Induction of Hibernation and Changes in Physiological and Metabolic Indices in Pelodiscus sinensis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Establishment of a Hibernation Induction Model
2.2. Sample Collection
2.3. Determination of Metabolism and Physiological Indicators
2.4. Analysis of Gene Transcriptional Activity
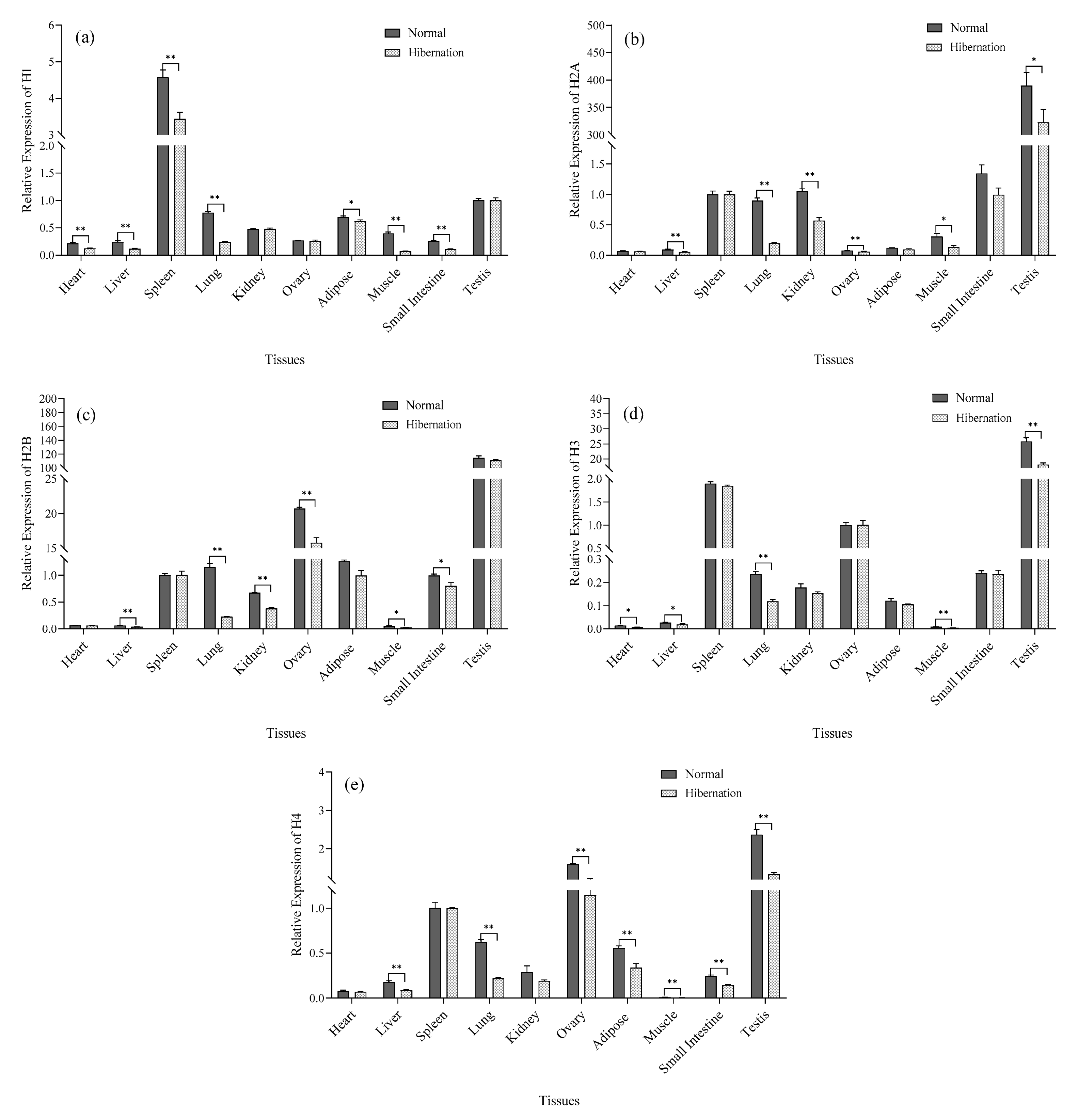
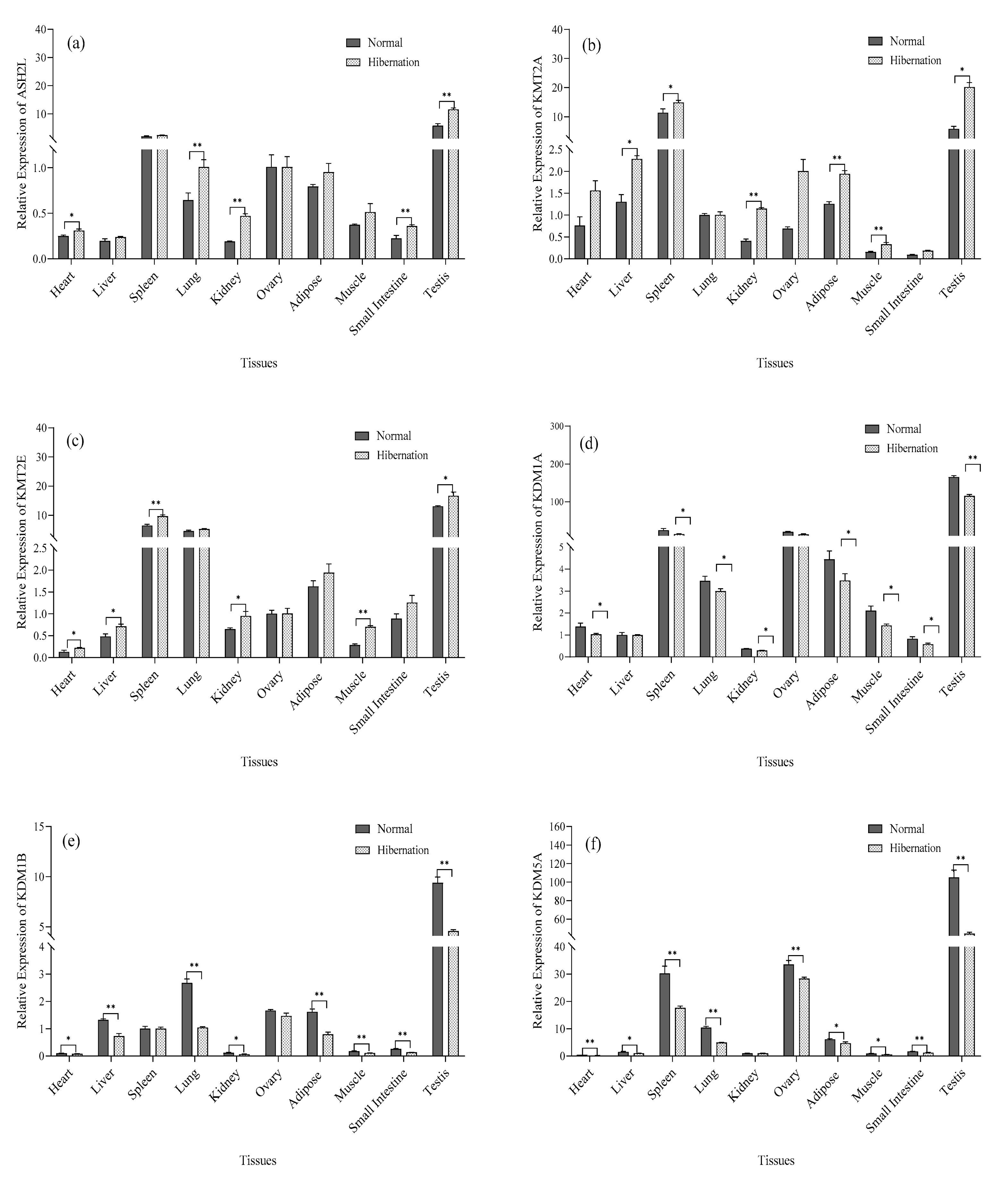
2.5. Analysis of mRNA Expression of Histones and (De)methylated Genes
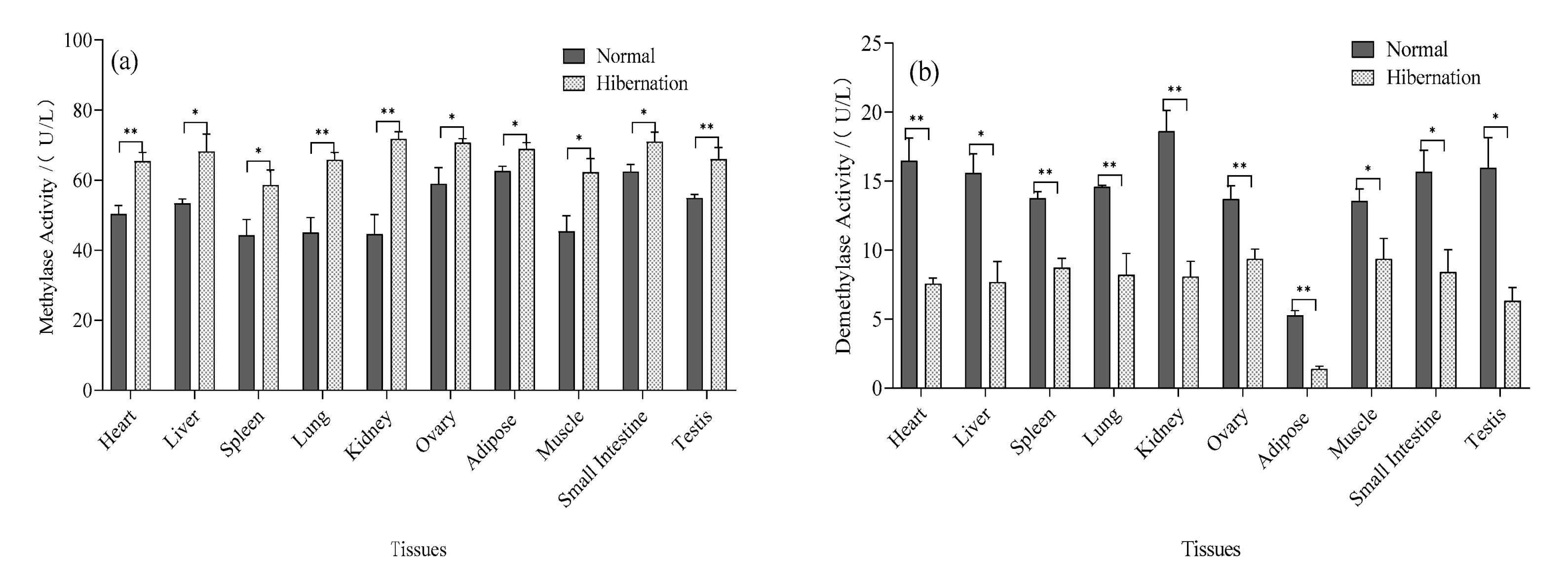
2.6. Analysis of Total HMT and DM Activity
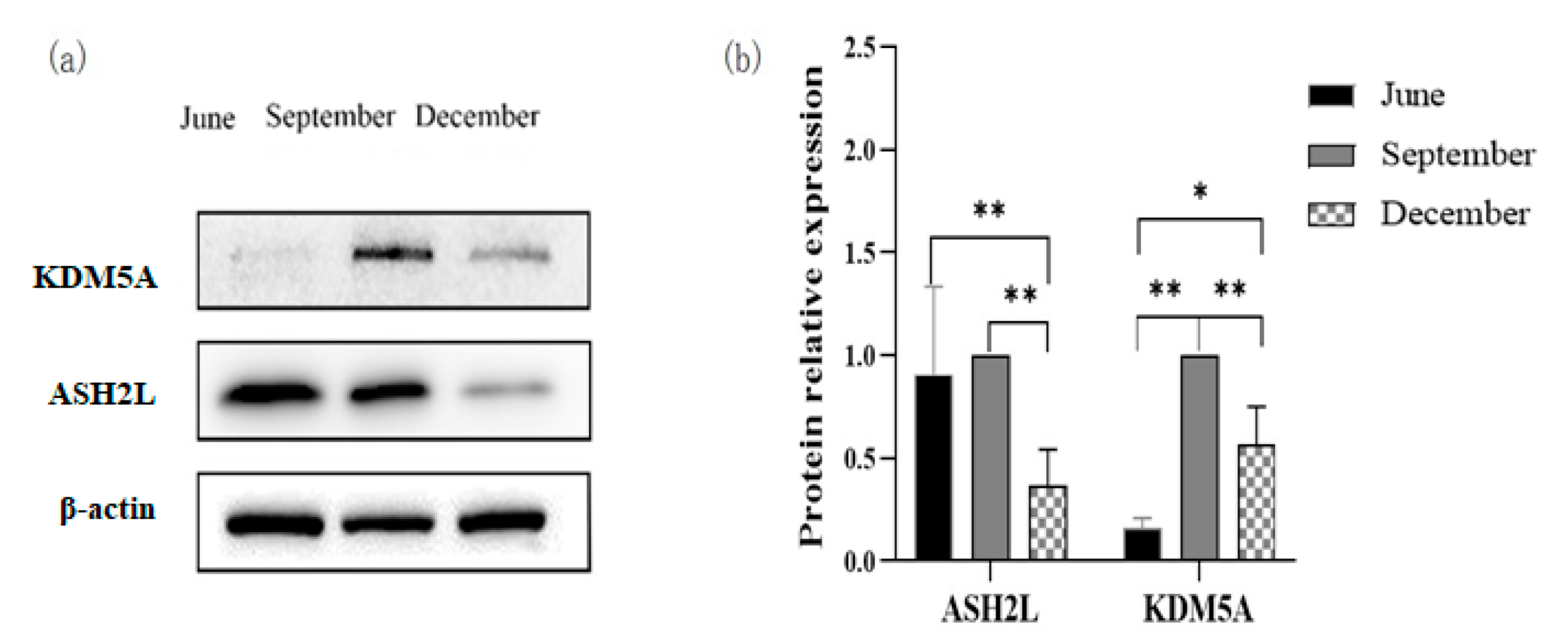
2.7. Analysis of ASH2L and KDM5A Protein Expression
2.8. Statistical Analysis
3. Results
3.1. Establishment of a Hibernation Induction Model
3.2. Measurement of Physiological Indices
3.3. Analysis of Transcriptional Activity
3.4. Analysis of Histone Gene Expression
3.5. Analysis of HMT and DM mRNA Expression
3.6. Determination of Total HMT and DM Activity
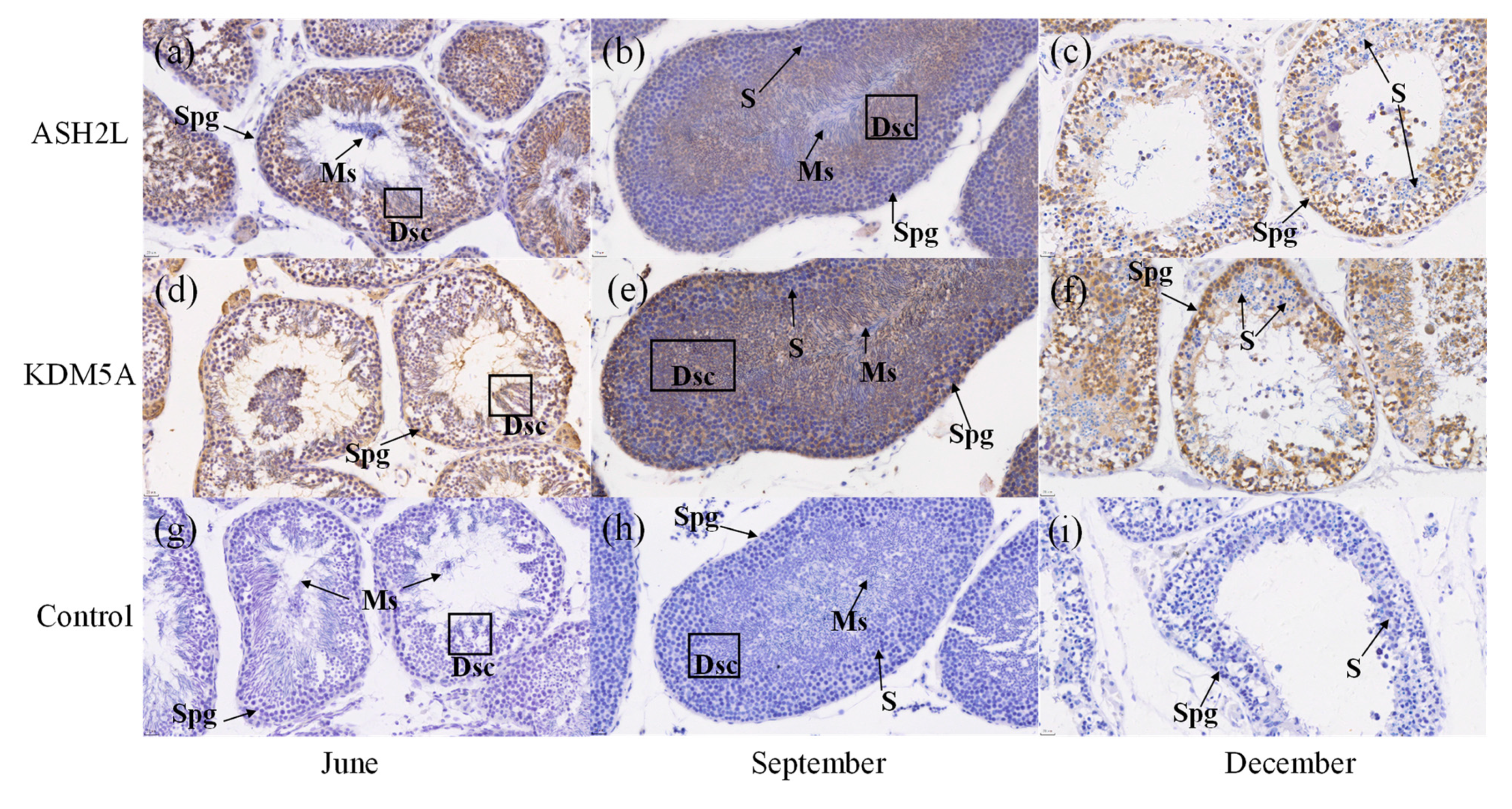
3.7. Expression of ASH2L and KDM5A
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| H3K4 | Histone H3 lysine 4 |
| HMT | Histone methylation |
| DM | Histone demethylation |
| CAT | Catalase |
| SOD | Superoxide dismutase |
| MDA | Malondialdehyde |
References
- Boyles, J.G.; Johnson, J.S.; Blomberg, A.; Lilley, T.M. Optimal hibernation theory. Mammal Rev. 2019, 50, 91–100. [Google Scholar] [CrossRef]
- Sprenger, R.; Milsom, W. Control of Breathing in Hibernation: Effect of Temperature on the Ventilatory Response to Co2. Cryobiology 2019, 133, 49–63. [Google Scholar] [CrossRef]
- Geiser, F. Hibernation. Curr. Biol. 2013, 23, R188–R193. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Kamata, T.; Nawa, H.; Sekijima, T.; Takei, N. AMPK activation, eEF2 inactivation, and reduced protein synthesis in the cerebral cortex of hibernating chipmunks. Sci. Rep. 2019, 9, 11904. [Google Scholar] [CrossRef] [PubMed]
- Christensen, M.; Hartmund, T.; Gesser, H. Creatine kinase, energy-rich phosphates and energy metabolism in heart muscle of different vertebrates. J. Comp. Physiol. B 1994, 164, 118–123. [Google Scholar] [CrossRef]
- Birkedal, R.; Gesser, H. Effects of hibernation on mitochondrial regulation and metabolic capacities in myocardium of painted turtle (Chrysemys picta). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2004, 139, 285–291. [Google Scholar] [CrossRef]
- Morin, P., Jr.; Storey, K.B. Evidence for a reduced transcriptional state during hibernation in ground squirrels. Cryobiology 2006, 53, 310–318. [Google Scholar] [CrossRef]
- Andrews, M.T.; Russeth, K.P.; Drewes, L.R.; Henry, P.G. Adaptive mechanisms regulate preferred utilization of ketones in the heart and brain of a hibernating mammal during arousal from torpor. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 296, R383–R393. [Google Scholar] [CrossRef]
- Zhang, J.; Kaasik, K.; Blackburn, M.R.; Lee, C.C. Constant darkness is a circadian metabolic signal in mammals. Nature 2006, 439, 340–343. [Google Scholar] [CrossRef]
- Toien, O.; Blake, J.; Edgar, D.M.; Grahn, D.A.; Heller, H.C.; Barnes, B.M. Hibernation in black bears: Independence of metabolic suppression from body temperature. Science 2011, 331, 906–909. [Google Scholar] [CrossRef]
- Gillen, A.E.; Fu, R.; Riemondy, K.A.; Jager, J.; Fang, B.; Lazar, M.A.; Martin, S.L. Liver Transcriptome Dynamics During Hibernation Are Shaped by a Shifting Balance between Transcription and RNA Stability. Front. Physiol. 2021, 12, 662132. [Google Scholar] [CrossRef]
- Jansen, H.T.; Trojahn, S.; Saxton, M.W.; Quackenbush, C.R.; Evans Hutzenbiler, B.D.; Nelson, O.L.; Cornejo, O.E.; Robbins, C.T.; Kelley, J.L. Hibernation induces widespread transcriptional remodeling in metabolic tissues of the grizzly bear. Commun. Biol. 2019, 2, 336. [Google Scholar] [CrossRef]
- Wu, C.-W.; Storey, K.B. Roles of the mTOR signaling pathway in hibernating ground squirrels, a differential suppression of active protein synthesis. Cryobiology 2013, 66, 356. [Google Scholar] [CrossRef]
- Kruman, I.I.; Ilyasova, E.N.; Rudchenko, S.A.; Khurkhulu, Z.S. The intestinal epithelial cells of ground squirrel (Citellus undulatus) accumulate at G2 phase of the cell cycle throughout a bout of hibernation. Comp. Biochem. Physiol. A Comp. Physiol. 1988, 90, 233–236. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Beisel, C.; Paro, R. Silencing chromatin: Comparing modes and mechanisms. Nat. Rev. Genet. 2011, 12, 123–135. [Google Scholar] [CrossRef]
- Martin, C.; Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005, 6, 838–849. [Google Scholar] [CrossRef]
- Tessier, S.N.; Luu, B.E.; Smith, J.C.; Storey, K.B. The role of global histone post-translational modifications during mammalian hibernation. Cryobiology 2017, 75, 28–36. [Google Scholar] [CrossRef]
- Shao, G.B.; Chen, J.C.; Zhang, L.P.; Huang, P.; Lu, H.Y.; Jin, J.; Gong, A.H.; Sang, J.R. Dynamic patterns of histone H3 lysine 4 methyltransferases and demethylases during mouse preimplantation development. In Vitro Cell. Dev. Biol. Anim. 2014, 50, 603–613. [Google Scholar] [CrossRef]
- Rouble, A.N.; Hawkins, L.J.; Storey, K.B. Roles for lysine acetyltransferases during mammalian hibernation. J. Therm. Biol. 2018, 74, 71–76. [Google Scholar] [CrossRef]
- Laurentin, A. A microtiter modification of the anthrone-sulfuric acid colorimetric assay for glucose-based carbohydrates. Anal. Biochem. 2003, 315, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.A.C.C. A simple colorimetric assay for muramic acid and lactic acid. Appl. Biochem. Biotechnol. 1996, 56, 49–58. [Google Scholar] [CrossRef]
- Sandri, F.; Danieli, M.; Zecca, M.; Centomo, P. Comparing Catalysts of the Direct Synthesis of Hydrogen Peroxide in Organic Solvent: Is the Measure of the Product an Issue? ChemCatChem 2021, 13, 2653–2663. [Google Scholar] [CrossRef]
- Lee, S.C.; Hsiao, J.K.; Yang, Y.C.; Haung, J.C.; Tien, L.Y.; Li, D.E.; Tsai, S.M. Insulin-like growth factor-1 positively associated with bone formation markers and creatine kinase in adults with general physical activity. J. Clin. Lab. Anal. 2021, 35, e23799. [Google Scholar] [CrossRef]
- Qian, J.; Deng, D. Stability comparison of three candidate internal reference genes in Chinese giant salamander (Andrias davidianus). Aquac. Res. 2019, 51, 362–369. [Google Scholar] [CrossRef]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. Biotechniques 1998, 24, 318–319. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Zakrajsek, B.A.; Mills, A.G.; Gorn, V.; Singer, M.J.; Reed, M.W. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: Comparison of endpoint and real-time methods. Anal. Biochem. 2000, 285, 194–204. [Google Scholar] [CrossRef]
- Starkings, S. Quantitative Data Analysis with IBM SPSS 17,18 & 19: A Guide for Social Scientists. Int. Stat. Rev. 2012, 80, 334–335. [Google Scholar]
- Ortmann, S.; Heldmaier, G. Regulation of body temperature and energy requirements of hibernating alpine marmots (Marmota marmota). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000, 278, R698–R704. [Google Scholar] [CrossRef]
- West, T.G.; Donohoe, P.H.; Staples, J.F.; Askew, G.N. Tribute to R. G. Boutilier: The role for skeletal muscle in the hypoxia-induced hypometabolic responses of submerged frogs. J. Exp. Biol. 2006, 209 Pt 7, 1159–1168. [Google Scholar] [CrossRef]
- Layne, J.R.; Kennedy, S.D. Cellular energetics of frozen wood frogs (Rana sylvatica) revealed via NMR spectroscopy. J. Therm. Biol. 2002, 27, 167–173. [Google Scholar] [CrossRef]
- Chen, B.J.; Zhang, W.Y.; Niu, C.J.; Li, W.J.; Jia, H.; Storey, K.B. Antioxidant response to acute cold exposure and during recovery in juvenile Chinese soft-shelled turtles (Pelodiscus sinensis). J. Exp. Biol. 2019, 222 Pt 4, jeb197863. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Regenstein, J.M.; Xie, D.; Lu, W.; Ren, X.; Yuan, J.; Mao, L. The oxidative stress and antioxidant responses of Litopenaeus vannamei to low temperature and air exposure. Fish Shellfish Immunol. 2018, 72, 564–571. [Google Scholar] [CrossRef]
- Chen, B.J.; Niu, C.J.; Yuan, L. Ascorbic acid regulation in stress responses during acute cold exposure and following recovery in juvenile Chinese soft-shelled turtle (Pelodiscus sinensis). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2015, 184, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Van Breukelen, F.; Martin, S.L. Reversible depression of transcription during hibernation. J. Comp. Physiol. B 2002, 172, 355–361. [Google Scholar] [CrossRef]
- Biggar, Y.; Storey, K.B. Global DNA modifications suppress transcription in brown adipose tissue during hibernation. Cryobiology 2014, 69, 333–338. [Google Scholar] [CrossRef]
- Roessler, T.Y.; Wirtz, A.; Slater, M.J.; Henjes, J. Growth performance and RNA/DNA ratio of noble crayfish (Astacus astacus) and narrow-clawed crayfish (Pontastacus leptodactylus) fed fish waste diets. Aquac. Res. 2020, 51, 3205–3215. [Google Scholar] [CrossRef]
- Doenecke, D.; Albig, W.; Bode, C.; Drabent, B.; Franke, K.; Gavenis, K.; Witt, O. Histones: Genetic diversity and tissue-specific gene expression. Histochem. Cell Biol. 1997, 107, 1–10. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, J.; Pan, Y.; Jin, J.; Sang, J.; Huang, P.; Shao, G. Expression of histone H3 lysine 4 methylation and its demethylases in the developing mouse testis. Cell Tissue Res. 2014, 358, 875–883. [Google Scholar] [CrossRef]
- Lüscher-Firzlaff, J.; Chatain, N.; Kuo, C.-C.; Braunschweig, T.; Bochyńska, A.; Ullius, A.; Denecke, B.; Costa, I.G.; Koschmieder, S.; Lüscher, B. Hematopoietic stem and progenitor cell proliferation and differentiation requires the trithorax protein Ash2l. Sci. Rep. 2019, 9, 8262. [Google Scholar] [CrossRef]
- Wan, M.; Liang, J.; Xiong, Y.; Shi, F.; Zhang, Y.; Lu, W.; He, Q.; Yang, D.; Chen, R.; Liu, D.; et al. The trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J. Biol. Chem. 2013, 288, 5039–5048. [Google Scholar] [CrossRef]
- Steward, M.M.; Lee, J.S.; O’Donovan, A.; Wyatt, M.; Bernstein, B.E.; Shilatifard, A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006, 13, 852–854. [Google Scholar] [CrossRef]
- Capell, B.C.; Drake, A.M.; Zhu, J.; Shah, P.P.; Dou, Z.; Dorsey, J.; Simola, D.F.; Donahue, G.; Sammons, M.; Rai, T.S.; et al. MLL1 is essential for the senescence-associated secretory phenotype. Genes Dev. 2016, 30, 321–336. [Google Scholar] [CrossRef]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef]
- Adamo, A.; Sese, B.; Boue, S.; Castano, J.; Paramonov, I.; Barrero, M.J.; Izpisua Belmonte, J.C. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat. Cell Biol. 2011, 13, 652–659. [Google Scholar] [CrossRef]
- Jin, Y.; Huo, B.; Fu, X.; Cheng, Z.; Zhu, J.; Zhang, Y.; Hao, T.; Hu, X. LSD1 knockdown reveals novel histone lysine methylation in human breast cancer MCF-7 cells. Biomed. Pharmacother. 2017, 92, 896–904. [Google Scholar] [CrossRef]
- Fang, R.; Barbera, A.J.; Xu, Y.; Rutenberg, M.; Leonor, T.; Bi, Q.; Lan, F.; Mei, P.; Yuan, G.C.; Lian, C.; et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol. Cell 2010, 39, 222–233. [Google Scholar] [CrossRef]
- Gaillard, S.; Charasson, V.; Ribeyre, C.; Salifou, K.; Pillaire, M.J.; Hoffmann, J.S.; Constantinou, A.; Trouche, D.; Vandromme, M. KDM5A and KDM5B histone-demethylases contribute to HU-induced replication stress response and tolerance. Biol. Open 2021, 10, bio057729. [Google Scholar] [CrossRef]
- Wijenayake, S.; Hawkins, L.J.; Storey, K.B. Dynamic regulation of six histone H3 lysine (K) methyltransferases in response to prolonged anoxia exposure in a freshwater turtle. Gene 2018, 649, 50–57. [Google Scholar] [CrossRef]
- Lee, S.; Lee, J.; Chae, S.; Moon, Y.; Lee, H.Y.; Park, B.; Yang, E.G.; Hwang, D.; Park, H. Multi-dimensional histone methylations for coordinated regulation of gene expression under hypoxia. Nucleic Acids Res. 2017, 45, 11643–11657. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Endo, D.; Koji, T. Roles of epigenome in mammalian spermatogenesis. Reprod. Med. Biol. 2014, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Wahlestedt, C. Epigenetic mechanisms of gene regulation during mammalian spermatogenesis. Epigenetics 2008, 3, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, M.; Simeone, A.; Hormanseder, E.; Teperek, M.; Gaggioli, V.; O’Doherty, A.; Falk, E.; Sporniak, M.; D’Santos, C.; Franklin, V.N.R.; et al. Epigenetic homogeneity in histone methylation underlies sperm programming for embryonic transcription. Nat. Commun. 2020, 11, 3491. [Google Scholar] [CrossRef]
- An, J.; Qin, J.; Wan, Y.; Zhang, Y.; Hu, Y.; Zhang, C.; Zeng, W. Histone lysine methylation exhibits a distinct distribution during spermatogenesis in pigs. Theriogenology 2015, 84, 1455–1462. [Google Scholar] [CrossRef]
- Godmann, M.; Auger, V.; Ferraroni-Aguiar, V.; Di Sauro, A.; Sette, C.; Behr, R.; Kimmins, S. Dynamic regulation of histone H3 methylation at lysine 4 in mammalian spermatogenesis. Biol. Reprod. 2007, 77, 754–764. [Google Scholar] [CrossRef]
- Weili, L.; Juliane, L.F.; Andrea, U.; Ursula, S.; Thomas, L.; Bernhard, L. Loss of the epigenetic regulator Ash2l results in desintegration of hepatocytes and liver failure. Int. J. Clin. Exp. Pathol. 2016, 9, 5167–5175. [Google Scholar]
- Nishio, H.; Hayashi, Y.; Moritoki, Y.; Kamisawa, H.; Mizuno, K.; Kojima, Y.; Kohri, K. Distinctive changes in histone H3K4 modification mediated via Kdm5a expression in spermatogonial stem cells of cryptorchid testes. J. Urol. 2014, 191, 1564–1572. [Google Scholar] [CrossRef]


| Target Gene | Primers | Primer Sequence(5′-3′) | Login ID |
|---|---|---|---|
| GAPDH | GAPDH-F GAPDH-R | CCTGGTATGACAATGAGTT GTGCCTGGTTTATTCCTT | NM_001286927.1 |
| H1 | H1-F H1-R | TCCTGCTGTGTCCGCTCCTG GGAAGACTTACGGGCTTTGGAACC | XM_006125889.3 |
| H2A | H2A-F H2A-R | AAGGTCAGTGGAAACTCTGGTTGC TGCCTTACTTGCTGGTCTGTGTTC | XM_006136395.2 |
| H2B | H2B-F H2B-R | GAGGGTCGGTCGAGATGTCTACG GACTCCTTGCGGCTCTTCTTACG | XM_025177927.1 |
| H3 | H3-F H3-R | GAAATCGCCCAGGACTTCAAGACC GGCATGATGGTGACTCGCTTAGC | XM_006125911.2 |
| H4 | H4-F H4-R | CTAAGGTGCCTTGAGTCTGCTGTC CGAGCCAAGCGACGAATAGCC | XM_006125900.3 |
| ASH2L | ASH2L-F ASH2L-R | ACTGACCGTTATTGGCGAGAAAGG CAAGTCTGGCTGCTGTGTCTGG | XM_006134041.3 |
| KMT2A | KMT2A-F KMT2A-R | TGCGATTCCGACACTTGAAGAAGAC TGAGGATGGAGCGAATGACATTGC | XM_006120948.3 |
| KMT2E | KMT2E-F KMT2E-R | AGTGTGGTAAGGCTGCTTGTAAGTG TGGTGAGGAGGATCAGGCTTCTATC | XM_014580823.2 |
| KDM1A | KDM1A-F KDM1A-R | AACGAAGAAGACTGGCAAGGTGATC CTTCCAGAAGCGTGACATCCATCC | XM_014568580.2 |
| KDM1B | KDM1B-F KDM1B-R | GAAGCAGCAGAGGATGATGATGAGG TAGCACACCTTTCAGCAGCACTTG | XM_006121822.3 |
| KDM5A | KDM5A-F KDM5A-R | CAAGGCTACAGGTGTGGTCTCAAG GCTGCTGATTGTAGGCTGGTATCC | XM_014579673.1 |
| RT (°C) | WT (°C) | CT (°C) | PT (°C) | OT (°C) | HHM | BOC (%) | TOT (s) | LNE (cm) |
|---|---|---|---|---|---|---|---|---|
| 26 | 20.1 | 20.6 ± 0.17 | 21.3 ± 0.83 | 18.7 ± 0.90 | 13.0 ± 0.82 | 93.3 ± 2.36 | 3.3 ± 0.47 | 15.0 ± 1.63 |
| 23 | 18.9 | 19.6 ± 0.21 | 21.4 ± 0.41 | 18.3 ± 0.73 | 11 ± 0.82 | 95 ± 0.00 | 6.3 ± 0.94 | 15 ± 0.82 |
| 20 | 16.1 | 18.0 ± 0.21 | 19.4 ± 0.50 | 17.5 ± 0.47 | 12.3 ± 2.62 | 95 ± 0.00 | 5.7 ± 0.94 | 16 ± 0.82 |
| 17 | 15.2 | 18.0 ± 0.21 | 19.4 ± 0.50 | 17.5 ± 0.47 | 12.3 ± 2.62 | 91.7 ± 2.36 | 6.7 ± 1.25 | 14.7 ± 1.25 |
| 14 | 14.3 | 15.2 ± 0.25 | 16.2 ± 0.33 | 16.7 ± 0.87 | 17.0 ± 5.10 | 88.3 ± 2.36 | 6.0 ± 0.82 | 14.7 ± 1.25 |
| 11 | 10.9 | 13.1 ± 0.29 | 13.2 ± 0.54 | 14.5 ± 0.24 | 6.7 ± 1.25 | 86.7 ± 2.36 | 7.3 ± 2.36 | 14.7 ± 1.25 |
| 8 * | 8.4 * | 9.6 ± 0.21 ** | 8.9 ± 0.47 * | 13.1 ± 0.34 * | 5.0 ± 0.82 | 81.7 ± 2.36 ** | 9.0 ± 2.45 | 8.0 ± 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, R.; Wu, J.; You, Z.; Xu, D.; Li, C.; Wang, W.; Qian, G. Induction of Hibernation and Changes in Physiological and Metabolic Indices in Pelodiscus sinensis. Biology 2023, 12, 720. https://doi.org/10.3390/biology12050720
Lin R, Wu J, You Z, Xu D, Li C, Wang W, Qian G. Induction of Hibernation and Changes in Physiological and Metabolic Indices in Pelodiscus sinensis. Biology. 2023; 12(5):720. https://doi.org/10.3390/biology12050720
Chicago/Turabian StyleLin, Runlan, Jiahao Wu, Ziyi You, Dongjie Xu, Caiyan Li, Wei Wang, and Guoying Qian. 2023. "Induction of Hibernation and Changes in Physiological and Metabolic Indices in Pelodiscus sinensis" Biology 12, no. 5: 720. https://doi.org/10.3390/biology12050720
APA StyleLin, R., Wu, J., You, Z., Xu, D., Li, C., Wang, W., & Qian, G. (2023). Induction of Hibernation and Changes in Physiological and Metabolic Indices in Pelodiscus sinensis. Biology, 12(5), 720. https://doi.org/10.3390/biology12050720








