Daily Rhythmicity of Muscle-Related and Rhythm Genes Expression in Mackerel Tuna (Euthynnus affinis)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal
2.2. Sampling Procedures
2.3. RNA Extraction and Reverse Transcription
2.4. Gene Expression Analyses
2.5. Statistics
3. Results
3.1. Analysis of the Rhythmicity of the Muscle Colck Gene in Mackerel Tuna
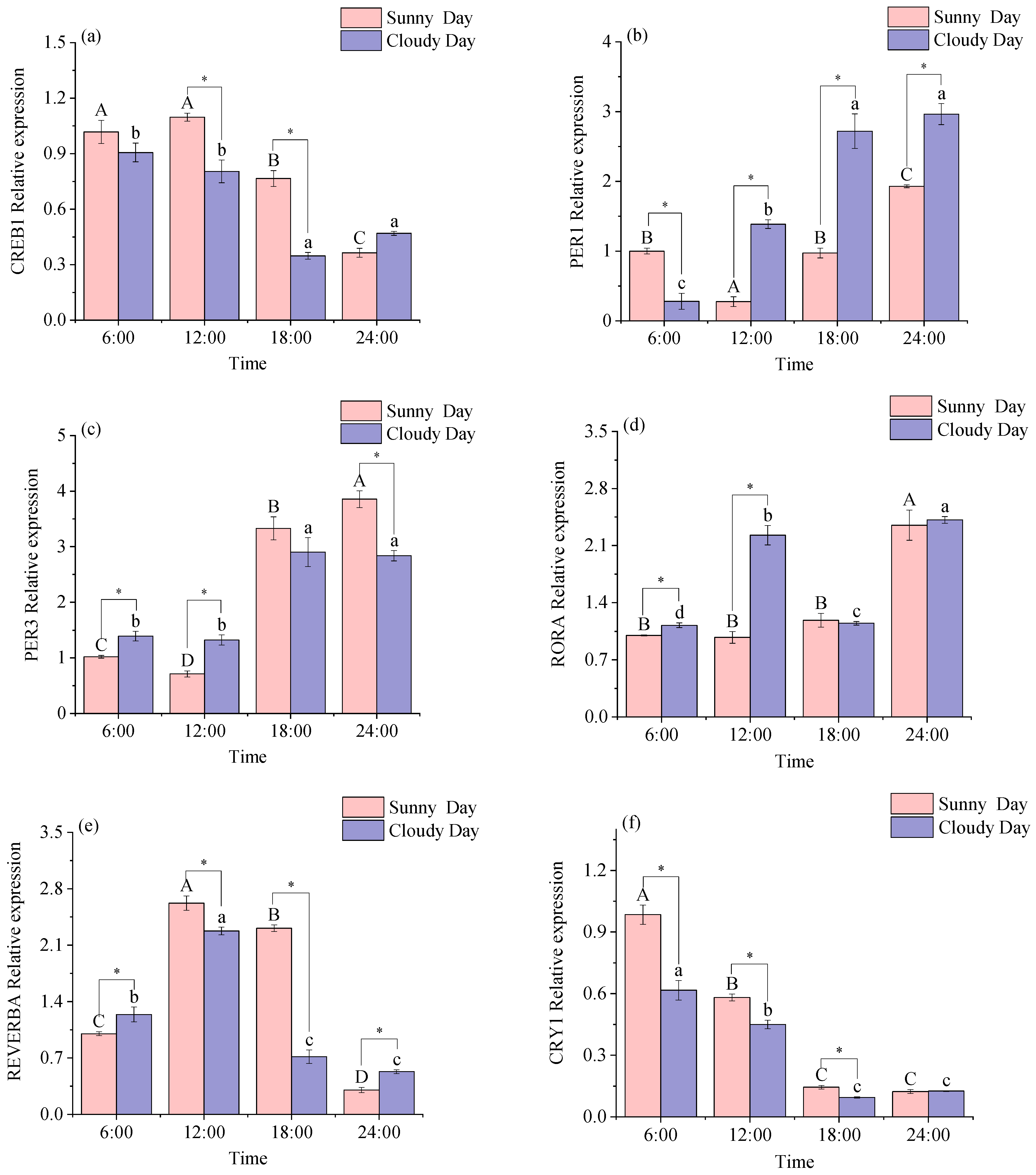
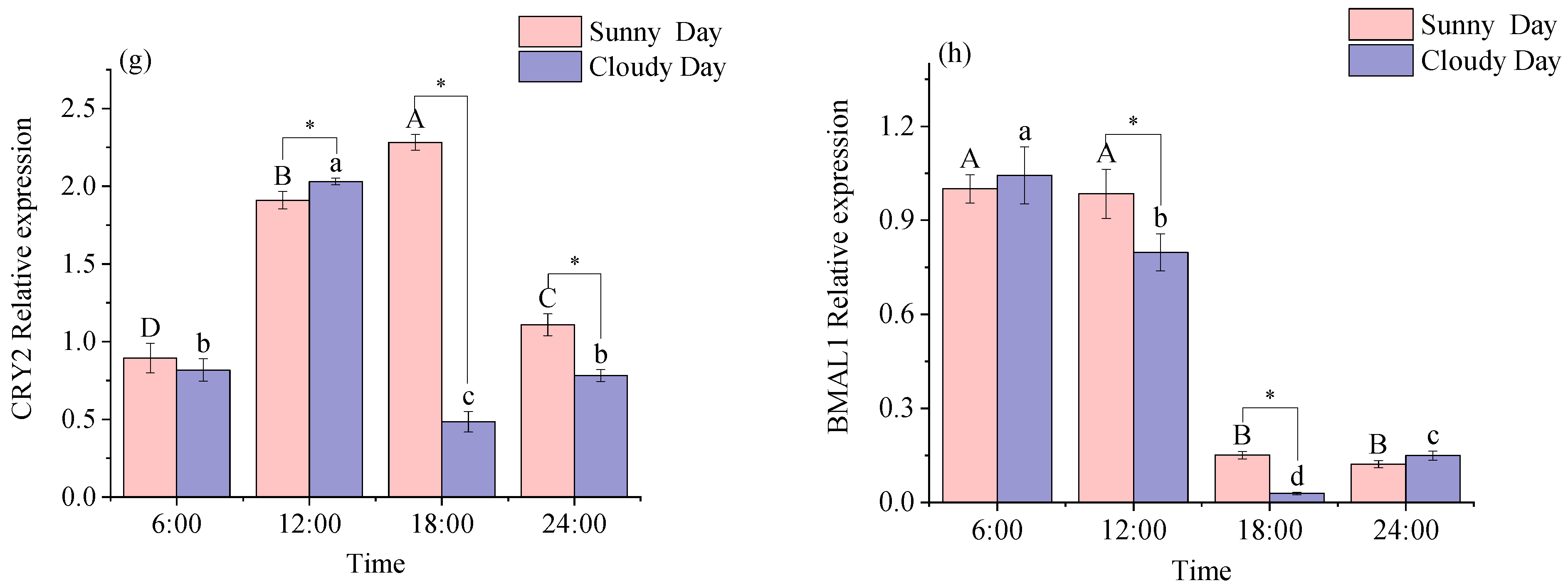
3.1.1. Rhythmical Analysis of the White Muscle Clock Gene
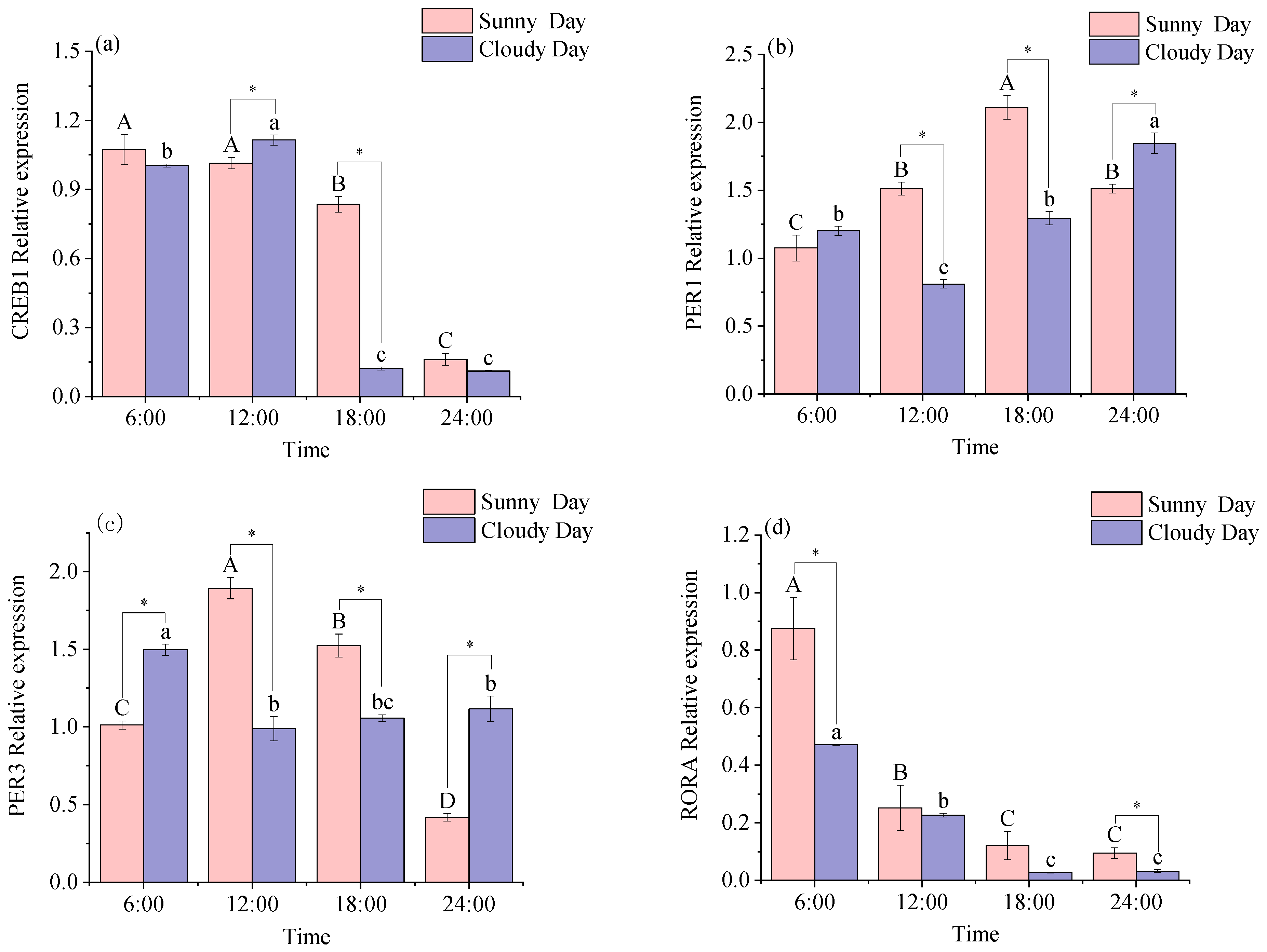
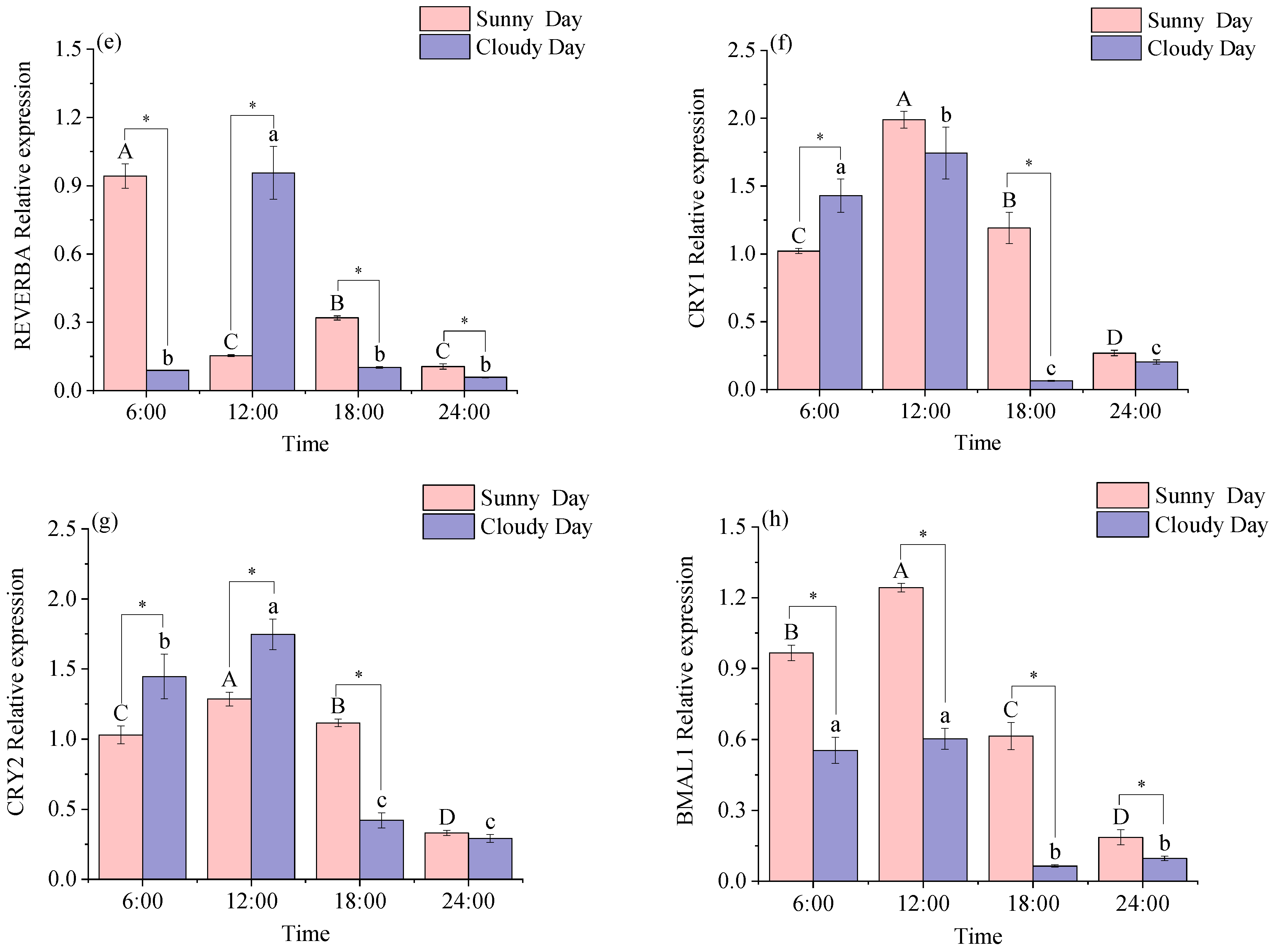
3.1.2. Rhythmical Analysis of the Red Muscle Clock Gene
3.2. Functional Genes Expression Levels in Mackerel Tuna Muscles
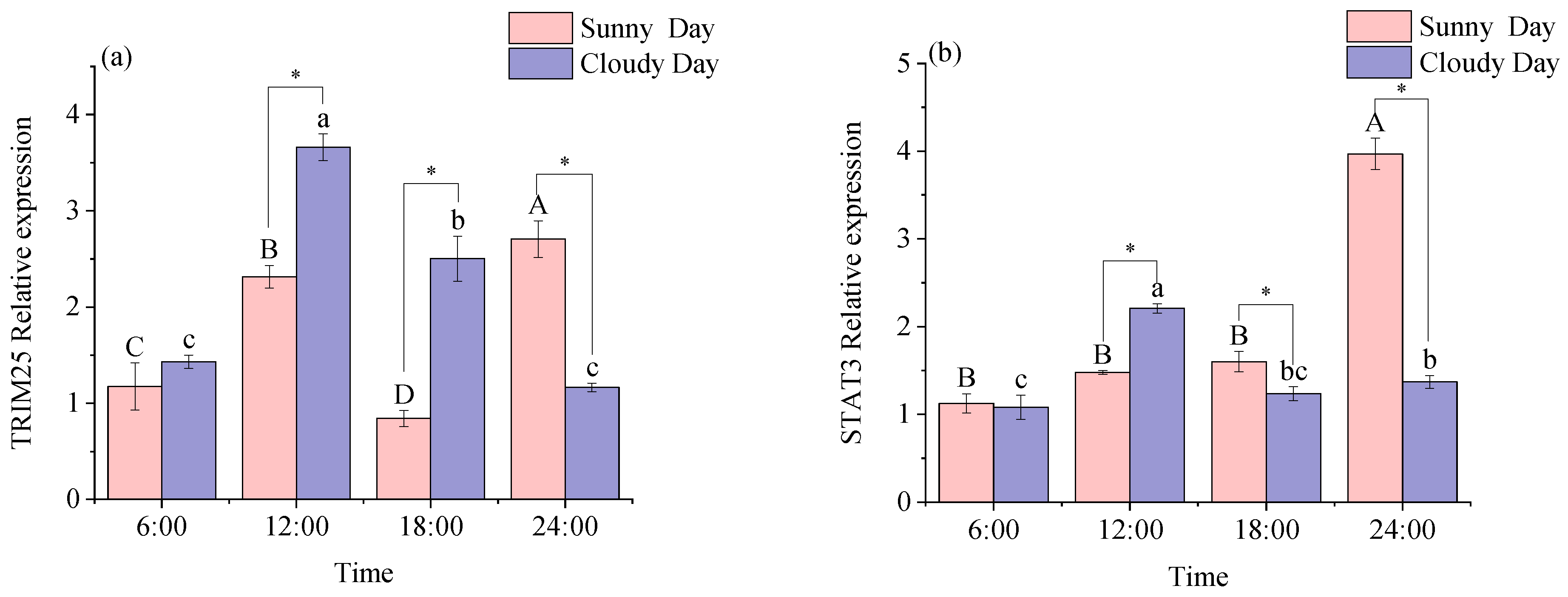
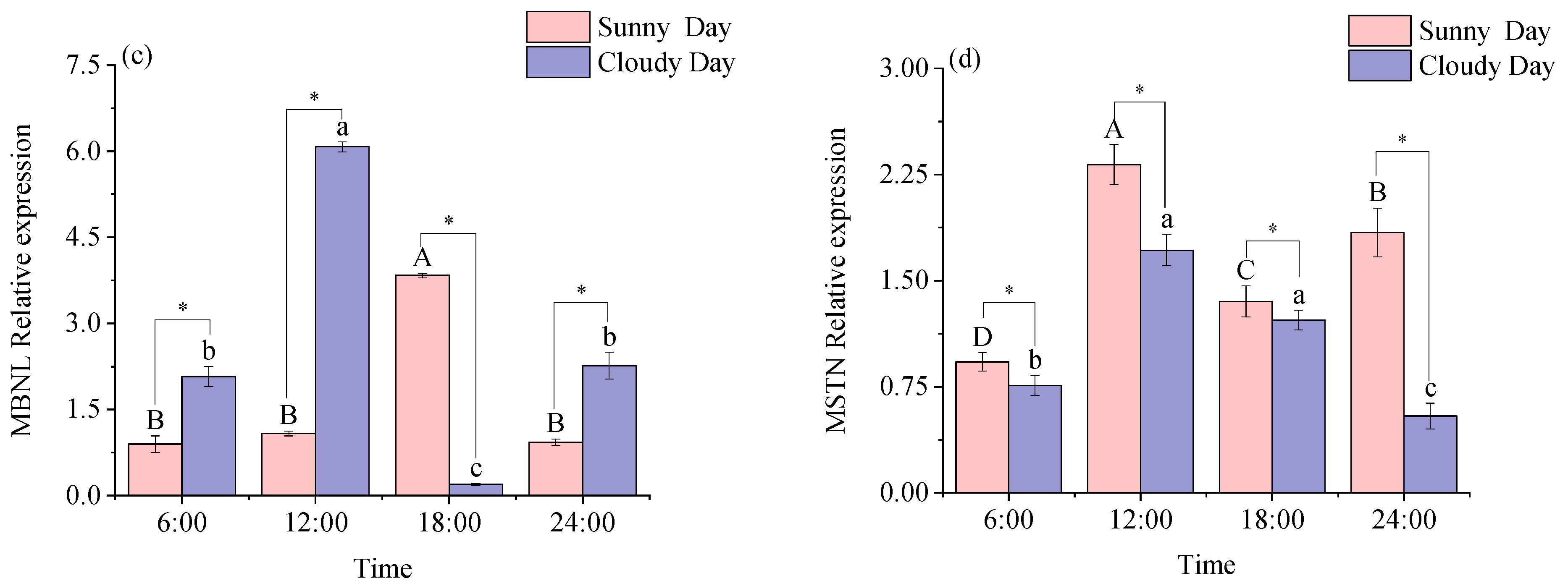
3.2.1. Functional Genes Expression Levels in White Muscle
3.2.2. Functional Genes Expression Levels in Red Muscle
4. Discussion
4.1. Muscle Rhythm Gene Expression in Mackerel Tuna
4.2. Functional Gene Expression in Mackerel Tuna Muscle
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Orem, J.M. Rhythms of Life: The Biological Clocks that Control the Daily Lives of Every Living Thing. Q. Rev. Biol. 2005, 80, 266–267. [Google Scholar] [CrossRef]
- Feng, N.Y.; Bass, A.H. Bass “Singing” Fish Rely on Circadian Rhythm and Melatonin for the Timing of Nocturnal Courtship Vocalization. Curr. Biol. 2016, 19, 2681–2689. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhu, W.; Wu, P. Analysis of the rhythms of gene expression in the liver and heart biological clocks of Siniperca chuatsi. Acta Hydrobiol. Sin. 2016, 40, 243–251. [Google Scholar]
- Shibata, S.; Tominaga, K. Brain neuronal mechanisms of circadian rhythms in mammalians. Yakugaku Zasshi J. Pharm. Soc. Jpn. 1991, 111, 270–283. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vatine, G.; Vallone, D.; Gothilf, Y.; Foulkes, N.S. It’s time to swim! Zebrafish and the circadian clock. EBS Lett. 2011, 585, 1485–1494. [Google Scholar] [CrossRef]
- Wang, W.; Hu, J.; Fu, Z.; Yu, G.; Ma, Z. Daily Rhythmicity of Hepatic Rhythm, Lipid Metabolism and Immune Gene Expression of Mackerel Tuna (Euthynnus affinis) under Different Weather. J. Mar. Sci. Eng. 2022, 10, 2. [Google Scholar] [CrossRef]
- Velarde, E.; Haque, R.; Iuvone, P.M.; Azpeleta, C.; Alonso-Gómez, A.L.; Delgado, M.J. Circadian clock genes of goldfish, Carassius auratus: cDNA cloning and rhythmic expression of period and cryptochrome transcripts in retina, liver, and gut. J. Biol. Rhythm. 2009, 24, 104–113. [Google Scholar] [CrossRef]
- Welsh, D.; Takahashi, J.; Kay, S. Suprachiasmatic nucleus: Cellautonomy and network properties. Annu. Rev. Physiol. 2010, 72, 551–577. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Chung, M. Genetics and epigenetics of circadian rhythms and their potential roles in neuropsychiatric disorders. Neurosci. Bull. 2015, 31, 141–159. [Google Scholar] [CrossRef]
- McCarthy, J.J.; Andrews, J.L.; McDearmon, E.L.; Campbell, K.S.; Barber, B.K.; Miller, B.H.; Walker, J.R.; Hogenesch, J.B.; Takahashi, J.S.; Esser, K.A.; et al. McDearmon Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol. Genom. 2007, 31, 86–95. [Google Scholar] [CrossRef]
- Wu, P.; Li, Y.L.; Cheng, J.; Chen, L.; Zhu, X.; Feng, Z.G.; Zhang, J.S.; Chu, W.Y. Daily rhythmicity of clock gene transcript levels in fast and slow muscle fibers from Chinese perch (Siniperca chuatsi). BMC Genom. 2016, 17, 1008. [Google Scholar] [CrossRef]
- Nagasawa, K.; Giannetto, A.; Fernandes, J.M.O. Photoperiod influences growth and MLL (mixed-lineage leukaemia) expression in Atlantic cod. PLoS ONE 2012, 7, e36908. [Google Scholar] [CrossRef]
- Lazado, C.C.; Kumaratunga, H.P.; Nagasawa, K.; Babiak, I.; Giannetto, A.; Fernandes, J.M. Daily Rhythmicity of clock Gene Transcripts in Atlantic Cod Fast Skeletal Muscle. PLoS ONE 2014, 9, e99172. [Google Scholar] [CrossRef]
- Zhang, X.; Patel, S.P.; McCarthy, J.J.; Rabchevsky, A.G.; Goldhamer, D.J.; Esser, K.A. A non-canonical E-box within the Myo D core enhancer is necessary for circadian expression in skeletal muscle. Nucleic Acids Res. 2012, 40, 3419–3430. [Google Scholar] [CrossRef] [PubMed]
- Cox, K.H.; Takahashi, J.S. Circandian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 2019, 63, R93–R102. [Google Scholar] [CrossRef] [PubMed]
- Heikel, G.; Choudhury, N.R.; Michlewski, G. The role of Trim25 in development, disease and RNA metabolism. Biochem. Soc. Trans. 2016, 44, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Sun, Y.M. The IL-6/JAK/STAT3 pathway: Potential therapeutic strategies in treating colorectal cancer (Review). Int. J. Oncol. 2014, 44, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Irimia, M.; Ross, P.J.; Sung, H.K.; Alipanahi, B.; David, L.; Golipour, A.; Gabut, M.; Michael, I.P.; Nachman, E.N.; et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 2013, 498, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Liu, X.N.; Wang, T.T.; Pan, W.; Tao, Q.F.; Zhou, W.P.; Wang, F.; Sun, S.H. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of IncRNA-PXN-ASI. Nat. Cell Biol. 2017, 19, 820–832. [Google Scholar] [CrossRef]
- Saina, M.; Technau, U. Characterization of myostatin/gdf8/11 in the Starlet Sea Anemone Nematostella vectensis. J. Exp. Zool. 2009, 312, 780–788. [Google Scholar] [CrossRef]
- Christoffels, A.; Koh, E.G.; Chia, J.M.; Brenner, S.; Aparicio, S.; Venkatesh, B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fish. Mol. Biol. Evol. 2004, 21, 1146–1151. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lawler, A.M.; Lee, S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Hadibarata, T.; Sathishkumar, P.; Prasetia, H.; Pusfitasari, E.D.; Tasfiyati, A.N.; Muzdalifah, D.; Waluyo, J.; Randy, A.; Ramadhaningtyas, D.P.; Zuas, O.; et al. Microplastic contamination in the Skipjack Tuna (Euthynnus affinis) collected from Southern Coast of Java, Indonesia. Chemosphere 2021, 276, 130185. [Google Scholar]
- Zhou, S.; Yang, R.; Yu, G.; Ma, Z.; Meng, X. Muscle composition and mutrition evaluation of three tuna species near Meiji Reef. South China Fish. Sci. 2020, 17, 51–59. [Google Scholar]
- Zhou, S.; Yang, R.; Yu, G.; Wu, Q.; Ma, Z. Growth of Longtail Tuna Thunnus tonggol and Small Tuna Euthymmus affinis in Circulating Water Ponds. Fish. Sci. 2021, 40, 339–346. [Google Scholar]
- Bravo, R.; Cubero, J.; Franco, L.; Mesa, M.; Galán, C.; Rodríguez, A.B.; Jarne, C.; Barriga, C. Body weight gain in rats by a high-fat diet produces chronodisruption in activity/inactivity circadian rhythm. Chronobiol. Int. 2014, 31, 363–370. [Google Scholar] [CrossRef]
- Wang, X. Bioinformatical Analysis of ATE/CREB Family and the Role of Creb1 on Gametogenesis in Nile Tilapia. Master Dissertation, Southwest University, Chongqing, China, 2020. [Google Scholar]
- Cuesta, I.H.; Lahiri, K.; Lopez-Olmeda, J.F.; Loosli, F.; Foulkes, N.S.; Vallone, D. Differential maturation of rhythmic clock gene expression during early development in medaka (Oryzias latipes). Chronobiol. Int. 2014, 31, 468–478. [Google Scholar] [CrossRef]
- Lazado, G.C.; Nagasawa, K.; Babiak, I.; Kumaratunga, H.P.S.; Fernandes, J.M.O. Circadian rhythmicity and photic plasticity of myosin gene transcription in fast skeletal muscle og Atlantic cod (Gadus morhua). Mar. Genom. 2014, 18 Pt A, 21–29. [Google Scholar] [CrossRef]
- Reppert, S.M.; Weaver, D.R.; Steven, M. Coordination of circadian timing in mammals. PLoS ONE 2002, 6, e17080. [Google Scholar] [CrossRef]
- Sardiello, M.; Cairo, S.; Fontanella, B.; Ballabio, A.; Meroni, G. Genomic analysis of the TRIM gamily reveals two groups of genes with distinct evolutionary properties. BMC Evol. Biol. 2008, 8, 225. [Google Scholar] [CrossRef]
- Manokaran, G.; Finol, E.; Wang, C.; Gunaratne, J.; Bahl, J.; Ong, E.Z.; Tan, H.C.; Sessions, O.M.; Ward, A.M.; Gubler, D.J.; et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 2015, 350, 217–221. [Google Scholar] [CrossRef]
- Zhou, Z.Z.; Jing, P.; Wei, K. Cloning and expression analysis of the TRIM25 gene from rhubarb fish. Acta Hydrobiol Sin. 2019, 43, 1189–1196. [Google Scholar]
- Rawlings, J.S.; Rosler, K.M.; Harrsion, D.A. The jak/stat signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef]
- Belletti, B.; Baldassarre, G. Stathmin: A protein with many tasks. New biomarker and potential target in cancer. Expert Opin. Ther. Targets 2011, 15, 1249–1266. [Google Scholar] [CrossRef]
- Liu, Y.Z.; Sepich, D.S.; Krezel, L.S. Stat3/Cdc25a-Dependent cell proliferation promotes embryonic axis extension during zebrafish gastrulation. PLoS Genet. 2017, 13, e1006564. [Google Scholar] [CrossRef]
- Oates, A.C.; Wollberg, P.; Pratt, S.J.; Paw, B.H.; Johnson, S.L.; Ho, R.K.; Postlethwait, J.H.; Zon, L.I.; Wilks, A.F. Zebrafish stat3 is expressed in restricted tissues during embryogenesis and stat1 rescues cytokine signaling in a STAT1-deficient human cell line. Dev. Dynam. 1999, 215, 352–370. [Google Scholar] [CrossRef]
- Guo, C.-J.; Zhang, Y.-F.; Yang, L.-S.; Yang, X.-B.; Wu, Y.-Y.; Liu, D.; Chen, W.-J.; Weng, S.-P.; Yu, X.-Q.; He, J.-G. The JAK and STAT family members of the Mandarin fish Siniperca chuatsi: Molecular cloning, tissues distribution and immunobiological activity. Fish Shellfish Immunol. 2009, 27, 349–359. [Google Scholar] [CrossRef]
- Jin, Y.L.; Zhou, T.; Li, N.; Liu, S.K.; Xu, X.Y.; Pan, Y.; Tan, S.X.; Shi, H.T.; Yang, Y.J.; Yuan, Z.H.; et al. JAK and STAT members in channel catfish: Identification, phylogenetic analysis and expression profiling after Edwardsiella ictalurid infection. Dev. Comp. Immunol. 2017, 81, 334–341. [Google Scholar] [CrossRef]
- Yamada EBastie, C.C.; Koga, H.; Wang, Y.; Cuervo, A.M.; Pessin, J.E. Mouse Skeletal Muscle Fiber-Type-Specific Macroautophagy and Muscle Wasting Are Regulated by a Fyn/STAT3/Vps34 Signaling Pathway. Cell Rep. 2012, 5, 557–569. [Google Scholar] [CrossRef]
- Fardaei, M.; Rogers, M.T.; Thorpe, H.M.; Larkin, K.; Hamshere, M.G.; Harper, P.S.; Brook, J.D. Thorpe Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002, 11, 805–814. [Google Scholar] [CrossRef]
- Squillace, R.M.; Chenault, D.M.; Wang, E.H. Inhibition of muscle differentiation by the novel muscleblind-related protein CHCR. J. Dev. Biol. 2002, 250, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Kanadia, R.N.; Urbinati, C.R.; Crusselle, V.J.; Luo, D.; Lee, Y.J.; Harrison, J.K.; Oh, S.P.; Swanson, M.S. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Gene Expr. Patterns 2003, 3, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Carlson, C.; Booth, F.W.; Cordon, S.E. Skeletal muscle myostatin mRNA expression is fiber-type specific and increase during hindtimb unloading. Am. J. Physiol. 1999, 277, 601–606. [Google Scholar]
- Chen, A.E.; Ginty, D.D.; Fan, C.M. Protein kinase A signalling via CREB controls myogenesis induced by Wnt proteins. Nature 2005, 433, 317–322. [Google Scholar] [CrossRef] [PubMed]

| Genes | Weather | Acro (p-Value) | Acrophase |
|---|---|---|---|
| CREB1 | S | <0.001 | 12 ± 0.82 |
| C | <0.001 | 6 ± 0.85 | |
| PER1 | S | <0.001 | 0 ± 1.00 |
| C | <0.02 | 18 ± 1.20 | |
| PER3 | S | n.s. | n/a |
| C | n.s. | n/a | |
| RORA | S | n.s. | n/a |
| C | n.s. | n/a | |
| REVERBA | S | <0.005 | 12 ± 0.98 |
| C | <0.001 | 12 ± 0.83 | |
| CRY1 | S | <0.001 | 6 ± 0.86 |
| C | <0.001 | 6 ± 0.83 | |
| CRY2 | S | <0.005 | 18 ± 1.03 |
| C | n.s. | n/a | |
| BMAL1 | S | n.s. | n/a |
| C | <0.001 | 6 ± 0.86 |
| Genes | Weather | Acro (p-Value) | Acrophase |
|---|---|---|---|
| CREB1 | S | <0.01 | 12 ± 1.07 |
| C | n.s. | n/a | |
| PER1 | S | <0.001 | 18 ± 0.53 |
| C | <0.001 | 0 ± 0.56 | |
| PER3 | S | <0.001 | 12 ± 0.68 |
| C | <0.05 | 6 ± 1.33 | |
| RORA | S | <0.005 | 6 ± 1.04 |
| C | <0.001 | 6 ± 0.76 | |
| REVERBA | S | n.s. | n/a |
| C | <0.02 | 12 ± 1.17 | |
| CRY1 | S | <0.001 | 12 ± 0.31 |
| C | n.s. | n/a | |
| CRY2 | S | <0.001 | 12 ± 0.69 |
| C | <0.05 | 12 ± 1.17 | |
| BMAL1 | S | <0.001 | 12 ± 0.66 |
| C | n.s. | n/a |
| Genes | Weather | Acro (p-Value) | Acrophase |
|---|---|---|---|
| TRIM25 | S | n.s. | n/a |
| C | <0.001 | 12 ± 0.93 | |
| STAT3 | S | <0.05 | 0 ± 1.29 |
| C | n.s. | n/a | |
| MBNL | S | <0.02 | 18 ± 1.16 |
| C | n.s. | n/a | |
| MSTN | S | n.s. | n/a |
| C | <0.02 | 12 ± 1.19 |
| Genes | Weather | Acro (p-Value) | Acrophase |
|---|---|---|---|
| TRIM25 | S | n.s. | n/a |
| C | n.s. | n/a | |
| STAT3 | S | <0.001 | 18 ± 0.67 |
| C | n.s. | n/a | |
| MBNL | S | n.s. | n/a |
| C | n.s. | n/a | |
| MSTN | S | <0.02 | 12 ± 1.2 |
| C | n.s. | n/a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Dai, S.; Liu, L.; Fu, Z.; Yang, R.; Yu, G.; Ma, Z.; Zong, H. Daily Rhythmicity of Muscle-Related and Rhythm Genes Expression in Mackerel Tuna (Euthynnus affinis). Biology 2023, 12, 1211. https://doi.org/10.3390/biology12091211
Wang W, Dai S, Liu L, Fu Z, Yang R, Yu G, Ma Z, Zong H. Daily Rhythmicity of Muscle-Related and Rhythm Genes Expression in Mackerel Tuna (Euthynnus affinis). Biology. 2023; 12(9):1211. https://doi.org/10.3390/biology12091211
Chicago/Turabian StyleWang, Wenwen, Shiming Dai, Longlong Liu, Zhengyi Fu, Rui Yang, Gang Yu, Zhenhua Ma, and Humin Zong. 2023. "Daily Rhythmicity of Muscle-Related and Rhythm Genes Expression in Mackerel Tuna (Euthynnus affinis)" Biology 12, no. 9: 1211. https://doi.org/10.3390/biology12091211
APA StyleWang, W., Dai, S., Liu, L., Fu, Z., Yang, R., Yu, G., Ma, Z., & Zong, H. (2023). Daily Rhythmicity of Muscle-Related and Rhythm Genes Expression in Mackerel Tuna (Euthynnus affinis). Biology, 12(9), 1211. https://doi.org/10.3390/biology12091211








