2 clustered separately in this phylogenetic tree analysis (Figure 5).
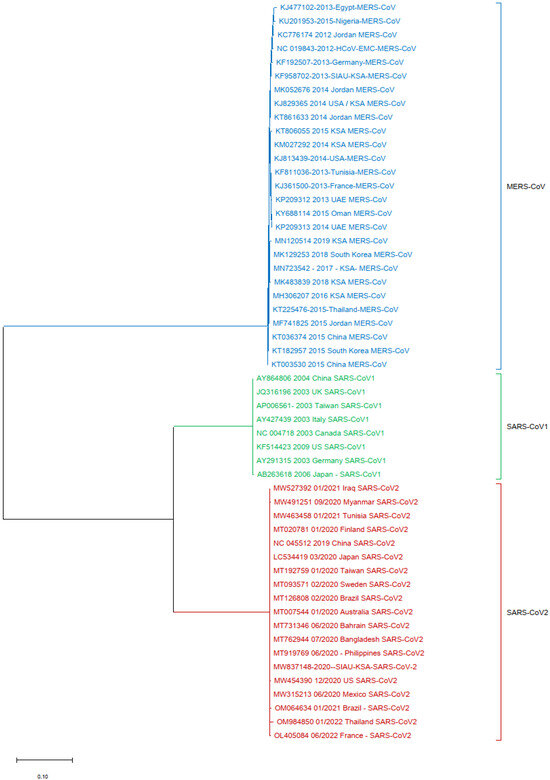
Figure 5.
Phylogeny tree based only on the nucleotide (NT) sequences of the S protein gene of MERS-CoV with SARS-CoV-1 and SARS-CoV-2.
3.3. Recombination Analyses
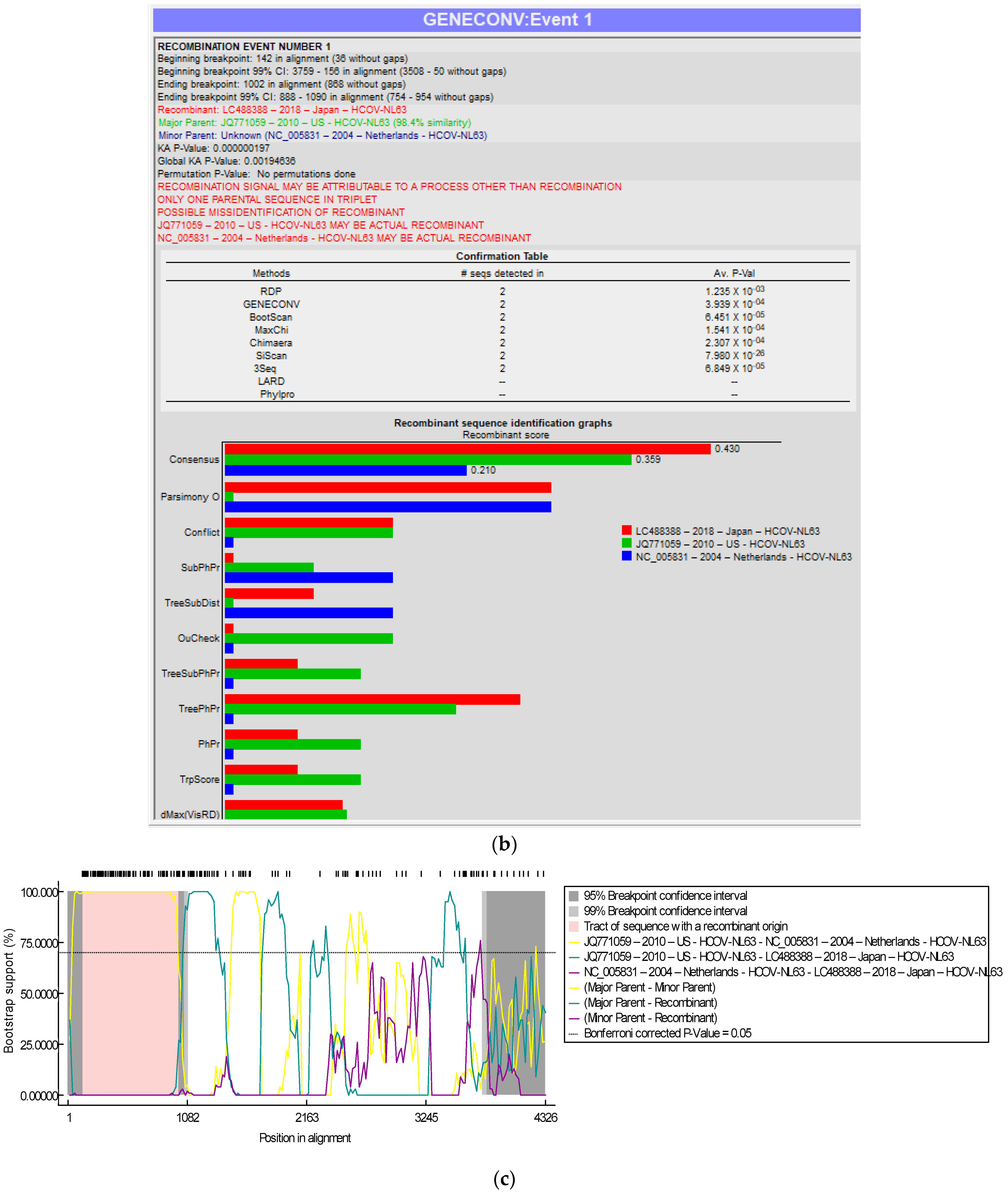
The genome of the SARS-CoV-2-S protein gene sequence (MW837148) was used as a reference sequence to elucidate the pattern of recombination with selected HCoVs, including MERS-CoV. Putatively, two recombination breakpoints were generated by using the RDP v. 5 program (Figure 6a). Respectively, for all of the sequences analyzed, HCoV-NL63-USA (Accession# JQ771059) was identified as a probable major parent with 98.4% similarity, and HCoV-NL63-Netherlands (Accession# NC_005831) was identified as a minor parent. HCoV-NL63-Japan (Accession# LC488388) was identified as a recombinant in GENCONV event number 1 (Figure 6b). The recombination breakpoints start at nucleotide position 142 in the alignment with a 95% confidence interval (CI) and end at the 1082 nucleotide position with a 99% CI and with a Bonferroni-corrected p-value of 0.05 (Figure 6c).

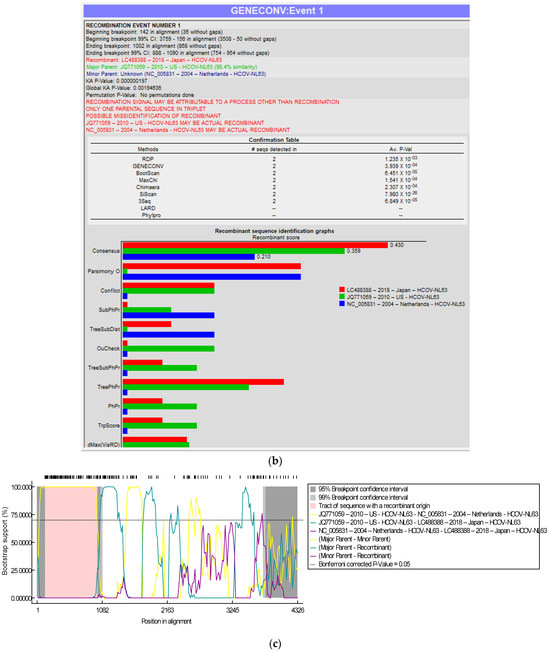
Figure 6.
(a). Recombination pattern and breakpoints of the SARS-CoV-2-S protein gene (MW837148) with selected HCoVs. (b). Recombination events of the SARS-CoV-2-S protein gene (MW837148) with selected HCoVs. (c). Recombination breakpoints of the SARS-CoV-2-S protein gene (MW837148) with selected HCoVs.
4. Discussion
HCoVs are serious pathogens associated with human and animal diseases, causing respiratory illnesses globally [3]. The monitoring of HCoV infections at a molecular level with an emphasis on the genome and phylogeny enables us to elucidate the emergence of new variants/strains that may infect and cause diseases to new hosts, including animals and humans at different geographic locations. Seasonal HCoVs such as HCoV-NL63, -229E, -OC43, and -HKU1 cause only seasonal infections, while SARS-CoV-1, MERS-CoV, and SARS-CoV-2 are known to cause respiratory illnesses throughout the year [2,22,47,48]. Genetic changes in the viral genomes lead to disruption of the virus-and-host cell interactions, as well as changes in virus reproduction, virulence, pathogenicity, gene expression, and ultimately determine the outcome of the severe infection [47]. Due to the favorable climatic conditions, frequent recombination and mutation occurs in the coronavirus genome and new virus variants/strains and isolates emerge, which results in interspecies transmission and infection. Based on globally published reports, there are many hosts that have been identified as coronavirus reservoirs, such as bats, palm civets, raccoon dogs, and camels [49,50]. They use different receptors such as ACE-2, DPP4, and APN for the host cell attachment and entry to the host cell [51]. SARS-CoV-1 emerged in 2002–2003 and caused epidemics. This coronaviral genome had seven NT and six AA variations in its S gene that resulted in a low pathogenicity identified in palm civets and raccoon dogs. In 2003, a global pandemic was caused by a highly pathogenic SARS-CoV-1 disease. Based on sequence analyses, fourteen single-nucleotide variations caused changes into eleven AA changes, which led to its high pathogenicity. Another six nucleotide variations resulted in four amino acid variations and caused a global epidemic in 2003. Just after the first epidemic, in 2004, due to interspecies transmission and viral adaptation, four new cases of human infection were reported in China [52].
There are many published reports available describing the comparative study of genetic determinants with high- and low-virulence properties and mortality rates caused by HCoVs like SARS-CoV-1, SARS-CoV-2, and MERS-CoV in comparison to other HCoVs like HCoV-NL63, -229E, -OC43, and -HKU1 [53]. Global viromics studies of more than 3000 viral genomes collected from both humans and animals (SARS-CoV-1, MERS-CoV, and SARS-CoV-2) confirmed variations in four locations situated in the nucleoprotein gene (N) and S protein gene as compared to HCoV-NL63, -229E, -OC43, and -HKU1 [47,53]. The WHO has kept MERS-CoV on the priority list for performing detailed studies because of its continuous infection and spread to humans and camels in different locations [22]. Recently, the whole genome of MERS-CoV isolated from humans and camels was used for a global analysis of genetic diversity, and the results showed that three clades (A, B, and C) were generated in the software, and it was concluded that MERS-CoV and its new variants are circulating in camels [22]. Additionally, one more study from Ethiopia reported that the MERS-CoV infecting Ethiopian camels phylogenetically belongs to clade C2 and forms closed clusters with East African strains [36]. Due to their continuous exposure to infected animals, animal handlers can facilitate the transmission and introduction of moderately to highly virulent HCoVs in a specific geographic location. High mutation rates result in efficient virus transmission, severe infection, and easy host adaptation, and can cause global epidemics and pandemics. A similar phenomenon and favorable conditions were also observed in the city of Wuhan, China, which resulted in the emergence of SARS-CoV-2. Changes in the nucleotides and amino acids favor the emergence of new isolates, strains, mutations, or recombinant viruses, as has been observed and reported earlier in many published papers from Saudi Arabia for MERS-CoV and South Korean mutants, as well as other HCoVs such as SARS-CoV-1 and SARS-CoV-2 from different geographic locations [20,32,54,55,56,57,58]. The S protein gene mutations in other HCoVs have favored the high rate of interspecies transmission towards human receptors [59,60,61]. In the present study, the genome sequences of MERS-CoV showed less identity with SARS-CoV-1 but higher genome similarity with SARS-CoV-2. In the phylogenetic tree relationship analyses, it was observed that most of the virus isolates formed a closed cluster with their similar isolates like MERS-CoV, SARS-CoV-1, and SARS-CoV-2, as well as other HCoVs. Our data and findings are supported by earlier published reports about genetic diversity, phylogenetic analyses, and recombination analyses based on the MERS-CoV-S gene with selected HCoVs from both humans and camels [20,22]. Based on the phylogeny, it has been observed that even after the continuous import of infected camels from African countries, the MERS-CoV-African isolate did not establish itself in the KSA as both isolates formed separate clusters, and the Arabian MERS-CoV isolate is still circulating in camels [21,22,34,36].
Recombination is very crucial and play an important role in the emergence of a recombinant virus, new virus isolates, and variants/strains with novel properties during the life cycle of an HCoV with other co-circulating viruses in different hosts and locations. The published reports suggest that coronaviruses undergo rapid and frequent recombination, which leads to the emergence of new virus strains or variants with altered virulence and serious effects on cytokine storms [2]. The genomic alterations and gene flow of both humans and pathogens significantly favor the spread of pathogenic organisms from one to another location, as well as interspecies and intraspecies transmission [62,63]. It has been observed that the rate of mutations in CoVs, including HCoVs, is high in comparison to other ssRNA and DNA viruses [64,65]. In 2015, an outbreak of MERS-CoV was reported due to the emergence of a recombinant virus isolate with the co-circulation of HCoVs and MERS-CoV in Saudi Arabia. The co-circulation of HCoVs favors genomic recombination with MERS-CoV, which infects both humans and camels, resulting in the emergence of a novel recombinant virus that was lethal to humans [4,22,34,58].
The recombination patterns, breakpoints, and events provide very useful information during viral outbreaks caused by one or more viruses or any other recombinant as well as variant viruses. The identification of recombination events leads to identifying new variants or recombinants that have other properties, such as altered transmission patterns, virus replication, and infectivity, as well as epidemiological fitness to sustain the virus isolates or variants in different environmental conditions. Recombination events may take place during the evolution and transmission of HCoVs. There are various published reports (in silico and in vivo) about recombination events in SARS-CoV-2 [66,67]. Recently, in one study, a Recombination Inference using Phylogenetic Patterns (RIPPLES) program was developed to detect recombination events in large mutation-annotated tree (MAT) files. This program breaks the sequences into two distinct fragments based on the recombination and mutation in the sequence, and two breakpoints are separated. By using this program, a total of six hundred and six events of recombination were identified, and it was concluded that SARS-CoV-2 genomes exhibit recombination in the S gene [68]. There are several reports about the recombination between Alpha and Delta, Beta and Delta, and Omicron BA.1 and BA.1–BA.2 recombinants [41]. Additionally, in another report published by Preska Steinberg in 2023, the ORF1ab and S genes showed a high frequency of recombination when analyzed in 191 SARS-CoV-2 and related genomes [69]. The positive selection site of the MERS-CoV-S gene in camels has been found to favor host jumping and human infection. As it has been observed in MERS-CoV, the frequent recombination breakpoint occurs in the ORF1b/S gene, while in SARS-CoV-2, the S gene shows major variations and recombination, which has led to the emergence of new variants globally [22,34]. The recombination pattern of MERS-CoV with other selected HCoVs indicates co-infections with different MERS-CoV variants in camels, while the SARS-CoV-2 recombination pattern indicates that HCoV-NL63-USA is the probable major parent and HCoV-NL63-Netherlands is the minor parent. HCoV-NL63-Japan was identified as a recombinant in GENCONV event number 1 [21,22,34].
In this study, we have discussed the S protein gene based on genetic diversity, phylogenetic relationships, and recombination patterns and breakpoints, which will enable us to identify the emergence and spread of new recombinant viruses, variants, and isolates with an extended host range and novel properties. These variants or strains may infect multiple hosts in new geographical regions globally. These findings suggest that more elaborate genetic analysis research is further required focusing on other geographical regions as well as the full genome of each HCoV. In the future, for the detailed study of HCoVs, genomic analyses are required to understand the emergence and spread of new variants/isolates/strains of HCoV in dromedaries, pangolins, bats, humans, and other unidentified alternative hosts. There are still many knowledge gaps left requiring detailed information on both MERS-CoV and SARS-CoV-2, and comprehensive genotypic studies and follow-up analyses are needed, which will provide a clue as to whether asymptomatic MERS-CoV and SARS-CoV-2 infections in camels and humans, as well as in other hosts, are currently developing locally and globally. The generated data will aid in understanding how genetic diversity, selection, and recombination play important roles in modifying and molding the genetic structure of a specific virus that may lead to the emergence of new pandemics or epidemics.
5. Conclusions
This study was based on the genetic diversity and recombination pattern analysis of the MERS-CoV and SARS-CoV-2-S protein genes compared with other HCoVs. Based on the data generated, it is concluded that these S protein genes have significant diversity and a different recombination pattern when compared to other HCoVs. Continuous monitoring and detailed genetic diversity and phylogenetic analysis studies are required to understand this virus’s evolution, interspecies transmission, host adaptation, virulence, disease severity, and genetic relationship with other HCoVs. This study will be highly helpful for combatting any further outbreaks. Our analyses of recombination patterns will be beneficial for understanding the emergence of any new recombinant viruses or variants. These genetic diversity analyses and the identification of any mutation in the viral genome can be used to design and develop an effective vaccine for broad-spectrum protection against HCoVs globally.
Author Contributions
Conceptualization and visualization, S.S.S. and E.I.A.; methodology, S.S.S. and F.A.; software, F.A., S.S.S. and E.I.A.; investigation, S.S.S. and E.I.A.; data curation, S.S.S. and F.A.; Figure 1, viral genome designing: A.M.H.; writing—original draft preparation, S.S.S. and F.A.; critical review and editing, S.S.S., E.I.A., F.A., L.H.B., A.M.H. and A.M.T.; funding acquisition, S.S.S.; formal analysis, S.S.S., F.A. and E.I.A.; resources: E.I.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Institutional Fund Projects under grant number IFPIP: 1700-141-1443.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors gratefully acknowledge the technical and financial support provided by the Ministry of Education and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Kirtipal, N.; Bharadwaj, S.; Kang, S.G. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. 2020, 85, 104502. [Google Scholar] [CrossRef] [PubMed]
- Cheng, V.C.; Lau, S.K.; Woo, P.C.; Yuen, K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007, 20, 660–694. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar] [CrossRef] [PubMed]
- Bolles, M.; Donaldson, E.; Baric, R. SARS-CoV and emergent coronaviruses: Viral determinants of interspecies transmission. Curr. Opin. Virol. 2011, 1, 624–634. [Google Scholar] [CrossRef]
- Lau, S.K.; Woo, P.C.; Yip, C.C.; Tse, H.; Tsoi, H.W.; Cheng, V.C.; Lee, P.; Tang, B.S.; Cheung, C.H.; Lee, R.A.; et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 2006, 44, 2063–2071. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Rasool, S.; Fielding, B.C. Understanding Human Coronavirus HCoV-NL63. Open Virol. J. 2010, 4, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.X.; Ng, Y.L.; Tam, J.P.; Liu, D.X. Human Coronaviruses: A Review of Virus-Host Interactions. Diseases 2016, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Che, X.Y.; Di, B.; Zhao, G.P.; Wang, Y.D.; Qiu, L.W.; Hao, W.; Wang, M.; Qin, P.Z.; Liu, Y.F.; Chan, K.H.; et al. A patient with asymptomatic severe acute respiratory syndrome (SARS) and antigenemia from the 2003–2004 community outbreak of SARS in Guangzhou, China. Clin. Infect. Dis. 2006, 43, e1–e5. [Google Scholar] [CrossRef] [PubMed]
- Tsang, K.W.; Ho, P.L.; Ooi, G.C.; Yee, W.K.; Wang, T.; Chan-Yeung, M.; Lam, W.K.; Seto, W.H.; Yam, L.Y.; Cheung, T.M.; et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003, 348, 1977–1985. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Peiris, M.; Perlman, S. Unresolved questions in the zoonotic transmission of MERS. Curr. Opin. Virol. 2022, 52, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; Hashem, A.M.; El-Kafrawy, S.A.; Sohrab, S.S.; Aburizaiza, A.S.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Jamjoom, G.A.; Madani, T.A. Detection of the Middle East respiratory syndrome coronavirus genome in an air sample originating from a camel barn owned by an infected patient. mBio 2014, 5, e01450-14. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef] [PubMed]
- Drosten, C.; Meyer, B.; Muller, M.A.; Corman, V.M.; Al-Masri, M.; Hossain, R.; Madani, H.; Sieberg, A.; Bosch, B.J.; Lattwein, E.; et al. Transmission of MERS-coronavirus in household contacts. N. Engl. J. Med. 2014, 371, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.S.; Azhar, E.I.; Kim, Y.J.; Memish, Z.A.; Oh, M.D.; Zumla, A. Middle East respiratory syndrome coronavirus: Risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect. Dis. 2018, 18, e217–e227. [Google Scholar] [CrossRef] [PubMed]
- Reusken, C.B.; Messadi, L.; Feyisa, A.; Ularamu, H.; Godeke, G.J.; Danmarwa, A.; Dawo, F.; Jemli, M.; Melaku, S.; Shamaki, D.; et al. Geographic distribution of MERS coronavirus among dromedary camels, Africa. Emerg. Infect. Dis. 2014, 20, 1370–1374. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.D.; Park, W.B.; Park, S.W.; Choe, P.G.; Bang, J.H.; Song, K.H.; Kim, E.S.; Kim, H.B.; Kim, N.J. Middle East respiratory syndrome: What we learned from the 2015 outbreak in the Republic of Korea. Korean J. Intern. Med. 2018, 33, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Sohrab, S.S.; Azhar, E.I. Genetic diversity of MERS-CoV spike protein gene in Saudi Arabia. J. Infect. Public. Health 2020, 13, 709–717. [Google Scholar] [CrossRef]
- Zhou, Z.; Hui, K.P.Y.; So, R.T.Y.; Lv, H.; Perera, R.; Chu, D.K.W.; Gelaye, E.; Oyas, H.; Njagi, O.; Abayneh, T.; et al. Phenotypic and genetic characterization of MERS coronaviruses from Africa to understand their zoonotic potential. Proc. Natl. Acad. Sci. USA 2021, 118, e2103984118. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; Velavan, T.P.; Rungsung, I.; Traore, T.; Hui, D.S.; McCloskey, B.; El-Kafrawy, S.A.; Zumla, A. Middle East respiratory syndrome coronavirus-a 10-year (2012–2022) global analysis of human and camel infections, genomic sequences, lineages, and geographical origins. Int. J. Infect. Dis. 2023, 131, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Parkhe, P.; Verma, S. Evolution, Interspecies Transmission, and Zoonotic Significance of Animal Coronaviruses. Front. Vet. Sci. 2021, 8, 719834. [Google Scholar] [CrossRef] [PubMed]
- Solomon, M.; Liang, C. Human coronaviruses: The emergence of SARS-CoV-2 and management of COVID-19. Virus Res. 2022, 319, 198882. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Giorgi, E.E.; Marichann, M.H.; Foley, B.; Xiao, C.; Kong, X.P.; Chen, Y.; Korber, B.; Gao, F.; Gnanakaran, S. Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci. Adv. 2020, 6, eabb9153. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Grunewald, M.; Perlman, S. Coronaviruses: An Updated Overview of Their Replication and Pathogenesis. In Coronaviruses; Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2203, pp. 1–29. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.Y.; Li, J.L.; Yang, X.L.; Chmura, A.A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef]
- Park, Y.J.; Walls, A.C.; Wang, Z.; Sauer, M.M.; Li, W.; Tortorici, M.A.; Bosch, B.J.; DiMaio, F.; Veesler, D. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat. Struct. Mol. Biol. 2019, 26, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Parra-Lucares, A.; Segura, P.; Rojas, V.; Pumarino, C.; Saint-Pierre, G.; Toro, L. Emergence of SARS-CoV-2 Variants in the World: How Could This Happen? Life 2022, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Raj, V.S.; Mou, H.H.; Smits, S.L.; Dekkers, D.H.W.; Muller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.A.; Zaki, A.; Fouchier, R.A.M.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Anthony, S.J.; Gilardi, K.; Menachery, V.D.; Goldstein, T.; Ssebide, B.; Mbabazi, R.; Navarrete-Macias, I.; Liang, E.; Wells, H.; Hicks, A.; et al. Further Evidence for Bats as the Evolutionary Source of Middle East Respiratory Syndrome Coronavirus. mBio 2017, 8, e00373-17. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.W.; Hui, K.P.Y.; Perera, R.; Miguel, E.; Niemeyer, D.; Zhao, J.; Channappanavar, R.; Dudas, G.; Oladipo, J.O.; Traore, A.; et al. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc. Natl. Acad. Sci. USA 2018, 115, 3144–3149. [Google Scholar] [CrossRef] [PubMed]
- El-Kafrawy, S.A.; Corman, V.M.; Tolah, A.M.; Al Masaudi, S.B.; Hassan, A.M.; Muller, M.A.; Bleicker, T.; Harakeh, S.M.; Alzahrani, A.A.; Alsaaidi, G.A.; et al. Enzootic patterns of Middle East respiratory syndrome coronavirus in imported African and local Arabian dromedary camels: A prospective genomic study. Lancet Planet Health 2019, 3, e521–e528. [Google Scholar] [CrossRef] [PubMed]
- Ngere, I.; Hunsperger, E.A.; Tong, S.; Oyugi, J.; Jaoko, W.; Harcourt, J.L.; Thornburg, N.J.; Oyas, H.; Muturi, M.; Osoro, E.M.; et al. Outbreak of Middle East Respiratory Syndrome Coronavirus in Camels and Probable Spillover Infection to Humans in Kenya. Viruses 2022, 14, 1743. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Ali, A.; Walelign, E.; Demissie, G.F.; El Masry, I.; Abayneh, T.; Getachew, B.; Krishnan, P.; Ng, D.Y.M.; Gardner, E.; et al. Genetic diversity and molecular epidemiology of Middle East Respiratory Syndrome Coronavirus in dromedaries in Ethiopia, 2017–2020. Emerg. Microbes Infect. 2023, 12, e2164218. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Meng, K.; Meng, G. Genomic recombination events may reveal the evolution of coronavirus and the origin of SARS-CoV-2. Sci. Rep. 2020, 10, 21617. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Yi, S.V. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021, 53, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.L.; Wu, C.C.; Li, X.; Song, Y.H.; Yao, X.M.; Wu, X.K.; Duan, Y.G.; Zhang, H.; Wang, Y.R.; Qian, Z.H.; et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020, 7, 1012–1023. [Google Scholar] [CrossRef] [PubMed]
- Scovino, A.M.; Dahab, E.C.; Vieira, G.F.; Freire-de-Lima, L.; Freire-de-Lima, C.G.; Morrot, A. SARS-CoV-2’s Variants of Concern: A Brief Characterization. Front. Immunol. 2022, 13, 834098. [Google Scholar] [CrossRef]
- Focosi, D.; Maggi, F. Recombination in Coronaviruses, with a Focus on SARS-CoV-2. Viruses 2022, 14, 1239. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Sharma, K.; Shaw, D.; Bhargava, A.; Negi, S.S. Mosaic Recombination Inflicted Various SARS-CoV-2 Lineages to Emerge into Novel Virus Variants: A Review Update. Indian J. Clin. Biochem 2022, 38, 418–425. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Koopmans, M. Tracking SARS-CoV-2 variants and resources. Nat. Methods 2023, 20, 489–490. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef] [PubMed]
- Nikonova, A.A.; Faizuloev, E.B.; Gracheva, A.V.; Isakov, I.Y.; Zverev, V.V. Genetic Diversity and Evolution of the Biological Features of the Pandemic SARS-CoV-2. Acta Naturae 2021, 13, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Liu, Z.; Chen, D. Human coronaviruses: Origin, host and receptor. J. Clin. Virol. 2022, 155, 105246. [Google Scholar] [CrossRef] [PubMed]
- Chathappady House, N.N.; Palissery, S.; Sebastian, H. Corona Viruses: A Review on SARS, MERS and COVID-19. Microbiol. Insights 2021, 14, 11786361211002481. [Google Scholar] [CrossRef] [PubMed]
- Jo, W.K.; de Oliveira-Filho, E.F.; Rasche, A.; Greenwood, A.D.; Osterrieder, K.; Drexler, J.F. Potential zoonotic sources of SARS-CoV-2 infections. Transbound. Emerg. Dis. 2021, 68, 1824–1834. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Kan, B.; Wang, M.; Jing, H.; Xu, H.; Jiang, X.; Yan, M.; Liang, W.; Zheng, H.; Wan, K.; Liu, Q.; et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005, 79, 11892–11900. [Google Scholar] [CrossRef] [PubMed]
- Gussow, A.B.; Auslander, N.; Faure, G.; Wolf, Y.I.; Zhang, F.; Koonin, E.V. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc. Natl. Acad. Sci. USA 2020, 117, 15193–15199. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Kim, Y.J.; Park, S.H.; Yun, M.R.; Yang, J.S.; Kang, H.J.; Han, Y.W.; Lee, H.S.; Kim, H.M.; Kim, H.; et al. Variations in Spike Glycoprotein Gene of MERS-CoV, South Korea, 2015. Emerg. Infect. Dis. 2016, 22, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Cheon, S.; Min, C.K.; Sohn, K.M.; Kang, Y.J.; Cha, Y.J.; Kang, J.I.; Han, S.K.; Ha, N.Y.; Kim, G.; et al. Spread of Mutant Middle East Respiratory Syndrome Coronavirus with Reduced Affinity to Human CD26 during the South Korean Outbreak. mBio 2016, 7, e00019. [Google Scholar] [CrossRef] [PubMed]
- Omrani, A.S.; Al-Tawfiq, J.A.; Memish, Z.A. Middle East respiratory syndrome coronavirus (MERS-CoV): Animal to human interaction. Pathog. Glob. Health 2015, 109, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Amer, H.; Alqahtani, A.S.; Alzoman, H.; Aljerian, N.; Memish, Z.A. Unusual presentation of Middle East respiratory syndrome coronavirus leading to a large outbreak in Riyadh during 2017. Am. J. Infect. Control 2018, 46, 1022–1025. [Google Scholar] [CrossRef] [PubMed]
- Assiri, A.M.; Midgley, C.M.; Abedi, G.R.; Bin Saeed, A.; Almasri, M.M.; Lu, X.; Al-Abdely, H.M.; Abdalla, O.; Mohammed, M.; Algarni, H.S.; et al. Epidemiology of a Novel Recombinant Middle East Respiratory Syndrome Coronavirus in Humans in Saudi Arabia. J. Infect. Dis. 2016, 214, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Huang, Y.; Yuen, K.Y. Coronavirus Diversity, Phylogeny and Interspecies Jumping. Exp. Biol. Med. 2009, 234, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Wang, Q.; Gao, G.F. Bat-to-human: Spike features determining host jump of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015, 23, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Dudas, G.; Carvalho, L.M.; Rambaut, A.; Bedford, T. MERS-CoV spillover at the camel-human interface. Elife 2018, 7, e31257. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.T.; Jia, N.; Zhang, Y.W.; Shum, M.H.; Jiang, J.F.; Zhu, H.C.; Tong, Y.G.; Shi, Y.X.; Ni, X.B.; Liao, Y.S.; et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X.; Zhong, W.; Hao, P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020, 63, 457–460. [Google Scholar] [CrossRef]
- Borucki, M.K.; Lao, V.; Hwang, M.; Gardner, S.; Adney, D.; Munster, V.; Bowen, R.; Allen, J.E. Middle East Respiratory Syndrome Coronavirus Intra-Host Populations Are Characterized by Numerous High Frequency Variants. PLoS ONE 2016, 11, e0146251. [Google Scholar] [CrossRef]
- Cotten, M.; Watson, S.J.; Zumla, A.I.; Makhdoom, H.Q.; Palser, A.L.; Ong, S.H.; Al Rabeeah, A.A.; Alhakeem, R.F.; Assiri, A.; Al-Tawfiq, J.A.; et al. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio 2014, 5, e01062-13. [Google Scholar] [CrossRef] [PubMed]
- Tonkin-Hill, G.; Martincorena, I.; Amato, R.; Lawson, A.R.J.; Gerstung, M.; Johnston, I.; Jackson, D.K.; Park, N.; Lensing, S.V.; Quail, M.A.; et al. Patterns of within-host genetic diversity in SARS-CoV-2. Elife 2021, 10, e66857. [Google Scholar] [CrossRef] [PubMed]
- Varabyou, A.; Pockrandt, C.; Salzberg, S.L.; Pertea, M. Rapid detection of inter-clade recombination in SARS-CoV-2 with Bolotie. Genetics 2021, 218, iyab074. [Google Scholar] [CrossRef] [PubMed]
- Turakhia, Y.; Thornlow, B.; Hinrichs, A.; McBroome, J.; Ayala, N.; Ye, C.; Smith, K.; De Maio, N.; Haussler, D.; Lanfear, R.; et al. Pandemic-scale phylogenomics reveals the SARS-CoV-2 recombination landscape. Nature 2022, 609, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Preska Steinberg, A.; Silander, O.K.; Kussell, E. Correlated substitutions reveal SARS-like coronaviruses recombine frequently with a diverse set of structured gene pools. Proc. Natl. Acad. Sci. USA 2023, 120, e2206945119. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).