RNA Interference in Insects: From a Natural Mechanism of Gene Expression Regulation to a Biotechnological Crop Protection Promise
Abstract
Simple Summary
Abstract
1. Introduction
2. RNAi Discovery
3. Three Different Pathways of RNA-Mediated Silencing
3.1. siRNAs
3.2. miRNAs
3.3. piRNAs
4. dsRNA Cell Uptake and Systemic Distribution of the Silencing Signal
5. Sources of dsRNAs with Insecticidal Effect
6. RNAi in Pest Control: Challenges and Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Culliney, T.W. Crop Losses to Arthropods. In Integrated Pest Management: Pesticide Problems; Springer: Amsterdam, The Netherlands, 2014; Volume 3, pp. 201–225. ISBN 9789400777965. [Google Scholar]
- Sharma, S.; Kooner, R.; Arora, R. Insect Pests and Crop Losses. In Breeding Insect Resistant Crops for Sustainable Agriculture; Springer: Singapore, 2017; pp. 45–66. ISBN 9789811060564. [Google Scholar]
- Escobar-Bravo, R.; Alba, J.M.; Pons, C.; Granell, A.; Kant, M.R.; Moriones, E.; Fernández-Muñoz, R. A Jasmonate-Inducible Defense Trait Transferred from Wild into Cultivated Tomato Establishes Increased Whitefly Resistance and Reduced Viral Disease Incidence. Front. Plant Sci. 2016, 7, 1732. [Google Scholar] [CrossRef] [PubMed]
- Monci, F.; García-Andrés, S.; Sánchez-Campos, S.; Fernández-Muñoz, R.; Díaz-Pendón, J.A.; Moriones, E. Use of Systemic Acquired Resistance and Whitefly Optical Barriers to Reduce Tomato Yellow Leaf Curl Disease Damage to Tomato Crops. Plant Dis. 2019, 103, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Fortes, I.M.; Fernández-Muñoz, R.; Moriones, E. Host Plant Resistance to Bemisia tabaci to Control Damage Caused in Tomato Plants by the Emerging Crinivirus Tomato Chlorosis Virus. Front. Plant Sci. 2020, 11, 585510. [Google Scholar] [CrossRef] [PubMed]
- Chala, B.; Hamde, F. Emerging and Re-Emerging Vector-Borne Infectious Diseases and the Challenges for Control: A Review. Front. Public. Health 2021, 9, 1466. [Google Scholar] [CrossRef] [PubMed]
- Deguine, J.P.; Atiama-Nurbel, T.; Aubertot, J.N.; Augusseau, X.; Atiama, M.; Jacquot, M.; Reynaud, B. Agroecological Management of Cucurbit-Infesting Fruit Fly: A Review. Agron. Sustain. Dev. 2015, 35, 937–965. [Google Scholar] [CrossRef]
- Tyagi, S.; Kesiraju, K.; Saakre, M.; Rathinam, M.; Raman, V.; Pattanayak, D.; Sreevathsa, R. Genome Editing for Resistance to Insect Pests: An Emerging Tool for Crop Improvement. ACS Omega 2020, 5, 20674–20683. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Izant, J.G.; Weintraub, H. Inhibition of Thymidine Kinase Gene Expression by Anti-Sense RNA: A Molecular Approach to Genetic Analysis. Cell 1984, 36, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Albertson, D.; Harrison, S.W.; Moerman, D.G. Production of Antisense RNA Leads to Effective and Specific Inhibition of Gene Expression in C. Elegans Muscle. Development 1991, 113, 503–514. [Google Scholar] [CrossRef]
- Guo, S.; Kemphues, K.J. Par-1, a Gene Required for Establishing Polarity in C. Elegans Embryos, Encodes a Putative Ser/Thr Kinase That Is Asymmetrically Distributed. Cell 1995, 81, 611–620. [Google Scholar] [CrossRef]
- Matzke, M.A.; Primig, M.; Trnovsky, J.; Matzke, A.J.M. Reversible Methylation and Inactivation of Marker Genes in Sequentially Transformed Tobacco Plants. EMBO J. 1989, 8, 643. [Google Scholar] [CrossRef]
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in Trans. Plant Cell 1990, 2, 279–289. [Google Scholar] [CrossRef]
- van der Krol, A.R.; Mur, L.A.; Beld, M.; Mol, J.N.M.; Stuitje, A.R. Flavonoid Genes in Petunia: Addition of a Limited Number of Gene Copies May Lead to a Suppression of Gene Expression. Plant Cell 1990, 2, 291–299. [Google Scholar] [CrossRef]
- Romano, N.; Macino, G. Quelling: Transient Inactivation of Gene Expression in Neurospora Crassa by Transformation with Homologous Sequences. Mol. Microbiol. 1992, 6, 3343–3353. [Google Scholar] [CrossRef] [PubMed]
- Cogoni, C.; Irelan, J.T.; Schumacher, M.; Schmidhauser, T.J.; Selker, E.U.; Macino, G. Transgene Silencing of the Al-1 Gene in Vegetative Cells of Neurospora Is Mediated by a Cytoplasmic Effector and Does Not Depend on DNA-DNA Interactions or DNA Methylation. EMBO J. 1996, 15, 3153–3163. [Google Scholar] [CrossRef] [PubMed]
- Kennerdell, J.R.; Carthew, R.W. Use of DsRNA-Mediated Genetic Interference to Demonstrate That Frizzled and Frizzled 2 Act in the Wingless Pathway. Cell 1998, 95, 1017–1026. [Google Scholar] [CrossRef]
- Hamilton, A.J.; Baulcombe, D.C. A Species of Small Antisense RNA in Posttranscriptional Gene Silencing in Plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef]
- Wianny, F.; Zernicka-Goetz, M. Specific Interference with Gene Function by Double-Stranded RNA in Early Mouse Development. Nat. Cell Biol. 1999, 2, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Drinnenberg, I.A.; Weinberg, D.E.; Xie, K.T.; Mower, J.P.; Wolfe, K.H.; Fink, G.R.; Bartel, D.P. RNAi in Budding Yeast. Science 2009, 326, 544–550. [Google Scholar] [CrossRef]
- Li, L.C.; Okino, S.T.; Zhao, H.; Pookot, D.; Place, R.F.; Urakami, S.; Enokida, H.; Dahiya, R. Small DsRNAs Induce Transcriptional Activation in Human Cells. Proc. Natl. Acad. Sci. USA 2006, 103, 17337. [Google Scholar] [CrossRef]
- de Hayr, L.; Asad, S.; Hussain, M.; Asgari, S. RNA Activation in Insects: The Targeted Activation of Endogenous and Exogenous Genes. Insect Biochem. Mol. Biol. 2020, 119, 103325. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Aliyari, R.; Li, W.X.; Li, H.W.; Kim, K.; Carthew, R.; Atkinson, P.; Ding, S.W. RNA Interference Directs Innate Immunity against Viruses in Adult Drosophila. Science 2006, 312, 452–454. [Google Scholar] [CrossRef] [PubMed]
- Galiana-Arnoux, D.; Dostert, C.; Schneemann, A.; Hoffmann, J.A.; Imler, J.L. Essential Function in Vivo for Dicer-2 in Host Defense against RNA Viruses in Drosophila. Nat. Immunol. 2006, 7, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.J.; Okamura, K.; Martin, R.; Lai, E.C. Endogenous RNA Interference Provides a Somatic Defense against Drosophila Transposons. Curr. Biol. 2008, 18, 795–802. [Google Scholar] [CrossRef]
- Czech, B.; Malone, C.D.; Zhou, R.; Stark, A.; Schlingeheyde, C.; Dus, M.; Perrimon, N.; Kellis, M.; Wohlschlegel, J.A.; Sachidanandam, R.; et al. An Endogenous Small Interfering RNA Pathway in Drosophila. Nature 2008, 453, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.W.; Zapp, M.L.; Weng, Z.; et al. Endogenous SiRNAs Derived from Transposons and MRNAs in Drosophila Somatic Cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef]
- Okamura, K.; Balla, S.; Martin, R.; Liu, N.; Lai, E.C. Two Distinct Mechanisms Generate Endogenous SiRNAs from Bidirectional Transcription in Drosophila Melanogaster. Nat. Struct. Mol. Biol. 2008, 15, 581–590. [Google Scholar] [CrossRef]
- Okamura, K.; Chung, W.J.; Ruby, J.G.; Guo, H.; Bartel, D.P.; Lai, E.C. The Drosophila Hairpin RNA Pathway Generates Endogenous Short Interfering RNAs. Nature 2008, 453, 803–806. [Google Scholar] [CrossRef]
- Lucchetta, E.M.; Carthew, R.W.; Ismagilov, R.F. The Endo-SiRNA Pathway Is Essential for Robust Development of the Drosophila Embryo. PLoS ONE 2009, 4, e7576. [Google Scholar] [CrossRef]
- Lim, D.H.; Oh, C.T.; Lee, L.; Hong, J.S.; Noh, S.H.; Hwang, S.; Kim, S.; Han, S.J.; Lee, Y.S. The Endogenous SiRNA Pathway in Drosophila Impacts Stress Resistance and Lifespan by Regulating Metabolic Homeostasis. FEBS Lett. 2011, 585, 3079–3085. [Google Scholar] [CrossRef]
- Lim, D.H.; Lee, L.; Oh, C.T.; Kim, N.H.; Hwang, S.; Han, S.J.; Lee, Y.S. Microarray Analysis of Drosophila Dicer-2 Mutants Reveals Potential Regulation of Mitochondrial Metabolism by Endogenous SiRNAs. J. Cell Biochem. 2013, 114, 418–427. [Google Scholar] [CrossRef]
- Zamore, P.D.; Tuschl, T.; Sharp, P.A.; Bartel, D.P. RNAi: Double-Stranded RNA Directs the ATP-Dependent Cleavage of MRNA at 21 to 23 Nucleotide Intervals. Cell 2000, 101, 25–33. [Google Scholar] [CrossRef]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-Directed Nuclease Mediates Post-Transcriptional Gene Silencing in Drosophila Cells. Nature 2000, 404, 293–296. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T. RNA Interference Is Mediated by 21- and 22-Nucleotide RNAs. Genes. Dev. 2001, 15, 188. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a Bidentate Ribonuclease in the Initiation Step of RNA Interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct Roles for Drosophila Dicer-1 and Dicer-2 in the SiRNA/MiRNA Silencing Pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef]
- Lau, P.W.; Guiley, K.Z.; De, N.; Potter, C.S.; Carragher, B.; MacRae, I.J. The Molecular Architecture of Human Dicer. Nat. Struct. Mol. Biol. 2012, 19, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Sinha, N.K.; Iwasa, J.; Shen, P.S.; Bass, B.L. Dicer Uses Distinct Modules for Recognizing DsRNA Termini. Science 2018, 359, 329–334. [Google Scholar] [CrossRef]
- Welker, N.C.; Maity, T.S.; Ye, X.; Aruscavage, P.J.; Krauchuk, A.A.; Liu, Q.; Bass, B.L. Dicer’s Helicase Domain Discriminates DsRNA Termini to Promote an Altered Reaction Mode. Mol. Cell 2011, 41, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Cenik, E.S.; Fukunaga, R.; Lu, G.; Dutcher, R.; Wang, Y.; Tanaka Hall, T.M.; Zamore, P.D. Phosphate and R2D2 Restrict the Substrate Specificity of Dicer-2, an ATP-Driven Ribonuclease. Mol. Cell 2011, 42, 172–184. [Google Scholar] [CrossRef]
- Sinha, N.K.; Trettin, K.D.; Aruscavage, P.J.; Bass, B.L. Drosophila Dicer-2 Cleavage Is Mediated by Helicase- and DsRNA Termini-Dependent States That Are Modulated by Loquacious-PD. Mol. Cell 2015, 58, 406–417. [Google Scholar] [CrossRef]
- Naganuma, M.; Tadakuma, H.; Tomari, Y. Single-Molecule Analysis of Processive Double-Stranded RNA Cleavage by Drosophila Dicer-2. Nat. Commun. 2021, 12, 4268. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Czech, B.; Brennecke, J.; Sachidanandam, R.; Wohlschlegel, J.A.; Perrimon, N.; Hannon, G.J. Processing of Drosophila Endo-SiRNAs Depends on a Specific Loquacious Isoform. RNA 2009, 15, 1886–1895. [Google Scholar] [CrossRef] [PubMed]
- Hartig, J.V.; Esslinger, S.; Böttcher, R.; Saito, K.; Förstemann, K. Endo-SiRNAs Depend on a New Isoform of Loquacious and Target Artificially Introduced, High-Copy Sequences. EMBO J. 2009, 28, 2932–2944. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, K.; Miyoshi, T.; Hartig, J.V.; Siomi, H.; Siomi, M.C. Molecular Mechanisms That Funnel RNA Precursors into Endogenous Small-Interfering RNA and MicroRNA Biogenesis Pathways in Drosophila. RNA 2010, 16, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Marques, J.T.; Wang, J.P.; Wang, X.; de Oliveira, K.P.V.; Gao, C.; Aguiar, E.R.G.R.; Jafari, N.; Carthew, R.W. Functional Specialization of the Small Interfering RNA Pathway in Response to Virus Infection. PLoS Pathog. 2013, 9, e1003579. [Google Scholar] [CrossRef]
- Trettin, K.D.; Sinha, N.K.; Eckert, D.M.; Apple, S.E.; Bass, B.L. Loquacious-PD Facilitates Drosophila Dicer-2 Cleavage through Interactions with the Helicase Domain and DsRNA. Proc. Natl. Acad. Sci. USA 2017, 114, E7939–E7948. [Google Scholar] [CrossRef] [PubMed]
- Jonely, M.; Singh, R.K.; Donelick, H.M.; Bass, B.L.; Noriega, R. Loquacious-PD Regulates the Terminus-Dependent Molecular Recognition of Dicer-2 toward Double-Stranded RNA. Chem. Commun. 2021, 57, 10879–10882. [Google Scholar] [CrossRef]
- MacRae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural Basis for Double-Stranded RNA Processing by Dicer. Science 2006, 311, 195–198. [Google Scholar] [CrossRef]
- MacRae, I.J.; Zhou, K.; Doudna, J.A. Structural Determinants of RNA Recognition and Cleavage by Dicer. Nat. Struct. Mol. Biol. 2007, 14, 934–940. [Google Scholar] [CrossRef]
- Kandasamy, S.K.; Fukunaga, R. Phosphate-Binding Pocket in Dicer-2 PAZ Domain for High-Fidelity SiRNA Production. Proc. Natl. Acad. Sci. USA 2016, 113, 14031–14036. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.; Mingels, L.; Vogel, E.; Wang, L.; Christiaens, O.; Cappelle, K.; Wynant, N.; Gansemans, Y.; van Nieuwerburgh, F.; Smagghe, G.; et al. Generation of Virus- and DsRNA-Derived SiRNAs with Species-Dependent Length in Insects. Viruses 2019, 11, 738. [Google Scholar] [CrossRef]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single Processing Center Models for Human Dicer and Bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Patkaniowska, A.; Urlaub, H.; Lührmann, R.; Tuschl, T. Single-Stranded Antisense SiRNAs Guide Target RNA Cleavage in RNAi. Cell 2002, 110, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Rand, T.A.; Petersen, S.; Du, F.; Wang, X. Argonaute2 Cleaves the Anti-Guide Strand of SiRNA during RISC Activation. Cell 2005, 123, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-Strand Cleavage Facilitates Assembly of SiRNA into Ago2-Containing RNAi Enzyme Complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, K.; Tsukumo, H.; Nagami, T.; Siomi, H.; Siomi, M.C. Slicer Function of Drosophila Argonautes and Its Involvement in RISC Formation. Genes. Dev. 2005, 19, 2837–2848. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional SiRNAs and MiRNAs Exhibit Strand Bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the Assembly of the RNAi Enzyme Complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, F.; Kalidas, S.; Smith, D.; Liu, Q. Dicer-2 and R2D2 Coordinately Bind SiRNA to Promote Assembly of the SiRISC Complexes. RNA 2006, 12, 1514–1520. [Google Scholar] [CrossRef]
- Tomari, Y.; Matranga, C.; Haley, B.; Martinez, N.; Zamore, P.D. A Protein Sensor for SiRNA Asymmetry. Science 2004, 306, 1377–1380. [Google Scholar] [CrossRef]
- Mirkovic-Hösle, M.; Förstemann, K. Transposon Defense by Endo-SiRNAs, PiRNAs and Somatic PilRNAs in Drosophila: Contributions of Loqs-PD and R2D2. PLoS ONE 2014, 9, e84994. [Google Scholar] [CrossRef]
- Tants, J.N.; Fesser, S.; Kern, T.; Stehle, R.; Geerlof, A.; Wunderlich, C.; Juen, M.; Hartlmüller, C.; Böttcher, R.; Kunzelmann, S.; et al. Molecular Basis for Asymmetry Sensing of SiRNAs by the Drosophila Loqs-PD/Dcr-2 Complex in RNA Interference. Nucleic Acids Res. 2017, 45, 12536–12550. [Google Scholar] [CrossRef]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a Link between Genetic and Biochemical Analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef]
- Song, J.J.; Liu, J.; Tolia, N.H.; Schneiderman, J.; Smith, S.K.; Martienssen, R.A.; Hannon, G.J.; Joshua-Tor, L. The Crystal Structure of the Argonaute2 PAZ Domain Reveals an RNA Binding Motif in RNAi Effector Complexes. Nat. Struct. Mol. Biol. 2003, 10, 1026–1032. [Google Scholar] [CrossRef]
- Kataoka, Y.; Takeichi, M.; Uemura, T. Developmental Roles and Molecular Characterization of a Drosophila Homologue of Arabidopsis Argonaute1, the Founder of a Novel Gene Superfamily. Genes. Cells 2001, 6, 313–325. [Google Scholar] [CrossRef]
- Rubio, M.; Maestro, J.L.; Piulachs, M.D.; Belles, X. Conserved Association of Argonaute 1 and 2 Proteins with MiRNA and SiRNA Pathways throughout Insect Evolution, from Cockroaches to Flies. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2018, 1861, 554–560. [Google Scholar] [CrossRef]
- Song, J.J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal Structure of Argonaute and Its Implications for RISC Slicer Activity. Science 2004, 305, 1434–1437. [Google Scholar] [CrossRef]
- Schirle, N.T.; MacRae, I.J. The Crystal Structure of Human Argonaute2. Science 2012, 336, 1037–1040. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Oe, A.; Nishida, K.M.; Yamashita, K.; Kajiya, A.; Hirano, S.; Matsumoto, N.; Dohmae, N.; Ishitani, R.; Saito, K.; et al. Crystal Structure of Drosophila Piwi. Nat. Commun. 2020, 11, 858. [Google Scholar] [CrossRef]
- Okamura, K.; Ishizuka, A.; Siomi, H.; Siomi, M.C. Distinct Roles for Argonaute Proteins in Small RNA-Directed RNA Cleavage Pathways. Genes. Dev. 2004, 18, 1655–1666. [Google Scholar] [CrossRef]
- van Rij, R.P.; Saleh, M.C.; Berry, B.; Foo, C.; Houk, A.; Antoniewski, C.; Andino, R. The RNA Silencing Endonuclease Argonaute 2 Mediates Specific Antiviral Immunity in Drosophila Melanogaster. Genes. Dev. 2006, 20, 2985–2995. [Google Scholar] [CrossRef]
- Förstemann, K.; Horwich, M.D.; Wee, L.M.; Tomari, Y.; Zamore, P.D. Drosophila MicroRNAs Are Sorted into Functionally Distinct Argonaute Complexes after Production by Dicer-1. Cell 2007, 130, 287–297. [Google Scholar] [CrossRef]
- Tomari, Y.; Du, T.; Zamore, P.D. Sorting of Drosophila Small Silencing RNAs. Cell 2007, 130, 299–308. [Google Scholar] [CrossRef]
- Czech, B.; Zhou, R.; Erlich, Y.; Brennecke, J.; Binari, R.; Villalta, C.; Gordon, A.; Perrimon, N.; Hannon, G.J. Hierarchical Rules for Argonaute Loading in Drosophila. Mol. Cell 2009, 36, 445–456. [Google Scholar] [CrossRef]
- Okamura, K.; Liu, N.; Lai, E.C. Distinct Mechanisms for MicroRNA Strand Selection by Drosophila Argonautes. Mol. Cell 2009, 36, 431–444. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Xu, J.; Seitz, H.; Weng, Z.; Zamore, P.D. Sorting of Drosophila Small Silencing RNAs Partitions MicroRNA* Strands into the RNA Interference Pathway. RNA 2010, 16, 43–56. [Google Scholar] [CrossRef]
- Ameres, S.L.; Hung, J.H.; Xu, J.; Weng, Z.; Zamore, P.D. Target RNA-Directed Tailing and Trimming Purifies the Sorting of Endo-SiRNAs between the Two Drosophila Argonaute Proteins. RNA 2011, 17, 54–63. [Google Scholar] [CrossRef]
- Nishida, K.M.; Miyoshi, K.; Ogino, A.; Miyoshi, T.; Siomi, H.; Siomi, M.C. Roles of R2D2, a Cytoplasmic D2 Body Component, in the Endogenous SiRNA Pathway in Drosophila. Mol. Cell 2013, 49, 680–691. [Google Scholar] [CrossRef]
- Liu, Q.; Rand, T.A.; Kalidas, S.; Du, F.; Kim, H.E.; Smith, D.P.; Wang, X. R2D2, a Bridge between the Initiation and Effector Steps of the Drosophila RNAi Pathway. Science 2003, 301, 1921–1925. [Google Scholar] [CrossRef]
- Iwasaki, S.; Sasaki, H.M.; Sakaguchi, Y.; Suzuki, T.; Tadakuma, H.; Tomari, Y. Defining Fundamental Steps in the Assembly of the Drosophila RNAi Enzyme Complex. Nature 2015, 521, 533–536. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, G.; Juranek, S.; Tuschl, T.; Patel, D.J. Structure of the Guide-Strand-Containing Argonaute Silencing Complex. Nature 2008, 456, 209–213. [Google Scholar] [CrossRef]
- Lingel, A.; Simon, B.; Izaurralde, E.; Sattler, M. Structure and Nucleic-Acid Binding of the Drosophila Argonaute 2 PAZ Domain. Nature 2003, 426, 465–469. [Google Scholar] [CrossRef]
- Kwak, P.B.; Tomari, Y. The N Domain of Argonaute Drives Duplex Unwinding during RISC Assembly. Nat. Struct. Mol. Biol. 2012, 19, 145–151. [Google Scholar] [CrossRef]
- Gu, S.; Jin, L.; Huang, Y.; Zhang, F.; Kay, M.A. Slicing-Independent RISC Activation Requires the Argonaute PAZ Domain. Curr. Biol. 2012, 22, 1536–1542. [Google Scholar] [CrossRef]
- Park, J.H.; Shin, C. Slicer-Independent Mechanism Drives Small-RNA Strand Separation during Human RISC Assembly. Nucleic Acids Res. 2015, 43, 9418–9433. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ye, X.; Jiang, F.; Liang, C.; Chen, D.; Peng, J.; Kinch, L.N.; Grishin, N.V.; Liu, Q. C3PO, an Endoribonuclease That Promotes RNAi by Facilitating RISC Activation. Science 2009, 325, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Horwich, M.D.; Li, C.; Matranga, C.; Vagin, V.; Farley, G.; Wang, P.; Zamore, P.D. The Drosophila RNA Methyltransferase, DmHen1, Modifies Germline PiRNAs and Single-Stranded SiRNAs in RISC. Curr. Biol. 2007, 17, 1265–1272. [Google Scholar] [CrossRef]
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/Hsp90 Chaperone Machinery Mediates ATP-Dependent RISC Loading of Small RNA Duplexes. Mol. Cell 2010, 39, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, T.; Takeuchi, A.; Siomi, H.; Siomi, M.C. A Direct Role for Hsp90 in Pre-RISC Formation in Drosophila. Nat. Struct. Mol. Biol. 2010, 17, 1024–1026. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 Is the Catalytic Engine of Mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Tuschl, T. RISC Is a 5′ Phosphomonoester-Producing RNA Endonuclease. Genes. Dev. 2004, 18, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Lima, W.F.; de Hoyos, C.L.; Liang, X.H.; Crooke, S.T. RNA Cleavage Products Generated by Antisense Oligonucleotides and SiRNAs Are Processed by the RNA Surveillance Machinery. Nucleic Acids Res. 2016, 44, 3351–3363. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. Bantam Encodes a Developmentally Regulated MicroRNA That Controls Cell Proliferation and Regulates the Proapoptotic Gene Hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Li, W.; Zhao, H.; Gao, L.; Fan, Y.; Zhou, S. The MicroRNAs Let 7 and Mir 278 Regulate Insect Metamorphosis and Oogenesis by Targeting the Juvenile Hormone Early Response Gene Krüppel Homolog. Development 2018, 145, dev.170670. [Google Scholar] [CrossRef]
- Kang, L.; Wang, M.; Cao, X.; Tang, S.; Xia, D.; Shen, X.; Zhao, Q. Inhibition of Expression of BmNPV Cg30 by Bmo-MiRNA-390 Is a Host Response to Baculovirus Invasion. Arch. Virol. 2018, 163, 2719–2725. [Google Scholar] [CrossRef]
- Ma, K.; Li, F.; Tang, Q.; Liang, P.; Liu, Y.; Zhang, B.; Gao, X. CYP4CJ1-Mediated Gossypol and Tannic Acid Tolerance in Aphis Gossypii Glover. Chemosphere 2019, 219, 961–970. [Google Scholar] [CrossRef]
- Aravin, A.A.; Lagos-Quintana, M.; Yalcin, A.; Zavolan, M.; Marks, D.; Snyder, B.; Gaasterland, T.; Meyer, J.; Tuschl, T. The Small RNA Profile during Drosophila Melanogaster Development. Dev. Cell 2003, 5, 337–350. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human MicroRNAs Are Processed from Capped, Polyadenylated Transcripts That Can Also Function as MRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- Church, V.A.; Pressman, S.; Isaji, M.; Truscott, M.; Cizmecioglu, N.T.; Buratowski, S.; Frolov, M.V.; Carthew, R.W. Microprocessor Recruitment to Elongating RNA Polymerase II Is Required for Differential Expression of MicroRNAs. Cell Rep. 2017, 20, 3123–3134. [Google Scholar] [CrossRef][Green Version]
- Xiong, H.; Qian, J.; He, T.; Li, F. Independent Transcription of MiR-281 in the Intron of ODA in Drosophila Melanogaster. Biochem. Biophys. Res. Commun. 2009, 378, 883–889. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA Maturation: Stepwise Processing and Subcellular Localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef]
- Landthaler, M.; Yalcin, A.; Tuschl, T. The Human DiGeorge Syndrome Critical Region Gene 8 and Its D. Melanogaster Homolog Are Required for MiRNA Biogenesis. Curr. Biol. 2004, 14, 2162–2167. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of Primary MicroRNAs by the Microprocessor Complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The Nuclear RNase III Drosha Initiates MicroRNA Processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Ma, H.; Wu, Y.; Choi, J.G.; Wu, H. Lower and Upper Stem-Single-Stranded RNA Junctions Together Determine the Drosha Cleavage Site. Proc. Natl. Acad. Sci. USA 2013, 110, 20687–20692. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.S. Functional Anatomy of the Human Microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef]
- Kwon, S.C.; Nguyen, T.A.; Choi, Y.G.; Jo, M.H.; Hohng, S.; Kim, V.N.; Woo, J.S. Structure of Human DROSHA. Cell 2016, 164, 81–90. [Google Scholar] [CrossRef]
- Partin, A.C.; Zhang, K.; Jeong, B.C.; Herrell, E.; Li, S.; Chiu, W.; Nam, Y. Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA. Mol. Cell 2020, 78, 411–422.e4. [Google Scholar] [CrossRef]
- Jin, W.; Wang, J.; Liu, C.P.; Wang, H.W.; Xu, R.M. Structural Basis for Pri-MiRNA Recognition by Drosha. Mol. Cell 2020, 78, 423–433.e5. [Google Scholar] [CrossRef]
- Herbert, K.M.; Sarkar, S.K.; Mills, M.; De La Herran, H.C.D.; Neuman, K.C.; Steitz, J.A. A Heterotrimer Model of the Complete Microprocessor Complex Revealed by Single-Molecule Subunit Counting. RNA 2016, 22, 175–183. [Google Scholar] [CrossRef]
- Fang, W.; Bartel, D.P. The Menu of Features That Define Primary MicroRNAs and Enable de Novo Design of MicroRNA Genes. Mol. Cell 2015, 60, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic MicroRNA Precursors That Bypass Drosha Processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The Mirtron Pathway Generates MicroRNA-Class Regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Shibata, S.; Sasaki, M.; Miki, T.; Shimamoto, A.; Furuichi, Y.; Katahira, J.; Yoneda, Y. Exportin-5 Orthologues Are Functionally Divergent among Species. Nucleic Acids Res. 2006, 34, 4711–4721. [Google Scholar] [CrossRef] [PubMed]
- Singh, C.P.; Singh, J.; Nagaraju, J. A Baculovirus-Encoded MicroRNA (MiRNA) Suppresses Its Host MiRNA Biogenesis by Regulating the Exportin-5 Cofactor Ran. J. Virol. 2012, 86, 7867–7879. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Cullen, B.R. Structural Requirements for Pre-MicroRNA Binding and Nuclear Export by Exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef] [PubMed]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A High-Resolution Structure of the Pre-Microrna Nuclear Export Machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, F.; Yang, F.; Meng, Z.; Zeng, Y. Selectivity of Exportin 5 Binding to Human Precursor MicroRNAs. RNA Biol. 2021, 18, 730–737. [Google Scholar] [CrossRef]
- Tsutsumi, A.; Kawamata, T.; Izumi, N.; Seitz, H.; Tomari, Y. Recognition of the Pre-MiRNA Structure by Drosophila Dicer-1. Nat. Struct. Mol. Biol. 2011, 18, 1153–1158. [Google Scholar] [CrossRef]
- Lim, M.Y.T.; Ng, A.W.T.; Chou, Y.; Lim, T.P.; Simcox, A.; Tucker-Kellogg, G.; Okamura, K. The Drosophila Dicer-1 Partner Loquacious Enhances MiRNA Processing from Hairpins with Unstable Structures at the Dicing Site. Cell Rep. 2016, 15, 1795–1808. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jiang, F.; Ye, X.; Liu, X.; Fincher, L.; McKearin, D.; Liu, Q. Dicer-1 and R3D1-L Catalyze MicroRNA Maturation in Drosophila. Genes. Dev. 2005, 19, 1674–1679. [Google Scholar] [CrossRef]
- Förstemann, K.; Tomari, Y.; Du, T.; Vagin, V.V.; Denli, A.M.; Bratu, D.P.; Klattenhoff, C.; Theurkauf, W.E.; Zamore, P.D. Normal MicroRNA Maturation and Germ-Line Stem Cell Maintenance Requires Loquacious, a Double-Stranded RNA-Binding Domain Protein. PLoS Biol. 2005, 3, e236. [Google Scholar] [CrossRef]
- Saito, K.; Ishizuka, A.; Siomi, H.; Siomi, M.C. Processing of Pre-MicroRNAs by the Dicer-1–Loquacious Complex in Drosophila Cells. PLoS Biol. 2005, 3, e235. [Google Scholar] [CrossRef]
- Liu, X.; Park, J.K.; Jiang, F.; Liu, Y.; Mckearin, D.; Liu, Q. Dicer-1, but Not Loquacious, Is Critical for Assembly of MiRNA-Induced Silencing Complexes. RNA 2007, 13, 2324–2329. [Google Scholar] [CrossRef]
- Kawamata, T.; Seitz, H.; Tomari, Y. Structural Determinants of MiRNAs for RISC Loading and Slicer-Independent Unwinding. Nat. Struct. Mol. Biol. 2009, 16, 953–960. [Google Scholar] [CrossRef]
- Chawla, G.; Sokol, N.S. ADAR Mediates Differential Expression of Polycistronic MicroRNAs. Nucleic Acids Res. 2014, 42, 5245–5255. [Google Scholar] [CrossRef]
- Reimão-Pinto, M.M.; Ignatova, V.; Burkard, T.R.; Hung, J.H.; Manzenreither, R.A.; Sowemimo, I.; Herzog, V.A.; Reichholf, B.; Fariña-Lopez, S.; Ameres, S.L. Uridylation of RNA Hairpins by Tailor Confines the Emergence of MicroRNAs in Drosophila. Mol. Cell 2015, 59, 203–216. [Google Scholar] [CrossRef]
- Agarwal, V.; Subtelny, A.O.; Thiru, P.; Ulitsky, I.; Bartel, D.P. Predicting MicroRNA Targeting Efficacy in Drosophila. Genome Biol. 2018, 19, 152. [Google Scholar] [CrossRef]
- Lai, E.C. Micro RNAs Are Complementary to 3′ UTR Sequence Motifs That Mediate Negative Post-Transcriptional Regulation. Nat. Genet. 2002, 30, 363–364. [Google Scholar] [CrossRef] [PubMed]
- Stark, A.; Brennecke, J.; Russell, R.B.; Cohen, S.M. Identification of Drosophila MicroRNA Targets. PLoS Biol. 2003, 1, e60. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Kuzin, A.; Kundu, M.; Brody, T.; Odenwald, W.F. The Drosophila Nerfin-1 MRNA Requires Multiple MicroRNAs to Regulate Its Spatial and Temporal Translation Dynamics in the Developing Nervous System. Dev. Biol. 2007, 310, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.J.; Hyun, S. Multiple Targets of the MicroRNA MiR-8 Contribute to Immune Homeostasis in Drosophila. Dev. Comp. Immunol. 2014, 45, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Zekri, L.; Huntzinger, E.; Heimstädt, S.; Izaurralde, E. The Silencing Domain of GW182 Interacts with PABPC1 to Promote Translational Repression and Degradation of MicroRNA Targets and Is Required for Target Release. Mol. Cell Biol. 2009, 29, 6220–6231. [Google Scholar] [CrossRef]
- Zdanowicz, A.; Thermann, R.; Kowalska, J.; Jemielity, J.; Duncan, K.; Preiss, T.; Darzynkiewicz, E.; Hentze, M.W. Drosophila MiR2 Primarily Targets the M7GpppN Cap Structure for Translational Repression. Mol. Cell 2009, 35, 881–888. [Google Scholar] [CrossRef]
- Fukaya, T.; Iwakawa, H.O.; Tomari, Y. MicroRNAs Block Assembly of EIF4F Translation Initiation Complex in Drosophila. Mol. Cell 2014, 56, 67–78. [Google Scholar] [CrossRef]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. MRNA Degradation by MiRNAs and GW182 Requires Both CCR4:NOT Deadenylase and DCP1:DCP2 Decapping Complexes. Genes. Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef]
- Eulalio, A.; Rehwinkel, J.; Stricker, M.; Huntzinger, E.; Yang, S.F.; Doerks, T.; Dorner, S.; Bork, P.; Boutros, M.; Izaurralde, E. Target-Specific Requirements for Enhancers of Decapping in MiRNA-Mediated Gene Silencing. Genes. Dev. 2007, 21, 2558–2570. [Google Scholar] [CrossRef]
- Braun, J.E.; Huntzinger, E.; Fauser, M.; Izaurralde, E. GW182 Proteins Directly Recruit Cytoplasmic Deadenylase Complexes to MiRNA Targets. Mol. Cell 2011, 44, 120–133. [Google Scholar] [CrossRef]
- Nishihara, T.; Zekri, L.; Braun, J.E.; Izaurralde, E. MiRISC Recruits Decapping Factors to MiRNA Targets to Enhance Their Degradation. Nucleic Acids Res. 2013, 41, 8692–8705. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Behm-Ansmant, I.; Gatfield, D.; Izaurralde, E. A Crucial Role for GW182 and the DCP1:DCP2 Decapping Complex in MiRNA-Mediated Gene Silencing. RNA 2005, 11, 1640–1647. [Google Scholar] [CrossRef]
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A Distinct Small RNA Pathway Silences Selfish Genetic Elements in the Germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Gonzalez, J.; Qi, H.; Liu, N.; Lin, H. Piwi Is a Key Regulator of Both Somatic and Germline Stem Cells in the Drosophila Testis. Cell Rep. 2015, 12, 150–161. [Google Scholar] [CrossRef]
- Klein, J.D.; Qu, C.; Yang, X.; Fan, Y.; Tang, C.; Peng, J.C. C-Fos Repression by Piwi Regulates Drosophila Ovarian Germline Formation and Tissue Morphogenesis. PLoS Genet. 2016, 12, e1006281. [Google Scholar] [CrossRef]
- Dietrich, I.; Shi, X.; McFarlane, M.; Watson, M.; Blomström, A.L.; Skelton, J.K.; Kohl, A.; Elliott, R.M.; Schnettler, E. The Antiviral RNAi Response in Vector and Non-Vector Cells against Orthobunyaviruses. PLoS Negl. Trop. Dis. 2017, 11, e0005272. [Google Scholar] [CrossRef]
- Katsuma, S.; Kawamoto, M.; Shoji, K.; Aizawa, T.; Kiuchi, T.; Izumi, N.; Ogawa, M.; Mashiko, T.; Kawasaki, H.; Sugano, S.; et al. Transcriptome Profiling Reveals Infection Strategy of an Insect Maculavirus. DNA Res. 2018, 25, 277–286. [Google Scholar] [CrossRef]
- Kotov, A.A.; Adashev, V.E.; Godneeva, B.K.; Ninova, M.; Shatskikh, A.S.; Bazylev, S.S.; Aravin, A.A.; Olenina, L.v. PiRNA Silencing Contributes to Interspecies Hybrid Sterility and Reproductive Isolation in Drosophila Melanogaster. Nucleic Acids Res. 2019, 47, 4255–4271. [Google Scholar] [CrossRef]
- Goriaux, C.; Desset, S.; Renaud, Y.; Vaury, C.; Brasset, E. Transcriptional Properties and Splicing of the Flamenco PiRNA Cluster. EMBO Rep. 2014, 15, 411–418. [Google Scholar] [CrossRef]
- Dennis, C.; Brasset, E.; Sarkar, A.; Vaury, C. Export of PiRNA Precursors by EJC Triggers Assembly of Cytoplasmic Yb-Body in Drosophila. Nat. Commun. 2016, 7, 13739. [Google Scholar] [CrossRef]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized PiRNA Pathways Act in Germline and Somatic Tissues of the Drosophila Ovary. Cell 2009, 137, 522–535. [Google Scholar] [CrossRef]
- Mohn, F.; Sienski, G.; Handler, D.; Brennecke, J. The Rhino-Deadlock-Cutoff Complex Licenses Noncanonical Transcription of Dual-Strand PiRNA Clusters in Drosophila. Cell 2014, 157, 1364–1379. [Google Scholar] [CrossRef]
- Klattenhoff, C.; Xi, H.; Li, C.; Lee, S.; Xu, J.; Khurana, J.S.; Zhang, F.; Schultz, N.; Koppetsch, B.S.; Nowosielska, A.; et al. The Drosophila HP1 Homolog Rhino Is Required for Transposon Silencing and PiRNA Production by Dual-Strand Clusters. Cell 2009, 138, 1137–1149. [Google Scholar] [CrossRef]
- Andersen, P.R.; Tirian, L.; Vunjak, M.; Brennecke, J. A Heterochromatin-Dependent Transcription Machinery Drives PiRNA Expression. Nature 2017, 549, 54–59. [Google Scholar] [CrossRef]
- Chen, Y.C.A.; Stuwe, E.; Luo, Y.; Ninova, M.; le Thomas, A.; Rozhavskaya, E.; Li, S.; Vempati, S.; Laver, J.D.; Patel, D.J.; et al. Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of PiRNA Precursors. Mol. Cell 2016, 63, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, J.; Schultz, N.; Zhang, F.; Parhad, S.S.; Tu, S.; Vreven, T.; Zamore, P.D.; Weng, Z.; Theurkauf, W.E. The HP1 Homolog Rhino Anchors a Nuclear Complex That Suppresses PiRNA Precursor Splicing. Cell 2014, 157, 1353–1363. [Google Scholar] [CrossRef]
- ElMaghraby, M.F.; Andersen, P.R.; Pühringer, F.; Hohmann, U.; Meixner, K.; Lendl, T.; Tirian, L.; Brennecke, J. A Heterochromatin-Specific RNA Export Pathway Facilitates PiRNA Production. Cell 2019, 178, 964–979.e20. [Google Scholar] [CrossRef]
- Kneuss, E.; Munafò, M.; Eastwood, E.L.; Deumer, U.S.; Preall, J.B.; Hannon, G.J.; Czech, B. Specialization of the Drosophila Nuclear Export Family Protein Nxf3 for PiRNA Precursor Export. Genes. Dev. 2019, 33, 1208–1220. [Google Scholar] [CrossRef]
- Ai, K.L.; Kai, T. Unique Germ-Line Organelle, Nuage, Functions to Repress Selfish Genetic Elements in Drosophila Melanogaster. Proc. Natl. Acad. Sci. USA 2007, 104, 6714–6719. [Google Scholar] [CrossRef]
- Qi, H.; Watanabe, T.; Ku, H.Y.; Liu, N.; Zhong, M.; Lin, H. The Yb Body, a Major Site for Piwi-Associated RNA Biogenesis and a Gateway for Piwi Expression and Transport to the Nucleus in Somatic Cells. J. Biol. Chem. 2011, 286, 3789–3797. [Google Scholar] [CrossRef] [PubMed]
- Szakmary, A.; Reedy, M.; Qi, H.; Lin, H. The Yb Protein Defines a Novel Organelle and Regulates Male Germline Stem Cell Self-Renewal in Drosophila Melanogaster. J. Cell Biol. 2009, 185, 613–627. [Google Scholar] [CrossRef]
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains PiRNA Production in Most Animals. Mol. Cell 2018, 71, 775–790.e5. [Google Scholar] [CrossRef]
- Nishida, K.M.; Sakakibara, K.; Iwasaki, Y.W.; Yamada, H.; Murakami, R.; Murota, Y.; Kawamura, T.; Kodama, T.; Siomi, H.; Siomi, M.C. Hierarchical Roles of Mitochondrial Papi and Zucchini in Bombyx Germline PiRNA Biogenesis. Nature 2018, 555, 260–264. [Google Scholar] [CrossRef]
- Izumi, N.; Shoji, K.; Suzuki, Y.; Katsuma, S.; Tomari, Y. Zucchini Consensus Motifs Determine the Mechanism of Pre-PiRNA Production. Nature 2020, 578, 311–316. [Google Scholar] [CrossRef]
- Han, B.W.; Wang, W.; Li, C.; Weng, Z.; Zamore, P.D. Noncoding RNA. PiRNA-Guided Transposon Cleavage Initiates Zucchini-Dependent, Phased PiRNA Production. Science 2015, 348, 817–821. [Google Scholar] [CrossRef]
- Mohn, F.; Handler, D.; Brennecke, J. Noncoding RNA. PiRNA-Guided Slicing Specifies Transcripts for Zucchini-Dependent, Phased PiRNA Biogenesis. Science 2015, 348, 812–817. [Google Scholar] [CrossRef] [PubMed]
- Kawaoka, S.; Izumi, N.; Katsuma, S.; Tomari, Y. 3′ End Formation of PIWI-Interacting RNAs in Vitro. Mol. Cell 2011, 43, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Izumi, N.; Shoji, K.; Sakaguchi, Y.; Honda, S.; Kirino, Y.; Suzuki, T.; Katsuma, S.; Tomari, Y. Identification and Functional Analysis of the Pre-PiRNA 3′ Trimmer in Silkworms. Cell 2016, 164, 962–973. [Google Scholar] [CrossRef]
- Hayashi, R.; Schnabl, J.; Handler, D.; Mohn, F.; Ameres, S.L.; Brennecke, J. Genetic and Mechanistic Diversity of PiRNA 3′-End Formation. Nature 2016, 539, 588–592. [Google Scholar] [CrossRef]
- Saito, K.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Siomi, H.; Siomi, M.C. Pimet, the Drosophila Homolog of HEN1, Mediates 2′-O-Methylation of Piwi- Interacting RNAs at Their 3′ Ends. Genes. Dev. 2007, 21, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Simanshu, D.K.; Ma, J.B.; Patel, D.J. Structural Basis for PiRNA 2′-O-Methylated 3′-End Recognition by Piwi PAZ (Piwi/Argonaute/Zwille) Domains. Proc. Natl. Acad. Sci. USA 2011, 108, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Gunawardane, L.S.; Saito, K.; Nishida, K.M.; Miyoshi, K.; Kawamura, Y.; Nagami, T.; Siomi, H.; Siomi, M.C. A Slicer-Mediated Mechanism for Repeat-Associated SiRNA 5′ End Formation in Drosophila. Science 2007, 315, 1587–1590. [Google Scholar] [CrossRef] [PubMed]
- Rouget, C.; Papin, C.; Boureux, A.; Meunier, A.C.; Franco, B.; Robine, N.; Lai, E.C.; Pelisson, A.; Simonelig, M. Maternal MRNA Deadenylation and Decay by the PiRNA Pathway in the Early Drosophila Embryo. Nature 2010, 467, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Ríos, P.; Chartier, A.; Pierson, S.; Simonelig, M. Aubergine and PiRNAs Promote Germline Stem Cell Self-Renewal by Repressing the Proto-Oncogene Cbl. EMBO J. 2017, 36, 3194–3211. [Google Scholar] [CrossRef] [PubMed]
- Ai, K.L.; Tao, L.; Kai, T. PiRNAs Mediate Posttranscriptional Retroelement Silencing and Localization to Pi-Bodies in the Drosophila Germline. J. Cell Biol. 2009, 186, 333–342. [Google Scholar] [CrossRef]
- Klenov, M.S.; Sokolova, O.A.; Yakushev, E.Y.; Stolyarenko, A.D.; Mikhaleva, E.A.; Lavrov, S.A.; Gvozdev, V.A. Separation of Stem Cell Maintenance and Transposon Silencing Functions of Piwi Protein. Proc. Natl. Acad. Sci. USA 2011, 108, 18760–18765. [Google Scholar] [CrossRef] [PubMed]
- Sienski, G.; Dönertas, D.; Brennecke, J. Transcriptional Silencing of Transposons by Piwi and Maelstrom and Its Impact on Chromatin State and Gene Expression. Cell 2012, 151, 964–980. [Google Scholar] [CrossRef]
- Shpiz, S.; Ryazansky, S.; Olovnikov, I.; Abramov, Y.; Kalmykova, A. Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-SiRNAs at the Target Sites in the Drosophila Germline. PLoS Genet. 2014, 10, e1004138. [Google Scholar] [CrossRef]
- Kordyukova, M.; Sokolova, O.; Morgunova, V.; Ryazansky, S.; Akulenko, N.; Glukhov, S.; Kalmykova, A. Nuclear Ccr4-Not Mediates the Degradation of Telomeric and Transposon Transcripts at Chromatin in the Drosophila Germline. Nucleic Acids Res. 2020, 48, 141–156. [Google Scholar] [CrossRef]
- Whangbo, J.S.; Hunter, C.P. Environmental RNA Interference. Trends Genet. 2008, 24, 297–305. [Google Scholar] [CrossRef]
- Hunter, C.P.; Winston, W.M.; Molodowitch, C.; Feinberg, E.H.; Shih, J.; Sutherlin, M.; Wright, A.J.; Fitzgerald, M.C. Systemic RNAi in Caenorhabditis Elegans. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 95–100. [Google Scholar] [CrossRef]
- Winston, W.M.; Sutherlin, M.; Wright, A.J.; Feinberg, E.H.; Hunter, C.P. Caenorhabditis Elegans SID-2 Is Required for Environmental RNA Interference. Proc. Natl. Acad. Sci. USA 2007, 104, 10565–10570. [Google Scholar] [CrossRef] [PubMed]
- Winston, W.M.; Molodowitch, C.; Hunter, C.P. Systemic RNAi in C. Elegans Requires the Putative Transmembrane Protein SID-1. Science 2002, 295, 2456–2459. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, E.H.; Hunter, C.P. Transport of DsRNA into Cells by the Transmembrane Protein SID-1. Science 2003, 301, 1545–1547. [Google Scholar] [CrossRef]
- Hinas, A.; Wright, A.J.; Hunter, C.P. SID-5 Is an Endosome-Associated Protein Required for Efficient Systemic RNAi in C. Elegans. Curr. Biol. 2012, 22, 1938–1943. [Google Scholar] [CrossRef]
- Jose, A.M.; Kim, Y.A.; Leal-Ekman, S.; Hunter, C.P. Conserved Tyrosine Kinase Promotes the Import of Silencing RNA into Caenorhabditis Elegans Cells. Proc. Natl. Acad. Sci. USA 2012, 109, 14520–14525. [Google Scholar] [CrossRef]
- Gao, J.; Zhao, L.; Luo, Q.; Liu, S.; Lin, Z.; Wang, P.; Fu, X.; Chen, J.; Zhang, H.; Lin, L.; et al. An EHBP-1-SID-3-DYN-1 Axis Promotes Membranous Tubule Fission during Endocytic Recycling. PLoS Genet. 2020, 16, e1008763. [Google Scholar] [CrossRef]
- Dowling, D.; Pauli, T.; Donath, A.; Meusemann, K.; Podsiadlowski, L.; Petersen, M.; Peters, R.S.; Mayer, C.; Liu, S.; Zhou, X.; et al. Phylogenetic Origin and Diversification of RNAi Pathway Genes in Insects. Genome Biol. Evol. 2016, 8, 3784–3793. [Google Scholar] [CrossRef]
- Tomoyasu, Y.; Miller, S.C.; Tomita, S.; Schoppmeier, M.; Grossmann, D.; Bucher, G. Exploring Systemic RNA Interference in Insects: A Genome-Wide Survey for RNAi Genes in Tribolium. Genome Biol. 2008, 9, R10. [Google Scholar] [CrossRef]
- Pinheiro, D.H.; Vélez, A.M.; Fishilevich, E.; Wang, H.; Carneiro, N.P.; Valencia-Jiménez, A.; Valicente, F.H.; Narva, K.E.; Siegfried, B.D. Clathrin-Dependent Endocytosis Is Associated with RNAi Response in the Western Corn Rootworm, Diabrotica virgifera virgifera LeConte. PLoS ONE 2018, 13, e0201849. [Google Scholar] [CrossRef]
- Dong, X.; Li, X.; Li, Q.; Jia, H.; Zhang, H. The Inducible Blockage of RNAi Reveals a Role for Polyunsaturated Fatty Acids in the Regulation of DsRNA-Endocytic Capacity in Bactrocera dorsalis. Sci. Rep. 2017, 7, 5584. [Google Scholar] [CrossRef]
- Cappelle, K.; de Oliveira, C.F.R.; van Eynde, B.; Christiaens, O.; Smagghe, G. The Involvement of Clathrin-Mediated Endocytosis and Two Sid-1-like Transmembrane Proteins in Double-Stranded RNA Uptake in the Colorado Potato Beetle Midgut. Insect Mol. Biol. 2016, 25, 315–323. [Google Scholar] [CrossRef]
- Yoon, J.S.; Shukla, J.N.; Gong, Z.J.; Mogilicherla, K.; Palli, S.R. RNA Interference in the Colorado Potato Beetle, Leptinotarsa decemlineata: Identification of Key Contributors. Insect Biochem. Mol. Biol. 2016, 78, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.C.; van Rij, R.P.; Hekele, A.; Gillis, A.; Foley, E.; O’Farrell, P.H.; Andino, R. The Endocytic Pathway Mediates Cell Entry of DsRNA to Induce RNAi Silencing. Nat. Cell Biol. 2006, 8, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Ulvila, J.; Parikka, M.; Kleino, A.; Sormunen, R.; Ezekowitz, R.A.; Kocks, C.; Rämet, M. Double-Stranded RNA Is Internalized by Scavenger Receptor-Mediated Endocytosis in Drosophila S2 Cells. J. Biol. Chem. 2006, 281, 14370–14375. [Google Scholar] [CrossRef] [PubMed]
- Aung, K.M.; Boldbaatar, D.; Umemiya-Shirafuji, R.; Liao, M.; Xuenan, X.; Suzuki, H.; Galay, R.; Tanaka, T.; Fujisaki, K. Scavenger Receptor Mediates Systemic RNA Interference in Ticks. PLoS ONE 2011, 6, e28407. [Google Scholar] [CrossRef] [PubMed]
- Wynant, N.; Santos, D.; van Wielendaele, P.; vanden Broeck, J. Scavenger Receptor-Mediated Endocytosis Facilitates RNA Interference in the Desert Locust, Schistocerca gregaria. Insect Mol. Biol. 2014, 23, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Gao, X.; Xu, J.; Liang, X.; Li, Q.; Yao, J.; Zhu, K.Y. Clathrin-Dependent Endocytosis Plays a Predominant Role in Cellular Uptake of Double-Stranded RNA in the Red Flour Beetle. Insect Biochem. Mol. Biol. 2015, 60, 68–77. [Google Scholar] [CrossRef]
- Meng, F.; Yang, M.; Li, Y.; Li, T.; Liu, X.; Wang, G.; Wang, Z.; Jin, X.; Li, W. Functional Analysis of RNA Interference-Related Soybean Pod Borer (Lepidoptera) Genes Based on Transcriptome Sequences. Front. Physiol. 2018, 9, 383. [Google Scholar] [CrossRef]
- Abbasi, R.; Heschuk, D.; Kim, B.; Whyard, S. A Novel Paperclip Double-Stranded RNA Structure Demonstrates Clathrin-Independent Uptake in the Mosquito Aedes Aegypti. Insect Biochem. Mol. Biol. 2020, 127, 103492. [Google Scholar] [CrossRef] [PubMed]
- Rocha, J.J.E.; Korolchuk, V.I.; Robinson, I.M.; O’Kane, C.J. A Phagocytic Route for Uptake of Double-Stranded RNA in RNAi. PLoS ONE 2011, 6, e19087. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, R.; Ramaseshadri, P.; Anderson, J.; Bachman, P.; Clinton, W.; Flannagan, R.; Ilagan, O.; Lawrence, C.; Levine, S.; Moar, W.; et al. Characterizing the Mechanism of Action of Double-Stranded RNA Activity against Western Corn Rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 2012, 7, e47534. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.C.; Miyata, K.; Brown, S.J.; Tomoyasu, Y. Dissecting Systemic RNA Interference in the Red Flour Beetle Tribolium castaneum: Parameters Affecting the Efficiency of RNAi. PLoS ONE 2012, 7, e47431. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.K.; Singh, S.; Mogilicherla, K.; Shukla, J.N.; Palli, S.R. Comparative Analysis of Double-Stranded RNA Degradation and Processing in Insects. Sci. Rep. 2017, 7, 17059. [Google Scholar] [CrossRef]
- Wang, K.; Peng, Y.; Pu, J.; Fu, W.; Wang, J.; Han, Z. Variation in RNAi Efficacy among Insect Species Is Attributable to DsRNA Degradation in Vivo. Insect Biochem. Mol. Biol. 2016, 77, 1–9. [Google Scholar] [CrossRef]
- Guan, R.B.; Li, H.C.; Fan, Y.J.; Hu, S.R.; Christiaens, O.; Smagghe, G.; Miao, X.X. A Nuclease Specific to Lepidopteran Insects Suppresses RNAi. J. Biol. Chem. 2018, 293, 6011–6021. [Google Scholar] [CrossRef]
- Lomate, P.R.; Bonning, B.C. Distinct Properties of Proteases and Nucleases in the Gut, Salivary Gland and Saliva of Southern Green Stink Bug, Nezara viridula. Sci. Rep. 2016, 6, 27587. [Google Scholar] [CrossRef]
- Shukla, J.N.; Kalsi, M.; Sethi, A.; Narva, K.E.; Fishilevich, E.; Singh, S.; Mogilicherla, K.; Palli, S.R. Reduced Stability and Intracellular Transport of DsRNA Contribute to Poor RNAi Response in Lepidopteran Insects. RNA Biol. 2016, 13, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.S.; Gurusamy, D.; Palli, S.R. Accumulation of DsRNA in Endosomes Contributes to Inefficient RNA Interference in the Fall Armyworm, Spodoptera frugiperda. Insect Biochem. Mol. Biol. 2017, 90, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Pressman, S.; Andress, A.P.; Kim, K.; White, J.L.; Cassidy, J.J.; Li, X.; Lubell, K.; Lim, D.H.; Cho, I.S.; et al. Silencing by Small RNAs Is Linked to Endosomal Trafficking. Nat. Cell Biol. 2009, 11, 1150–1156. [Google Scholar] [CrossRef]
- Shi, X.; Liu, X.; Cooper, A.M.W.; Silver, K.; Merzendorfer, H.; Zhu, K.Y.; Zhang, J. Vacuolar (H+)-ATPase Subunit c Is Essential for the Survival and Systemic RNA Interference Response in Locusta migratoria. Pest. Manag. Sci. 2022, 78, 1555–1566. [Google Scholar] [CrossRef]
- Karlikow, M.; Goic, B.; Mongelli, V.; Salles, A.; Schmitt, C.; Bonne, I.; Zurzolo, C.; Saleh, M.C. Drosophila Cells Use Nanotube-like Structures to Transfer DsRNA and RNAi Machinery between Cells. Sci. Rep. 2016, 6, 27085. [Google Scholar] [CrossRef] [PubMed]
- Valiunas, V.; Polosina, Y.Y.; Miller, H.; Potapova, I.A.; Valiuniene, L.; Doronin, S.; Mathias, R.T.; Robinson, R.B.; Rosen, M.R.; Cohen, I.S.; et al. Connexin-Specific Cell-to-Cell Transfer of Short Interfering RNA by Gap Junctions. J. Physiol. 2005, 568, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Garbutt, J.S.; Bellés, X.; Richards, E.H.; Reynolds, S.E. Persistence of Double-Stranded RNA in Insect Hemolymph as a Potential Determiner of RNA Interference Success: Evidence from Manduca sexta and Blattella germanica. J. Insect Physiol. 2013, 59, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zheng, Y.; Chao, Z.; Chen, H.; Zhang, Y.; Yin, M.; Shen, J.; Yan, S. Visualization of the Process of a Nanocarrier-Mediated Gene Delivery: Stabilization, Endocytosis and Endosomal Escape of Genes for Intracellular Spreading. J. Nanobiotechnol. 2022, 20, 124. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Du, J.; Li, S.; Wang, X. Identification and Characterization of a Double-Stranded RNA Degrading Nuclease Influencing RNAi Efficiency in the Rice Leaf Folder Cnaphalocrocis medinalis. Int. J. Mol. Sci. 2022, 23, 3961. [Google Scholar] [CrossRef] [PubMed]
- Sakashita, K.; Tatsuke, T.; Masaki, Y.; Lee, M.; Kawaguch, Y.; Kusakabe, T. DsRNA Binding Activity of Silkworm Larval Hemolymph Is Mediated by Lipophorin Complex. J. Fac. Agric. Kyushu Univ. 2009, 54, 401–406. [Google Scholar] [CrossRef]
- Wynant, N.; Duressa, T.F.; Santos, D.; van Duppen, J.; Proost, P.; Huybrechts, R.; vanden Broeck, J. Lipophorins Can Adhere to DsRNA, Bacteria and Fungi Present in the Hemolymph of the Desert Locust: A Role as General Scavenger for Pathogens in the Open Body Cavity. J. Insect Physiol. 2014, 64, 7–13. [Google Scholar] [CrossRef] [PubMed]
- van den Brande, S.; Gijbels, M.; Wynant, N.; Santos, D.; Mingels, L.; Gansemans, Y.; van Nieuwerburgh, F.; vanden Broeck, J. The Presence of Extracellular MicroRNAs in the Media of Cultured Drosophila Cells. Sci. Rep. 2018, 8, 17312. [Google Scholar] [CrossRef]
- Dhahbi, J.M.; Atamna, H.; Li, R.; Yamakawa, A.; Guerrero, N.; Lam, H.T.; Mote, P.; Spindler, S.R. MicroRNAs Circulate in the Hemolymph of Drosophila and Accumulate Relative to Tissue Micrornas in an Age-Dependent Manner. Genom. Insights 2016, 9, 29–39. [Google Scholar] [CrossRef]
- Tassetto, M.; Kunitomi, M.; Andino, R. Circulating Immune Cells Mediate a Systemic RNAi-Based Adaptive Antiviral Response in Drosophila. Cell 2017, 169, 314–325.e13. [Google Scholar] [CrossRef]
- Mingels, L.; Wynant, N.; Santos, D.; Peeters, P.; Gansemans, Y.; Billen, J.; van Nieuwerburgh, F.; vanden Broeck, J. Extracellular Vesicles Spread the RNA Interference Signal of Tribolium castaneum TcA Cells. Insect Biochem. Mol. Biol. 2020, 122, 103377. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.S.; Kim, K.; Palli, S.R. Double-Stranded RNA in Exosomes: Potential Systemic RNA Interference Pathway in the Colorado Potato Beetle, Leptinotarsa decemlineata. J. Asia Pac. Entomol. 2020, 23, 1160–1164. [Google Scholar] [CrossRef]
- Bucher, G.; Scholten, J.; Klingler, M. Parental RNAi in Tribolium (Coleoptera). Curr. Biol. 2002, 12, R85–R86. [Google Scholar] [CrossRef] [PubMed]
- Horn, T.; Narov, K.D.; Panfilio, K.A. Persistent Parental RNAi in the Beetle Tribolium castaneum Involves Maternal Transmission of Long Double-Stranded RNA. Adv. Genet. 2022, 3, 2100064. [Google Scholar] [CrossRef] [PubMed]
- Khajuria, C.; Vélez, A.M.; Rangasamy, M.; Wang, H.; Fishilevich, E.; Frey, M.L.F.; Carneiro, N.P.; Gandra, P.; Narva, K.E.; Siegfried, B.D. Parental RNA Interference of Genes Involved in Embryonic Development of the Western Corn Rootworm, Diabrotica virgifera virgifera LeConte. Insect Biochem. Mol. Biol. 2015, 63, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Vicari, P.E.; Chang, E.S.; Perondini, A.L.P.; Selivon, D. Parental RNAi Silencing of the Transformer-2 Gene in a Species of the Anastrepha Fraterculus Complex of Cryptic Species (Diptera, Tephritidae). J. Agric. Sci. 2021, 13, p70. [Google Scholar] [CrossRef]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of Coleopteran Insect Pests through RNA Interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.B.; Cai, W.J.; Wang, J.W.; Hong, G.J.; Tao, X.Y.; Wang, L.J.; Huang, Y.P.; Chen, X.Y. Silencing a Cotton Bollworm P450 Monooxygenase Gene by Plant-Mediated RNAi Impairs Larval Tolerance of Gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef]
- Head, G.P.; Carroll, M.W.; Evans, S.P.; Rule, D.M.; Willse, A.R.; Clark, T.L.; Storer, N.P.; Flannagan, R.D.; Samuel, L.W.; Meinke, L.J. Evaluation of SmartStax and SmartStax PRO Maize against Western Corn Rootworm and Northern Corn Rootworm: Efficacy and Resistance Management. Pest. Manag. Sci. 2017, 73, 1883–1899. [Google Scholar] [CrossRef]
- AgbioInvestor. Time and Cost to Develop a New GM Trait; AgbioInvestor: Pathhead, UK, 2022. [Google Scholar]
- Sikora, D.; Rzymski, P. Public Acceptance of GM Foods: A Global Perspective (1999–2019). In Policy Issues in Genetically Modified Crops; Academic Press: Cambridge, MA, USA, 2021; pp. 293–315. [Google Scholar]
- Gordon, K.H.J.; Waterhouse, P.M. RNAi for Insect-Proof Plants. Nat. Biotechnol. 2007, 25, 1231–1232. [Google Scholar] [CrossRef]
- Jin, S.; Singh, N.D.; Li, L.; Zhang, X.; Daniell, H. Engineered Chloroplast DsRNA Silences Cytochrome P450 Monooxygenase, V-ATPase and Chitin synthase Genes in the Insect Gut and Disrupts Helicoverpa armigera Larval Development and Pupation. Plant Biotechnol. J. 2015, 13, 435–446. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, S.A.; Hasse, C.; Ruf, S.; Heckel, D.G.; Bock, R. Pest Control. Full Crop Protection from an Insect Pest by Expression of Long Double-Stranded RNAs in Plastids. Science 2015, 347, 991–994. [Google Scholar] [CrossRef]
- Bally, J.; McIntyre, G.J.; Doran, R.L.; Lee, K.; Perez, A.; Jung, H.; Naim, F.; Larrinua, I.M.; Narva, K.E.; Waterhouse, P.M. In-Plant Protection against Helicoverpa armigera by Production of Long HpRNA in Chloroplasts. Front. Plant Sci. 2016, 7, 1453. [Google Scholar] [CrossRef]
- Burke, W.G.; Kaplanoglu, E.; Kolotilin, I.; Menassa, R.; Donly, C. RNA Interference in the Tobacco Hornworm, Manduca Sexta, Using Plastid-Encoded Long Double-Stranded RNA. Front. Plant Sci. 2019, 10, 313. [Google Scholar] [CrossRef]
- He, W.; Xu, W.; Xu, L.; Fu, K.; Guo, W.; Bock, R.; Zhang, J. Length-Dependent Accumulation of Double-Stranded RNAs in Plastids Affects RNA Interference Efficiency in the Colorado Potato Beetle. J. Exp. Bot. 2020, 71, 2670–2677. [Google Scholar] [CrossRef] [PubMed]
- Kaplanoglu, E.; Kolotilin, I.; Menassa, R.; Donly, C. Plastid Transformation of Micro-Tom Tomato with a Hemipteran Double-Stranded RNA Results in RNA Interference in Multiple Insect Species. Int. J. Mol. Sci. 2022, 23, 3918. [Google Scholar] [CrossRef] [PubMed]
- Bally, J.; Fishilevich, E.; Doran, R.L.; Lee, K.; de Campos, S.B.; German, M.A.; Narva, K.E.; Waterhouse, P.M. Plin-AmiR, a Pre-MicroRNA-Based Technology for Controlling Herbivorous Insect Pests. Plant Biotechnol. J. 2020, 18, 1925–1932. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Pandit, S.S.; Baldwin, I.T. Tobacco Rattle Virus Vector: A Rapid and Transient Means of Silencing Manduca sexta Genes by Plant Mediated RNA Interference. PLoS ONE 2012, 7, e31347. [Google Scholar] [CrossRef] [PubMed]
- Wuriyanghan, H.; Falk, B.W. RNA Interference towards the Potato Psyllid, Bactericera cockerelli, Is Induced in Plants Infected with Recombinant Tobacco mosaic Virus (TMV). PLoS ONE 2013, 8, e66050. [Google Scholar] [CrossRef]
- Ramos, J.E.; Jain, R.G.; Powell, C.A.; Dawson, W.O.; Gowda, S.; Borovsky, D.; Shatters, R.G., Jr. Crowdsourced Identification of Potential Target Genes for CTV Induced Gene Silencing for Controlling the Citrus Greening Vector Diaphorina citri. Front Physiol. 2021, 12, 571826. [Google Scholar] [CrossRef]
- Khan, A.M.; Ashfaq, M.; Khan, A.A.; Naseem, M.T.; Mansoor, S. Evaluation of Potential RNA-Interference-Target Genes to Control Cotton Mealybug, Phenacoccus solenopsis (Hemiptera: Pseudococcuidae). Insect Sci. 2018, 25, 778–786. [Google Scholar] [CrossRef]
- Valentine, T.A.; Randall, E.; Wypijewski, K.; Chapman, S.; Jones, J.; Oparka, K.J. Delivery of Macromolecules to Plant Parasitic Nematodes Using a Tobacco Rattle Virus Vector. Plant Biotechnol. J. 2007, 5, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, G.; Magliano, M.; Dubrana, M.P.; Lozano, J.; Lecomte, P.; Favery, B.; Abad, P.; Rosso, M.N. Tobacco Rattle Virus Mediates Gene Silencing in a Plant Parasitic Root-Knot Nematode. J. Exp. Bot. 2009, 60, 4041–4050. [Google Scholar] [CrossRef]
- Hajós, J.P.; Vermunt, A.M.W.; Zuidema, D.; Kulcsár, P.; Varjas, L.; de Kort, C.A.D.; Závodszky, P.; Vlak, J.M. Dissecting Insect Development: Baculovirus-Mediated Gene Silencing in Insects. Insect Mol. Biol. 1999, 8, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Uhlirova, M.; Foy, B.D.; Beaty, B.J.; Olson, K.E.; Riddiford, L.M.; Jindra, M. Use of Sindbis Virus-Mediated RNA Interference to Demonstrate a Conserved Role of Broad-Complex in Insect Metamorphosis. Proc. Natl. Acad. Sci. USA 2003, 100, 15607–15612. [Google Scholar] [CrossRef]
- Huang, Y.; Deng, F.; Hu, Z.; Vlak, J.M.; Wang, H. Baculovirus-Mediated Gene Silencing in Insect Cells Using Intracellularly Produced Long Double-Stranded RNA. J. Biotechnol. 2007, 128, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Liu, M.; Deng, Y.; Peng, H.; Chen, X. Development of an Efficient Recombinant Mosquito Densovirus-Mediated RNA Interference System and Its Preliminary Application in Mosquito Control. PLoS ONE 2011, 6, e21329. [Google Scholar] [CrossRef]
- Kontogiannatos, D.; Swevers, L.; Maenaka, K.; Park, E.Y.; Iatrou, K.; Kourti, A. Functional Characterization of a Juvenile Hormone Esterase Related Gene in the Moth Sesamia nonagrioides through RNA Interference. PLoS ONE 2013, 8, e73834. [Google Scholar] [CrossRef]
- Taning, C.N.T.; Christiaens, O.; Li, X.X.; Swevers, L.; Casteels, H.; Maes, M.; Smagghe, G. Engineered Flock House Virus for Targeted Gene Suppression through RNAi in Fruit Flies (Drosophila melanogaster) in Vitro and in Vivo. Front. Physiol. 2018, 9, 805. [Google Scholar] [CrossRef] [PubMed]
- Selling, B.H.; Allison, R.F.; Kaesberg, P. Genomic RNA of an Insect Virus Directs Synthesis of Infectious Virions in Plants. Proc. Natl. Acad. Sci. USA 1990, 87, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, R.; Garcia, B.H.; Goodman, R.M. Systemic Spread of an RNA Insect Virus in Plants Expressing Plant Viral Movement Protein Genes. Proc. Natl. Acad. Sci. USA 2001, 98, 4910–4915. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Maharaj, P.D.; Mallajosyula, J.K.; McCormick, A.A.; Kearney, C.M. In Planta Production of Flock House Virus Transencapsidated RNA and Its Potential Use as a Vaccine. Mol. Biotechnol. 2015, 57, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, W.X.; Ding, S.W. Induction and Suppression of RNA Silencing by an Animal Virus. Science 2002, 296, 1319–1321. [Google Scholar] [CrossRef] [PubMed]
- Hajeri, S.; Killiny, N.; El-Mohtar, C.; Dawson, W.O.; Gowda, S. Citrus tristeza Virus-Based RNAi in Citrus Plants Induces Gene Silencing in Diaphorina citri, a Phloem-Sap Sucking Insect Vector of Citrus Greening Disease (Huanglongbing). J. Biotechnol. 2014, 176, 42–49. [Google Scholar] [CrossRef]
- Niehl, A.; Soininen, M.; Poranen, M.M.; Heinlein, M. Synthetic Biology Approach for Plant Protection Using DsRNA. Plant Biotechnol. J. 2018, 16, 1679–1687. [Google Scholar] [CrossRef]
- Xue, Q.; Samakovli, D.; Swevers, L.; Taning, C.N.T. Drosophila X Virus-like Particles as Efficient DsRNA Carriers for Improved RNAi against the Invasive Species, Drosophila suzukii. J. Pest Sci. 2023, 97, 429–443. [Google Scholar] [CrossRef]
- Taracena, M.L.; Oliveira, P.L.; Almendares, O.; Umaña, C.; Lowenberger, C.; Dotson, E.M.; Paiva-Silva, G.O.; Pennington, P.M. Genetically Modifying the Insect Gut Microbiota to Control Chagas Disease Vectors through Systemic RNAi. PLoS Negl. Trop. Dis. 2015, 9, e0003358. [Google Scholar] [CrossRef]
- Whitten, M.M.A.; Facey, P.D.; del Sol, R.; Fernández-Martínez, L.T.; Evans, M.C.; Mitchell, J.J.; Bodger, O.G.; Dyson, P.J. Symbiont-Mediated RNA Interference in Insects. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160042. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, L.; Hu, Q.; Zhang, B.; Wu, W.; Jin, F.; Jiang, J. Expression of DsRNA in Recombinant isaria fumosorosea Strain Targets the TLR7 Gene in Bemisia tabaci. BMC Biotechnol. 2015, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Wu, W. Recombinant Fungal Entomopathogen RNAi Target Insect Gene. Bioengineered 2016, 7, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.A.; Tabuloc, C.A.; Cervantes, K.R.; Chiu, J.C. Ingestion of Genetically Modified Yeast Symbiont Reduces Fitness of an Insect Pest via RNA Interference. Sci. Rep. 2016, 6, 22587. [Google Scholar] [CrossRef] [PubMed]
- Tenllado, F.; Martínez-García, B.; Vargas, M.; Díaz-Ruíz, J.R. Crude Extracts of Bacterially Expressed DsRNA Can Be Used to Protect Plants against Virus Infections. BMC Biotechnol. 2003, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- San Miguel, K.; Scott, J.G. The next Generation of Insecticides: DsRNA Is Stable as a Foliar-Applied Insecticide. Pest. Manag. Sci. 2016, 72, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.B.; Glick, E.; Paldi, N.; Bextine, B.R. Advances in RNA Interference: DsRNA Treatment in Trees and Grapevines for Insect Pest Suppression. Southwest. Entomol. 2012, 37, 85–87. [Google Scholar] [CrossRef]
- Li, H.; Guan, R.; Guo, H.; Miao, X. New Insights into an RNAi Approach for Plant Defence against Piercing-Sucking and Stem-Borer Insect Pests. Plant Cell Environ. 2015, 38, 2277–2285. [Google Scholar] [CrossRef] [PubMed]
- Dalakouras, A.; Jarausch, W.; Buchholz, G.; Bassler, A.; Braun, M.; Manthey, T.; Krczal, G.; Wassenegger, M. Delivery of Hairpin Rnas and Small Rnas into Woody and Herbaceous Plants by Trunk Injection and Petiole Absorption. Front. Plant Sci. 2018, 9, 1253. [Google Scholar] [CrossRef]
- Pampolini, F.; Rodrigues, T.B.; Leelesh, R.S.; Kawashima, T.; Rieske, L.K. Confocal Microscopy Provides Visual Evidence and Confirms the Feasibility of DsRNA Delivery to Emerald Ash Borer through Plant Tissues. J. Pest Sci. 2020, 93, 1143–1153. [Google Scholar] [CrossRef]
- Dalakouras, A.; Wassenegger, M.; McMillan, J.N.; Cardoza, V.; Maegele, I.; Dadami, E.; Runne, M.; Krczal, G.; Wassenegger, M. Induction of Silencing in Plants by High-Pressure Spraying of in Vitro-Synthesized Small RNAs. Front. Plant Sci. 2016, 7, 1327. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-Based Control of Fusarium graminearum Infections through Spraying of Long DsRNAs Involves a Plant Passage and Is Controlled by the Fungal Silencing Machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef] [PubMed]
- Biedenkopf, D.; Will, T.; Knauer, T.; Jelonek, L.; Furch, A.C.U.; Busche, T.; Koch, A. Systemic Spreading of Exogenous Applied Rna Biopesticides in the Crop Plant Hordeum Vulgare. ExRNA 2020, 2, 12. [Google Scholar] [CrossRef]
- Dalakouras, A.; Ganopoulos, I. Induction of Promoter DNA Methylation upon High-Pressure Spraying of Double-Stranded RNA in Plants. Agronomy 2021, 11, 789. [Google Scholar] [CrossRef]
- Das, P.R.; Sherif, S.M. Application of Exogenous DsRNAs-Induced RNAi in Agriculture: Challenges and Triumphs. Front. Plant Sci. 2020, 11, 946. [Google Scholar] [CrossRef] [PubMed]
- Taning, C.N.T.; Arpaia, S.; Christiaens, O.; Dietz-Pfeilstetter, A.; Jones, H.; Mezzetti, B.; Sabbadini, S.; Sorteberg, H.G.; Sweet, J.; Ventura, V.; et al. RNA-Based Biocontrol Compounds: Current Status and Perspectives to Reach the Market. Pest. Manag. Sci. 2020, 76, 841–845. [Google Scholar] [CrossRef]
- Timmons, L.; Court, D.L.; Fire, A. Ingestion of Bacterially Expressed DsRNAs Can Produce Specific and Potent Genetic Interference in Caenorhabditis Elegans. Gene 2001, 263, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Timmons, L.; Fire, A. Specific Interference by Ingested DsRNA. Nature 1998, 395, 854. [Google Scholar] [CrossRef] [PubMed]
- Papić, L.; Rivas, J.; Toledo, S.; Romero, J. Double-Stranded RNA Production and the Kinetics of Recombinant Escherichia coli HT115 in Fed-Batch Culture. Biotechnol. Rep. 2018, 20, e00292. [Google Scholar] [CrossRef]
- Ahn, S.-J.; Donahue, K.; Koh, Y.; Martin, R.R.; Choi, M.-Y. Microbial-Based Double-Stranded RNA Production to Develop Cost-Effective RNA Interference Application for Insect Pest Management. Int. J. Insect Sci. 2019, 11, 1179543319840323. [Google Scholar] [CrossRef]
- Ma, Z.Z.; Zhou, H.; Wei, Y.L.; Yan, S.; Shen, J. A Novel Plasmid-Escherichia coli System Produces Large Batch DsRNAs for Insect Gene Silencing. Pest. Manag. Sci. 2020, 76, 2505–2512. [Google Scholar] [CrossRef]
- Park, M.G.; Kim, W.J.; Choi, J.Y.; Kim, J.H.; Park, D.H.; Kim, J.Y.; Wang, M.; Je, Y.H. Development of a Bacillus Thuringiensis Based DsRNA Production Platform to Control Sacbrood Virus in Apis Cerana. Pest. Manag. Sci. 2020, 76, 1699–1704. [Google Scholar] [CrossRef] [PubMed]
- Hashiro, S.; Chikami, Y.; Kawaguchi, H.; Krylov, A.A.; Niimi, T.; Yasueda, H. Efficient Production of Long Double-Stranded RNAs Applicable to Agricultural Pest Control by Corynebacterium Glutamicum Equipped with Coliphage T7-Expression System. Appl. Microbiol. Biotechnol. 2021, 105, 4987–5000. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T.; Davis, Z.; Chen, L.; Englaender, J.; Zomorodi, S.; Frank, J.; Bartlett, K.; Somers, E.; Carballo, S.M.; Kester, M.; et al. Minicell-Based Fungal RNAi Delivery for Sustainable Crop Protection. Microb. Biotechnol. 2021, 14, 1847–1856. [Google Scholar] [CrossRef]
- Figueiredo Prates, L.H.; Merlau, M.; Rühl-Teichner, J.; Schetelig, M.F.; Häcker, I. An Optimized/Scale Up-Ready Protocol for Extraction of Bacterially Produced DsRNA at Good Yield and Low Costs. Int. J. Mol. Sci. 2023, 24, 9266. [Google Scholar] [CrossRef]
- Cordero, T.; Aragonés, V.; Daròs, J.A. Large-Scale Production of Recombinant RNAs on a Circular Scaffold Using a Viroid-Derived System in Escherichia coli. J. Vis. Exp. 2018, 141, e58472. [Google Scholar] [CrossRef]
- Daròs, J.A.; Aragonés, V.; Cordero, T. A Viroid-Derived System to Produce Large Amounts of Recombinant RNA in Escherichia coli. Sci. Rep. 2018, 8, 1904. [Google Scholar] [CrossRef]
- Ortolá, B.; Daròs, J.-A. Production of Recombinant RNA in Escherichia coli Using Eggplant Latent Viroid as a Scaffold. In Viroids: Methods and Protocols; Rao Ayala, L.N., Lavagi-Craddock, I., Georgios, V., Eds.; Springer: New York, NY, USA, 2021; ISBN 978-1-0716-1463-1. [Google Scholar]
- Ortolá, B.; Cordero, T.; Hu, X.; Daròs, J.A. Intron-Assisted, Viroid-Based Production of Insecticidal Circular Double-Stranded RNA in Escherichia coli. RNA Biol. 2021, 18, 1846–1857. [Google Scholar] [CrossRef]
- Ortolá, B.; Urbaneja, A.; Eiras, M.; Pérez-Hedo, M.; Daròs, J.A. RNAi-Mediated Silencing of Mediterranean Fruit Fly (Ceratitis capitata) Endogenous Genes Using Orally-Supplied Double-Stranded RNAs Produced in Escherichia coli. Pest. Manag. Sci. 2024, 80, 1087–1098. [Google Scholar] [CrossRef]
- Muerdter, F.; Olovnikov, I.; Molaro, A.; Rozhkov, N.V.; Czech, B.; Gordon, A.; Hannon, G.J.; Aravin, A.A. Production of Artificial PiRNAs in Flies and Mice. RNA 2012, 18, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Liu, X.; Guo, J.; Sun, X.; Sun, Y.; Shen, B.; Zhou, D.; Zhu, C. PiRNA-3878 Targets P450 (CpCYP307B1) to Regulate Pyrethroid Resistance in Culex Pipiens Pallens. Parasitol. Res. 2017, 116, 2489–2497. [Google Scholar] [CrossRef] [PubMed]
- Mondal, M.; Brown, J.K.; Flynt, A. Exploiting Somatic PiRNAs in Bemisia tabaci Enables Novel Gene Silencing through RNA Feeding. Life Sci. Alliance 2020, 3, e202000731. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Dou, W.; Taning, C.N.T.; Smagghe, G.; Wang, J.J. Regulatory Roles of MicroRNAs in Insect Pests: Prospective Targets for Insect Pest Control. Curr. Opin. Biotechnol. 2021, 70, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Mitchell, M.J.; Nie, G. Nanomaterials for Therapeutic RNA Delivery. Matter 2020, 3, 1948–1975. [Google Scholar] [CrossRef]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested Double-Stranded RNAs Can Act as Species-Specific Insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Taning, C.N.T.; Christiaens, O.; Berkvens, N.; Casteels, H.; Maes, M.; Smagghe, G. Oral RNAi to Control Drosophila suzukii: Laboratory Testing against Larval and Adult Stages. J. Pest Sci. 2016, 89, 803–814. [Google Scholar] [CrossRef]
- Castellanos, N.L.; Smagghe, G.; Sharma, R.; Oliveira, E.E.; Christiaens, O. Liposome Encapsulation and EDTA Formulation of DsRNA Targeting Essential Genes Increase Oral RNAi-Caused Mortality in the Neotropical Stink Bug Euschistus Heros. Pest. Manag. Sci. 2019, 75, 537–548. [Google Scholar] [CrossRef]
- Gurusamy, D.; Mogilicherla, K.; Shukla, J.N.; Palli, S.R. Lipids Help Double-Stranded RNA in Endosomal Escape and Improve RNA Interference in the Fall Armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol. 2020, 104, e21678. [Google Scholar] [CrossRef]
- Wang, K.; Peng, Y.; Chen, J.; Peng, Y.; Wang, X.; Shen, Z.; Han, Z. Comparison of Efficacy of RNAi Mediated by Various Nanoparticles in the Rice Striped Stem Borer (Chilo Suppressalis). Pestic. Biochem. Physiol. 2020, 165, 104467. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Zhu, K.Y. Chitosan/Double-Stranded RNA Nanoparticle-Mediated RNA Interference to Silence Chitin Synthase Genes through Larval Feeding in the African Malaria Mosquito (Anopheles Gambiae). Insect Mol. Biol. 2010, 19, 683–693. [Google Scholar] [CrossRef]
- Das, S.; Debnath, N.; Cui, Y.; Unrine, J.; Palli, S.R. Chitosan, Carbon Quantum Dot, and Silica Nanoparticle Mediated DsRNA Delivery for Gene Silencing in Aedes Aegypti: A Comparative Analysis. ACS Appl. Mater. Interfaces 2015, 7, 19530–19535. [Google Scholar] [CrossRef]
- Gurusamy, D.; Mogilicherla, K.; Palli, S.R. Chitosan Nanoparticles Help Double-Stranded RNA Escape from Endosomes and Improve RNA Interference in the Fall Armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol. 2020, 104, e21677. [Google Scholar] [CrossRef]
- Dhandapani, R.K.; Gurusamy, D.; Howell, J.L.; Palli, S.R. Development of CS-TPP-DsRNA Nanoparticles to Enhance RNAi Efficiency in the Yellow Fever Mosquito, Aedes Aegypti. Sci. Rep. 2019, 9, 8775. [Google Scholar] [CrossRef]
- Dou, T.; Wang, J.; Han, C.; Shao, X.; Zhang, J.; Lu, W. Cellular Uptake and Transport Characteristics of Chitosan Modified Nanoparticles in Caco-2 Cell Monolayers. Int. J. Biol. Macromol. 2019, 138, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Chen, Q.; Wu, W.; Wang, J.; Chu, P.K.; Bai, H.; Tang, G. Reconstructed Chitosan with Alkylamine for Enhanced Gene Delivery by Promoting Endosomal Escape. Carbohydr. Polym. 2020, 227, 115339. [Google Scholar] [CrossRef] [PubMed]
- Elhaj Baddar, Z.; Gurusamy, D.; Laisney, J.; Tripathi, P.; Palli, S.R.; Unrine, J.M. Polymer-Coated Hydroxyapatite Nanocarrier for Double-Stranded RNA Delivery. J. Agric. Food Chem. 2020, 68, 6811–6818. [Google Scholar] [CrossRef]
- Edwards, C.H.; Christie, C.R.; Masotti, A.; Celluzzi, A.; Caporali, A.; Campbell, E.M. Dendrimer-Coated Carbon Nanotubes Deliver DsRNA and Increase the Efficacy of Gene Knockdown in the Red Flour Beetle Tribolium castaneum. Sci. Rep. 2020, 10, 12422. [Google Scholar] [CrossRef] [PubMed]
- Henry, D.; Lacarriere, V.C. Alkali-Resistant Calcium Phosphate/Nucleic Acids Hybrid Carrier for Pest Control and Method to Produce the Particles. U.S. Patent Application US20210169083A1, 10 June 2021. [Google Scholar]
- He, B.; Chu, Y.; Yin, M.; Müllen, K.; An, C.; Shen, J. Fluorescent Nanoparticle Delivered DsRNA toward Genetic Control of Insect Pests. Adv. Mater. 2013, 25, 4580–4584. [Google Scholar] [CrossRef]
- Christiaens, O.; Tardajos, M.G.; Reyna, Z.L.M.; Dash, M.; Dubruel, P.; Smagghe, G. Increased RNAi Efficacy in Spodoptera Exigua via the Formulation of DsRNA with Guanylated Polymers. Front. Physiol. 2018, 9, 316. [Google Scholar] [CrossRef]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay Nanosheets for Topical Delivery of RNAi for Sustained Protection against Plant Viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef]
- Worrall, E.A.; Bravo-Cazar, A.; Nilon, A.T.; Fletcher, S.J.; Robinson, K.E.; Carr, J.P.; Mitter, N. Exogenous Application of RNAi-Inducing Double-Stranded RNA Inhibits Aphid-Mediated Transmission of a Plant Virus. Front. Plant Sci. 2019, 10, 265. [Google Scholar] [CrossRef]
- Sundaresha, S.; Sharma, S.; Bairwa, A.; Tomar, M.; Kumar, R.; Bhardwaj, V.; Jeevalatha, A.; Bakade, R.; Salaria, N.; Thakur, K.; et al. Spraying of DsRNA Molecules Derived from Phytophthora infestans, along with Nanoclay Carriers as a Proof of Concept for Developing Novel Protection Strategy for Potato Late Blight. Pest. Manag. Sci. 2022, 78, 3183–3192. [Google Scholar] [CrossRef]
- Jain, R.G.; Fletcher, S.J.; Manzie, N.; Robinson, K.E.; Li, P.; Lu, E.; Brosnan, C.A.; Xu, Z.P.; Mitter, N. Foliar Application of Clay-Delivered RNA Interference for Whitefly Control. Nat. Plants 2022, 8, 535–548. [Google Scholar] [CrossRef]
- Gudlur, S.; Sukthankar, P.; Gao, J.; Avila, L.A.; Hiromasa, Y.; Chen, J.; Iwamoto, T.; Tomich, J.M. Peptide Nanovesicles Formed by the Self-Assembly of Branched Amphiphilic Peptides. PLoS ONE 2012, 7, e45374. [Google Scholar] [CrossRef]
- Avila, L.A.; Chandrasekar, R.; Wilkinson, K.E.; Balthazor, J.; Heerman, M.; Bechard, J.; Brown, S.; Park, Y.; Dhar, S.; Reeck, G.R.; et al. Delivery of Lethal DsRNAs in Insect Diets by Branched Amphiphilic Peptide Capsules. J. Control. Release 2018, 273, 139–146. [Google Scholar] [CrossRef]
- McGraw, E.; Roberts, J.D.; Kunte, N.; Westerfield, M.; Streety, X.; Held, D.; Avila, L.A. Insight into Cellular Uptake and Transcytosis of Peptide Nanoparticles in Spodoptera frugiperda Cells and Isolated Midgut. ACS Omega 2022, 7, 10933–10943. [Google Scholar] [CrossRef] [PubMed]
- Falato, L.; Gestin, M.; Langel, Ü. Cell-Penetrating Peptides Delivering SiRNAs: An Overview. In Design and Delivery of SiRNA Therapeutics; Humana Press Inc.: Totowa, NJ, USA, 2021; Volume 2282, pp. 329–352. [Google Scholar]
- Gillet, F.X.; Garcia, R.A.; Macedo, L.L.P.; Albuquerque, E.V.S.; Silva, M.C.M.; Grossi-de-Sa, M.F. Investigating Engineered Ribonucleoprotein Particles to Improve Oral RNAi Delivery in Crop Insect Pests. Front. Physiol. 2017, 8, 256. [Google Scholar] [CrossRef]
- Martinez, Z.; de Schutter, K.; van Damme, E.J.M.; Vogel, E.; Wynant, N.; vanden Broeck, J.; Christiaens, O.; Smagghe, G. Accelerated Delivery of DsRNA in Lepidopteran Midgut Cells by a Galanthus Nivalis Lectin (GNA)-DsRNA-Binding Domain Fusion Protein. Pestic. Biochem. Physiol. 2021, 175, 104853. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.M.W.; Song, H.; Yu, Z.; Biondi, M.; Bai, J.; Shi, X.; Ren, Z.; Weerasekara, S.M.; Hua, D.H.; Silver, K.; et al. Comparison of Strategies for Enhancing RNA Interference Efficiency in Ostrinia nubilalis. Pest. Manag. Sci. 2021, 77, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Howard, J.D.; Beghyn, M.; Dewulf, N.; De Vos, Y.; Philips, A.; Portwood, D.; Kilby, P.M.; Oliver, D.; Maddelein, W.; Brown, S.; et al. Chemically Modified DsRNA Induces RNAi Effects in Insects in Vitro and in Vivo: A Potential New Tool for Improving RNA-Based Plant Protection. J. Biol. Chem. 2022, 298, 102311. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Dong, X.; Zou, C.; Zhang, H. Endocytic Pathway Mediates Refractoriness of Insect Bactrocera dorsalis to RNA Interference. Sci. Rep. 2015, 5, 8700. [Google Scholar] [CrossRef] [PubMed]
- Cedden, D.; Güney, G.; Scholten, S.; Rostás, M. Lethal and Sublethal Effects of Orally Delivered Double-Stranded RNA on the Cabbage Stem Flea Beetle, Psylliodes chrysocephala. Pest. Manag. Sci. 2023; early view. [Google Scholar] [CrossRef]
- Ullah, F.; Gul, H.; Tariq, K.; Hafeez, M.; Desneux, N.; Gao, X.; Song, D. RNA Interference-Mediated Silencing of Ecdysone Receptor (EcR) Gene Causes Lethal and Sublethal Effects on Melon Aphid, Aphis Gossypii. Entomol. Gen. 2022, 42, 791–797. [Google Scholar] [CrossRef]
- Khajuria, C.; Ivashuta, S.; Wiggins, E.; Flagel, L.; Moar, W.; Pleau, M.; Miller, K.; Zhang, Y.; Ramaseshadri, P.; Jiang, C.; et al. Development and Characterization of the First DsRNA-Resistant Insect Population from Western Corn Rootworm, Diabrotica virgifera virgifera LeConte. PLoS ONE 2018, 13, e0197059. [Google Scholar] [CrossRef]
- Zhang, X.; Fan, Z.; Wang, Q.; Kong, X.; Liu, F.; Fang, J.; Zhang, S.; Zhang, Z. RNAi Efficiency through DsRNA Injection Is Enhanced by Knockdown of DsRNA Nucleases in the Fall Webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int. J. Mol. Sci. 2022, 23, 6182. [Google Scholar] [CrossRef]
- Prentice, K.; Smagghe, G.; Gheysen, G.; Christiaens, O. Nuclease Activity Decreases the RNAi Response in the Sweetpotato Weevil Cylas Puncticollis. Insect Biochem. Mol. Biol. 2019, 110, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Tayler, A.; Heschuk, D.; Giesbrecht, D.; Park, J.Y.; Whyard, S. Efficiency of RNA Interference Is Improved by Knockdown of DsRNA Nucleases in Tephritid Fruit Flies. Open Biol. 2019, 9, 190198. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.H.; Song, H.F.; Abbas, M.; Wang, Y.L.; Li, T.; Ma, E.B.; Cooper, A.M.W.; Silver, K.; Zhu, K.Y.; Zhang, J.Z. A DsRNA-Degrading Nuclease (DsRNase2) Limits RNAi Efficiency in the Asian Corn Borer (Ostrinia furnacalis). Insect Sci. 2021, 28, 1677–1689. [Google Scholar] [CrossRef]
- Kim, J.S.; Park, J.; Choi, J.H.; Kang, S.; Park, N. RNA–DNA Hybrid Nano-Materials for Highly Efficient and Long Lasting RNA Interference Effect. RSC Adv. 2023, 13, 3139–3146. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, J.S.; Lee, J.B. Generation of SiRNA Nanosheets for Efficient RNA Interference. Sci. Rep. 2016, 6, 25146. [Google Scholar] [CrossRef]
- Abe, N.; Abe, H.; Nagai, C.; Harada, M.; Hatakeyama, H.; Harashima, H.; Ohshiro, T.; Nishihara, M.; Furukawa, K.; Maeda, M.; et al. Synthesis, Structure, and Biological Activity of Dumbbell-Shaped Nanocircular RNAs for RNA Interference. Bioconjug Chem. 2011, 22, 2082–2092. [Google Scholar] [CrossRef]
- Grabow, W.W.; Zakrevsky, P.; Afonin, K.A.; Chworos, A.; Shapiro, B.A.; Jaeger, L. Self-Assembling RNA Nanorings Based on RNAI/II Inverse Kissing Complexes. Nano Lett. 2011, 11, 878–887. [Google Scholar] [CrossRef]
- Lund, V.K.; Madsen, K.L.; Kjaerulff, O. Drosophila Rab2 Controls Endosome-Lysosome Fusion and LAMP Delivery to Late Endosomes. Autophagy 2018, 14, 1520–1542. [Google Scholar] [CrossRef]
- Johannes, L.; Lucchino, M. Current Challenges in Delivery and Cytosolic Translocation of Therapeutic RNAs. Nucleic Acid. Ther. 2018, 28, 178–193. [Google Scholar] [CrossRef]
- Pinzón, N.; Bertrand, S.; Subirana, L.; Busseau, I.; Escrivá, H.; Seitz, H. Functional Lability of RNA-Dependent RNA Polymerases in Animals. PLoS Genet. 2019, 15, e1007915. [Google Scholar] [CrossRef]
- Hu, X.; Richtman, N.M.; Zhao, J.Z.; Duncan, K.E.; Niu, X.; Procyk, L.A.; Oneal, M.A.; Kernodle, B.M.; Steimel, J.P.; Crane, V.C.; et al. Discovery of Midgut Genes for the RNA Interference Control of Corn Rootworm. Sci. Rep. 2016, 6, 30542. [Google Scholar] [CrossRef]
- Wynant, N.; Verlinden, H.; Breugelmans, B.; Simonet, G.; Vanden Broeck, J. Tissue-Dependence and Sensitivity of the Systemic RNA Interference Response in the Desert Locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2012, 42, 911–917. [Google Scholar] [CrossRef]
- Cooper, A.M.W.; Song, H.; Shi, X.; Yu, Z.; Lorenzen, M.; Silver, K.; Zhang, J.; Zhu, K.Y. Characterization, Expression Patterns, and Transcriptional Responses of Three Core RNA Interference Pathway Genes from Ostrinia nubilalis. J. Insect Physiol. 2021, 129, 104181. [Google Scholar] [CrossRef]
- Guo, W.C.; Fu, K.Y.; Yang, S.; Li, X.X.; Li, G.Q. Instar-Dependent Systemic RNA Interference Response in Leptinotarsa decemlineata Larvae. Pestic. Biochem. Physiol. 2015, 123, 64–73. [Google Scholar] [CrossRef]
- Bansal, R.; Michel, A.P. Core RNAi Machinery and Sid1, a Component for Systemic RNAi, in the Hemipteran Insect, Aphis glycines. Int. J. Mol. Sci. 2013, 14, 3786–3801. [Google Scholar] [CrossRef] [PubMed]
- Arraes, F.B.M.; Martins-de-Sa, D.; Noriega Vasquez, D.D.; Melo, B.P.; Faheem, M.; de Macedo, L.L.P.; Morgante, C.V.; Barbosa, J.A.R.G.; Togawa, R.C.; Moreira, V.J.V.; et al. Dissecting Protein Domain Variability in the Core RNA Interference Machinery of Five Insect Orders. RNA Biol. 2021, 18, 1653–1681. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Hu, S.; Li, H.; Shi, Z.; Miao, X. The in Vivo DsRNA Cleavage Has Sequence Preference in Insects. Front. Physiol. 2018, 9, 1768. [Google Scholar] [CrossRef] [PubMed]
- Swevers, L.; Broeck, J.V.; Smagghe, G. The Possible Impact of Persistent Virus Infection on the Function of the RNAi Machinery in Insects: A Hypothesis. Front. Physiol. 2013, 4, 66388. [Google Scholar] [CrossRef] [PubMed]
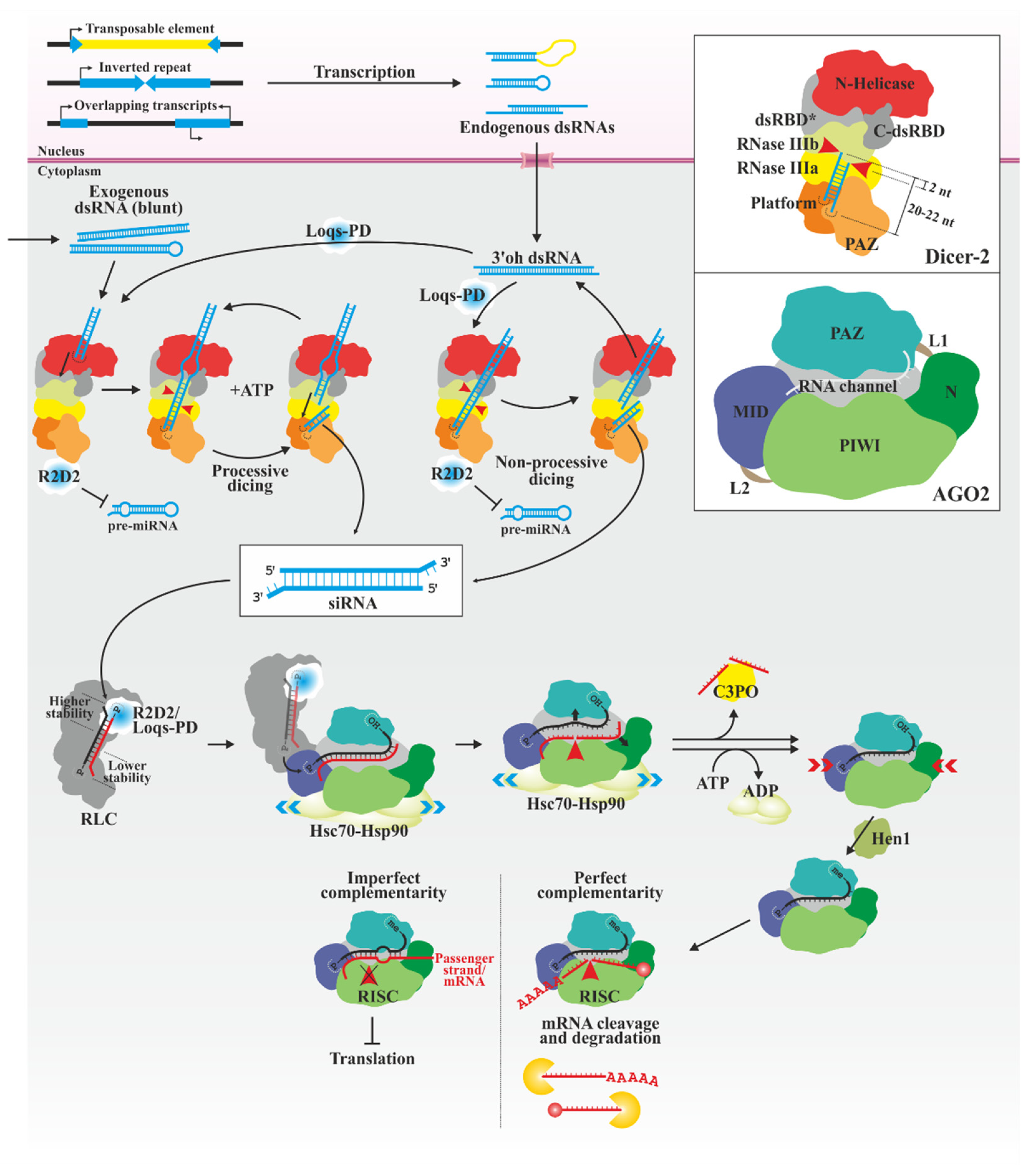
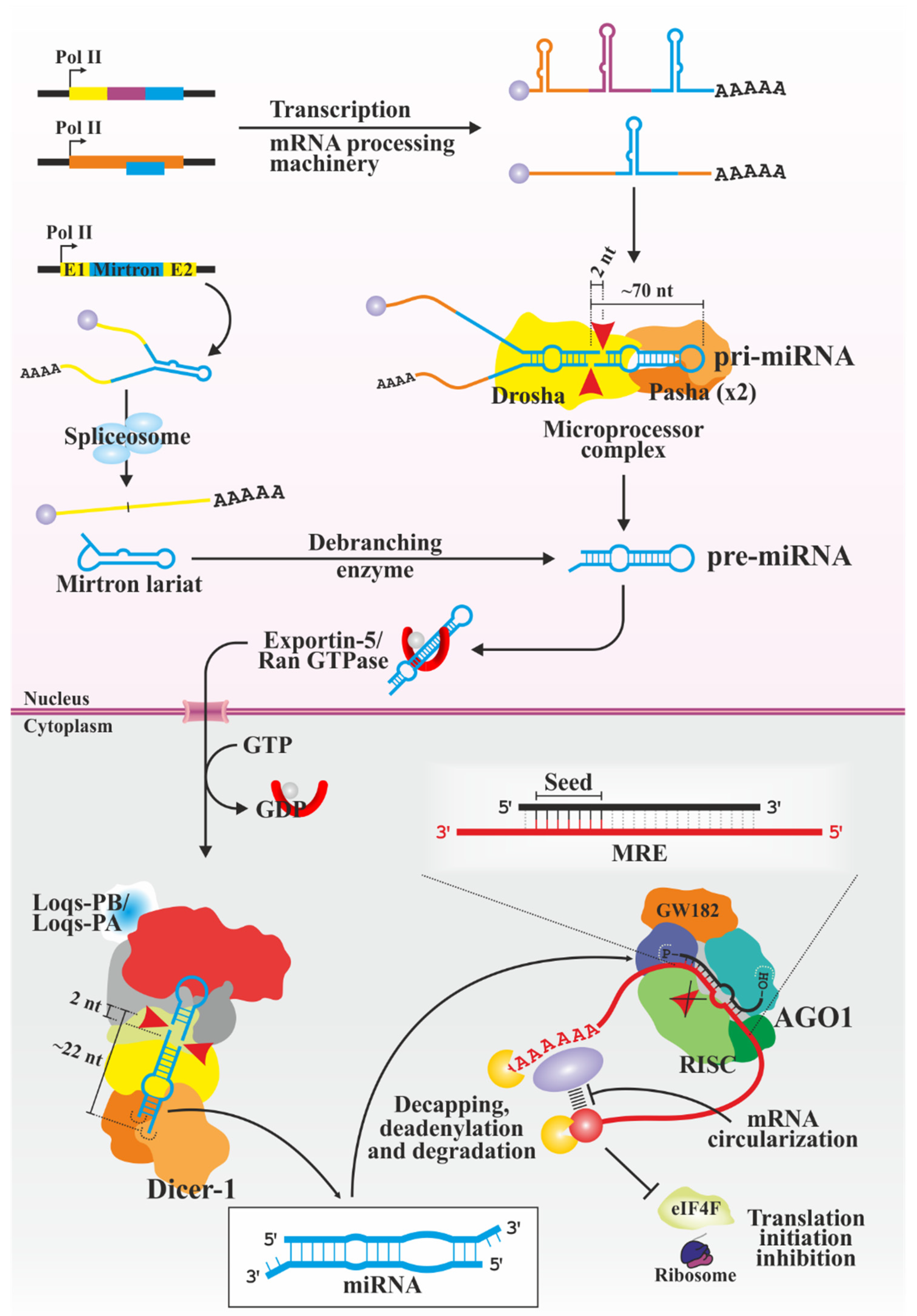
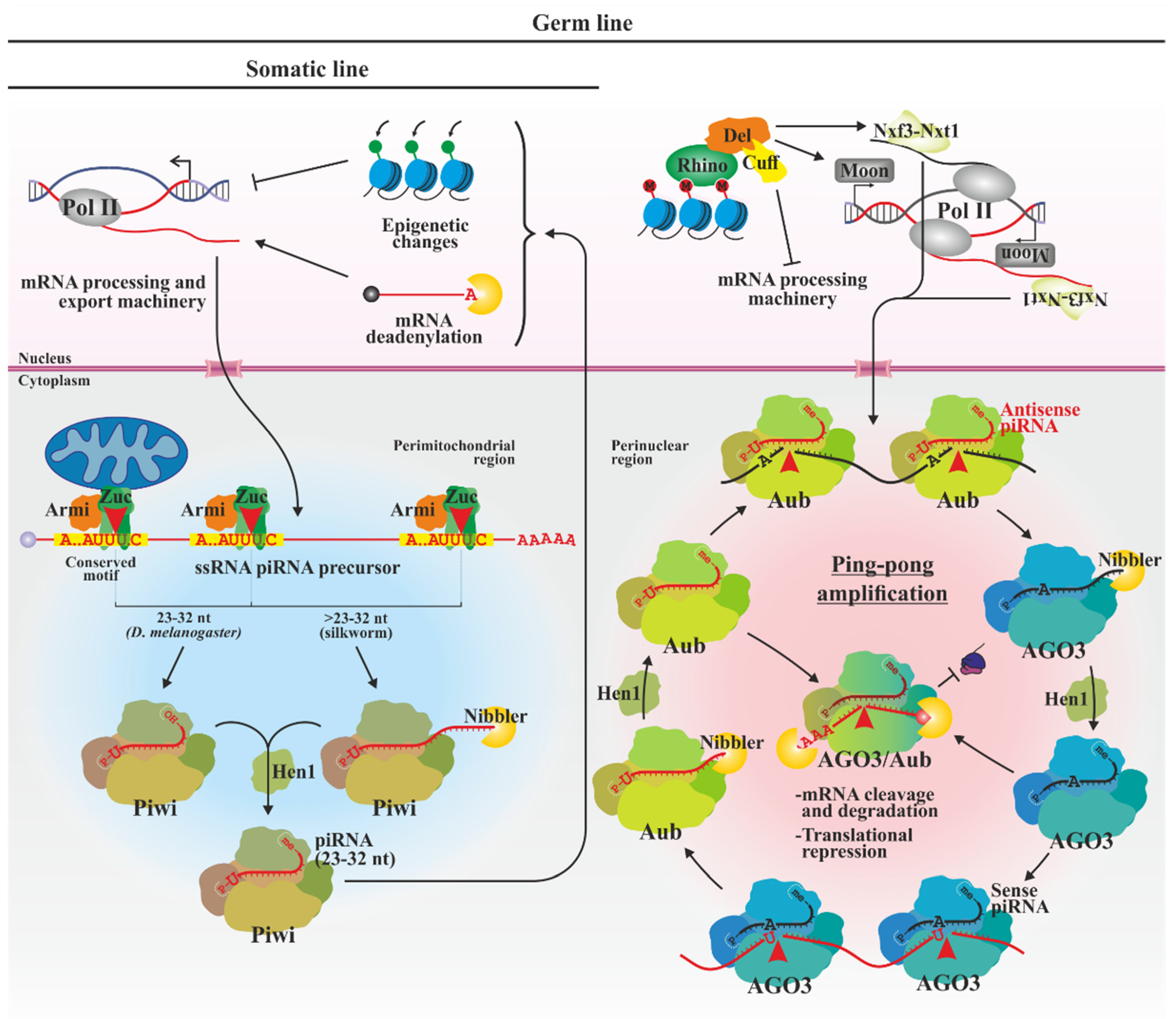
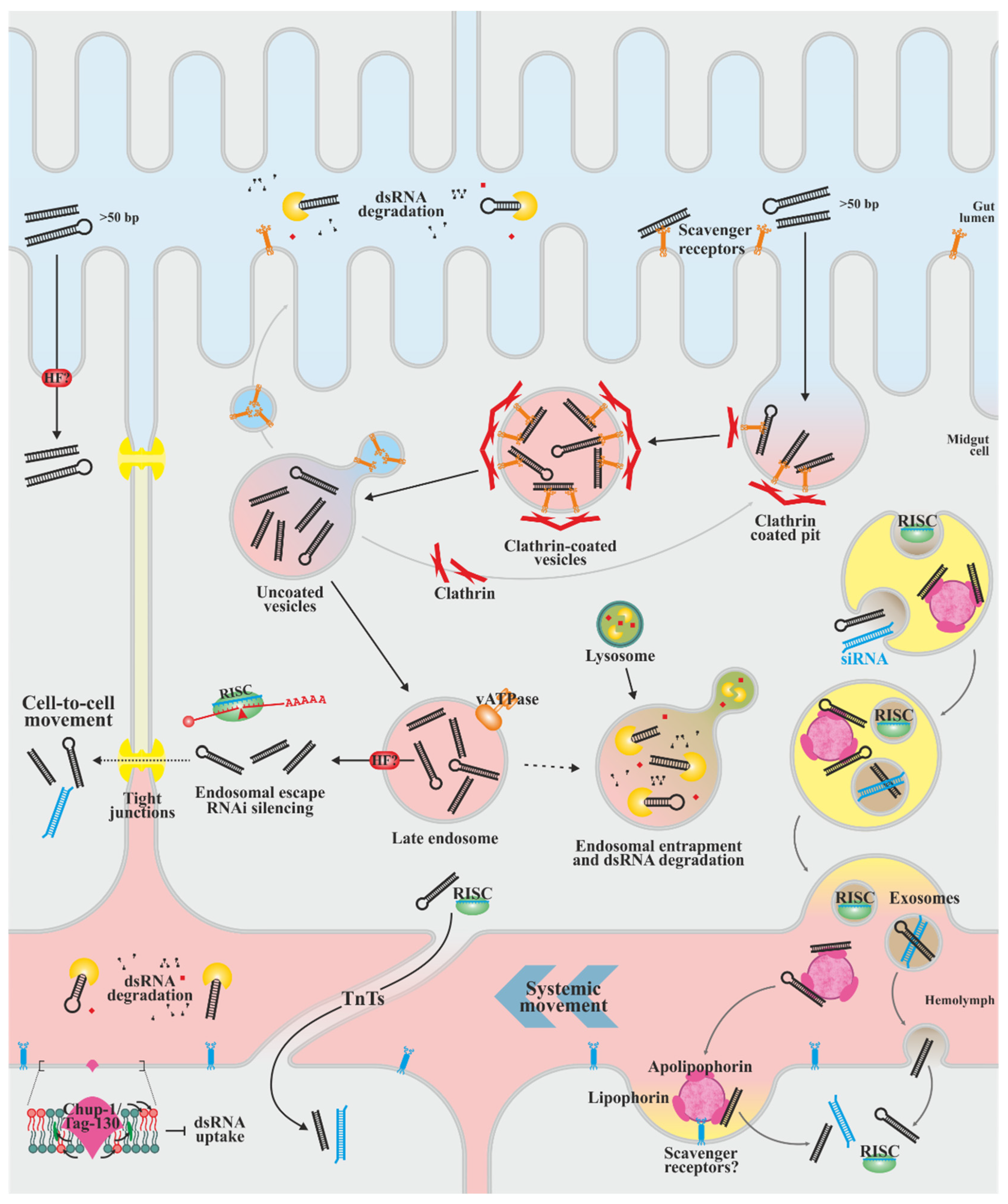
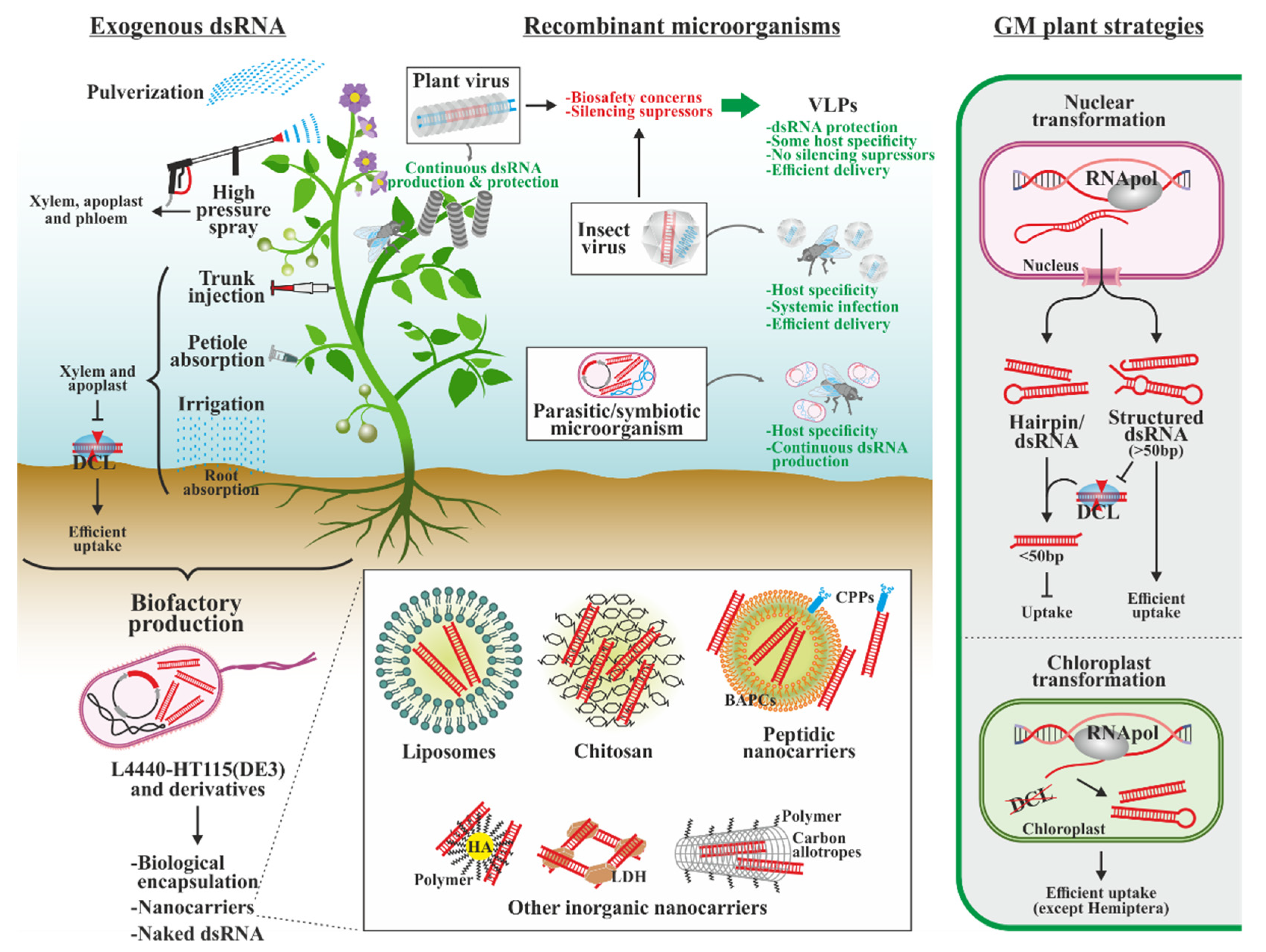
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortolá, B.; Daròs, J.-A. RNA Interference in Insects: From a Natural Mechanism of Gene Expression Regulation to a Biotechnological Crop Protection Promise. Biology 2024, 13, 137. https://doi.org/10.3390/biology13030137
Ortolá B, Daròs J-A. RNA Interference in Insects: From a Natural Mechanism of Gene Expression Regulation to a Biotechnological Crop Protection Promise. Biology. 2024; 13(3):137. https://doi.org/10.3390/biology13030137
Chicago/Turabian StyleOrtolá, Beltrán, and José-Antonio Daròs. 2024. "RNA Interference in Insects: From a Natural Mechanism of Gene Expression Regulation to a Biotechnological Crop Protection Promise" Biology 13, no. 3: 137. https://doi.org/10.3390/biology13030137
APA StyleOrtolá, B., & Daròs, J.-A. (2024). RNA Interference in Insects: From a Natural Mechanism of Gene Expression Regulation to a Biotechnological Crop Protection Promise. Biology, 13(3), 137. https://doi.org/10.3390/biology13030137





