Reproductive Ability Disparity in the Pacific Whiteleg Shrimp (Penaeus vannamei): Insights from Ovarian Cellular and Molecular Levels
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Induction of Ovary Development and Spawning Frequency Statistics
2.3. Sample Collection
2.4. Transmission Electron Microscopy Sample Preparation
2.5. RNA-Seq and Bioinformatic Analysis
2.5.1. RNA-Seq of Ovarian Samples
2.5.2. Data Quality Control and Sequence Alignment
2.5.3. Expression Level Statistics and Inter-Group Analysis
2.6. qRT-PCR and Statistical Analysis
2.7. Fluorescence In Situ Hybridization (FISH)
3. Results
3.1. Maturation Frequency of 632 Female Shrimp in a Breeding Cycle
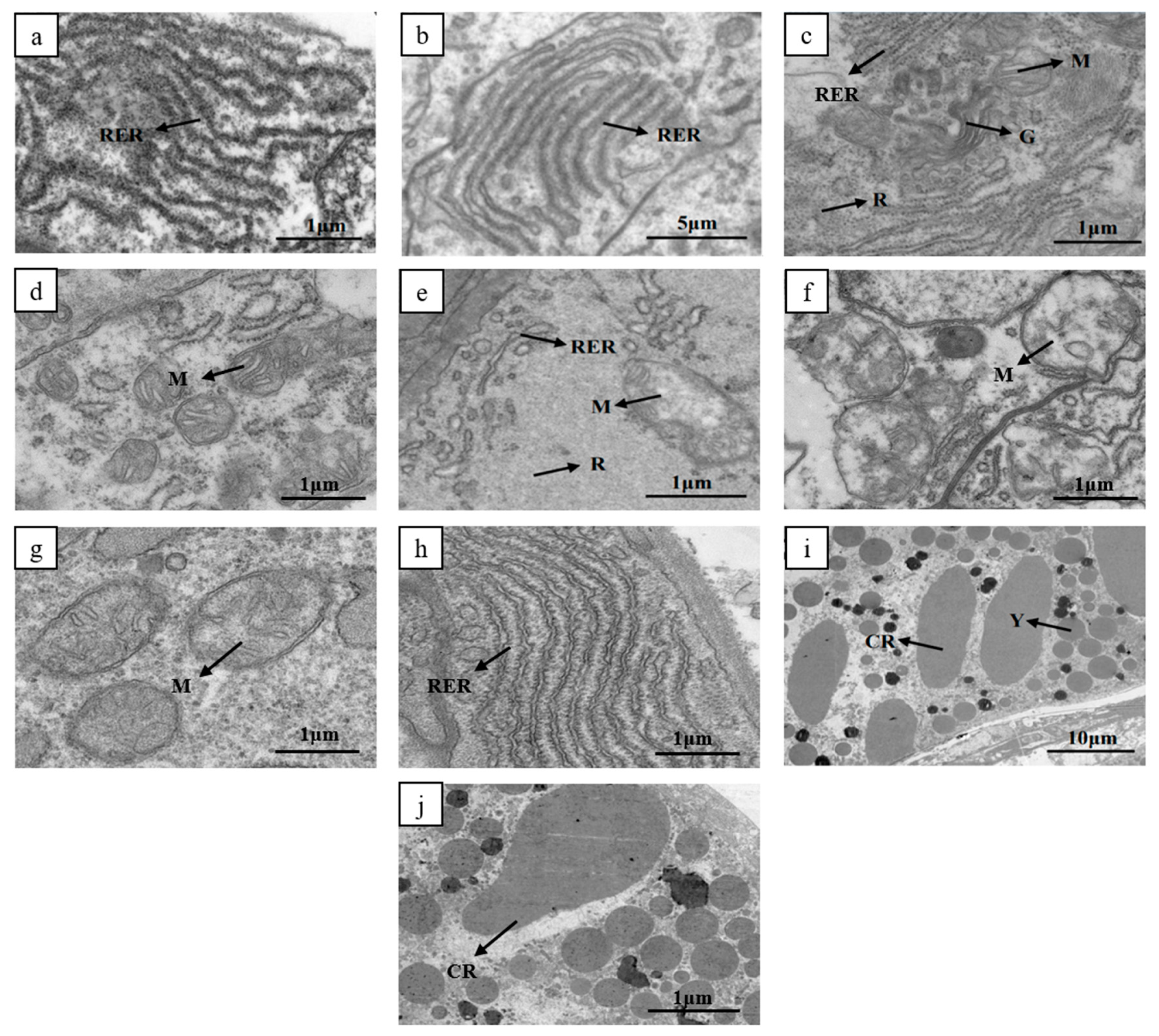
3.2. Comparison of Ovarian Ultrastructure between High and Low Reproductive Female Shrimp
3.3. Transcriptome Data
3.3.1. Overview of RNA Sequencing Data
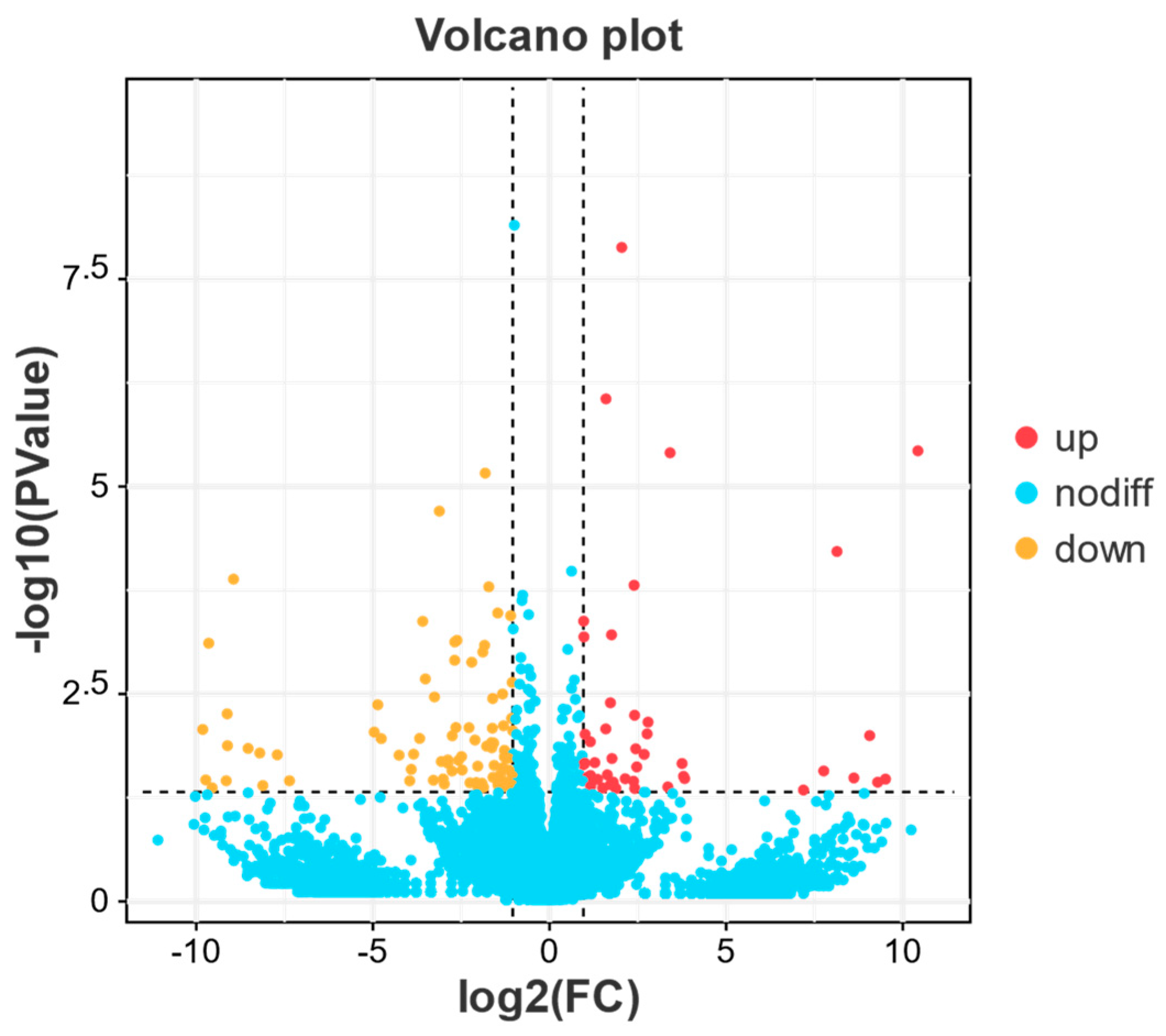
3.3.2. Differential Gene Expression Analysis
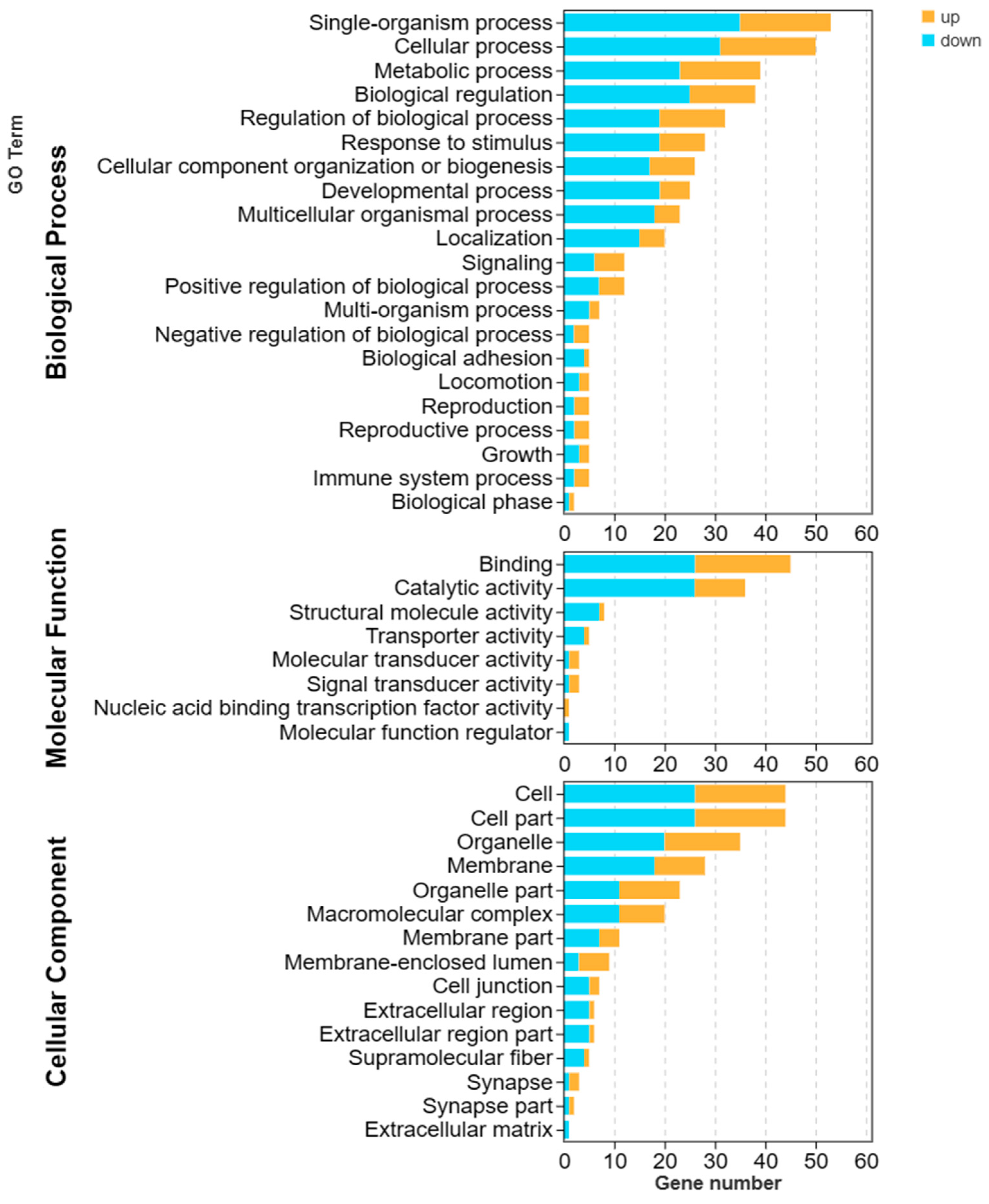
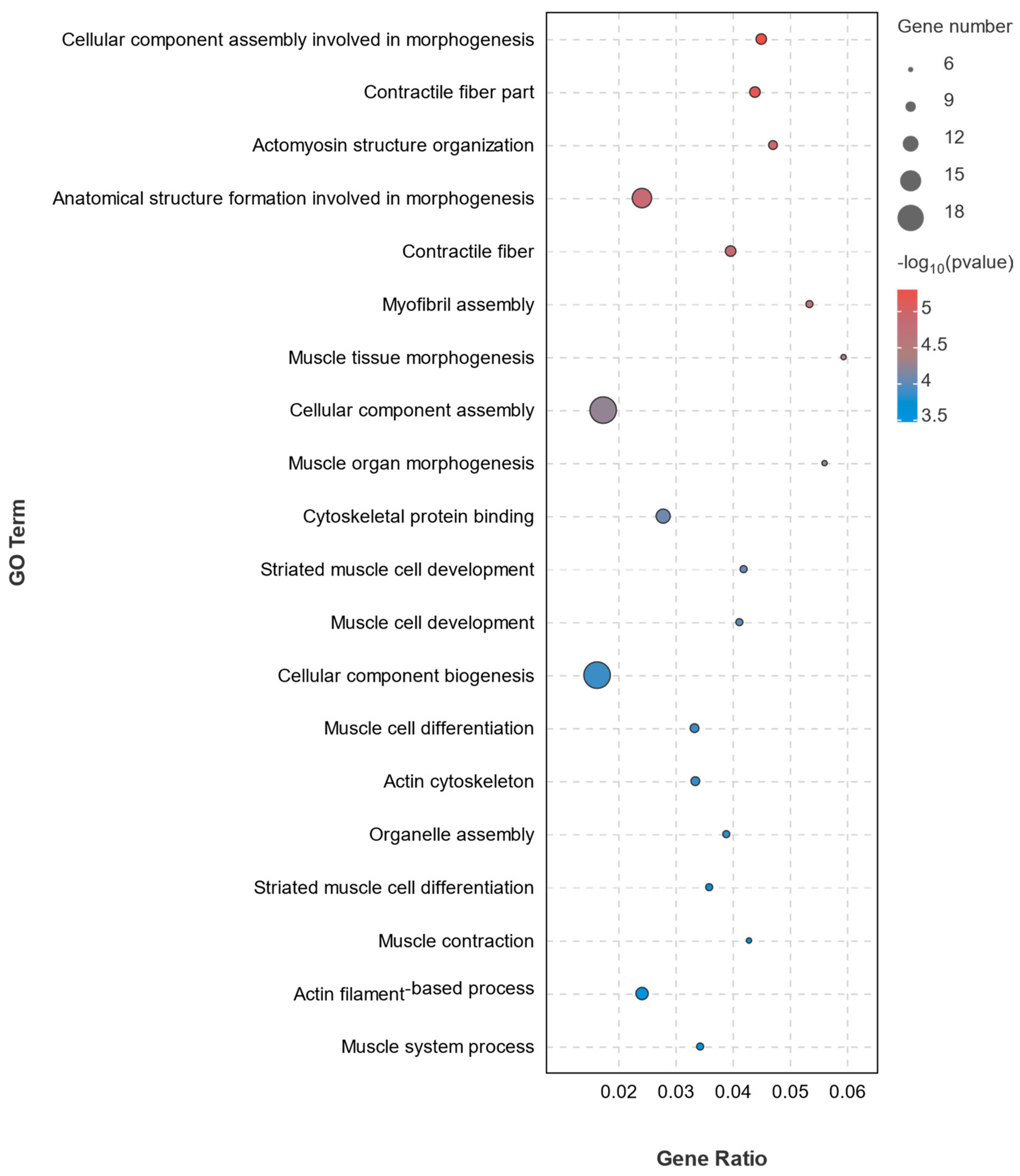
3.3.3. GO Enrichment Analysis of DEGs
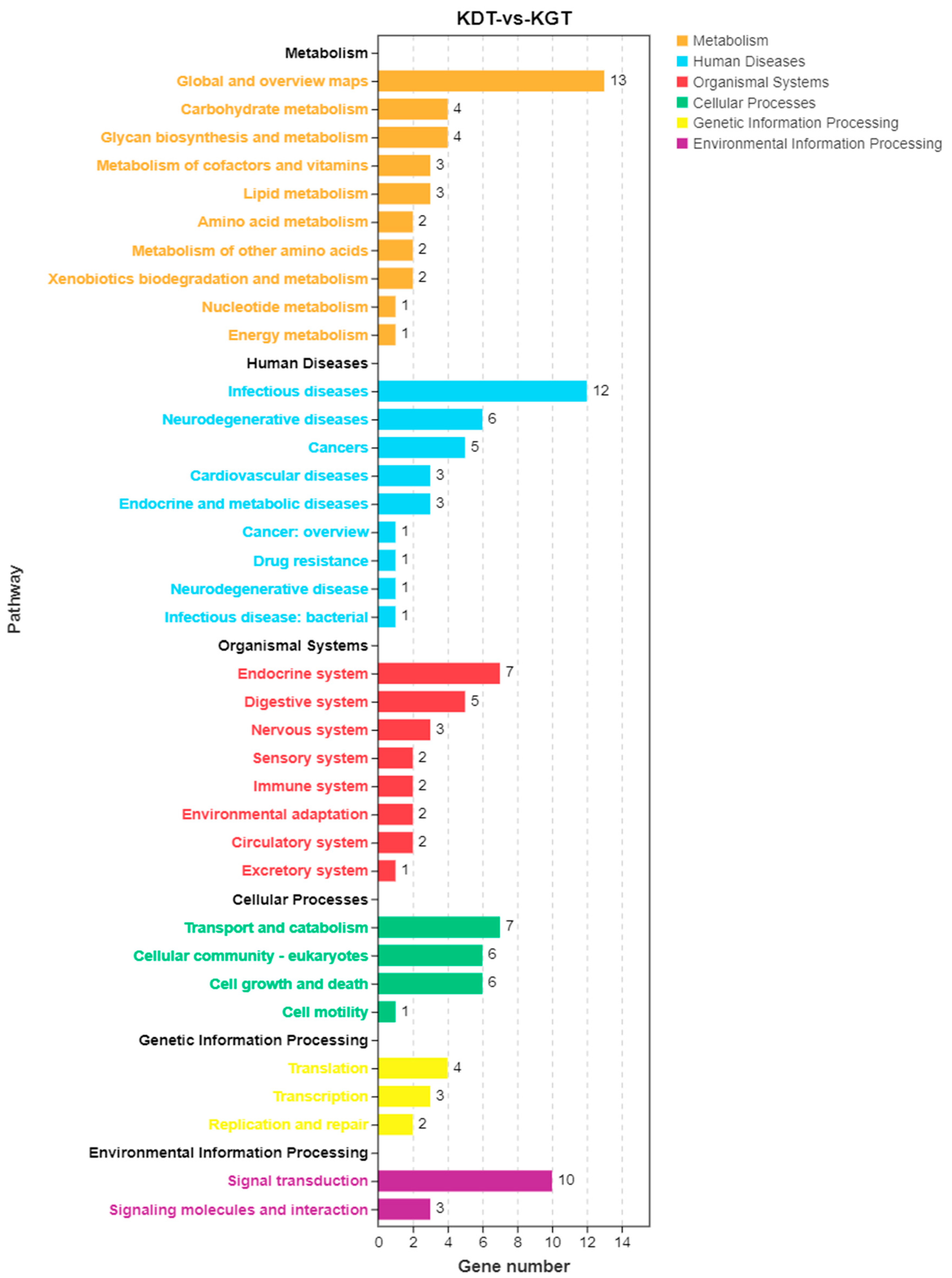
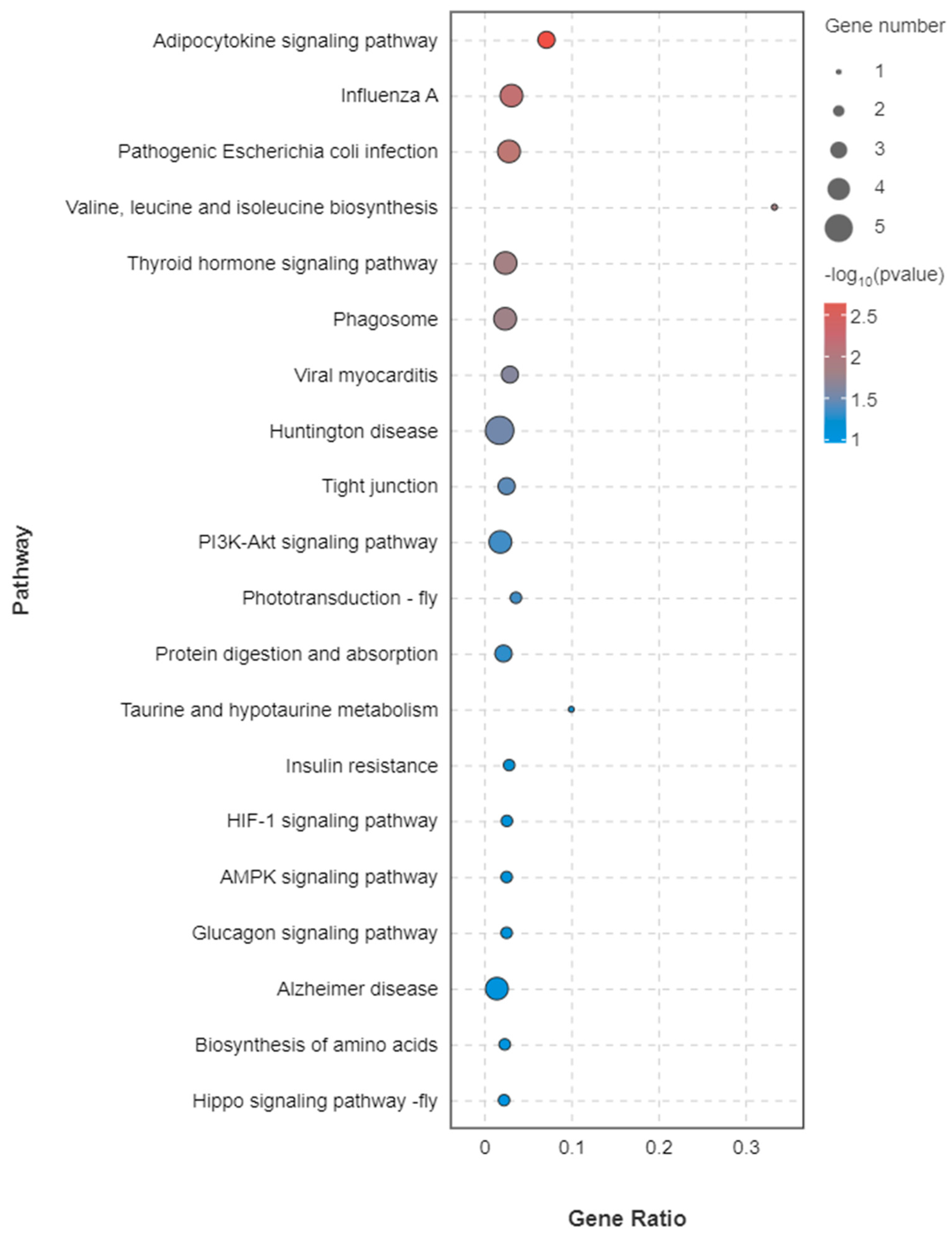
3.3.4. KEGG Annotation of DEGs
3.4. Analysis of EHF and PRDM9 Genes in DEGs
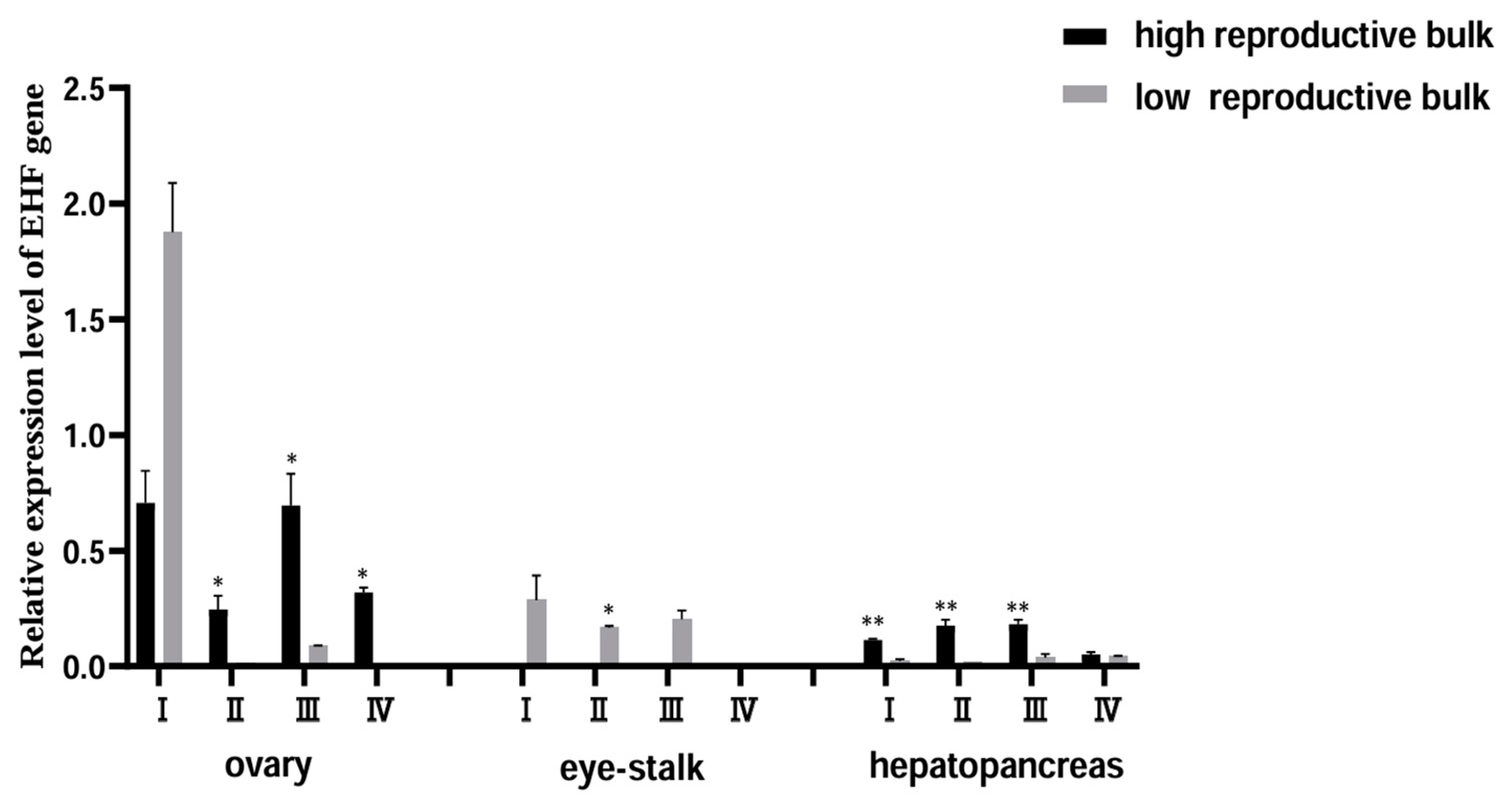
3.4.1. Expression Pattern of EHF Gene and Localization of Ovarian Cells at Different Mature Stages
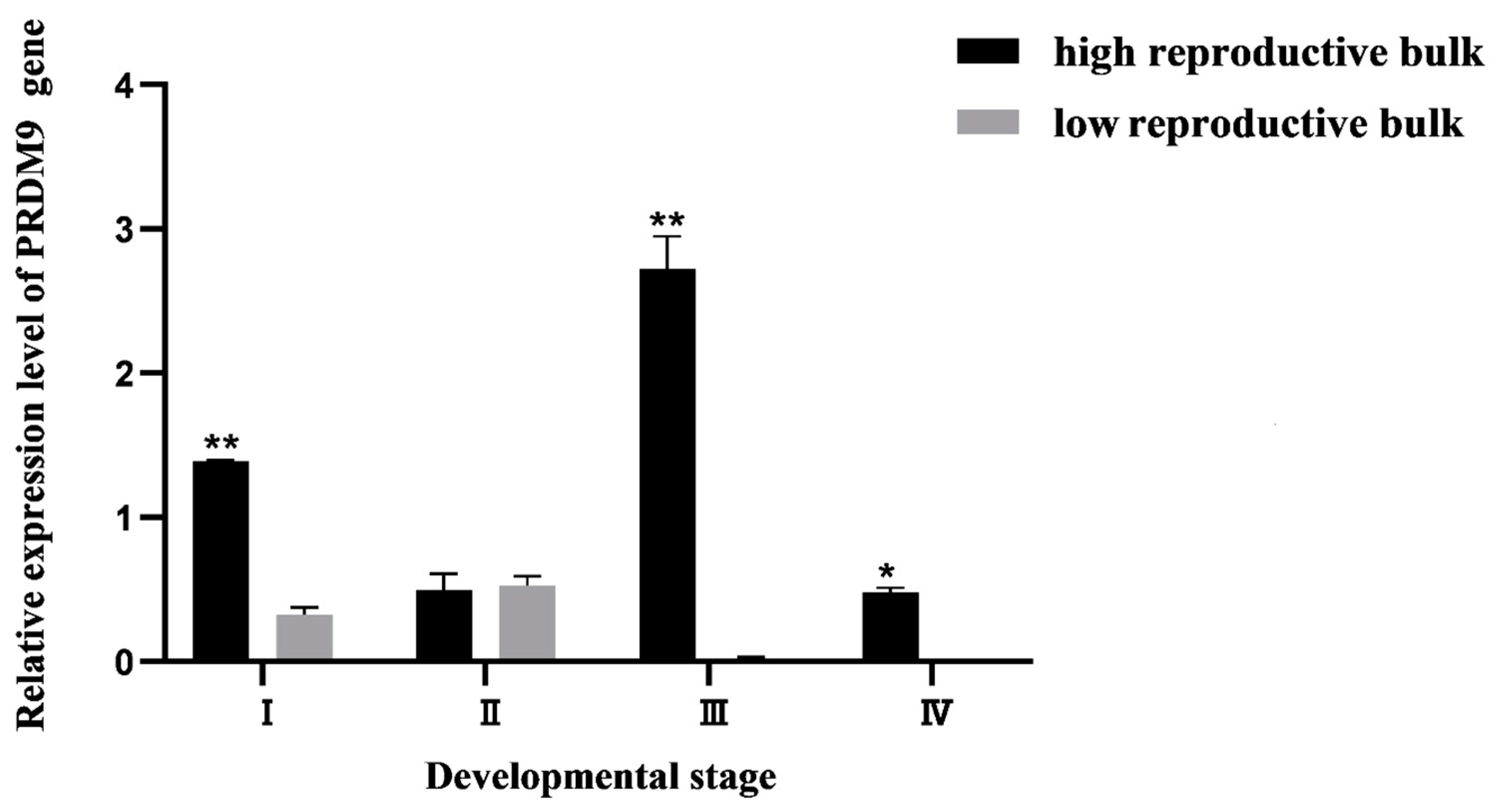
3.4.2. Expression Pattern Analysis of PRDM9 Gene and Localization of Ovarian Cells at Different Mature Stages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. FishStat Database. 2024. Available online: https://www.fao.org/fishery/statistics-query/en/global_production/global_production_quantity (accessed on 24 March 2024).
- Uawisetwathana, U.; Leelatanawit, R.; Klanchui, A.; Prommoon, J.; Klinbunga, S.; Karoonuthaisiri, N. Insights into eyestalk ablation mechanism to induce ovarian maturation in the black tiger shrimp. PLoS ONE 2011, 6, e24427. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.; Wang, W.; Shi, L.; Liu, J.; Zhou, T.; Wang, C.; Amankwah, B.K. Eyestalk Ablation, a Prerequisite for Crustacean Reproduction: A review. Isr. J. Aquac.-Bamidgeh 2019, 71. [Google Scholar] [CrossRef]
- Liu, J.H.; Zhou, T.T.; Wang, C.G.; Wang, W.; Chan, S.M. Comparative transcriptomics reveals eyestalk ablation induced responses of the neuroendocrine-immune system in the Pacific white shrimp Litopenaeus vannamei. Fish. Shellfish. Immun. 2020, 106, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.L.; Zhang, M.Q.; Gao, B.Q.; Lv, J.J.; Li, J.; Liu, P. Integrative Proteomic and MicroRNA Analysis: Insights into Mechanisms of Eyestalk Ablation-Induced Ovarian Maturation in the Swimming Crab Portunus trituberculatus. Front. Endocrinol. 2020, 11, 557324. [Google Scholar] [CrossRef] [PubMed]
- Arcos, F.G.; Ibarra, A.M.; Palacios, E.; Vazquez-Boucard, C.; Racotta, I.S. Feasible predictive criteria for reproductive performance of white shrimp Litopenaeus vannamei: Egg quality and female physiological condition. Aquaculture 2003, 228, 335–349. [Google Scholar] [CrossRef]
- Fan, Y.P.; Tan, J.; Luan, S.; Meng, X.H.; Luo, K.; Sui, J.; Chen, B.L.; Cao, J.W.; Kong, J. Comparative analysis of breeding characteristics of different strains of Litopenaeus vannamei. J. Fish. Sci. China 2021, 28, 1141–1151. [Google Scholar]
- Lee, S.G.; Mykles, D.L. Proteomics and signal transduction in the crustacean molting gland. Integr. Comp. Biol. 2006, 46, 965–977. [Google Scholar] [CrossRef]
- Mykles, D.L. Signaling Pathways That Regulate the Crustacean Molting Gland. Front. Endocrinol. 2021, 12, 674711. [Google Scholar] [CrossRef] [PubMed]
- Fanjul-Moles, M.L. Biochemical and functional aspects of crustacean hyperglycemic hormone in decapod crustaceans: Review and update. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2006, 142, 390–400. [Google Scholar] [CrossRef]
- Jayasankar, V.; Tomy, S.; Wilder, M.N. Insights on Molecular Mechanisms of Ovarian Development in Decapod Crustacea: Focus on Vitellogenesis-Stimulating Factors and Pathways. Front. Endocrinol. 2020, 11, 577925. [Google Scholar] [CrossRef]
- Chen, H.Y.; Toullec, J.Y.; Lee, C.Y. The Crustacean Hyperglycemic Hormone Superfamily: Progress Made in the Past Decade. Front. Endocrinol. 2020, 11, 578958. [Google Scholar] [CrossRef] [PubMed]
- Ohira, T. Chapter 76—Crustacean hyperglycemic hormone. In Handbook of Hormones, 2nd ed.; Ando, H., Ukena, K., Nagata, S., Eds.; Academic Press: San Diego, CA, USA, 2021; pp. 731–733. [Google Scholar]
- Nagaraju, G.P.C. Reproductive regulators in decapod crustaceans: An overview. J. Exp. Biol. 2011, 214, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Ozhan-Kizil, G.; Havemann, J.; Gerberding, M. Germ cells in the crustacean Parhyale hawaiensis depend on Vasa protein for their maintenance but not for their formation. Dev. Biol. 2009, 327, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Clemm, V.H.K.; Müller, S.; Schleich, S.; Meister, M.; Bohlen, J.; Hofmann, T.G.; Teleman, A.A. Cyclin B/CDK1 and Cyclin A/CDK2 phosphorylate DENR to promote mitotic protein translation and faithful cell division. Nat. Commun. 2022, 13, 668. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, K.; Yamagata, H.; Shiga, Y. Exploring embryonic germ line development in the water flea, Daphnia magna, by zinc-finger-containing VASA as a marker. Gene Expr. Patterns 2005, 5, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.H.; Yan, S.F.; Wang, C.G.; Song, Z.R.; Zou, W.Z. Ultrastructure on oogenesis of Litopenaeus vannamel. J. Fish. China 2005, 29, 7. [Google Scholar]
- Sui, J.; Luan, S.; Cao, J.W.; Dai, P.; Meng, X.H.; Luo, K.; Chen, B.L.; Tan, J.; Fu, Q.; Kong, J. Genomic signatures of artificial selection in fecundity of Pacific white shrimp, Penaeus vannamei. Front. Genet. 2022, 13, 929889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.C.; Kong, J.; Cao, J.W.; Tan, J.; Dai, P.; Meng, X.H.; Luo, K.; Fu, Q.; Chen, B.L.; LIU, D.Y.; et al. Histological Observation of Ovary and Expression Analysis of Related Candidate Genes in the High- and Low- Fecundity Populations of Litopenaeus vannamei. Prog. Fish. Sci. 2023, 1–13. [Google Scholar] [CrossRef]
- Tinikul, Y.; Poljaroen, J.; Kornthong, N.; Chotwiwatthanakun, C.; Anuracpreeda, P.; Poomtong, T.; Hanna, P.J.; Sobhon, P. Distribution and changes of serotonin and dopamine levels in the central nervous system and ovary of the Pacific white shrimp, Litopenaeus vannamei, during ovarian maturation cycle. Cell Tissue Res. 2011, 345, 103–124. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.L.; Kong, J.; Meng, X.H.; Luan, S.; Luo, K.; Cao, B.X.; Liu, N.; Lu, X.; Deng, K.Y.; Cao, J.W.; et al. Comparative microarray profile of the hepatopancreas in the response of “Huanghai No. 2” Fenneropenaeus chinensis to white spot syndrome virus. Fish Shellfish. Immunol. 2016, 58, 210–219. [Google Scholar] [CrossRef]
- Giacomello, M.; Pyakurel, A.; Glytsou, C.; Scorrano, L. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Bio. 2020, 21, 204–224. [Google Scholar] [CrossRef]
- Abate, M.; Festa, A.; Falco, M.; Lombardi, A.; Luce, A.; Grimaldi, A.; Zappavigna, S.; Sperlongano, P.; Irace, C.; Caraglia, M.; et al. Mitochondria as playmakers of apoptosis, autophagy and senescence. Semin. Cell Dev. Biol. 2020, 98, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ratchford, A.M.; Chi, M.M.Y.; Schoeller, E.; Frolova, A.; Schedl, T.; Moley, K.H. Maternal Diabetes Causes Mitochondrial Dysfunction and Meiotic Defects in Murine Oocytes. Mol. Endocrinol. 2009, 23, 1603–1612. [Google Scholar] [CrossRef]
- Dalton, C.M.; Szabadkai, G.; Carroll, J. Measurement of ATP in Single Oocytes: Impact of Maturation and Cumulus Cells on Levels and Consumption. J. Cell Physiol. 2014, 229, 353–361. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Lin, M.; Zhuan, Q.R.; Wang, Z.C.; Li, J.J.; Liu, K.X.; Fu, X.W.; Hou, Y.P. Mitochondrial Calcium uniporters are essential for meiotic progression in mouse oocytes by controlling Ca2+ entry. Cell Proliferat 2021, 54, e13127. [Google Scholar] [CrossRef]
- Beams, H.W. Electron microscope studies on developing crayfish oocytes with special reference to the origin of yolk. J. Cell Biol. 1963, 18, 621–649. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Yan, S.F.; Song, Z.R. Ultrastructure of vitellogenesis in the oocytes of Litopenaeus vannamei. Acta Zool. Sin. 2005, 51, 133–141. [Google Scholar]
- Kang, X.; Wang, J.; Yan, L.Y. Endoplasmic reticulum in oocytes: Spatiotemporal distribution and function. J. Assist. Reprod. Gen. 2023, 40, 1255–1263. [Google Scholar] [CrossRef]
- Udagawa, O.; Ishihara, N. Mitochondrial dynamics and interorganellar communication in the development and dysmorphism of mammalian oocytes. J. Biochem. 2020, 167, 257–266. [Google Scholar] [CrossRef]
- Fanibunda, S.E.; Deb, S.; Maniyadath, B.; Tiwari, P.; Ghai, U.; Gupta, S.; Figueiredo, D.; Weisstaub, N.; Gingrich, J.A.; Vaidya, A.; et al. Serotonin regulates mitochondrial biogenesis and function in rodent cortical neurons via the 5-HT(2A) receptor and SIRT1-PGC-1α axis. Proc. Natl. Acad. Sci. USA 2019, 116, 11028–11037. [Google Scholar] [CrossRef]
- Simmons, E.C.; Scholpa, N.E.; Cleveland, K.H.; Schnellmann, R.G. 5-hydroxytryptamine 1F Receptor Agonist Induces Mitochondrial Biogenesis and Promotes Recovery from Spinal Cord Injury. J. Pharmacol. Exp. Ther. 2020, 372, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Valenti, D.; de Bari, L.; Vigli, D.; Lacivita, E.; Leopoldo, M.; Laviola, G.; Vacca, R.A.; De Filippis, B. Stimulation of the brain serotonin receptor 7 rescues mitochondrial dysfunction in female mice from two models of Rett syndrome. Neuropharmacology 2017, 121, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Sola-Penna, M.; Paixão, L.P.; Branco, J.R.; Ochioni, A.C.; Albanese, J.M.; Mundim, D.M.; Baptista-de-Souza, D.; Figueiredo, C.P.; Coelho, W.S.; Marcondes, M.C.; et al. Serotonin activates glycolysis and mitochondria biogenesis in human breast cancer cells through activation of the Jak1/STAT3/ERK1/2 and adenylate cyclase/PKA, respectively. Br. J. Cancer 2020, 122, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Vaca, A.A.; Alfaro, J. Ovarian maturation and spawning in the white shrimp, Penaeus vannamei, by serotonin injection. Aquaculture 2000, 182, 373–385. [Google Scholar] [CrossRef]
- Wongprasert, K.; Asuvapongpatana, S.; Poltana, P.; Tiensuwan, M.; Withyachumnarnkul, B. Serotonin stimulates ovarian maturation and spawning in the black tiger shrimp Penaeus monodon. Aquaculture 2006, 261, 1447–1454. [Google Scholar] [CrossRef]
- Tinikul, Y.; Mercier, A.J.; Soonklang, N.; Sobhon, P. Changes in the levels of serotonin and dopamine in the central nervous system and ovary, and their possible roles in the ovarian development in the giant freshwater prawn, Macrobrachium rosenbergii. Gen. Comp. Endocr. 2008, 158, 250–258. [Google Scholar] [CrossRef]
- Tomy, S.; Saikrithi, P.; James, N.; Balasubramanian, C.P.; Panigrahi, A.; Otta, S.K.; Subramoniam, T.; Ponniah, A.G. Serotonin induced changes in the expression of ovarian gene network in the Indian white shrimp, Penaeus indicus. Aquaculture 2016, 452, 239–246. [Google Scholar] [CrossRef]
- Ganglia, C.; Mattson, M.; Spaziani, E. 5-Hydroxytryptamine Mediates Release of Molt-Inhibiting Hormone Activity from Isolated Crab Eyestalk Ganglia. Biol. Bull. 1985, 169, 246–255. [Google Scholar]
- Kornthong, N.; Cummins, S.F.; Chotwiwatthanakun, C.; Khornchatri, K.; Engsusophon, A.; Hanna, P.J.; Sobhon, P. Identification of Genes Associated with Reproduction in the Mud Crab (Scylla olivacea) and Their Differential Expression following Serotonin Stimulation. PLoS ONE 2014, 9, e115867. [Google Scholar] [CrossRef]
- Kornthong, N.; Chotwiwatthanakun, C.; Chansela, P.; Tinikul, Y.; Cummins, S.F.; Hanna, P.J.; Sobhon, P. Characterization of red pigment concentrating hormone (RPCH) in the female mud crab (Scylla olivacea) and the effect of 5-HT on its expression. Gen. Comp. Endocr. 2013, 185, 28–36. [Google Scholar] [CrossRef]
- John, S.; Russell, L.; Chin, S.S.; Luo, W.; Oshima, R.; Garrett-Sinha, L.A. Transcription factor Ets1, but not the closely related factor Ets2, inhibits antibody-secreting cell differentiation. Mol. Cell Biol. 2014, 34, 522–532. [Google Scholar] [CrossRef]
- Schulz, R.A.; The, S.M.; Hogue, D.A.; Galewsky, S.; Guo, Q. Ets oncogene-related gene Elg functions in Drosophila oogenesis. Proc. Natl. Acad. Sci. USA 1993, 90, 10076–10080. [Google Scholar] [CrossRef]
- Yan, R.; Cheng, X.; Gu, C.; Xu, Y.; Long, X.; Zhai, J.; Sun, F.; Qian, J.; Du, Y.; Wang, H.; et al. Dynamics of DNA hydroxymethylation and methylation during mouse embryonic and germline development. Nat. Genet. 2023, 55, 130–143. [Google Scholar] [CrossRef]
- Eram, M.S.; Bustos, S.P.; Lima-Fernandes, E.; Siarheyeva, A.; Senisterra, G.; Hajian, T.; Chau, I.; Duan, S.; Wu, H.; Dombrovski, L. Trimethylation of histone H3 lysine 36 by human methyltransferase PRDM9 protein. J. Biol. Chem. 2014, 289, 12177–12188. [Google Scholar] [CrossRef]
- Di Tullio, F.; Schwarz, M.; Zorgati, H.; Mzoughi, S.; Guccione, E. The duality of PRDM proteins: Epigenetic and structural perspectives. FEBS J. 2022, 289, 1256–1275. [Google Scholar] [CrossRef]
- Hohenauer, T.; Moore, A.W. The Prdm family: Expanding roles in stem cells and development. Development 2012, 139, 2267. [Google Scholar] [CrossRef] [PubMed]
- Mihola, O.; Pratto, F.; Brick, K.; Linhartova, E.; Kobets, T.; Flachs, P.; Baker, C.L.; Sedlacek, R.; Paigen, K.; Petkov, P.M.; et al. Histone methyltransferase PRDM9 is not essential for meiosis in male mice. Genome Res. 2019, 29, 1078–1086. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Yoshida, K.; Matsui, Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005, 438, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.Q. Research on Hybrid Sterility Gene Prdm9 of Yak. Master’s Thesis, Southwest University for Nationalities, Chengdu, China, 2014. [Google Scholar]
- Yamaji, M.; Seki, Y.; Kurimoto, K.; Yabuta, Y.; Yuasa, M.; Shigeta, M.; Yamanaka, K.; Ohinata, Y.; Saitou, M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008, 40, 1016–1022. [Google Scholar] [CrossRef]



| Primer Name | Sequence (5′-3′) | Fragment Length (bp) | TM/°C | Reference |
|---|---|---|---|---|
| 18S-F | TATACGCTAGTGGAGCTGGAA | 147 | 59 | Shi et al. (2016) [27] |
| 18S-R | GGGGAGGTAGTGACGAAAAAT | |||
| EHF-F | CAAGGTCTATGGCGATGGAGAATGG | 115 | 60 | The present study |
| EHF-R | TGGTAGTGATTGGAAGGCGAGGTAG | |||
| PRDM9-F | GCCAGGAGTGAGAACTACGATGATG | 142 | 59 | The present study |
| PRDM9-R | CTCCACTTGACCTGTCGGCTATTG |
| Gene Name | Probe Sequence |
|---|---|
| EHF | 5′-GATGCCGATTTGTCTTTTTTTGGTC-3′ |
| 5′-ACCATCTGGCAACACTCCTGTCCT-3′ | |
| 5′-GAAACCCGAAGAACACTGTAATCCG-3′ | |
| PRDM9 | 5′-GTTGCGTCGCTTCCTCTGTTGTT-3′ |
| 5′-CCGCCTACGCTTCCATTTTTGT-3 | |
| 5′-CACTCTGCCTTCCCCTTATGACC-3′ |
| Group | Raw Data | Clean Data | AF-Q20 (%) | AF-Q30 (%) | AF-GC |
|---|---|---|---|---|---|
| KDT1 | 46,520,442 | 46,288,750 (99.50%) | 6,715,262,029 (97.44%) | 6,399,880,267 (92.87%) | 3,395,451,947 (49.27%) |
| KDT2 | 41,907,276 | 41,722,290 (99.56%) | 6,040,934,649 (97.28%) | 5,742,949,875 (92.49%) | 3,063,680,490 (49.34%) |
| KDT3 | 44,090,614 | 43,907,530 (99.58%) | 6,372,805,328 (97.53%) | 6,077,816,927 (93.02%) | 3,238,788,915 (49.57%) |
| KGT1 | 47,324,512 | 47,103,908 (99.53%) | 6,831,597,125 (97.54%) | 6,521,213,357 (93.11%) | 3,482,699,644 (49.73%) |
| KGT2 | 47,507,396 | 47,263,324 (99.49%) | 6,862,474,739 (97.58%) | 6,550,690,414 (93.15%) | 3,453,633,923 (49.11%) |
| KGT3 | 45,450,430 | 45,294,532 (99.66%) | 6,585,079,758 (97.59%) | 6,288,860,139 (93.20%) | 3,334,162,581 (49.41%) |
| Group | Total | Unmapped (%) | Unique_ Mapped (%) | Multiple_ Mapped (%) | Total_ Mapped (%) |
|---|---|---|---|---|---|
| KDT1 | 46,076,744 | 2,956,233 (6.42%) | 30,466,203 (66.12%) | 12,654,308 (27.46%) | 43,120,511 (93.58%) |
| KDT2 | 41,505,912 | 2,804,291 (6.76%) | 27,175,879 (65.47%) | 11,525,742 (27.77%) | 38,701,621 (93.24%) |
| KDT3 | 43,747,662 | 2,772,509 (6.34%) | 28,830,719 (65.90%) | 12,144,434 (27.76%) | 40,975,153 (93.66%) |
| KGT1 | 46,969,794 | 3,032,178 (6.46%) | 30,984,340 (65.97%) | 12,953,276 (27.58%) | 43,937,616 (93.54%) |
| KGT2 | 46,992,062 | 3,140,999 (6.68%) | 30,799,565 (65.54%) | 13,051,498 (27.77%) | 43,851,063 (93.32%) |
| KGT3 | 45,165,458 | 2,808,346 (6.22%) | 29,754,848 (65.88%) | 12,602,264 (27.90%) | 42,357,112 (93.78%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Kong, J.; Cao, J.; Dai, P.; Chen, B.; Tan, J.; Meng, X.; Luo, K.; Fu, Q.; Wei, P.; et al. Reproductive Ability Disparity in the Pacific Whiteleg Shrimp (Penaeus vannamei): Insights from Ovarian Cellular and Molecular Levels. Biology 2024, 13, 218. https://doi.org/10.3390/biology13040218
Zhang J, Kong J, Cao J, Dai P, Chen B, Tan J, Meng X, Luo K, Fu Q, Wei P, et al. Reproductive Ability Disparity in the Pacific Whiteleg Shrimp (Penaeus vannamei): Insights from Ovarian Cellular and Molecular Levels. Biology. 2024; 13(4):218. https://doi.org/10.3390/biology13040218
Chicago/Turabian StyleZhang, Jianchun, Jie Kong, Jiawang Cao, Ping Dai, Baolong Chen, Jian Tan, Xianhong Meng, Kun Luo, Qiang Fu, Peiming Wei, and et al. 2024. "Reproductive Ability Disparity in the Pacific Whiteleg Shrimp (Penaeus vannamei): Insights from Ovarian Cellular and Molecular Levels" Biology 13, no. 4: 218. https://doi.org/10.3390/biology13040218
APA StyleZhang, J., Kong, J., Cao, J., Dai, P., Chen, B., Tan, J., Meng, X., Luo, K., Fu, Q., Wei, P., Luan, S., & Sui, J. (2024). Reproductive Ability Disparity in the Pacific Whiteleg Shrimp (Penaeus vannamei): Insights from Ovarian Cellular and Molecular Levels. Biology, 13(4), 218. https://doi.org/10.3390/biology13040218





