Tubulin Post-Translational Modifications: The Elusive Roles of Acetylation
Abstract
Simple Summary
Abstract
1. Introduction
1.1. The Dynamic Nature of Microtubules
1.2. The Functional Diversity of Microtubules and Microtubule-Based Structures
1.3. In Vivo Microtubule Dynamics Is Modulated by Microtubule-Associated Proteins
1.4. The In Vivo Diversity of Tubulin Pools and Microtubule Functional Diversity
2. The Tubulin Acetyltransferases and Deacetylases
2.1. Tubulin Acetyltransferases
2.2. Tubulin Deacetylases
3. Tubulin Acetylation: Structural and Functional Implications
3.1. Structural Implications of Tubulin Acetylation
3.2. Functional Impact of Tubulin Acetylation
3.2.1. Cilia Microtubules
3.2.2. Centrioles
3.2.3. Cytoplasmic Microtubules Arrays
3.3. Tubulin Acetylation beyond Lys40
4. Tubulin Acetylation and Stress Conditions
4.1. Mechanisms of Microtubules α-Tubulin Acetylation Regulation under Stress
4.2. Dysregulation of Tubulin Acetylation and Disease
4.2.1. Neurodegenerative Diseases
4.2.2. Cancer
4.2.3. Cardiac Diseases
4.2.4. Innate Immunity and Virus Infections
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Tavana, O.; Gu, W. p53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 2019, 11, 564–577. [Google Scholar] [CrossRef]
- Millán-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone post-translational modifications—Cause and consequence of genome function. Nat. Rev. Genet. 2022, 23, 563–580. [Google Scholar] [CrossRef]
- Wloga, D.; Joachimiak, E.; Fabczak, H. Tubulin Post-Translational Modifications and Microtubule Dynamics. Int. J. Mol. Sci. 2017, 18, 2207. [Google Scholar] [CrossRef] [PubMed]
- Gudimchuk, N.B.; McIntosh, J.R. Regulation of microtubule dynamics, mechanics and function through the growing tip. Nat. Rev. Mol. Cell Biol. 2021, 22, 777–795. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, J.; Tavares, A.; Carvalhal, S.; Soares, H. Revisiting the tubulin folding pathway: New roles in centrosomes and cilia. Biomol. Concepts 2010, 1, 423–434. [Google Scholar] [CrossRef]
- Cowan, N.J.; Lewis, S.A. Type II chaperonns, prefoldin, and the tubulin-specific chaperones. Adv. Protein Chem. 2001, 59, 73–104. [Google Scholar] [CrossRef]
- Lopez, T.; Dalton, K.; Frydman, J. The Mechanism and Function of Group II Chaperonins. J. Mol. Biol. 2015, 427, 2919–2930. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.A.; Tian, G.; Cowan, N.J. The α- and β-tubulin folding pathways. Trends Cell Biol. 1997, 7, 479–484. [Google Scholar] [CrossRef]
- Lopez-Fanarraga, M.; Avila, J.; Guasch, A.; Coll, M.; Zabala, J.C. Review: Postchaperonin Tubulin Folding Cofactors and Their Role in Microtubule Dynamics. J. Struct. Biol. 2001, 135, 219–229. [Google Scholar] [CrossRef]
- Kortazar, D.; Fanarraga, M.; Carranza, G.; Bellido, J.; Villegas, J.; Avila, J.; Zabala, J. Role of cofactors B (TBCB) and E (TBCE) in tubulin heterodimer dissociation. Exp. Cell Res. 2007, 313, 425–436. [Google Scholar] [CrossRef]
- Voloshin, O.; Gocheva, Y.; Gutnick, M.; Movshovich, N.; Bakhrat, A.; Baranes-Bachar, K.; Bar-Zvi, D.; Parvari, R.; Gheber, L.; Raveh, D. Tubulin chaperone E binds microtubules and proteasomes and protects against misfolded protein stress. Cell. Mol. Life Sci. 2010, 67, 2025–2038. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Mitchison, T.J. Microtubule Polymerization Dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83–117. [Google Scholar] [CrossRef] [PubMed]
- Serna, M.; Zabala, J.C. Tubulin Folding and Degradation. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016; pp. 1–9. [Google Scholar] [CrossRef]
- Nolasco, S.; Bellido, J.; Serna, M.; Carmona, B.; Soares, H.; Zabala, J.C. Colchicine Blocks Tubulin Heterodimer Recycling by Tubulin Cofactors TBCA, TBCB, and TBCE. Front. Cell Dev. Biol. 2021, 9, 950. [Google Scholar] [CrossRef] [PubMed]
- Roll-Mecak, A. The Tubulin Code in Microtubule Dynamics and Information Encoding. Dev. Cell 2020, 54, 7–20. [Google Scholar] [CrossRef]
- Camelo, C.; Peneda, C.; Carmona, B.; Soarets, H. TBCC. In Encyclopedia of Signaling Molecules; Choi, S., Ed.; Springer: New York, NY, USA, 2016; pp. 1–8. [Google Scholar] [CrossRef]
- Lomakin, A.J.; Semenova, I.; Zaliapin, I.; Kraikivski, P.; Nadezhdina, E.; Slepchenko, B.M.; Akhmanova, A.; Rodionov, V. CLIP-170-Dependent Capture of Membrane Organelles by Microtubules Initiates Minus-End Directed Transport. Dev. Cell 2009, 17, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Lomakin, A.J.; Kraikivski, P.; Semenova, I.; Ikeda, K.; Zaliapin, I.; Tirnauer, J.S.; Akhmanova, A.; Rodionov, V. Stimulation of the CLIP-170–dependent capture of membrane organelles by microtubules through fine tuning of microtubule assembly dynamics. Mol. Biol. Cell 2011, 22, 4029–4037. [Google Scholar] [CrossRef]
- Kanfer, G.; Peterka, M.; Arzhanik, V.K.; Drobyshev, A.L.; Ataullakhanov, F.I.; Volkov, V.A.; Kornmann, B. CENP-F couples cargo to growing and shortening microtubule ends. Mol. Biol. Cell 2017, 28, 2400–2409. [Google Scholar] [CrossRef]
- Seetharaman, S.; Etienne-Manneville, S. Microtubules at focal adhesions—A double-edged sword. J. Cell Sci. 2019, 132, jcs232843. [Google Scholar] [CrossRef]
- Stehbens, S.; Wittmann, T. Targeting and transport: How microtubules control focal adhesion dynamics. J. Cell Biol. 2012, 198, 481–489. [Google Scholar] [CrossRef]
- Stehbens, S.J.; Paszek, M.; Pemble, H.; Ettinger, A.; Gierke, S.; Wittmann, T. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nature 2014, 16, 558–570. [Google Scholar] [CrossRef]
- Kopf, A.; Kiermaier, E. Dynamic Microtubule Arrays in Leukocytes and Their Role in Cell Migration and Immune Synapse Formation. Front. Cell Dev. Biol. 2021, 9, 635511. [Google Scholar] [CrossRef]
- Seetharaman, S.; Etienne-Manneville, S. Cytoskeletal Crosstalk in Cell Migration. Trends Cell Biol. 2020, 30, 720–735. [Google Scholar] [CrossRef]
- Mayor, R.; Etienne-Manneville, S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 2016, 17, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Bayless, K.J.; Johnson, G.A. Role of the Cytoskeleton in Formation and Maintenance of Angiogenic Sprouts. J. Vasc. Res. 2011, 48, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, T.; Dehghani, F. The Cytoskeleton—A Complex Interacting Meshwork. Cells 2019, 8, 362. [Google Scholar] [CrossRef]
- Bornens, M. Organelle positioning and cell polarity. Nat. Rev. Mol. Cell Biol. 2008, 9, 874–886. [Google Scholar] [CrossRef] [PubMed]
- Budovsky, A.; Fraifeld, V.E.; Aronov, S. Linking cell polarity, aging and rejuvenation. Biogerontology 2010, 12, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Soares, H.; Marinho, H.S.; Real, C.; Antunes, F. Cellular polarity in aging: Role of redox regulation and nutrition. Genes Nutr. 2013, 9, 371. [Google Scholar] [CrossRef]
- Vasileva, E.; Citi, S. The role of microtubules in the regulation of epithelial junctions. Tissue Barriers 2018, 6, 1–20. [Google Scholar] [CrossRef]
- Matis, M. The Mechanical Role of Microtubules in Tissue Remodeling. Bioessays 2020, 42, e1900244. [Google Scholar] [CrossRef]
- Singh, A.; Saha, T.; Begemann, I.; Ricker, A.; Nüsse, H.; Thorn-Seshold, O.; Klingauf, J.; Galic, M.; Matis, M. Polarized microtubule dynamics directs cell mechanics and coordinates forces during epithelial morphogenesis. Nat. Cell Biol. 2018, 20, 1126–1133. [Google Scholar] [CrossRef]
- Ghalloussi, D.; Dhenge, A.; Bergmeier, W. New insights into cytoskeletal remodeling during platelet production. J. Thromb. Haemost. 2019, 17, 1430–1439. [Google Scholar] [CrossRef]
- Patel, S.R.; Richardson, J.L.; Schulze, H.; Kahle, E.; Galjart, N.; Drabek, K.; Shivdasani, R.A.; Hartwig, J.H.; Italiano, J.J.E. Differential roles of microtubule assembly and sliding in proplatelet formation by megakaryocytes. Blood 2005, 106, 4076–4085. [Google Scholar] [CrossRef]
- Thon, J.N.; Macleod, H.; Begonja, A.J.; Zhu, J.; Lee, K.-C.; Mogilner, A.; Hartwig, J.H.; Jr, J.E.I. Microtubule and cortical forces determine platelet size during vascular platelet production. Nat. Commun. 2012, 3, 852. [Google Scholar] [CrossRef]
- Sánchez, I.; Dynlacht, B.D. Cilium assembly and disassembly. Nat. Cell Biol. 2016, 18, 711–717. [Google Scholar] [CrossRef]
- Pitaval, A.; Senger, F.; Letort, G.; Gidrol, X.; Guyon, L.; Sillibourne, J.; Théry, M. Microtubule stabilization drives 3D centrosome migration to initiate primary ciliogenesis. J. Cell Biol. 2017, 216, 3713–3728. [Google Scholar] [CrossRef]
- van der Vaart, B.; Akhmanova, A.; Straube, A. Regulation of microtubule dynamic instability. Biochem. Soc. Trans. 2009, 37, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Bodakuntla, S.; Jijumon, A.; Villablanca, C.; Gonzalez-Billault, C.; Janke, C. Microtubule-Associated Proteins: Structuring the Cytoskeleton. Trends Cell Biol. 2019, 29, 804–819. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, T.M. Metaphase Spindle Assembly. Biology 2017, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Starr, D.A. A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 2009, 122, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Dogterom, M.; Kerssemakers, J.W.J.; Romet-Lemonne, G.; E Janson, M. Force generation by dynamic microtubules. Curr. Opin. Cell Biol. 2005, 17, 67–74. [Google Scholar] [CrossRef]
- Kimura, A.; Onami, S. Modeling Microtubule-Mediated Forces and Centrosome Positioning in Caenorhabditis elegans Embryos. Methods Cell Biol. 2010, 97, 437–453. [Google Scholar] [CrossRef]
- Burakov, A.; Nadezhdina, E.; Slepchenko, B.; Rodionov, V. Centrosome positioning in interphase cells. J. Cell Biol. 2003, 162, 963–969. [Google Scholar] [CrossRef]
- Zhu, J.; Burakov, A.; Rodionov, V.; Mogilner, A. Finding the Cell Center by a Balance of Dynein and Myosin Pulling and Microtubule Pushing: A Computational Study. Mol. Biol. Cell 2010, 21, 4418–4427. [Google Scholar] [CrossRef]
- Wu, J.; Misra, G.; Russell, R.J.; Ladd, A.J.C.; Lele, T.P.; Dickinson, R.B. Effects of dynein on microtubule mechanics and centrosome positioning. Mol. Biol. Cell 2011, 22, 4834–4841. [Google Scholar] [CrossRef]
- Tanimoto, H.; Sallé, J.; Dodin, L.; Minc, N. Physical forces determining the persistency and centring precision of microtubule asters. Nat. Phys. 2018, 14, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Odell, J.; Sikirzhytski, V.; Tikhonenko, I.; Cobani, S.; Khodjakov, A.; Koonce, M. Force balances between interphase centrosomes as revealed by laser ablation. Mol. Biol. Cell 2019, 30, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Daga, R.R.; Yonetani, A.; Chang, F. Asymmetric Microtubule Pushing Forces in Nuclear Centering. Curr. Biol. 2006, 16, 1544–1550. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.; Marsh, L.; Doye, V.; Inoué, S.; Chang, F. A Mechanism for Nuclear Positioning in Fission Yeast Based on Microtubule Pushing. J. Cell Biol. 2001, 153, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Graham, O.S.; Raposo, A.; Johnston, D.S. Growing Microtubules Push the Oocyte Nucleus to Polarize the Drosophila Dorsal-Ventral Axis. Science 2012, 336, 999–1003. [Google Scholar] [CrossRef]
- Letort, G.; Nedelec, F.; Blanchoin, L.; Théry, M. Centrosome centering and decentering by microtubule network rearrangement. Mol. Biol. Cell 2016, 27, 2833–2843. [Google Scholar] [CrossRef]
- Théry, M.; Racine, V.; Piel, M.; Pépin, A.; Dimitrov, A.; Chen, Y.; Sibarita, J.-B.; Bornens, M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA 2006, 103, 19771–19776. [Google Scholar] [CrossRef] [PubMed]
- Komarova, Y.; De Groot, C.O.; Grigoriev, I.; Gouveia, S.M.; Munteanu, E.L.; Schober, J.M.; Honnappa, S.; Buey, R.M.; Hoogenraad, C.C.; Dogterom, M.; et al. Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 2009, 184, 691–706. [Google Scholar] [CrossRef] [PubMed]
- Akhmanova, A.; Steinmetz, M.O. Tracking the ends: A dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 2008, 9, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Lansbergen, G.; Akhmanova, A. Microtubule Plus End: A Hub of Cellular Activities. Traffic 2006, 7, 499–507. [Google Scholar] [CrossRef]
- Honnappa, S.; Gouveia, S.; Weisbrich, A.; Damberger, F.; Bhavesh, N.S.; Jawhari, H.; Grigoriev, I.; van Rijssel, F.J.; Martinez-Buey, R.; Lawera, A.; et al. An EB1-Binding Motif Acts as a Microtubule Tip Localization Signal. Cell 2009, 138, 366–376. [Google Scholar] [CrossRef]
- Vitre, B.; Coquelle, F.M.; Heichette, C.; Garnier, C.; Chrétien, D.; Arnal, I. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 2008, 10, 415–421. [Google Scholar] [CrossRef]
- Bieling, P.; Laan, L.; Schek, H.; Munteanu, E.L.; Sandblad, L.; Dogterom, M.; Brunner, D.; Surrey, T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature 2007, 450, 1100–1105. [Google Scholar] [CrossRef]
- Maurer, S.P.; Fourniol, F.J.; Bohner, G.; Moores, C.A.; Surrey, T. EBs Recognize a Nucleotide-Dependent Structural Cap at Growing Microtubule Ends. Cell 2012, 149, 371–382. [Google Scholar] [CrossRef]
- Seetapun, D.; Castle, B.T.; McIntyre, A.J.; Tran, P.T.; Odde, D.J. Estimating the Microtubule GTP Cap Size In Vivo. Curr. Biol. 2012, 22, 1681–1687. [Google Scholar] [CrossRef]
- Zhang, R.; Alushin, G.M.; Brown, A.; Nogales, E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell 2015, 162, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Duellberg, C.; Cade, N.I.; Holmes, D.; Surrey, T. The size of the EB cap determines instantaneous microtubule stability. Elife 2016, 5, e13470. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Lyle, K.S.; Gierke, S.; Matov, A.; Danuser, G.; Wittmann, T. GSK3β phosphorylation modulates CLASP–microtubule association and lamella microtubule attachment. J. Cell Biol. 2009, 184, 895–908. [Google Scholar] [CrossRef]
- Lansbergen, G.; Grigoriev, I.; Mimori-Kiyosue, Y.; Ohtsuka, T.; Higa, S.; Kitajima, I.; Demmers, J.; Galjart, N.; Houtsmuller, A.B.; Grosveld, F.; et al. CLASPs Attach Microtubule Plus Ends to the Cell Cortex through a Complex with LL5β. Dev. Cell 2006, 11, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Galjart, N. CLIPs and CLASPs and cellular dynamics. Nat. Rev. Mol. Cell Biol. 2005, 6, 487–498. [Google Scholar] [CrossRef]
- Walczak, C.E.; Gayek, S.; Ohi, R. Microtubule-Depolymerizing Kinesins. Annu. Rev. Cell Dev. Biol. 2013, 29, 417–441. [Google Scholar] [CrossRef]
- Cassimeris, L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 2002, 14, 18–24. [Google Scholar] [CrossRef]
- Gupta, K.K.; Li, C.; Duan, A.; Alberico, E.O.; Kim, O.V.; Alber, M.S.; Goodson, H.V. Mechanism for the catastrophe-promoting activity of the microtubule destabilizer Op18/stathmin. Proc. Natl. Acad. Sci. USA 2013, 110, 20449–20454. [Google Scholar] [CrossRef]
- Roostalu, J.; Surrey, T. Microtubule nucleation: Beyond the template. Nat. Rev. Mol. Cell Biol. 2017, 18, 702–710. [Google Scholar] [CrossRef]
- McNally, F.J.; Vale, R.D. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 1993, 75, 419–429. [Google Scholar] [CrossRef]
- Evans, K.J.; Gomes, E.; Reisenweber, S.M.; Gundersen, G.G.; Lauring, B.P. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J. Cell Biol. 2005, 168, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Roll-Mecak, A.; Vale, R.D. The Drosophila Homologue of the Hereditary Spastic Paraplegia Protein, Spastin, Severs and Disassembles Microtubules. Curr. Biol. 2005, 15, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Valencia, J.D.D.; Stewman, S.; Metz, J.; Monnier, S.; Rath, U.; Asenjo, A.B.; Charafeddine, R.A.; Sosa, H.J.; Ross, J.L.; et al. Human Fidgetin is a microtubule severing the enzyme and minus-end depolymerase that regulates mitosis. Cell Cycle 2012, 11, 2359–2366. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.-W.; Howard, J. Cutting, Amplifying, and Aligning Microtubules with Severing Enzymes. Trends Cell Biol. 2020, 31, 50–61. [Google Scholar] [CrossRef]
- McNally, F.J.; Roll-Mecak, A. Microtubule-severing enzymes: From cellular functions to molecular mechanism. J. Cell Biol. 2018, 217, 4057–4069. [Google Scholar] [CrossRef]
- Zhang, D.; Grode, K.D.; Stewman, S.F.; Diaz-Valencia, J.D.; Liebling, E.; Rath, U.; Riera, T.; Currie, J.D.; Buster, D.W.; Asenjo, A.B.; et al. Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat. Cell Biol. 2011, 13, 361–369. [Google Scholar] [CrossRef]
- E Mains, P.; Kemphues, K.J.; A Sprunger, S.; A Sulston, I.; Wood, W.B. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics 1990, 126, 593–605. [Google Scholar] [CrossRef]
- Ahmad, F.; Yu, W.; McNally, F.J.; Baas, P.W. An Essential Role for Katanin in Severing Microtubules in the Neuron. J. Cell Biol. 1999, 145, 305–315. [Google Scholar] [CrossRef]
- Kapitein, L.C.; Hoogenraad, C.C. Building the Neuronal Microtubule Cytoskeleton. Neuron 2015, 87, 492–506. [Google Scholar] [CrossRef]
- Baas, P.W.; Rao, A.N.; Matamoros, A.J.; Leo, L. Stability properties of neuronal microtubules. Cytoskeleton 2016, 73, 442–460. [Google Scholar] [CrossRef]
- Lohret, T.A.; McNally, F.J.; Quarmby, L.M. A Role for Katanin-mediated Axonemal Severing during Chlamydomonas Deflagellation. Mol. Biol. Cell 1998, 9, 1195–1207. [Google Scholar] [CrossRef] [PubMed]
- Vemu, A.; Szczesna, E.; Zehr, E.A.; Spector, J.O.; Grigorieff, N.; Deaconescu, A.M.; Roll-Mecak, A. Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science 2018, 361, eaau1504. [Google Scholar] [CrossRef]
- Komarova, Y.A.; Akhmanova, A.S.; Kojima, S.-I.; Galjart, N.; Borisy, G.G. Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 2002, 159, 589–599. [Google Scholar] [CrossRef]
- Chen, J.; Kanai, Y.; Cowan, N.J.; Hirokawa, N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature 1992, 360, 674–677. [Google Scholar] [CrossRef]
- Harada, A.; Oguchi, K.; Okabe, S.; Kuno, J.; Terada, S.; Ohshima, T.; Sato-Yoshitake, R.; Takei, Y.; Noda, T.; Hirokawa, N. Altered microtubule organization in small-calibre axons of mice lacking tau protein. Nature 1994, 369, 488–491. [Google Scholar] [CrossRef] [PubMed]
- Shahpasand, K.; Uemura, I.; Saito, T.; Asano, T.; Hata, K.; Shibata, K.; Toyoshima, Y.; Hasegawa, M.; Hisanaga, S.-I. Regulation of Mitochondrial Transport and Inter-Microtubule Spacing by Tau Phosphorylation at the Sites Hyperphosphorylated in Alzheimer’s Disease. J. Neurosci. 2012, 32, 2430–2441. [Google Scholar] [CrossRef] [PubMed]
- Fourniol, F.J.; Sindelar, C.V.; Amigues, B.; Clare, D.K.; Thomas, G.; Perderiset, M.; Francis, F.; Houdusse, A.; Moores, C.A. Template-free 13-protofilament microtubule–MAP assembly visualized at 8 Å resolution. J. Cell Biol. 2010, 191, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Hooikaas, P.J.; Martin, M.; Mühlethaler, T.; Kuijntjes, G.-J.; Peeters, C.A.; Katrukha, E.A.; Ferrari, L.; Stucchi, R.; Verhagen, D.G.; van Riel, W.E.; et al. MAP7 family proteins regulate kinesin-1 recruitment and activation. J. Cell Biol. 2019, 218, 1298–1318. [Google Scholar] [CrossRef]
- Subramanian, R.; Wilson-Kubalek, E.M.; Arthur, C.P.; Bick, M.J.; Campbell, E.A.; Darst, S.A.; Milligan, R.A.; Kapoor, T.M. Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein. Cell 2010, 142, 433–443. [Google Scholar] [CrossRef]
- Bieling, P.; Telley, I.A.; Surrey, T. A Minimal Midzone Protein Module Controls Formation and Length of Antiparallel Microtubule Overlaps. Cell 2010, 142, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, M.; Bui, K.H. Microtubule Inner Proteins: A Meshwork of Luminal Proteins Stabilizing the Doublet Microtubule. Bioessays 2018, 40, 1700209. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, A.A.Z.; Ichikawa, M.; Dai, D.; Kubo, S.; Black, C.S.; Peri, K.; McAlear, T.S.; Veyron, S.; Yang, S.K.; Vargas, J.; et al. The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications. Elife 2020, 9, 52760. [Google Scholar] [CrossRef]
- Ma, M.; Stoyanova, M.; Rademacher, G.; Dutcher, S.K.; Brown, A.; Zhang, R. Structure of the Decorated Ciliary Doublet Microtubule. Cell 2019, 179, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Goodson, H.V.; Jonasson, E.M. Microtubules and Microtubule-Associated Proteins. Cold Spring Harb. Perspect. Biol. 2018, 10, a022608. [Google Scholar] [CrossRef] [PubMed]
- Dumont, E.L.P.; Do, C.; Hess, H. Molecular wear of microtubules propelled by surface-adhered kinesins. Nat. Nanotechnol. 2015, 10, 166–169. [Google Scholar] [CrossRef]
- Triclin, S.; Inoue, D.; Gaillard, J.; Htet, Z.M.; DeSantis, M.E.; Portran, D.; Derivery, E.; Aumeier, C.; Schaedel, L.; John, K.; et al. Self-repair protects microtubules from destruction by molecular motors. Nat. Mater. 2021, 20, 883–891. [Google Scholar] [CrossRef]
- Janke, C. The tubulin code: Molecular components, readout mechanisms, and functions. J. Cell Biol. 2014, 206, 461–472. [Google Scholar] [CrossRef]
- Wloga, D.; Joachimiak, E.; Louka, P.; Gaertig, J. Posttranslational Modifications of Tubulin and Cilia. Cold Spring Harb. Perspect. Biol. 2016, 9, a028159. [Google Scholar] [CrossRef]
- Ludueña, R.F. A Hypothesis on the Origin and Evolution of Tubulin. Int. Rev. Cell Mol. Biol. 2013, 302, 41–185. [Google Scholar] [CrossRef]
- Breuss, M.W.; Leca, I.; Gstrein, T.; Hansen, A.H.; Keays, D.A. Tubulins and brain development—The origins of functional specification. Mol. Cell. Neurosci. 2017, 84, 58–67. [Google Scholar] [CrossRef]
- Kemphues, K.J.; Raff, E.C.; Raff, R.A.; Kaufman, T.C. Mutation in a testis-specific β-tubulin in Drosophila: Analysis of its effects on meiosis and map location of the gene. Cell 1980, 21, 445–451. [Google Scholar] [CrossRef]
- Nielsen, M.G.; Turner, F.; Hutchens, J.A.; Raff, E.C. Axoneme-specific β-tubulin specialization: A Conserved C-Terminal Motif Specifies the Central Pair. Curr. Biol. 2001, 11, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Fukushige, T.; Siddiqui, Z.; Chou, M.; Culotti, J.; Gogonea, C.; Siddiqui, S.; Hamelin, M. MEC-12, an alpha-tubulin required for touch sensitivity in C. elegans. J. Cell Sci. 1999, 112, 395–403. [Google Scholar] [CrossRef]
- Savage, C.; Hamelin, M.; Culotti, J.G.; Coulson, A.; Albertson, D.G.; Chalfie, M. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 1989, 3, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Schwer, H.D.; Lecine, P.; Tiwari, S.; E Italiano, J.; Hartwig, J.H.; A Shivdasani, R. A lineage-restricted and divergent β-tubulin isoform is essential for the biogenesis, structure and function of blood platelets. Curr. Biol. 2001, 11, 579–586. [Google Scholar] [CrossRef]
- Janke, C.; Magiera, M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 307–326. [Google Scholar] [CrossRef]
- Song, Y.; Brady, S.T. Post-translational modifications of tubulin: Pathways to functional diversity of microtubules. Trends Cell Biol. 2014, 25, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Fourest-Lieuvin, A.; Peris, L.; Gache, V.; Garcia-Saez, I.; Juillan-Binard, C.; Lantez, V.; Job, D. Microtubule Regulation in Mitosis: Tubulin Phosphorylation by the Cyclin-dependent Kinase Cdk1. Mol. Biol. Cell 2006, 17, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Ori-McKenney, K.M.; McKenney, R.J.; Huang, H.H.; Li, T.; Meltzer, S.; Jan, L.Y.; Vale, R.D.; Wiita, A.P.; Jan, Y.N. Phosphorylation of β-Tubulin by the Down Syndrome Kinase, Minibrain/DYRK1a, Regulates Microtubule Dynamics and Dendrite Morphogenesis. Neuron 2016, 90, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Ludueña, R.F.; Zimmermann, H.-P.; Little, M. Identification of the phosphorylated β-tubulin isotype in differentiated neuroblastoma cells. FEBS Lett. 1988, 230, 142–146. [Google Scholar] [CrossRef]
- Peters, J.D.; Furlong, M.T.; Asai, D.J.; Harrison, M.L.; Geahlen, R.L. Syk, Activated by Cross-linking the B-cell Antigen Receptor, Localizes to the Cytosol Where It Interacts with and Phosphorylates α-Tubulin on Tyrosine. J. Biol. Chem. 1996, 271, 4755–4762. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, T.; Kadowaki, T.; Nishida, E.; Kadooka, T.; Ogawara, H.; Fukami, Y.; Sakai, H.; Takaku, F.; Kasuga, M. Substrate specificities of tyrosine-specific protein kinases toward cytoskeletal proteins in vitro. J. Biol. Chem. 1986, 261, 14797–14803. [Google Scholar] [CrossRef]
- Matten, W.T.; Aubry, M.; West, J.; Maness, P.F. Tubulin is phosphorylated at tyrosine by pp60c-src in nerve growth cone membranes. J. Cell Biol. 1990, 111, 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Powell, R.T.; Tripathi, D.N.; Dere, R.; Ho, T.H.; Blasius, T.L.; Chiang, Y.-C.; Davis, I.J.; Fahey, C.C.; Hacker, K.E.; et al. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell 2016, 166, 950–962. [Google Scholar] [CrossRef]
- Chin, H.G.; Esteve, P.-O.; Ruse, C.; Lee, J.; Schaus, S.E.; Pradhan, S.; Hansen, U. The microtubule-associated histone methyltransferase SET8, facilitated by transcription factor LSF, methylates α-tubulin. J. Biol. Chem. 2020, 295, 4748–4759. [Google Scholar] [CrossRef] [PubMed]
- Ozols, J.; Caron, J.M. Posttranslational modification of tubulin by palmitoylation: II. Identification of sites of palmitoylation. Mol. Biol. Cell 1997, 8, 637–645. [Google Scholar] [CrossRef]
- Song, Y.; Kirkpatrick, L.L.; Schilling, A.B.; Helseth, D.L.; Chabot, N.; Keillor, J.W.; Johnson, G.V.; Brady, S.T. Transglutaminase and Polyamination of Tubulin: Posttranslational Modification for Stabilizing Axonal Microtubules. Neuron 2013, 78, 109–123. [Google Scholar] [CrossRef]
- Hallak, M.E.; Rodriguez, J.; Barra, H.; Caputto, R. Release of tyrosine from tyrosinated tubulin. Some common factors that affect this process and the assembly of tubulin. FEBS Lett. 1977, 73, 147–150. [Google Scholar] [CrossRef]
- Barra, H.S.; Arce, C.A.; Rodriguez, J.A.; Caputto, R. Incorporation of phenylalanine as a single unit into rat brain protein: Reciprocal inhibition by phenylalanine and tyrosine of their respective incorporations. J. Neurochem. 1973, 21, 1241–1251. [Google Scholar] [CrossRef]
- Arce, C.A.; Rodriguez, J.A.; Barra, H.S.; Caputto, R. Incorporation of l-Tyrosine, l-Phenylalanine and l-3,4-Dihydroxyphenylalanine as Single Units into Rat Brain Tubulin. JBIC J. Biol. Inorg. Chem. 1975, 59, 145–149. [Google Scholar] [CrossRef]
- Redeker, V.; Levilliers, N.; Schmitter, J.-M.; Le Caer, J.-P.; Rossier, J.; Adoutte, A.; Bré, M.-H. Polyglycylation of Tubulin: A Posttranslational Modification in Axonemal Microtubules. Science 1994, 266, 1688–1691. [Google Scholar] [CrossRef]
- Bre, M.; Redeker, V.; Quibell, M.; Darmanaden-Delorme, J.; Bressac, C.; Cosson, J.; Huitorel, P.; Schmitter, J.; Rossler, J.; Johnson, T.; et al. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: Widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J. Cell Sci. 1996, 109, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Eddé, B.; Rossier, J.; Le Caer, J.-P.; Desbruyères, E.; Gros, F.; Denoulet, P. Posttranslational Glutamylation of α-tubulin. Science 1990, 247, 83–85. [Google Scholar] [CrossRef] [PubMed]
- E Alexander, J.; Hunt, D.F.; Lee, M.K.; Shabanowitz, J.; Michel, H.; Berlin, S.C.; MacDonald, T.L.; Sundberg, R.J.; I Rebhun, L.; Frankfurter, A. Characterization of posttranslational modifications in neuron-specific class III beta-tubulin by mass spectrometry. Proc. Natl. Acad. Sci. USA 1991, 88, 4685–4689. [Google Scholar] [CrossRef]
- Rüdiger, M.; Plessman, U.; Klöppel, K.-D.; Wehland, J.; Weber, K. Class II tubulin, the major brain β tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS Lett. 1992, 308, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhao, J.; Feng, J. Parkin Binds to α/β Tubulin and Increases their Ubiquitination and Degradation. J. Neurosci. 2003, 23, 3316–3324. [Google Scholar] [CrossRef]
- Wang, Q.; Peng, Z.; Long, H.; Deng, X.; Huang, K. Poly-ubiquitylation of α-tubulin at K304 is required for flagellar disassembly in Chlamydomonas. J. Cell Sci. 2019, 132, jcs.229047. [Google Scholar] [CrossRef]
- Rosas-Acosta, G.; Russell, W.K.; Deyrieux, A.; Russell, D.H.; Wilson, V.G. A Universal Strategy for Proteomic Studies of SUMO and Other Ubiquitin-like Modifiers. Mol. Cell. Proteom. 2005, 4, 56–72. [Google Scholar] [CrossRef]
- Paturle, L.; Wehland, J.; Margolis, R.L.; Job, D. Complete separation of tyrosinated, detyrosinated, and nontyrosinatable brain tubulin subpopulations using affinity chromatography. Biochemistry 1989, 28, 2698–2704. [Google Scholar] [CrossRef]
- Paturle-Lafanechere, L.; Edde, B.; Denoulet, P.; Van Dorsselaer, A.; Mazarguil, H.; Le Caer, J.P.; Wehland, J.; Job, D. Characterization of a major brain tubulin variant which cannot be tyrosinated. Biochemistry 1991, 30, 10523–10528. [Google Scholar] [CrossRef]
- Aillaud, C.; Bosc, C.; Saoudi, Y.; Denarier, E.; Peris, L.; Sago, L.; Taulet, N.; Cieren, A.; Tort, O.; Magiera, M.M.; et al. Evidence for new C-terminally truncated variants of α- and β-tubulins. Mol. Biol. Cell 2016, 27, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.K.; Gupta, R.; Baldus, L.; Lyon, D.; Narita, T.; Lammers, M.; Choudhary, C.; Weinert, B.T. Analysis of human acetylation stoichiometry defines mechanistic constraints on protein regulation. Nat. Commun. 2019, 10, 1055. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Xiong, Y.; Li, S.; Ren, Y.; He, Q.; Gao, S.; Zhou, J.; Shui, W. New HDAC6-mediated deacetylation sites of tubulin in the mouse brain identified by quantitative mass spectrometry. Sci. Rep. 2015, 5, 16869. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Xiong, Y.; Ren, Y.; Zhang, L.; He, X.; Wang, X.; Liu, M.; Li, D.; Shui, W.; Zhou, J. Proteomic Profiling and Functional Characterization of Multiple Post-Translational Modifications of Tubulin. J. Proteome Res. 2015, 14, 3292–3304. [Google Scholar] [CrossRef]
- Lundby, A.; Lage, K.; Weinert, B.T.; Bekker-Jensen, D.B.; Secher, A.; Skovgaard, T.; Kelstrup, C.D.; Dmytriyev, A.; Choudhary, C.; Lundby, C.; et al. Proteomic Analysis of Lysine Acetylation Sites in Rat Tissues Reveals Organ Specificity and Subcellular Patterns. Cell Rep. 2012, 2, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Weinert, B.T.; Wagner, S.A.; Horn, H.; Henriksen, P.; Liu, W.R.; Olsen, J.V.; Jensen, L.J.; Choudhary, C. Proteome-Wide Mapping of the Drosophila Acetylome Demonstrates a High Degree of Conservation of Lysine Acetylation. Sci. Signal. 2011, 4, ra48. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef]
- Piroli, G.G.; Manuel, A.M.; Walla, M.D.; Jepson, M.J.; Brock, J.W.C.; Rajesh, M.P.; Tanis, R.M.; Cotham, W.E.; Frizzell, N. Identification of protein succination as a novel modification of tubulin. Biochem. J. 2014, 462, 231–245. [Google Scholar] [CrossRef]
- Ji, S.; Kang, J.G.; Park, S.Y.; Lee, J.; Oh, Y.J.; Cho, J.W. O-GlcNAcylation of tubulin inhibits its polymerization. Amino Acids 2010, 40, 809–818. [Google Scholar] [CrossRef]
- Verhey, K.J.; Gaertig, J. The Tubulin Code. Cell Cycle 2007, 6, 2152–2160. [Google Scholar] [CrossRef]
- Bär, J.; Popp, Y.; Bucher, M.; Mikhaylova, M. Direct and indirect effects of tubulin post-translational modifications on microtubule stability: Insights and regulations. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2022, 15, 119241. [Google Scholar] [CrossRef]
- Kumar, N.; Flavin, M. Preferential action of a brain detyrosinolating carboxypeptidase on polymerized tubulin. J. Biol. Chem. 1981, 256, 7678–7686. [Google Scholar] [CrossRef]
- Shida, T.; Cueva, J.G.; Xu, Z.; Goodman, M.B.; Nachury, M.V. The major α-tubulin K40 acetyltransferase αTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. USA 2010, 107, 21517–21522. [Google Scholar] [CrossRef] [PubMed]
- Regnard, C.; Audebert, S.; Desbruyères, É.; Denoulet, P.; Eddé, B. Tubulin Polyglutamylase: Partial Purification and Enzymatic Properties. Biochemistry 1998, 37, 8395–8404. [Google Scholar] [CrossRef] [PubMed]
- Audebert, S.; Desbruyères, E. Reversible Polyglutamylation of Alpha- and Beta-Tubulin and Microtubule Dynamics in Mouse Brain Neurons. Mol. Biol. Cell 1993, 4, 615–626. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, N.; Caron, C.; Matthias, G.; Hess, D.; Khochbin, S.; Matthias, P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003, 22, 1168–1179. [Google Scholar] [CrossRef]
- Szyk, A.; Deaconescu, A.M.; Piszczek, G.; Roll-Mecak, A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 2011, 18, 1250–1258. [Google Scholar] [CrossRef] [PubMed]
- Prota, A.E.; Magiera, M.M.; Kuijpers, M.; Bargsten, K.; Frey, D.; Wieser, M.; Jaussi, R.; Hoogenraad, C.C.; Kammerer, R.A.; Janke, C.; et al. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J. Cell Biol. 2013, 200, 259–270. [Google Scholar] [CrossRef]
- He, K.; Ling, K.; Hu, J. The emerging role of tubulin posttranslational modifications in cilia and ciliopathies. Biophys. Rep. 2020, 6, 1–16. [Google Scholar] [CrossRef]
- Gadadhar, S.; Dadi, H.; Bodakuntla, S.; Schnitzler, A.; Bièche, I.; Rusconi, F.; Janke, C. Tubulin glycylation controls primary cilia length. J. Cell Biol. 2017, 216, 2701–2713. [Google Scholar] [CrossRef]
- Rogowski, K.; Juge, F.; van Dijk, J.; Wloga, D.; Strub, J.-M.; Levilliers, N.; Thomas, D.; Bré, M.-H.; Van Dorsselaer, A.; Gaertig, J.; et al. Evolutionary Divergence of Enzymatic Mechanisms for Posttranslational Polyglycylation. Cell 2009, 137, 1076–1087. [Google Scholar] [CrossRef]
- Wloga, D.; Webster, D.M.; Rogowski, K.; Bré, M.-H.; Levilliers, N.; Jerka-Dziadosz, M.; Janke, C.; Dougan, S.T.; Gaertig, J. TTLL3 Is a Tubulin Glycine Ligase that Regulates the Assembly of Cilia. Dev. Cell 2009, 16, 867–876. [Google Scholar] [CrossRef]
- Koenning, M.; Wang, X.; Karki, M.; Jangid, R.K.; Kearns, S.; Tripathi, D.N.; Cianfrocco, M.; Verhey, K.J.; Jung, S.Y.; Coarfa, C.; et al. Neuronal SETD2 activity links microtubule methylation to an anxiety-like phenotype in mice. Brain 2021, 144, 2527–2540. [Google Scholar] [CrossRef]
- Xie, X.; Wang, S.; Li, M.; Diao, L.; Pan, X.; Chen, J.; Zou, W.; Zhang, X.; Feng, W.; Bao, L. α-TubK40me3 is required for neuronal polarization and migration by promoting microtubule formation. Nat. Commun. 2021, 12, 4113. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Schaedel, L.; Portran, D.; Aguilar, A.; Gaillard, J.; Marinkovich, M.P.; Théry, M.; Nachury, M.V. Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 2017, 356, 328–332. [Google Scholar] [CrossRef]
- Martínez-Hernández, J.; Parato, J.; Sharma, A.; Soleilhac, J.-M.; Qu, X.; Tein, E.; Sproul, A.; Andrieux, A.; Goldberg, Y.; Moutin, M.-J.; et al. Crosstalk between acetylation and the tyrosination/detyrosination cycle of α-tubulin in Alzheimer’s disease. Front. Cell Dev. Biol. 2022, 10, 926914. [Google Scholar] [CrossRef] [PubMed]
- Rogowski, K.; van Dijk, J.; Magiera, M.M.; Bosc, C.; Deloulme, J.-C.; Bosson, A.; Peris, L.; Gold, N.D.; Lacroix, B.; Grau, M.B.; et al. A Family of Protein-Deglutamylating Enzymes Associated with Neurodegeneration. Cell 2010, 143, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Magiera, M.M.; Singh, P.; Gadadhar, S.; Janke, C. Tubulin Posttranslational Modifications and Emerging Links to Human Disease. Cell 2018, 173, 1323–1327. [Google Scholar] [CrossRef]
- Yu, I.; Garnham, C.P.; Roll-Mecak, A. Writing and Reading the Tubulin Code. J. Biol. Chem. 2015, 290, 17163–17172. [Google Scholar] [CrossRef]
- Kubo, T.; Yanagisawa, H.-A.; Yagi, T.; Hirono, M.; Kamiya, R. Tubulin Polyglutamylation Regulates Axonemal Motility by Modulating Activities of Inner-Arm Dyneins. Curr. Biol. 2010, 20, 441–445. [Google Scholar] [CrossRef]
- Suryavanshi, S.; Eddé, B.; Fox, L.A.; Guerrero, S.; Hard, R.; Hennessey, T.; Kabi, A.; Malison, D.; Pennock, D.; Sale, W.S.; et al. Tubulin Glutamylation Regulates Ciliary Motility by Altering Inner Dynein Arm Activity. Curr. Biol. 2010, 20, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K. The axonemal microtubules of the Chlamydomonas flagellum differ in tubulin isoform content. J. Cell Sci. 1998, 111, 313–320. [Google Scholar] [CrossRef]
- Akella, J.S.; Wloga, D.; Kim, J.; Starostina, N.G.; Lyons-Abbott, S.; Morrissette, N.S.; Dougan, S.T.; Kipreos, E.T.; Gaertig, J. MEC-17 is an α-tubulin acetyltransferase. Nature 2010, 467, 218–222. [Google Scholar] [CrossRef]
- Kim, G.-W.; Li, L.; Gorbani, M.; You, L.; Yang, X.-J. Mice Lacking α-Tubulin Acetyltransferase 1 Are Viable but Display α-Tubulin Acetylation Deficiency and Dentate Gyrus Distortion. J. Biol. Chem. 2013, 288, 20334–20350. [Google Scholar] [CrossRef] [PubMed]
- Kalebic, N.; Sorrentino, S.; Perlas, E.; Bolasco, G.; Martinez, C.; Heppenstall, P.A. αTAT1 is the major α-tubulin acetyltransferase in mice. Nat. Commun. 2013, 4, 1962. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, A.; Becker, L.; Tedeschi, T.; Heller, S.; Iomini, C.; Nachury, M.V. α-Tubulin K40 acetylation is required for contact inhibition of proliferation and cell–substrate adhesion. Mol. Biol. Cell 2014, 25, 1854–1866. [Google Scholar] [CrossRef]
- Steczkiewicz, K.; Kinch, L.; Grishin, N.V.; Rychlewski, L.; Ginalski, K. Eukaryotic Domain of Unknown Function DUF738 Belongs to Gcn5-related N-acetyltransferase Superfamily. Cell Cycle 2006, 5, 2927–2930. [Google Scholar] [CrossRef]
- Friedmann, D.R.; Aguilar, A.; Fan, J.; Nachury, M.V.; Marmorstein, R. Structure of the α-tubulin acetyltransferase, αTAT1, and implications for tubulin-specific acetylation. Proc. Natl. Acad. Sci. USA 2012, 109, 19655–19660. [Google Scholar] [CrossRef]
- Kormendi, V.; Szyk, A.; Piszczek, G.; Roll-Mecak, A. Crystal Structures of Tubulin Acetyltransferase Reveal a Conserved Catalytic Core and the Plasticity of the Essential N Terminus. J. Biol. Chem. 2012, 287, 41569–41575. [Google Scholar] [CrossRef]
- Li, W.; Zhong, C.; Li, L.; Sun, B.; Wang, W.; Xu, S.; Zhang, T.; Wang, C.; Bao, L.; Ding, J. Molecular basis of the acetyltransferase activity of MEC-17 towards α-tubulin. Cell Res. 2012, 22, 1707–1711. [Google Scholar] [CrossRef]
- Taschner, M.; Vetter, M.; Lorentzen, E. Atomic resolution structure of human α-tubulin acetyltransferase bound to acetyl-CoA. Proc. Natl. Acad. Sci. USA 2012, 109, 19649–19654. [Google Scholar] [CrossRef]
- Vetting, M.W.; de Carvalho, L.P.S.; Yu, M.; Hegde, S.S.; Magnet, S.; Roderick, S.L.; Blanchard, J.S. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2004, 433, 212–226. [Google Scholar] [CrossRef]
- Saunders, H.A.; Johnson-Schlitz, D.M.; Jenkins, B.V.; Volkert, P.J.; Yang, S.Z.; Wildonger, J. Acetylated α-tubulin K394 regulates microtubule stability to shape the growth of axon terminals. Curr. Biol. 2022, 32, 614–630. [Google Scholar] [CrossRef]
- Chu, C.-W.; Hou, F.; Zhang, J.; Phu, L.; Loktev, A.V.; Kirkpatrick, D.; Jackson, P.K.; Zhao, Y.; Zou, H. A novel acetylation of β-tubulin by San modulates microtubule polymerization via down-regulating tubulin incorporation. Mol. Biol. Cell 2011, 22, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Davenport, A.M.; Collins, L.N.; Chiu, H.; Minor, P.J.; Sternberg, P.W.; Hoelz, A. Structural and Functional Characterization of the α-Tubulin Acetyltransferase MEC-17. J. Mol. Biol. 2014, 426, 2605–2616. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, N.; Sugisaki, S.; Tokunaga, E.; Fujitani, K.; Hayasaka, T.; Setou, M.; Inokuchi, K. N-acetyltransferase ARD1-NAT1 regulates neuronal dendritic development. Genes Cells 2008, 13, 1171–1183. [Google Scholar] [CrossRef]
- Conacci-Sorrell, M.; Ngouenet, C.; Eisenman, R.N. Myc-Nick: A Cytoplasmic Cleavage Product of Myc that Promotes α-Tubulin Acetylation and Cell Differentiation. Cell 2010, 142, 480–493. [Google Scholar] [CrossRef]
- Creppe, C.; Malinouskaya, L.; Volvert, M.-L.; Gillard, M.; Close, P.; Malaise, O.; Laguesse, S.; Cornez, I.; Rahmouni, S.; Ormenese, S.; et al. Elongator Controls the Migration and Differentiation of Cortical Neurons through Acetylation of α-Tubulin. Cell 2009, 136, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zheng, X.; McNutt, M.A.; Guang, L.; Sun, Y.; Wang, J.; Gong, Y.; Hou, L.; Zhang, B. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp. Cell Res. 2009, 315, 1653–1667. [Google Scholar] [CrossRef]
- Williams, B.C.; Garrett-Engele, C.M.; Li, Z.; Williams, E.V.; Rosenman, E.D.; Goldberg, M.L. Two Putative Acetyltransferases, San and Deco, Are Required for Establishing Sister Chromatid Cohesion in Drosophila. Curr. Biol. 2003, 13, 2025–2036. [Google Scholar] [CrossRef]
- Hou, F.; Chu, C.-W.; Kong, X.; Yokomori, K.; Zou, H. The acetyltransferase activity of San stabilizes the mitotic cohesin at the centromeres in a shugoshin-independent manner. J. Cell Biol. 2007, 177, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Gaertig, J.; A Cruz, M.; Bowen, J.; Gu, L.; Pennock, D.G.; Gorovsky, M.A. Acetylation of lysine 40 in alpha-tubulin is not essential in Tetrahymena thermophila. J. Cell Biol. 1995, 129, 1301–1310. [Google Scholar] [CrossRef]
- Varberg, J.M.; Padgett, L.R.; Arrizabalaga, G.; Sullivan, W.J. TgATAT-Mediated α-Tubulin Acetylation Is Required for Division of the Protozoan Parasite Toxoplasma gondii. Msphere 2016, 1, e00088-15. [Google Scholar] [CrossRef]
- Castro-Castro, A.; Janke, C.; Montagnac, G.; Paul-Gilloteaux, P.; Chavrier, P. ATAT1/MEC-17 acetyltransferase and HDAC6 deacetylase control a balance of acetylation of alpha-tubulin and cortactin and regulate MT1-MMP trafficking and breast tumor cell invasion. Eur. J. Cell Biol. 2012, 91, 950–960. [Google Scholar] [CrossRef] [PubMed]
- Kalebic, N.; Martinez, C.; Perlas, E.; Hublitz, P.; Bilbao-Cortes, D.; Fiedorczuk, K.; Andolfo, A.; Heppenstall, P.A. Tubulin Acetyltransferase αTAT1 Destabilizes Microtubules Independently of Its Acetylation Activity. Mol. Cell. Biol. 2013, 33, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Chalfie, M.; Au, M. Genetic Control of Differentiation of the Caenorhabditis elegans Touch Receptor Neurons. Science 1989, 243, 1027–1033. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, C.; Delohery, T.; Nasipak, B.; Foat, B.C.; Bounoutas, A.; Bussemaker, H.J.; Kim, S.K.; Chalfie, M. Identification of genes expressed in C. elegans touch receptor neurons. Nature 2002, 418, 331–335. [Google Scholar] [CrossRef]
- Topalidou, I.; Keller, C.; Kalebic, N.; Nguyen, K.C.; Somhegyi, H.; Politi, K.A.; Heppenstall, P.; Hall, D.H.; Chalfie, M. Genetically Separable Functions of the MEC-17 Tubulin Acetyltransferase Affect Microtubule Organization. Curr. Biol. 2012, 22, 1057–1065. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, D.; Song, J. Cortactin is involved in transforming growth factor-β1-induced epithelial-mesenchymal transition in AML-12 cells. Acta Biochim. Biophys. Sin. 2009, 41, 839–845. [Google Scholar] [CrossRef]
- Montagnac, G.; Meas-Yedid, V.; Irondelle, M.; Castro-Castro, A.; Franco, M.; Shida, T.; Nachury, M.V.; Benmerah, A.; Olivo-Marin, J.-C.; Chavrier, P. αTAT1 catalyses microtubule acetylation at clathrin-coated pits. Nature 2013, 502, 567–570. [Google Scholar] [CrossRef]
- Nogales, E.; Whittaker, M.; Milligan, R.A.; Downing, K.H. High-Resolution Model of the Microtubule. Cell 1999, 96, 79–88. [Google Scholar] [CrossRef]
- Soppina, V.; Herbstman, J.F.; Skiniotis, G.; Verhey, K.J. Luminal Localization of α-tubulin K40 Acetylation by Cryo-EM Analysis of Fab-Labeled Microtubules. PLoS ONE 2012, 7, e48204. [Google Scholar] [CrossRef]
- Szyk, A.; Deaconescu, A.M.; Spector, J.; Goodman, B.; Valenstein, M.L.; Ziolkowska, N.E.; Kormendi, V.; Grigorieff, N.; Roll-Mecak, A. Molecular Basis for Age-Dependent Microtubule Acetylation by Tubulin Acetyltransferase. Cell 2014, 157, 1405–1415. [Google Scholar] [CrossRef]
- Coombes, C.; Yamamoto, A.; McClellan, M.; Reid, T.A.; Plooster, M.; Luxton, G.W.G.; Alper, J.; Howard, J.; Gardner, M.K. Mechanism of microtubule lumen entry for the α-tubulin acetyltransferase enzyme αTAT1. Proc. Natl. Acad. Sci. USA 2016, 113, E7176–E7184. [Google Scholar] [CrossRef]
- Ly, N.; Elkhatib, N.; Bresteau, E.; Piétrement, O.; Khaled, M.; Magiera, M.M.; Janke, C.; Le Cam, E.; Rutenberg, A.D.; Montagnac, G. αTAT1 controls longitudinal spreading of acetylation marks from open microtubules extremities. Sci. Rep. 2016, 6, 35624. [Google Scholar] [CrossRef]
- Odde, D. Diffusion inside microtubules. Eur. Biophys. J. 1998, 27, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Akhmanova, A.; Steinmetz, M.O. Control of microtubule organization and dynamics: Two ends in the limelight. Nat. Rev. Mol. Cell Biol. 2015, 16, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Chrétien, D.; Metoz, F.; Verde, F.; Karsenti, E.; Wade, R.H. Lattice defects in microtubules: Protofilament numbers vary within individual microtubules. J. Cell Biol. 1992, 117, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Schaedel, L.; John, K.; Gaillard, J.; Nachury, M.V.; Blanchoin, L.; Théry, M. Microtubules self-repair in response to mechanical stress. Nat. Mater. 2015, 14, 1156–1163. [Google Scholar] [CrossRef]
- Howes, S.C.; Alushin, G.M.; Shida, T.; Nachury, M.V.; Nogales, E. Effects of tubulin acetylation and tubulin acetyltransferase binding on microtubule structure. Mol. Biol. Cell 2014, 25, 257–266. [Google Scholar] [CrossRef]
- Janke, C.; Montagnac, G. Causes and Consequences of Microtubule Acetylation. Curr. Biol. 2017, 27, R1287–R1292. [Google Scholar] [CrossRef] [PubMed]
- Hubbert, C.; Guardiola, A.; Shao, R.; Kawaguchi, Y.; Ito, A.; Nixon, A.; Yoshida, M.; Wang, X.-F.; Yao, T.-P. HDAC6 is a microtubule-associated deacetylase. Nature 2002, 417, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kwon, S.; Yamaguchi, T.; Cubizolles, F.; Rousseaux, S.; Kneissel, M.; Cao, C.; Li, N.; Cheng, H.-L.; Chua, K.; et al. Mice Lacking Histone Deacetylase 6 Have Hyperacetylated Tubulin but Are Viable and Develop Normally. Mol. Cell. Biol. 2008, 28, 1688–1701. [Google Scholar] [CrossRef]
- North, B.J.; Marshall, B.L.; Borra, M.T.; Denu, J.M.; Verdin, E. The Human Sir2 Ortholog, SIRT2, Is an NAD+-Dependent Tubulin Deacetylase. Mol. Cell 2003, 11, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Toro, T.B.; Watt, T.J. Critical review of non-histone human substrates of metal-dependent lysine deacetylases. FASEB J. 2020, 34, 13140. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.-L.; Yang, W.-M. Beyond Histone and Deacetylase: An Overview of Cytoplasmic Histone Deacetylases and Their Nonhistone Substrates. J. Biomed. Biotechnol. 2010, 2011, 146493. [Google Scholar] [CrossRef]
- Vaquero, A.; Scher, M.B.; Lee, D.H.; Sutton, A.; Cheng, H.-L.; Alt, F.W.; Serrano, L.; Sternglanz, R.; Reinberg, D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006, 20, 1256–1261. [Google Scholar] [CrossRef]
- North, B.J.; Verdin, E. Interphase Nucleo-Cytoplasmic Shuttling and Localization of SIRT2 during Mitosis. PLoS ONE 2007, 2, e784. [Google Scholar] [CrossRef]
- Dryden, S.C.; Nahhas, F.A.; Nowak, J.E.; Goustin, A.-S.; Tainsky, M.A. Role for Human SIRT2 NAD-Dependent Deacetylase Activity in Control of Mitotic Exit in the Cell Cycle. Mol. Cell. Biol. 2003, 23, 3173–3185. [Google Scholar] [CrossRef]
- Kim, H.-S.; Vassilopoulos, A.; Wang, R.-H.; Lahusen, T.; Xiao, Z.; Xu, X.; Li, C.; Veenstra, T.D.; Li, B.; Yu, H.; et al. SIRT2 Maintains Genome Integrity and Suppresses Tumorigenesis through Regulating APC/C Activity. Cancer Cell 2011, 20, 487–499. [Google Scholar] [CrossRef]
- Zhang, H.; Park, S.-H.; Pantazides, B.G.; Karpiuk, O.; Warren, M.D.; Hardy, C.W.; Duong, D.M.; Park, S.-J.; Kim, H.-S.; Vassilopoulos, A.; et al. SIRT2 directs the replication stress response through CDK9 deacetylation. Proc. Natl. Acad. Sci. USA 2013, 110, 13546–13551. [Google Scholar] [CrossRef] [PubMed]
- Pandithage, R.; Lilischkis, R.; Harting, K.; Wolf, A.; Jedamzik, B.; Lüscher-Firzlaff, J.; Vervoorts, J.; Lasonder, E.; Kremmer, E.; Knöll, B.; et al. The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. J. Cell Biol. 2008, 180, 915–929. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, R.M.; Vicente Miranda, H.; Francelle, L.; Pinho, R.; Szegö, É.M.; Martinho, R.; Munari, F.; Lázaro, D.F.; Moniot, S.; Guerreiro, P.; et al. The mechanism of sirtuin 2–mediated exacerbation of alpha-synuclein toxicity in models of Parkinson disease. PLoS Biol. 2017, 15, e2000374. [Google Scholar] [CrossRef]
- Quinti, L.; Casale, M.; Moniot, S.; Pais, T.F.; Van Kanegan, M.J.; Kaltenbach, L.S.; Pallos, J.; Lim, R.G.; Naidu, S.D.; Runne, H.; et al. SIRT2- and NRF2-Targeting Thiazole-Containing Compound with Therapeutic Activity in Huntington’s Disease Models. Cell Chem. Biol. 2016, 23, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Biella, G.; Fusco, F.; Nardo, E.; Bernocchi, O.; Colombo, A.; Lichtenthaler, S.F.; Forloni, G.; Albani, D. Sirtuin 2 Inhibition Improves Cognitive Performance and Acts on Amyloid-β Protein Precursor Processing in Two Alzheimer’s Disease Mouse Models. J. Alzheimer’s Dis. 2016, 53, 1193–1207. [Google Scholar] [CrossRef]
- Garske, A.L.; Smith, B.C.; Denu, J.M. Linking SIRT2 to Parkinson’s Disease. ACS Chem. Biol. 2007, 2, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Peng, L.; Seto, E.; Huang, S.; Qiu, Y. Modulation of Histone Deacetylase 6 (HDAC6) Nuclear Import and Tubulin Deacetylase Activity through Acetylation. J. Biol. Chem. 2012, 287, 29168–29174. [Google Scholar] [CrossRef]
- Li, Y.; Shin, D.; Kwon, S.H. Histone deacetylase 6 plays a role as a distinct regulator of diverse cellular processes. FEBS J. 2012, 280, 775–793. [Google Scholar] [CrossRef]
- Fusco, C.; Micale, L.; Augello, B.; Mandriani, B.; Pellico, M.T.; De Nittis, P.; Calcagnì, A.; Monti, M.; Cozzolino, F.; Pucci, P.; et al. HDAC6 mediates the acetylation of TRIM50. Cell. Signal. 2014, 26, 363–369. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Kovacs, J.J.; McLaurin, A.; Vance, J.M.; Ito, A.; Yao, T.-P. The Deacetylase HDAC6 Regulates Aggresome Formation and Cell Viability in Response to Misfolded Protein Stress. Cell 2003, 115, 727–738. [Google Scholar] [CrossRef]
- Boyault, C.; Sadoul, K.; Pabion, M.; Khochbin, S. HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene 2007, 26, 5468–5476. [Google Scholar] [CrossRef]
- Chen, S.; Owens, G.C.; Makarenkova, H.; Edelman, D.B. HDAC6 Regulates Mitochondrial Transport in Hippocampal Neurons. PLoS ONE 2010, 5, e10848. [Google Scholar] [CrossRef]
- Simões-Pires, C.; Zwick, V.; Nurisso, A.; Schenker, E.; Carrupt, P.-A.; Cuendet, M. HDAC6 as a target for neurodegenerative diseases: What makes it different from the other HDACs? Mol. Neurodegener. 2013, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Svoboda, M.; Zhang, G.; Cavasin, M.A.; Motlová, L.; McKinsey, T.A.; Eubanks, J.H.; Bařinka, C.; Kozikowski, A.P. Structural and in Vivo Characterization of Tubastatin A, a Widely Used Histone Deacetylase 6 Inhibitor. ACS Med. Chem. Lett. 2020, 11, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, C.; Hassan, S.; Liu, X.; Song, F.; Chen, K.; Zhang, W.; Yang, J. Histone deacetylase 6 in cancer. J. Hematol. Oncol. 2018, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Seigneurin-Berny, D.; Verdel, A.; Curtet, S.; Lemercier, C.; Garin, J.; Rousseaux, S.; Khochbin, S. Identification of Components of the Murine Histone Deacetylase 6 Complex: Link between Acetylation and Ubiquitination Signaling Pathways. Mol. Cell. Biol. 2001, 21, 8035–8044. [Google Scholar] [CrossRef]
- Boyault, C.; Zhang, Y.; Fritah, S.; Caron, C.; Gilquin, B.; Kwon, S.H.; Garrido, C.; Yao, T.-P.; Vourc’H, C.; Matthias, P.; et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007, 21, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Piperno, G.; Ledizet, M.; Chang, X.J. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J. Cell Biol. 1987, 104, 289–302. [Google Scholar] [CrossRef]
- Matsuyama, A.; Shimazu, T.; Sumida, Y.; Saito, A.; Yoshimatsu, Y.; Seigneurin-Berny, D.; Osada, H.; Komatsu, Y.; Nishino, N.; Khochbin, S.; et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002, 21, 6820–6831. [Google Scholar] [CrossRef]
- Zhao, Z.; Xu, H.; Gong, W. Histone Deacetylase 6 (HDAC6) Is an Independent Deacetylase for α-Tubulin. Protein Pept. Lett. 2010, 17, 555–558. [Google Scholar] [CrossRef]
- Miyake, Y.; Keusch, J.J.; Wang, L.; Saito, M.; Hess, D.; Wang, X.; Melancon, B.J.; Helquist, P.; Gut, H.; Matthias, P. Structural insights into HDAC6 tubulin deacetylation and its selective inhibition. Nat. Chem. Biol. 2016, 12, 748–754. [Google Scholar] [CrossRef]
- Zilberman, Y.; Ballestrem, C.; Carramusa, L.; Mazitschek, R.; Khochbin, S.; Bershadsky, A. Regulation of microtubule dynamics by inhibition of the tubulin deacetylase HDAC6. J. Cell Sci. 2009, 122, 3531–3541. [Google Scholar] [CrossRef]
- Bobrowska, A.; Donmez, G.; Weiss, A.; Guarente, L.P.; Bates, G.P. SIRT2 Ablation Has No Effect on Tubulin Acetylation in Brain, Cholesterol Biosynthesis or the Progression of Huntington’s Disease Phenotypes In Vivo. PLoS ONE 2012, 7, e34805. [Google Scholar] [CrossRef]
- Skoge, R.H.; Ziegler, M. SIRT2 inactivation reveals a subset of hyperacetylated perinuclear microtubules inaccessible to HDAC6. J. Cell Sci. 2016, 129, 2972–2982. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.; Marmo, T.P.; Salam, A.A.; Che, S.; Finkelstein, E.; Kabarriti, R.; Xenias, H.S.; Mazitschek, R.; Hubbert, C.; Kawaguchi, Y.; et al. HDAC6 deacetylation of tubulin modulates dynamics of cellular adhesions. J. Cell Sci. 2007, 120, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Cabrero, J.R.; Serrador, J.M.; Barreiro, O.; Mittelbrunn, M.; Naranjo-Suárez, S.; Martin-Cofreces, N.; Vicente-Manzanares, M.; Mazitschek, R.; Bradner, J.E.; Ávila, J.; et al. Lymphocyte Chemotaxis Is Regulated by Histone Deacetylase 6, Independently of Its Deacetylase Activity. Mol. Biol. Cell 2006, 17, 3435–3445. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, Z.; Zhang, Y.; Yong, S.; Salas-Burgos, A.; Koomen, J.; Olashaw, N.; Parsons, J.T.; Yang, X.-J.; Dent, S.R.; et al. HDAC6 Modulates Cell Motility by Altering the Acetylation Level of Cortactin. Mol. Cell 2007, 27, 197–213. [Google Scholar] [CrossRef]
- Pugacheva, E.N.; Jablonski, S.A.; Hartman, T.R.; Henske, E.P.; Golemis, E.A. HEF1-Dependent Aurora A Activation Induces Disassembly of the Primary Cilium. Cell 2007, 129, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ran, J.; Liu, M.; Li, D.; Li, Y.; Shi, X.; Meng, D.; Pan, J.; Ou, G.; Aneja, R.; et al. CYLD mediates ciliogenesis in multiple organs by deubiquitinating Cep70 and inactivating HDAC6. Cell Res. 2014, 24, 1342–1353. [Google Scholar] [CrossRef]
- Zhou, X.; Fan, L.X.; Li, K.; Ramchandran, R.; Calvet, J.P.; Li, X. SIRT2 regulates ciliogenesis and contributes to abnormal centrosome amplification caused by loss of polycystin-1. Hum. Mol. Genet. 2013, 23, 1644–1655. [Google Scholar] [CrossRef]
- Bangs, F.K.; Schrode, N.; Hadjantonakis, A.-K.; Anderson, K.V. Lineage specificity of primary cilia in the mouse embryo. Nat. Cell Biol. 2015, 17, 113–122. [Google Scholar] [CrossRef]
- Ran, J.; Yang, Y.; Li, D.; Liu, M.; Zhou, J. Deacetylation of α-tubulin and cortactin is required for HDAC6 to trigger ciliary disassembly. Sci. Rep. 2015, 5, 12917. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Meng, D.; Zhu, B.; Pan, J. Mechanism of ciliary disassembly. Cell. Mol. Life Sci. 2016, 73, 1787–1802. [Google Scholar] [CrossRef] [PubMed]
- Mackeh, R.; Lorin, S.; Ratier, A.; Mejdoubi-Charef, N.; Baillet, A.; Bruneel, A.; Hamaï, A.; Codogno, P.; Poüs, C.; Perdiz, D. Reactive Oxygen Species, AMP-activated Protein Kinase, and the Transcription Cofactor p300 Regulate α-Tubulin Acetyltransferase-1 (αTAT-1/MEC-17)-dependent Microtubule Hyperacetylation during Cell Stress. J. Biol. Chem. 2014, 289, 11816–11828. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Jeong, H.M.; Jin, Y.-H.; Kim, Y.-J.; Jeong, H.G.; Yeo, C.-Y.; Lee, K.-Y. Acetylation of histone deacetylase 6 by p300 attenuates its deacetylase activity. Biochem. Biophys. Res. Commun. 2009, 383, 88–92. [Google Scholar] [CrossRef]
- Even, A.; Morelli, G.; Turchetto, S.; Shilian, M.; Le Bail, R.; Laguesse, S.; Krusy, N.; Brisker, A.; Brandis, A.; Inbar, S.; et al. ATP-citrate lyase promotes axonal transport across species. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Cantó, C.; Wanders, R.J.; Auwerx, J. The Secret Life of NAD+: An Old Metabolite Controlling New Metabolic Signaling Pathways. Endocr. Rev. 2010, 31, 194–223. [Google Scholar] [CrossRef]
- Cantó, C.; Menzies, K.J.; Auwerx, J. NAD+ Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef]
- Covarrubias, A.J.; Perrone, R.; Grozio, A.; Verdin, E. NAD+ metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2020, 22, 119–141. [Google Scholar] [CrossRef]
- Shi, L.; Tu, B.P. Acetyl-CoA and the regulation of metabolism: Mechanisms and consequences. Curr. Opin. Cell Biol. 2015, 33, 125–131. [Google Scholar] [CrossRef]
- Piperno, G.; Fuller, M.T. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 1985, 101, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- LeDizet, M.; Piperno, G. Identification of an acetylation site of Chlamydomonas alpha-tubulin. Proc. Natl. Acad. Sci. USA 1987, 84, 5720–5724. [Google Scholar] [CrossRef] [PubMed]
- Kull, F.J.; Sloboda, R.D. A Slow Dance for Microtubule Acetylation. Cell 2014, 157, 1255–1256. [Google Scholar] [CrossRef] [PubMed]
- Maruta, H.; Greer, K.; Rosenbaum, J.L. The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J. Cell Biol. 1986, 103, 571–579. [Google Scholar] [CrossRef]
- Palazzo, A.; Ackerman, B.; Gundersen, G.G. Tubulin acetylation and cell motility. Nature 2003, 421, 230. [Google Scholar] [CrossRef]
- Webster, D.; Borisy, G. Microtubules are acetylated in domains that turn over slowly. J. Cell Sci. 1989, 92, 57–65. [Google Scholar] [CrossRef]
- Haggarty, S.J.; Koeller, K.M.; Wong, J.C.; Grozinger, C.M.; Schreiber, S.L. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. USA 2003, 100, 4389–4394. [Google Scholar] [CrossRef]
- Cueva, J.G.; Hsin, J.; Huang, K.C.; Goodman, M.B. Posttranslational Acetylation of α-Tubulin Constrains Protofilament Number in Native Microtubules. Curr. Biol. 2012, 22, 1066–1074. [Google Scholar] [CrossRef]
- Chrétien, D.; Flyvbjerg, H.; Fuller, S.D. Limited flexibility of the inter-protofilament bonds in microtubules assembled from pure tubulin. Eur. Biophys. J. 1998, 27, 490–500. [Google Scholar] [CrossRef]
- Wang, C.; Guo, Z.; Wang, R.; Luo, Y. Role of the inter-protofilament sliding in the bending of protein microtubules. J. Biomech. 2016, 49, 3803–3807. [Google Scholar] [CrossRef]
- Eshun-Wilson, L.; Zhang, R.; Portran, D.; Nachury, M.V.; Toso, D.B.; Löhr, T.; Vendruscolo, M.; Bonomi, M.; Fraser, J.S.; Nogales, E. Effects of α-tubulin acetylation on microtubule structure and stability. Proc. Natl. Acad. Sci. USA 2019, 116, 10366–10371. [Google Scholar] [CrossRef]
- Portran, D.; Schaedel, L.; Xu, Z.; Théry, M.; Nachury, M.V. Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nature 2017, 19, 391–398. [Google Scholar] [CrossRef]
- Neumann, B.; Hilliard, M.A. Loss of MEC-17 Leads to Microtubule Instability and Axonal Degeneration. Cell Rep. 2014, 6, 93–103. [Google Scholar] [CrossRef]
- Moutin, M.; Bosc, C.; Peris, L.; Andrieux, A. Tubulin post-translational modifications control neuronal development and functions. Dev. Neurobiol. 2020, 81, 253–272. [Google Scholar] [CrossRef]
- Mao, C.-X.; Xiong, Y.; Xiong, Z.; Wang, Q.; Zhang, Y.Q.; Jin, S. Microtubule-severing protein Katanin regulates neuromuscular junction development and dendritic elaboration in Drosophila. Development 2014, 141, 1064–1074. [Google Scholar] [CrossRef] [PubMed]
- Sudo, H.; Baas, P.W. Acetylation of Microtubules Influences Their Sensitivity to Severing by Katanin in Neurons and Fibroblasts. J. Neurosci. 2010, 30, 7215–7226. [Google Scholar] [CrossRef] [PubMed]
- Leo, L.; Yu, W.; D’Rozario, M.; Waddell, E.A.; Marenda, D.R.; Baird, M.A.; Davidson, M.W.; Zhou, B.; Wu, B.; Baker, L.; et al. Vertebrate Fidgetin Restrains Axonal Growth by Severing Labile Domains of Microtubules. Cell Rep. 2015, 12, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Nekooki-Machida, Y.; Nakakura, T.; Nishijima, Y.; Tanaka, H.; Arisawa, K.; Kiuchi, Y.; Miyashita, T.; Hagiwara, H. Dynamic localization of α-tubulin acetyltransferase ATAT1 through the cell cycle in human fibroblastic KD cells. Med. Mol. Morphol. 2018, 51, 217–226. [Google Scholar] [CrossRef]
- L’Hernault, S.W.; Rosenbaum, J.L. Chlamydomonas. alpha.-tubulin is posttranslationally modified by acetylation on the.epsilon.-amino group of a lysine. Biochemistry 1985, 24, 473–478. [Google Scholar] [CrossRef]
- Nakakura, T.; Suzuki, T.; Nemoto, T.; Tanaka, H.; Asano-Hoshino, A.; Arisawa, K.; Nishijima, Y.; Kiuchi, Y.; Hagiwara, H. Intracellular localization of α-tubulin acetyltransferase ATAT1 in rat ciliated cells. Med. Mol. Morphol. 2015, 49, 133–143. [Google Scholar] [CrossRef]
- Nakakura, T.; Asano-Hoshino, A.; Suzuki, T.; Arisawa, K.; Tanaka, H.; Sekino, Y.; Kiuchi, Y.; Kawai, K.; Hagiwara, H. The elongation of primary cilia via the acetylation of α-tubulin by the treatment with lithium chloride in human fibroblast KD cells. Med. Mol. Morphol. 2014, 48, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Keryer, G.; Pineda, J.R.; Liot, G.; Kim, J.; Dietrich, P.; Benstaali, C.; Smith, K.; Cordelières, F.; Spassky, N.; Ferrante, R.J.; et al. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J. Clin. Investig. 2011, 121, 4372–4382. [Google Scholar] [CrossRef]
- Loktev, A.V.; Zhang, Q.; Beck, J.S.; Searby, C.C.; Scheetz, T.E.; Bazan, J.F.; Slusarski, D.C.; Sheffield, V.C.; Jackson, P.K.; Nachury, M.V. A BBSome Subunit Links Ciliogenesis, Microtubule Stability, and Acetylation. Dev. Cell 2008, 15, 854–865. [Google Scholar] [CrossRef]
- Kozminski, K.G.; Diener, D.R.; Rosenbaum, J.L. High level expression of nonacetylatable ?-tubulin inChlamydomonas reinhardtii. Cell Motil. Cytoskelet. 1993, 25, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Alper, J.D.; Decker, F.; Agana, B.; Howard, J. The Motility of Axonemal Dynein Is Regulated by the Tubulin Code. Biophys. J. 2014, 107, 2872–2880. [Google Scholar] [CrossRef]
- Reed, N.A.; Cai, D.; Blasius, T.L.; Jih, G.T.; Meyhofer, E.; Gaertig, J.; Verhey, K.J. Microtubule Acetylation Promotes Kinesin-1 Binding and Transport. Curr. Biol. 2006, 16, 2166–2172. [Google Scholar] [CrossRef]
- Kaul, N.; Soppina, V.; Verhey, K.J. Effects of α-Tubulin K40 Acetylation and Detyrosination on Kinesin-1 Motility in a Purified System. Biophys. J. 2014, 106, 2636–2643. [Google Scholar] [CrossRef] [PubMed]
- Walter, W.J.; Beránek, V.; Fischermeier, E.; Diez, S. Tubulin Acetylation Alone Does Not Affect Kinesin-1 Velocity and Run Length In Vitro. PLoS ONE 2012, 7, e42218. [Google Scholar] [CrossRef]
- Sullenberger, C.; Vasquez-Limeta, A.; Kong, D.; Loncarek, J. With Age Comes Maturity: Biochemical and Structural Transformation of a Human Centriole in the Making. Cells 2020, 9, 1429. [Google Scholar] [CrossRef]
- Guichard, P.; Laporte, M.H.; Hamel, V. The centriolar tubulin code. Semin. Cell Dev. Biol. 2023, 137, 16–25. [Google Scholar] [CrossRef]
- Sahabandu, N.; Kong, D.; Magidson, V.; Nanjundappa, R.; Sullenberger, C.; Mahjoub, M.; Loncarek, J. Expansion microscopy for the analysis of centrioles and cilia. J. Microsc. 2019, 276, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Nekooki-Machida, Y.; Hagiwara, H. Role of tubulin acetylation in cellular functions and diseases. Med. Mol. Morphol. 2020, 53, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Pienkowski, T.P.; Baas, P.W. Regional differences in microtubule dynamics in the axon. J. Neurosci. 1993, 13, 856–866. [Google Scholar] [CrossRef] [PubMed]
- A Cambray-Deakin, M.; Burgoyne, R.D. Posttranslational modifications of alpha-tubulin: Acetylated and detyrosinated forms in axons of rat cerebellum. J. Cell Biol. 1987, 104, 1569–1574. [Google Scholar] [CrossRef]
- Brown, A.; Li, Y.; Slaughter, T.; Black, M. Composite microtubules of the axon: Quantitative analysis of tyrosinated and acetylated tubulin along individual axonal microtubules. J. Cell Sci. 1993, 104, 339–352. [Google Scholar] [CrossRef]
- Morley, S.J.; Qi, Y.; Iovino, L.; Andolfi, L.; Guo, D.; Kalebic, N.; Castaldi, L.; Tischer, C.; Portulano, C.; Bolasco, G.; et al. Acetylated tubulin is essential for touch sensation in mice. Elife 2016, 5, e20813. [Google Scholar] [CrossRef]
- Bounoutas, A.; O’Hagan, R.; Chalfie, M. The Multipurpose 15-Protofilament Microtubules in C. elegans Have Specific Roles in Mechanosensation. Curr. Biol. 2009, 19, 1362–1367. [Google Scholar] [CrossRef]
- Yan, C.; Wang, F.; Peng, Y.; Williams, C.; Jenkins, B.; Wildonger, J.; Kim, H.-J.; Perr, J.B.; Vaughan, J.C.; Kern, M.E.; et al. Microtubule Acetylation Is Required for Mechanosensation in Drosophila. Cell Rep. 2018, 25, 1051–1065. [Google Scholar] [CrossRef]
- Tas, R.P.; Chazeau, A.; Cloin, B.M.; Lambers, M.L.; Hoogenraad, C.C.; Kapitein, L.C. Differentiation between Oppositely Oriented Microtubules Controls Polarized Neuronal Transport. Neuron 2017, 96, 1264–1271. [Google Scholar] [CrossRef]
- Bhuwania, R.; Castro-Castro, A.; Linder, S. Microtubule acetylation regulates dynamics of KIF1C-powered vesicles and contact of microtubule plus ends with podosomes. Eur. J. Cell Biol. 2014, 93, 424–437. [Google Scholar] [CrossRef]
- Cai, D.; McEwen, D.P.; Martens, J.R.; Meyhofer, E.; Verhey, K.J. Single Molecule Imaging Reveals Differences in Microtubule Track Selection Between Kinesin Motors. PLOS Biol. 2009, 7, e1000216. [Google Scholar] [CrossRef]
- Dompierre, J.P.; Godin, J.D.; Charrin, B.C.; Cordelières, F.P.; King, S.J.; Humbert, S.; Saudou, F. Histone Deacetylase 6 Inhibition Compensates for the Transport Deficit in Huntington’s Disease by Increasing Tubulin Acetylation. J. Neurosci. 2007, 27, 3571–3583. [Google Scholar] [CrossRef] [PubMed]
- Godena, V.K.; Brookes-Hocking, N.; Moller, A.; Shaw, G.; Oswald, M.; Sancho, R.M.; Miller, C.C.J.; Whitworth, A.J.; De Vos, K.J. Increasing microtubule acetylation rescues axonal transport and locomotor deficits caused by LRRK2 Roc-COR domain mutations. Nat. Commun. 2014, 5, 5245. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.R.; Webster, B.M.; Mastronarde, D.N.; Verhey, K.J.; Voeltz, G.K. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 2010, 190, 363–375. [Google Scholar] [CrossRef]
- Misawa, T.; Takahama, M.; Kozaki, T.; Lee, H.; Zou, J.; Saitoh, T.; Akira, S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 2013, 14, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Even, A.; Morelli, G.; Broix, L.; Scaramuzzino, C.; Turchetto, S.; Gladwyn-Ng, I.; Le Bail, R.; Shilian, M.; Freeman, S.; Magiera, M.M.; et al. ATAT1-enriched vesicles promote microtubule acetylation via axonal transport. Sci. Adv. 2019, 5, eaax2705. [Google Scholar] [CrossRef]
- Sadoul, K.; Khochbin, S. The growing landscape of tubulin acetylation: Lysine 40 and many more. Biochem. J. 2016, 473, 1859–1868. [Google Scholar] [CrossRef]
- Lessard, D.V.; Zinder, O.J.; Hotta, T.; Verhey, K.J.; Ohi, R.; Berger, C.L. Polyglutamylation of tubulin’s C-terminal tail controls pausing and motility of kinesin-3 family member KIF1A. J. Biol. Chem. 2019, 294, 6353–6363. [Google Scholar] [CrossRef]
- Semenova, I.; Ikeda, K.; Resaul, K.; Kraikivski, P.; Aguiar, M.; Gygi, S.; Zaliapin, I.; Cowan, A.; Rodionov, V. Regulation of microtubule-based transport by MAP4. Mol. Biol. Cell 2014, 25, 3119–3132. [Google Scholar] [CrossRef]
- Tymanskyj, S.R.; Yang, B.H.; Verhey, K.J.; Ma, L. MAP7 regulates axon morphogenesis by recruiting kinesin-1 to microtubules and modulating organelle transport. Elife 2018, 7, e36374. [Google Scholar] [CrossRef]
- Monroy, B.Y.; Sawyer, D.L.; Ackermann, B.E.; Borden, M.M.; Tan, T.C.; Ori-McKenney, K.M. Competition between microtubule-associated proteins directs motor transport. Nat. Commun. 2018, 9, 1487. [Google Scholar] [CrossRef] [PubMed]
- Siahaan, V.; Krattenmacher, J.; Hyman, A.A.; Diez, S.; Hernández-Vega, A.; Lansky, Z.; Braun, M. Kinetically distinct phases of tau on microtubules regulate kinesin motors and severing enzymes. Nature 2019, 21, 1086–1092. [Google Scholar] [CrossRef]
- Takemura, R.; Okabe, S.; Umeyama, T.; Kanai, Y.; Cowan, N.; Hirokawa, N. Increased microtubule stability and alpha tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or tau. J. Cell Sci. 1992, 103, 953–964. [Google Scholar] [CrossRef]
- Lee, G.; Rook, S. Expression of tau protein in non-neuronal cells: Microtubule binding and stabilization. J. Cell Sci. 1992, 102, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Saragoni, L.; Hernandez, P.; Maccioni, R.B. Differential Association of Tau With Subsets of Microtubules Containing Posttranslationally-Modified Tubulin Variants in Neuroblastoma Cells. Neurochem. Res. 2000, 25, 59–70. [Google Scholar] [CrossRef]
- Perez, M.; Santa-Maria, I.; de Barreda, E.G.; Zhu, X.; Cuadros, R.; Cabrero, J.R.; Sanchez-Madrid, F.; Dawson, H.N.; Vitek, M.P.; Perry, G.; et al. Tau—An inhibitor of deacetylase HDAC6 function. J. Neurochem. 2009, 109, 1756–1766. [Google Scholar] [CrossRef]
- Mao, C.-X.; Wen, X.; Jin, S.; Zhang, Y.Q. Increased acetylation of microtubules rescues human tau-induced microtubule defects and neuromuscular junction abnormalities in Drosophila. Dis. Model. Mech. 2017, 10, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Esteves, A.; Palma, A.; Gomes, R.; Santos, D.; Silva, D.; Cardoso, S. Acetylation as a major determinant to microtubule-dependent autophagy: Relevance to Alzheimer’s and Parkinson disease pathology. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2018, 1865, 2008–2023. [Google Scholar] [CrossRef] [PubMed]
- Barisic, M.; e Sousa, R.S.; Tripathy, S.K.; Magiera, M.M.; Zaytsev, A.V.; Pereira, A.L.; Janke, C.; Grishchuk, E.L.; Maiato, H. Microtubule detyrosination guides chromosomes during mitosis. Science 2015, 348, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.J.; Forer, A. Acetylated ?-tubulin in spermatogenic cells of the crane flyNephrotoma suturalis: Kinetochore microtubules are selectively acetylated. Cell Motil. Cytoskelet. 1989, 14, 237–250. [Google Scholar] [CrossRef]
- Akera, T. Tubulin post-translational modifications in meiosis. Semin. Cell Dev. Biol. 2023, 137, 38–45. [Google Scholar] [CrossRef]
- Schatten, G.; Simerly, C.; Asai, D.J.; Szöke, E.; Cooke, P.; Schatten, H. Acetylated α-tubulin in microtubules during mouse fertilization and early development. Dev. Biol. 1988, 130, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K.W. Acetylation of α-tubilin in different bovine cell types: Implications for microtubule dynamics in interphase and mitosis. Cell Biol. Int. 1995, 19, 43–52. [Google Scholar] [CrossRef]
- Chu, D.T.; Klymkowsky, M. The appearance of acetylated α-tubulin during early development and cellular differentiation in Xenopus. Dev. Biol. 1989, 136, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Hiratsuka, M.; Osaki, M.; Oshimura, M. The Molecular Biology of Mammalian SIRT Proteins: SIRT2 Functions on Cell Cycle Regulation. Cell Cycle 2007, 6, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Ikeda, M.; Chiba, S.; Kanno, S.-I.; Mizuno, K. Furry promotes acetylation of microtubules in the mitotic spindle by inhibition of SIRT2 tubulin deacetylase. J. Cell Sci. 2013, 126, 4369–4380. [Google Scholar] [CrossRef]
- A Wickström, S.; Masoumi, K.C.; Khochbin, S.; Fässler, R.; Massoumi, R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2009, 29, 131–144. [Google Scholar] [CrossRef]
- Patel-Hett, S.; Richardson, J.L.; Schulze, H.; Drabek, K.; Isaac, N.A.; Hoffmeister, K.; Shivdasani, R.A.; Bulinski, J.C.; Galjart, N.; Hartwig, J.H.; et al. Visualization of microtubule growth in living platelets reveals a dynamic marginal band with multiple microtubules. Blood 2008, 111, 4605–4616. [Google Scholar] [CrossRef]
- Sadoul, K.; Wang, J.; Diagouraga, B.; Vitte, A.-L.; Buchou, T.; Rossini, T.; Polack, B.; Xi, X.; Matthias, P.; Khochbin, S. HDAC6 controls the kinetics of platelet activation. Blood 2012, 120, 4215–4218. [Google Scholar] [CrossRef]
- Deakin, N.O.; Turner, C.E. Paxillin inhibits HDAC6 to regulate microtubule acetylation, Golgi structure, and polarized migration. J. Cell Biol. 2014, 206, 395–413. [Google Scholar] [CrossRef]
- Gu, S.; Liu, Y.; Zhu, B.; Ding, K.; Yao, T.-P.; Chen, F.; Zhan, L.; Xu, P.; Ehrlich, M.; Liang, T.; et al. Loss of α-Tubulin Acetylation Is Associated with TGF-β-induced Epithelial-Mesenchymal Transition. J. Biol. Chem. 2016, 291, 5396–5405. [Google Scholar] [CrossRef] [PubMed]
- Boggs, A.E.; Vitolo, M.I.; Whipple, R.A.; Charpentier, M.S.; Goloubeva, O.G.; Ioffe, O.B.; Tuttle, K.C.; Slovic, J.; Lu, Y.; Mills, G.B.; et al. α-Tubulin Acetylation Elevated in Metastatic and Basal-like Breast Cancer Cells Promotes Microtentacle Formation, Adhesion, and Invasive Migration. Cancer Res. 2015, 75, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Bance, B.; Seetharaman, S.; Leduc, C.; Boëda, B.; Etienne-Manneville, S. Microtubule acetylation but not detyrosination promotes focal adhesion dynamics and astrocyte migration. J. Cell Sci. 2019, 132, jcs.225805. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.; Kim, S.K.; Ventrella, R.; Carruzzo, H.M.; Wortman, J.C.; Han, H.; Suva, E.E.; Mitchell, J.W.; Yu, C.C.; Mitchell, B.J. Tubulin acetylation promotes penetrative capacity of cells undergoing radial intercalation. Cell Rep. 2021, 36, 109556. [Google Scholar] [CrossRef]
- Seetharaman, S.; Vianay, B.; Roca, V.; Farrugia, A.J.; De Pascalis, C.; Boëda, B.; Dingli, F.; Loew, D.; Vassilopoulos, S.; Bershadsky, A.; et al. Microtubules tune mechanosensitive cell responses. Nat. Mater. 2021, 21, 366–377. [Google Scholar] [CrossRef]
- Yang, J.; Song, C.; Zhan, X. The role of protein acetylation in carcinogenesis and targeted drug discovery. Front. Endocrinol. 2022, 13, 2228. [Google Scholar] [CrossRef] [PubMed]
- Black, M.M.; Keyser, P. Acetylation of alpha-tubulin in cultured neurons and the induction of alpha-tubulin acetylation in PC12 cells by treatment with nerve growth factor. J. Neurosci. 1987, 7, 1833–1842. [Google Scholar] [CrossRef]
- Knossow, M.; Campanacci, V.; Khodja, L.A.; Gigant, B. The Mechanism of Tubulin Assembly into Microtubules: Insights from Structural Studies. iScience 2020, 23, 101511. [Google Scholar] [CrossRef]
- Geeraert, C.; Ratier, A.; Pfisterer, S.G.; Perdiz, D.; Cantaloube, I.; Rouault, A.; Pattingre, S.; Proikas-Cezanne, T.; Codogno, P.; Poüs, C. Starvation-induced Hyperacetylation of Tubulin Is Required for the Stimulation of Autophagy by Nutrient Deprivation. J. Biol. Chem. 2010, 285, 24184–24194. [Google Scholar] [CrossRef]
- Xie, R.; Nguyen, S.; McKeehan, W.L.; Liu, L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol. 2010, 11, 89. [Google Scholar] [CrossRef]
- Li, W.; Zhao, Y.; Chou, I.-N. Nickel (Ni2+) Enhancement of α-Tubulin Acetylation in Cultured 3T3 Cells. Toxicol. Appl. Pharmacol. 1996, 140, 461–470. [Google Scholar] [CrossRef]
- Giustiniani, J.; Daire, V.; Cantaloube, I.; Durand, G.; Poüs, C.; Perdiz, D.; Baillet, A. Tubulin acetylation favors Hsp90 recruitment to microtubules and stimulates the signaling function of the Hsp90 clients Akt/PKB and p53. Cell. Signal. 2009, 21, 529–539. [Google Scholar] [CrossRef]
- Kratzer, E.; Tian, Y.; Sarich, N.; Wu, T.; Meliton, A.; Leff, A.; Birukova, A.A. Oxidative Stress Contributes to Lung Injury and Barrier Dysfunction via Microtubule Destabilization. Am. J. Respir. Cell Mol. Biol. 2012, 47, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Oakhill, J.S.; Steel, R.; Chen, Z.-P.; Scott, J.W.; Ling, N.; Tam, S.; Kemp, B.E. AMPK Is a Direct Adenylate Charge-Regulated Protein Kinase. Science 2011, 332, 1433–1435. [Google Scholar] [CrossRef] [PubMed]
- Galdieri, L.; Gatla, H.; Vancurova, I.; Vancura, A. Activation of AMP-activated Protein Kinase by Metformin Induces Protein Acetylation in Prostate and Ovarian Cancer Cells. J. Biol. Chem. 2016, 291, 25154–25166. [Google Scholar] [CrossRef]
- Vancura, A.; Nagar, S.; Kaur, P.; Bu, P.; Bhagwat, M.; Vancurova, I. Reciprocal Regulation of AMPK/SNF1 and Protein Acetylation. Int. J. Mol. Sci. 2018, 19, 3314. [Google Scholar] [CrossRef]
- Currais, A.; Huang, L.; Goldberg, J.; Petrascheck, M.; Ates, G.; Pinto-Duarte, A.; Shokhirev, M.N.; Schubert, D.; Maher, P. Elevating acetyl-CoA levels reduces aspects of brain aging. Elife 2019, 8, e47866. [Google Scholar] [CrossRef] [PubMed]
- Tarasiuk, O.; Miceli, M.; Di Domizio, A.; Nicolini, G. AMPK and Diseases: State of the Art Regulation by AMPK-Targeting Molecules. Biology 2022, 11, 1041. [Google Scholar] [CrossRef]
- Zmijewski, J.W.; Banerjee, S.; Bae, H.; Friggeri, A.; Lazarowski, E.R.; Abraham, E. Exposure to Hydrogen Peroxide Induces Oxidation and Activation of AMP-activated Protein Kinase*. J. Biol. Chem. 2010, 285, 33154–33164. [Google Scholar] [CrossRef]
- Chen, Z.; Shen, X.; Shen, F.; Zhong, W.; Wu, H.; Liu, S.; Lai, J. TAK1 activates AMPK-dependent cell death pathway in hydrogen peroxide-treated cardiomyocytes, inhibited by heat shock protein-70. Mol. Cell. Biochem. 2013, 377, 35–44. [Google Scholar] [CrossRef]
- Hinchy, E.C.; Gruszczyk, A.V.; Willows, R.; Navaratnam, N.; Hall, A.R.; Bates, G.; Bright, T.P.; Krieg, T.; Carling, D.; Murphy, M.P. Mitochondria-derived ROS activate AMP-activated protein kinase (AMPK) indirectly. J. Biol. Chem. 2018, 293, 17208–17217. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Kaarniranta, K. AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res. Rev. 2012, 11, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.D.; Gross, E.G.; Pillai, G.S.; Seetharaman, S.; Etienne-Manneville, S.; Inoue, T. Non-catalytic allostery in α-TAT1 by a phospho-switch drives dynamic microtubule acetylation. J. Cell Biol. 2022, 221, e202202100. [Google Scholar] [CrossRef]
- Shah, N.; Kumar, S.; Zaman, N.; Pan, C.C.; Bloodworth, J.C.; Lei, W.; Streicher, J.M.; Hempel, N.; Mythreye, K.; Lee, N.Y. TAK1 activation of alpha-TAT1 and microtubule hyperacetylation control AKT signaling and cell growth. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Yaciuk, P.; Moran, E. Analysis with Specific Polyclonal Antiserum Indicates That the E1A-Associated 300-KDa Product Is a Stable Nuclear Phosphoprotein That Undergoes Cell Cycle Phase-Specific Modification. Mol. Cell. Biol. 1991, 11, 5389–5397. [Google Scholar] [PubMed]
- Yang, W.; Hong, Y.H.; Shen, X.-Q.; Frankowski, C.; Camp, H.S.; Leff, T. Regulation of Transcription by AMP-activated Protein Kinase: Phosphorylation of P300 Blocks Its Interaction with Nuclear Receptors. J. Biol. Chem. 2001, 276, 38341–38344. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiu, J.; Wang, X.; Zhang, Y.; Xia, M. AMP-Activated Protein Kinase Suppresses Endothelial Cell Inflammation Through Phosphorylation of Transcriptional Coactivator p300. Arter. Thromb. Vasc. Biol. 2011, 31, 2897–2908. [Google Scholar] [CrossRef]
- A Caporizzo, M.; Chen, C.Y.; Prosser, B.L. Cardiac microtubules in health and heart disease. Exp. Biol. Med. 2019, 244, 1255–1272. [Google Scholar] [CrossRef]
- Mackeh, R.; Perdiz, D.; Lorin, S.; Codogno, P.; Poüs, C. Autophagy and microtubules—New story, old players. J. Cell Sci. 2013, 126, 1071–1080. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Chun, Y.; Kim, J. Autophagy: An Essential Degradation Program for Cellular Homeostasis and Life. Cells 2018, 7, 278. [Google Scholar] [CrossRef]
- Hać, A.; Pierzynowska, K.; Herman-Antosiewicz, A. S6K1 Is Indispensible for Stress-Induced Microtubule Acetylation and Autophagic Flux. Cells 2021, 10, 929. [Google Scholar] [CrossRef]
- Hempen, B.; Brion, J.-P. Reduction of Acetylated α-Tubulin Immunoreactivity in Neurofibrillary Tangle-bearing Neurons in Alzheimerʼs Disease. J. Neuropathol. Exp. Neurol. 1996, 55, 964–972. [Google Scholar] [CrossRef]
- Silva, D.F.; Esteves, A.R.; Oliveira, C.R.; Cardoso, S.M. Mitochondrial Metabolism Power SIRT2-Dependent Deficient Traffic Causing Alzheimer’s-Disease Related Pathology. Mol. Neurobiol. 2017, 54, 4021–4040. [Google Scholar] [CrossRef] [PubMed]
- D’Ydewalle, C.; Krishnan, J.; Chiheb, D.M.; Van Damme, P.; Irobi, J.; Kozikowski, A.P.; Van den Berghe, P.; Timmerman, V.; Robberecht, W.; Van den Bosch, L. HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1–induced Charcot-Marie-Tooth disease. Nat. Med. 2011, 17, 968–974. [Google Scholar] [CrossRef]
- Benoy, V.; Vanden Berghe, P.V.; Jarpe, M.; Van Damme, P.; Robberecht, W.; Van Den Bosch, L. Development of Improved HDAC6 Inhibitors as Pharmacological Therapy for Axonal Charcot–Marie–Tooth Disease. Neurotherapeutics 2017, 14, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Adalbert, R.; Kaieda, A.; Antoniou, C.; Loreto, A.; Yang, X.; Gilley, J.; Hoshino, T.; Uga, K.; Makhija, M.T.; Coleman, M.P. Novel HDAC6 Inhibitors Increase Tubulin Acetylation and Rescue Axonal Transport of Mitochondria in a Model of Charcot–Marie–Tooth Type 2F. ACS Chem. Neurosci. 2019, 11, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Picci, C.; Wong, V.; Costa, C.; McKinnon, M.C.; Goldberg, D.C.; Swift, M.; Alam, N.M.; Prusky, G.T.; Shen, S.; Kozikowski, A.P.; et al. HDAC6 inhibition promotes α-tubulin acetylation and ameliorates CMT2A peripheral neuropathy in mice. Exp. Neurol. 2020, 328, 113281. [Google Scholar] [CrossRef] [PubMed]
- Taes, I.; Timmers, M.; Hersmus, N.; Bento-Abreu, A.; Van Den Bosch, L.; Van Damme, P.; Auwerx, J.; Robberecht, W. Hdac6 deletion delays disease progression in the SOD1G93A mouse model of ALS. Hum. Mol. Genet. 2013, 22, 1783–1790. [Google Scholar] [CrossRef]
- Govindarajan, N.; Rao, P.; Burkhardt, S.; Sananbenesi, F.; Schlüter, O.M.; Bradke, F.; Lu, J.; Fischer, A. Reducing HDAC6 ameliorates cognitive deficits in a mouse model for Alzheimer’s disease. EMBO Mol. Med. 2012, 5, 52–63. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, C.; Wu, J.; Tao, J.-J.; Sui, X.-L.; Yao, Z.-G.; Xu, Y.-F.; Huang, L.; Zhu, H.; Sheng, S.-L.; et al. Tubastatin A/ACY-1215 Improves Cognition in Alzheimer’s Disease Transgenic Mice. J. Alzheimer’s Dis. 2014, 41, 1193–1205. [Google Scholar] [CrossRef]
- Cappelletti, G.; Calogero, A.M.; Rolando, C. Microtubule acetylation: A reading key to neural physiology and degeneration. Neurosci. Lett. 2021, 755, 135900. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.-Y.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. α-Synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef]
- Zimprich, A. Genetics of Parkinson’s disease and essential tremor. Curr. Opin. Neurol. 2011, 24, 318–323. [Google Scholar] [CrossRef]
- Alim, M.A.; Ma, Q.-L.; Takeda, K.; Aizawa, T.; Matsubara, M.; Nakamura, M.; Asada, A.; Saito, T.; Xkaji, M.; Yoshii, M.; et al. Demonstration of a role for α-synuclein as a functional microtubule-associated protein. J. Alzheimer’s Dis. 2004, 6, 435–442. [Google Scholar] [CrossRef]
- Cartelli, D.; Aliverti, A.; Barbiroli, A.; Santambrogio, C.; Ragg, E.M.; Casagrande, F.V.; Cantele, F.; Beltramone, S.; Marangon, J.; De Gregorio, C.; et al. α-Synuclein is a Novel Microtubule Dynamase. Sci. Rep. 2016, 6, 33289. [Google Scholar] [CrossRef] [PubMed]
- Outeiro, T.F.; Kontopoulos, E.; Altmann, S.M.; Kufareva, I.; Strathearn, K.E.; Amore, A.M.; Volk, C.B.; Maxwell, M.M.; Rochet, J.-C.; McLean, P.J.; et al. Sirtuin 2 Inhibitors Rescue α-Synuclein-Mediated Toxicity in Models of Parkinson’s Disease. Science 2007, 317, 516–519. [Google Scholar] [CrossRef]
- Kluss, J.H.; Mamais, A.; Cookson, M.R. LRRK2 links genetic and sporadic Parkinson’s disease. Biochem. Soc. Trans. 2019, 47, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Simón-Sánchez, J.; Schulte, C.; Bras, J.M.; Sharma, M.; Gibbs, J.R.; Berg, D.; Paisan-Ruiz, C.; Lichtner, P.; Scholz, S.W.; Hernandez, D.G.; et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009, 41, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Snead, D.M.; Matyszewski, M.; Dickey, A.M.; Lin, Y.X.; Leschziner, A.E.; Reck-Peterson, S.L. Structural basis for Parkinson’s disease-linked LRRK2′s binding to microtubules. Nat. Struct. Mol. Biol. 2022, 29, 1196–1207. [Google Scholar] [CrossRef]
- Law, B.M.H.; Spain, V.A.; Leinster, V.H.L.; Chia, R.; Beilina, A.; Cho, H.J.; Taymans, J.-M.; Urban, M.K.; Sancho, R.M.; Ramírez, M.B.; et al. A Direct Interaction between Leucine-rich Repeat Kinase 2 and Specific β-Tubulin Isoforms Regulates Tubulin Acetylation. J. Biol. Chem. 2014, 289, 895–908. [Google Scholar] [CrossRef]
- Leschziner, A.E.; Reck-Peterson, S.L. Structural Biology of LRRK2 and its Interaction with Microtubules. Mov. Disord. 2021, 36, 2494–2504. [Google Scholar] [CrossRef] [PubMed]
- Deniston, C.K.; Salogiannis, J.; Mathea, S.; Snead, D.M.; Lahiri, I.; Matyszewski, M.; Donosa, O.; Watanabe, R.; Böhning, J.; Shiau, A.K.; et al. Structure of LRRK2 in Parkinson’s disease and model for microtubule interaction. Nature 2020, 588, 344–349. [Google Scholar] [CrossRef]
- Lloret, A.; Fuchsberger, T.; Giraldo, E.; Viña, J. Molecular mechanisms linking amyloid β toxicity and Tau hyperphosphorylation in Alzheimer׳s disease. Free. Radic. Biol. Med. 2015, 83, 186–191. [Google Scholar] [CrossRef]
- Ding, H.; Dolan, P.J.; Johnson, G.V.W. Histone deacetylase 6 interacts with the microtubule-associated protein tau. J. Neurochem. 2008, 106, 2119–2130. [Google Scholar] [CrossRef] [PubMed]
- Selenica, M.-L.; Benner, L.; Housley, S.B.; Manchec, B.; Lee, D.C.; Nash, K.R.; Kalin, J.; Bergman, J.A.; Kozikowski, A.; Gordon, M.N.; et al. Histone deacetylase 6 inhibition improves memory and reduces total tau levels in a mouse model of tau deposition. Alzheimer’s Res. Ther. 2014, 6, 12. [Google Scholar] [CrossRef]
- Tseng, J.-H.; Xie, L.; Song, S.; Xie, Y.; Allen, L.; Ajit, D.; Hong, J.-S.; Chen, X.; Meeker, R.B.; Cohen, T.J. The Deacetylase HDAC6 Mediates Endogenous Neuritic Tau Pathology. Cell Rep. 2017, 20, 2169–2183. [Google Scholar] [CrossRef] [PubMed]
- Onishi, T.; Maeda, R.; Terada, M.; Sato, S.; Fujii, T.; Ito, M.; Hashikami, K.; Kawamoto, T.; Tanaka, M. A novel orally active HDAC6 inhibitor T-518 shows a therapeutic potential for Alzheimer’s disease and tauopathy in mice. Sci. Rep. 2021, 11, 15423. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.; Ross, J.L.; Goldman, Y.E.; Holzbaur, E.L.F. Differential Regulation of Dynein and Kinesin Motor Proteins by Tau. Science 2008, 319, 1086–1089. [Google Scholar] [CrossRef]
- Cohen, T.J.; Guo, J.L.; Hurtado, D.E.; Kwong, L.K.; Mills, I.P.; Trojanowski, J.Q.; Lee, V.M.Y. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat. Commun. 2011, 2, 252. [Google Scholar] [CrossRef]
- Zhang, F.; Su, B.; Wang, C.; Siedlak, S.L.; Mondragon-Rodriguez, S.; Lee, H.-G.; Wang, X.; Perry, G.; Zhu, X. Posttranslational modifications of α-tubulin in alzheimer disease. Transl. Neurodegener. 2015, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Saba, N.F.; Magliocca, K.R.; Kim, S.; Muller, S.; Chen, Z.; Owonikoko, T.K.; Sarlis, N.J.; Eggers, C.; Phelan, V.; Grist, W.J.; et al. Acetylated Tubulin (AT) as a Prognostic Marker in Squamous Cell Carcinoma of the Head and Neck. Head Neck Pathol. 2013, 8, 66–72. [Google Scholar] [CrossRef]
- Seeley, E.S.; Carrière, C.; Goetze, T.; Longnecker, D.S.; Korc, M. Pancreatic Cancer and Precursor Pancreatic Intraepithelial Neoplasia Lesions Are Devoid of Primary Cilia. Cancer Res 2009, 69, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Del Bufalo, D.; Desideri, M.; De Luca, T.; Di Martile, M.; Gabellini, C.; Monica, V.; Busso, S.; Eramo, A.; De Maria, R.; Milella, M.; et al. Histone deacetylase inhibition synergistically enhances pemetrexed cytotoxicity through induction of apoptosis and autophagy in non-small cell lung cancer. Mol. Cancer 2014, 13, 230. [Google Scholar] [CrossRef]
- Rey, M.; Irondelle, M.; Waharte, F.; Lizarraga, F.; Chavrier, P. HDAC6 is required for invadopodia activity and invasion by breast tumor cells. Eur. J. Cell Biol. 2011, 90, 128–135. [Google Scholar] [CrossRef]
- Lee, C.-C.; Cheng, Y.-C.; Chang, C.-Y.; Lin, C.-M.; Chang, J.-Y. Alpha-tubulin acetyltransferase/MEC-17 regulates cancer cell migration and invasion through epithelial–mesenchymal transition suppression and cell polarity disruption. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Depetter, Y.; Geurs, S.; De Vreese, R.; Goethals, S.; Vandoorn, E.; Laevens, A.; Steenbrugge, J.; Meyer, E.; De Tullio, P.; Bracke, M.; et al. Selective pharmacological inhibitors of HDAC6 reveal biochemical activity but functional tolerance in cancer models. Int. J. Cancer 2019, 145, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Lopes, D.; Seabra, A.L.; Orr, B.; Maiato, H. α-Tubulin detyrosination links the suppression of MCAK activity with taxol cytotoxicity. J. Cell Biol. 2022, 222, e202205092. [Google Scholar] [CrossRef] [PubMed]
- Herczenik, E.; Gebbink, M.F.B.G. Molecular and cellular aspects of protein misfolding and disease. FASEB J. 2008, 22, 2115–2133. [Google Scholar] [CrossRef]
- Willis, M.S.; Patterson, C. Proteotoxicity and Cardiac Dysfunction—Alzheimer’s Disease of the Heart? N. Engl. J. Med. 2013, 368, 455–464. [Google Scholar] [CrossRef]
- Folger, A.; Wang, Y. The Cytotoxicity and Clearance of Mutant Huntingtin and Other Misfolded Proteins. Cells 2021, 10, 2835. [Google Scholar] [CrossRef]
- Wang, X.; Osinska, H.; Klevitsky, R.; Gerdes, A.M.; Nieman, M.; Lorenz, J.; Hewett, T.; Robbins, J. Expression of R120G-αB-Crystallin Causes Aberrant Desmin and αB-Crystallin Aggregation and Cardiomyopathy in Mice. Circ. Res. 2001, 89, 84–91. [Google Scholar] [CrossRef]
- Maloyan, A.; Sayegh, J.; Osinska, H.; Chua, B.H.; Robbins, J. Manipulation of Death Pathways in Desmin-Related Cardiomyopathy. Circ. Res. 2010, 106, 1524–1532. [Google Scholar] [CrossRef]
- Henning, R.; Brundel, B.J.J.M. Proteostasis in cardiac health and disease. Nat. Rev. Cardiol. 2017, 14, 637–653. [Google Scholar] [CrossRef] [PubMed]
- Sanbe, A.; Osinska, H.; Saffitz, J.E.; Glabe, C.G.; Kayed, R.; Maloyan, A.; Robbins, J. Desmin-related cardiomyopathy in transgenic mice: A cardiac amyloidosis. Proc. Natl. Acad. Sci. USA 2004, 101, 10132–10136. [Google Scholar] [CrossRef]
- McLendon, P.M.; Ferguson, B.S.; Osinska, H.; Bhuiyan, S.; James, J.; McKinsey, T.A.; Robbins, J. Tubulin hyperacetylation is adaptive in cardiac proteotoxicity by promoting autophagy. Proc. Natl. Acad. Sci. USA 2014, 111, E5178-86. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, C.-T.; Qi, X.; Meijering, R.A.; Hoogstra-Berends, F.; Tadevosyan, A.; Deniz, G.C.; Durdu, S.; Akar, A.R.; Sibon, O.C.; et al. Activation of Histone Deacetylase-6 Induces Contractile Dysfunction Through Derailment of α-Tubulin Proteostasis in Experimental and Human Atrial Fibrillation. Circulation 2014, 129, 346–358. [Google Scholar] [CrossRef]
- Naghavi, M.H.; Walsh, D. Microtubule Regulation and Function during Virus Infection. J. Virol. 2017, 91, e00538-17. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.; Gammon, D.B. Manipulation of Host Microtubule Networks by Viral Microtubule-Associated Proteins. Viruses 2022, 14, 979. [Google Scholar] [CrossRef]
- Valenzuela-Fernández, A.; Álvarez, S.; Gordon, M.; Barrero, M.; Ursa, A.; Cabrero, J.R.; Fernández, G.; Naranjo-Suárez, S.; Yáñez-Mó, M.; Serrador, J.M.; et al. Histone Deacetylase 6 Regulates Human Immunodeficiency Virus Type 1 Infection. Mol. Biol. Cell 2005, 16, 5445–5454. [Google Scholar] [CrossRef] [PubMed]
- Xuan, C.; Qiao, W.; Gao, J.; Liu, M.; Zhang, X.; Cao, Y.; Chen, Q.; Geng, Y.; Zhou, J. Regulation of Microtubule Assembly and Stability by the Transactivator of Transcription Protein of Jembrana Disease Virus. J. Biol. Chem. 2007, 282, 28800–28806. [Google Scholar] [CrossRef]
- Kannan, H.; Fan, S.; Patel, D.; Bossis, I.; Zhang, Y.-J. The Hepatitis E Virus Open Reading Frame 3 Product Interacts with Microtubules and Interferes with Their Dynamics. J. Virol. 2009, 83, 6375–6382. [Google Scholar] [CrossRef]
- Husain, M.; Harrod, K.S. Enhanced acetylation of alpha-tubulin in influenza A virus infected epithelial cells. FEBS Lett. 2010, 585, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Jiang, Y.; He, Z.; Kitazato, K.; Wang, Y. Cellular defence or viral assist: The dilemma of HDAC6. J. Gen. Virol. 2017, 98, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Husain, M.; Cheung, C.-Y. Histone Deacetylase 6 Inhibits Influenza A Virus Release by Downregulating the Trafficking of Viral Components to the Plasma Membrane via Its Substrate, Acetylated Microtubules. J. Virol. 2014, 88, 11229–11239. [Google Scholar] [CrossRef] [PubMed]
- Naranatt, P.P.; Krishnan, H.H.; Smith, M.S.; Chandran, B. Kaposi’s Sarcoma-Associated Herpesvirus Modulates Microtubule Dynamics via RhoA-GTP-Diaphanous 2 Signaling and Utilizes the Dynein Motors To Deliver Its DNA to the Nucleus. J. Virol. 2005, 79, 1191–1206. [Google Scholar] [CrossRef] [PubMed]
- Sabo, Y.; Walsh, D.; Barry, D.S.; Tinaztepe, S.; Santos, K.D.L.; Goff, S.P.; Gundersen, G.G.; Naghavi, M.H. HIV-1 Induces the Formation of Stable Microtubules to Enhance Early Infection. Cell Host Microbe 2013, 14, 535–546. [Google Scholar] [CrossRef]
- Zhang, S.; Jiang, Y.; Cheng, Q.; Zhong, Y.; Qin, Y.; Chen, M. Inclusion Body Fusion of Human Parainfluenza Virus Type 3 Regulated by Acetylated α-Tubulin Enhances Viral Replication. J. Virol. 2017, 91, e01802-16. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, S.; Ma, J.; Jin, D.; Qin, Y.; Chen, M. Vimentin inhibits α-tubulin acetylation via enhancing α-TAT1 degradation to suppress the replication of human parainfluenza virus type 3. PLoS Pathog. 2022, 18, e1010856. [Google Scholar] [CrossRef]
- Glon, D.; Vilmen, G.; Perdiz, D.; Hernandez, E.; Beauclair, G.; Quignon, F.; Berlioz-Torrent, C.; Maréchal, V.; Poüs, C.; Lussignol, M.; et al. Essential role of hyperacetylated microtubules in innate immunity escape orchestrated by the EBV-encoded BHRF1 protein. PLoS Pathog. 2022, 18, e1010371. [Google Scholar] [CrossRef]
- Destaing, O.; Saltel, F.; Gilquin, B.; Chabadel, A.; Khochbin, S.; Ory, S.; Jurdic, P. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J. Cell Sci. 2005, 118, 2901–2911. [Google Scholar] [CrossRef] [PubMed]
- Chabin-Brion, K.; Marceiller, J.; Perez, F.; Settegrana, C.; Drechou, A.; Durand, G.; Poüs, C. The Golgi Complex Is a Microtubule-organizing Organelle. Mol. Biol. Cell 2001, 12, 2047–2060. [Google Scholar] [CrossRef] [PubMed]
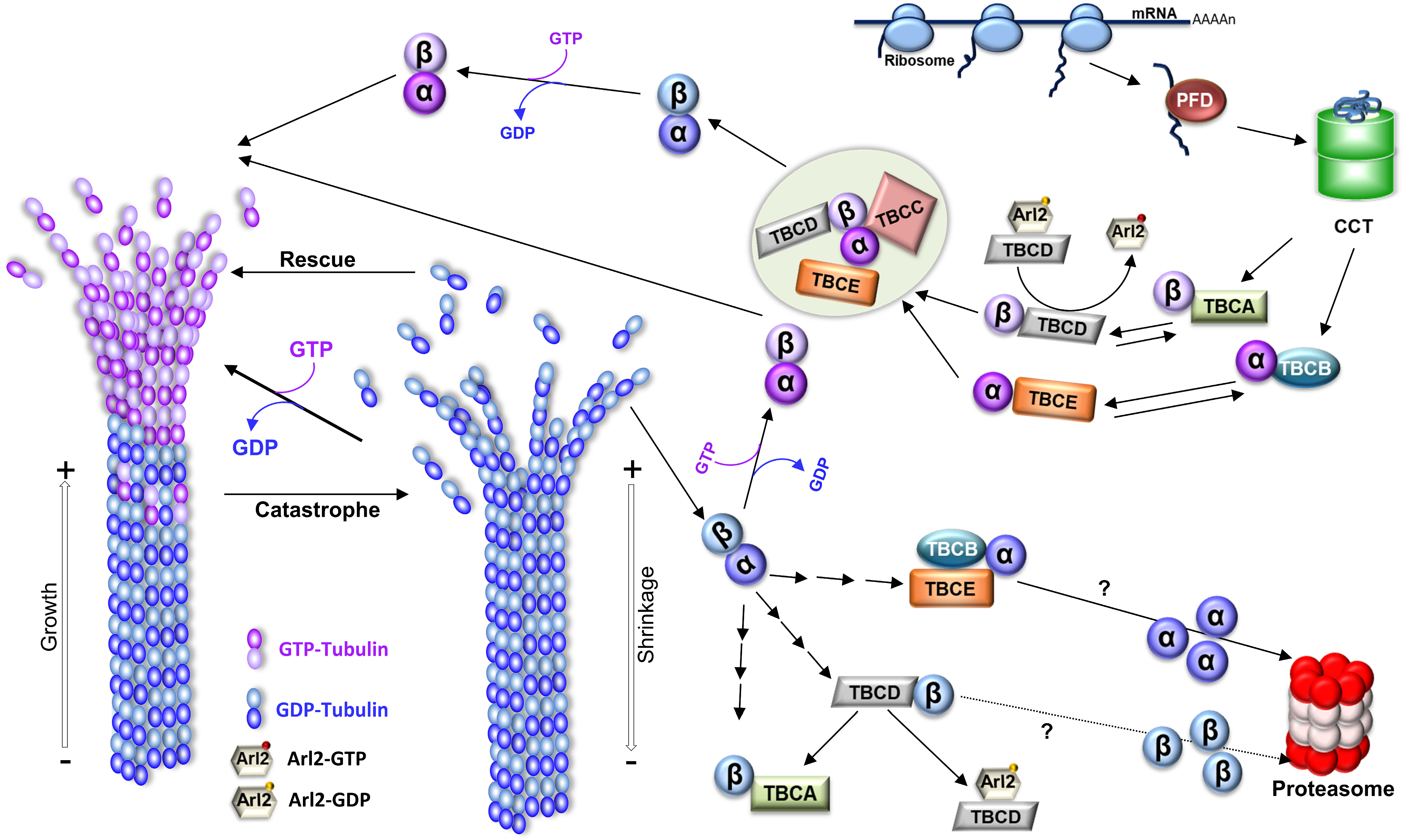

| Acetylated Tubulin and Amino Acid Residue | Acetylases/Deacetylases | Structural Localization | Functions | References |
|---|---|---|---|---|
| α-tubulin lysine 40 (Lys40) residue | αTAT1/HDAC6 and SIRT2 | Unstructured loop of α-tubulin of polymerized MTs. | Mechanical properties of MTs and prevention from mechanical aging - Establishing salt bridges between adjacent α-tubulins, leading to a rearrangement in the inter-protofilament angle. - Reducing interactions between protofilaments, potentially promoting protofilament sliding and enhancing MT flexibility and controlling the protofilament number. - Modulation of lateral, but not longitudinal, interactions of the protofilaments. - Avoiding long-lived MTs to lose their rigidity due to repetitive bending and stopping MT breakage and extending the MTs’ lifespan. | [157,167,187,231,237,257,259,269] |
| Regulation of MT dynamics - Self-assembly rate of acetylated tubulin was much slower than that of deacetylated tubulin. - No differences were found in polymerization rates for acetylated and deacetylated tubulin - When the measurements were made with the MTs already formed - acetylated MTs had a disassembly rate threefold faster than deacetylated MTs | [157,260,263,264] | |||
| Acetylation in centrioles - Allow centrioles to bear more mechanical stress and rendering them more flexible. | [282] | |||
| Cilia assembly - Lys40 acetylation is the most abundant acetylation site in cilia MTs - It is controversial that Lys40 acetylation is critical for correct cilia assembly and structure maintenance and stability. This may depend on cell type. - Controversial if it is required for sperm motility - Controversial in controlling ciliary motor proteins | [145,165,167,187,244,273,277,278,279,280] | |||
| Cargo transport - Cytoplasmic MT acetylation is more heterogeneous than that of cilia and centrioles. - The abundance and distribution patterns vary according to cell type, cell life cycle, and cell region; neurons illustrate this complexity. - A relationship between the MT acetylation status and the binding of molecular motors dynein and kinesin, especially in neurons, has been suggested. - Regulation of intracellular trafficking | [166,205,253,279,280,284,285,286,287,288,292,293,294,295,296,297,298,299] | |||
| Cell division - The acetylation of α-tubulin Lys40 is highly abundant in the mitotic spindle, midbody, and kinetochore MTs. - It is controversial that cell cycle progression is affected by MT acetylation/deacetylation. - Enzymes αTAT1, HDAC6, and SIRT2 present a regulation dependent on the cell cycle. | [166,167,235,270,311,312,316,317,318,319] | |||
| Cell shape and migration - MT acetylation is required for change in platelets’ shapes. - Tubulin acetylation in migration is involved in intracellular rearrangement of MTs accompanied by organelles re-localization. - It is controversial that loss of either HDAC6 or αTAT1 is associated with an increase in the number of focal adhesions. - It is proposed that MT acetylation is critical for the fine tuning of the mechanosensitive cell adhesion and migration. - Acetylation of MTs is critical for the penetrative capacity of cells undergoing radial intercalation in epithelia. | [186,192,204,237,259,320,321,322,323,324,325,326,327] | |||
| Stress response and autophagy - Increased levels of MT acetylation have been found in cells exposed to several cellular stresses, and this leads to increased cell survival. - MT α-tubulin hyperacetylation favors cell survival during stress through the induction of autophagy. | [227,246,331,332] | |||
| β-tubulin Lys252 residue | SAN/? | Interface between α/β-tubulins in the heterodimer | Stabilization of tubulin heterodimer - Affect the α/β tubulin interaction with impact in MT polymerization - Low levels of San acetylase, after cold-shock depolymerization, leads to MT depolymerization at a normal rate, but MT regrowth is faster. | [176] |
| α-tubulin Lys394 residue | ?/HDAC6 | Interface of the α/β-tubulin heterodimer in the MT surface, a region that suffers a conformational change as the dimers are added to the MT | Microtubule polymerization - This is required for heterodimer incorporation during MT assembly. | [135,175,330] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carmona, B.; Marinho, H.S.; Matos, C.L.; Nolasco, S.; Soares, H. Tubulin Post-Translational Modifications: The Elusive Roles of Acetylation. Biology 2023, 12, 561. https://doi.org/10.3390/biology12040561
Carmona B, Marinho HS, Matos CL, Nolasco S, Soares H. Tubulin Post-Translational Modifications: The Elusive Roles of Acetylation. Biology. 2023; 12(4):561. https://doi.org/10.3390/biology12040561
Chicago/Turabian StyleCarmona, Bruno, H. Susana Marinho, Catarina Lopes Matos, Sofia Nolasco, and Helena Soares. 2023. "Tubulin Post-Translational Modifications: The Elusive Roles of Acetylation" Biology 12, no. 4: 561. https://doi.org/10.3390/biology12040561
APA StyleCarmona, B., Marinho, H. S., Matos, C. L., Nolasco, S., & Soares, H. (2023). Tubulin Post-Translational Modifications: The Elusive Roles of Acetylation. Biology, 12(4), 561. https://doi.org/10.3390/biology12040561






