Simple Summary
The delimitation of species as the most important rank in biological classification is an essential contribution of taxonomy to biodiversity research, with all of its evolutionary, ecological, political, and legislative ramifications. Species delimitation is extremely tricky in plant groups evolving by polyploidisation (multiplication of chromosome sets) because the rapid formation of new, reproductively isolated lineages (species) is often not paralleled by conspicuous genetic, morphological, physiological, and/or ecological differentiation. Having clarified the taxonomy of diploid (2x) representatives of the genus Leucanthemum (marguerites, ox-eye daisies) in a previous contribution, the present study aims at an objective and reproducible delimitation of evolutionarily significant units (species) at the tetraploid (4x) level. We used DNA-based fingerprinting and statistical analyses of leaf shapes, ecological niches, and distribution ranges for eight predefined morphotaxa to judge their ranks as species or subspecies and propose a taxonomical treatment for the surveyed group with six species (two of them with two subspecies). Having clarified the taxonomic structure of the ancestral diploid (the ‘warps and wefts’) and the subsequent tetraploid layer (the ‘picks of the fabric’), we will be able to provide a taxonomy for the remainder of this well-known plant group and study its reticulate evolutionary history.
Abstract
Based on the results of a preceding species-delimitation analysis for the diploid representatives of the genus Leucanthemum (Compositae, Anthemideae), the present study aims at the elaboration of a specific and subspecific taxonomic treatment of the tetraploid members of the genus. Following an integrative taxonomic approach, species-level decisions on eight predefined morphotaxon hypotheses were based on genetic/genealogical, morphological, ecological, and geographical differentiation patterns. ddRADseq fingerprinting and SNP-based clustering revealed genetic integrity for six of the eight morphotaxa, with no clear differentiation patterns observed between the widespread L. ircutianum subsp. ircutianum and the N Spanish (Cordillera Cantábrica) L. cantabricum and the S French L. delarbrei subsp. delabrei (northern Massif Central) and L. meridionale (western Massif Central). The inclusion of differentiation patterns in morphological (leaf dissection and shape), ecological (climatological and edaphic niches), and geographical respects (pair-wise tests of sympatry vs. allopatry) together with the application of a procedural protocol for species-rank decisions (the ‘Wettstein tesseract’) led to the proposal of an acknowledgement of the eight predefined morphotaxon hypotheses as six species (two of them with two subspecies). Nomenclatural consequences following from these results are drawn and lead to the following new combinations: Leucanthemum delarbrei subsp. meridionale (Legrand) Oberpr., T.Ott & Vogt, comb. nov. and Leucanthemum ruscinonense (Jeanb. & Timb.-Lagr.) Oberpr., T.Ott & Vogt, comb. et stat. nov.
Keywords:
Asteraceae; ecology; ddRADseq; geography; leaf morphology; nomenclature; polyploidy; taxonomy 1. Introduction
The delimitation of species as the most paramount rank in biological classification is an essential contribution of taxonomy to biodiversity research, with all of its evolutionary, ecological, political, and legislative ramifications and corollaries. Following Stuessy [1] and Zachos et al. [2], this alpha-taxonomy procedure has a twofold nature: while in the first step (the ‘grouping’ step), ‘species discovery’ and ‘species validation’ methods are used to infer and subsequently test species-group hypotheses (the ‘species taxa’ of Zachos et al. [2]) by detecting genealogical, morphological, or ecological discontinuities [3,4], the second step (the ‘ranking’ step) constitutes ‘an executive decision that the species taxon warrants recognition at the species level’ [2]. However, the latter—the decision whether ‘species taxa’ should be ranked as species under the Linnaean classification system—is clearly subjective, due to its dependence on the acceptance of a species concept. Of these, a broad array exists, though without any cognisable chance for the unrestricted applicability of a single one throughout the realm of organismic diversity.
The ‘unified species concept’ proposed by De Queiroz [5] was a game-changer in the futile search for a generally applicable species concept because it altered the perspective that the properties entertained by the plethora of concepts (e.g., reproductive isolation, genealogy, morphology, ecology, geography, etc.) are helpful in species conceptualisation. Instead, the ‘unified species concept’ defined species as hypothetical independently evolving metapopulation lineages, for which the above-mentioned properties could be made to subserve as indicators or proxies. This conceptual shift paved the way for the renaissance of ‘biosystematics’ or ‘experimental taxonomy’ approaches to species delimitation used in the second half of the 20th century as ‘integrative taxonomy’ [6,7], which makes use of all available sources of empirical evidence for the conceptualisation of species rank. This is nowadays carried out either by using computational tools, e.g., Geneland [8] for the joint analysis of morphology, genetics, and geography; “multivariate normal mixtures and tolerance regions” analysis [9,10] for morphology and geography; iBPP [11] for genealogy and morphology; regression analysis [12] for genetics and geography; or by entertaining procedural protocols, e.g., [13,14,15,16]. Most recent approaches also try to incorporate the speciation process itself into species-delimitation software programs (Delineate [17]).
Species formation by polyploidy—a speciation mode realised in significant numbers of pteridophyte and angiosperm groups—poses considerable problems for species delimitation [18]: while polyploidisation will instantly lead to postzygotic reproductive isolation between parental taxa and their polyploid derivatives (biological species concept), detectable trait differences entertained by a morphological or a physiological/ecological species concept may only be realised in an allopolyploid speciation scenario and not (or at least not immediately or obviously) in an autopolyploid one. Additionally, a strict phylogenetic species concept (with species defined as monophyletic evolutionary units) is violated both in the auto- and allopolyploid speciation mode due to (a) the parental species becoming paraphyletic relative to the newly formed polyploid species and (b) the potentially polyphyletic nature of a polyploid species caused by its multiple (and sometimes reciprocally parented) origin or (c) gene flow between independently formed allo- and/or autopolyploid populations/lineages. Finally, at least in some plant groups, the switch of polyploid lineages toward asexual reproduction (agamospermy) makes the application of a biological species concept senseless. As a consequence, these peculiarities of polyploid species formation make the application of a ‘unified species concept’ sensu De Queiroz [5] (species as metapopulation lineages) and the integrative approach to species delimitation indispensable for plant groups diversifying through this speciation mode.
The genus Leucanthemum Mill. (Compositae, Anthemideae; marguerites or ox-eye daisies) comprises 39 [19] to 42 species [20], with ploidy levels ranging from diploid (2x) to dodecaploid (12x), and one species [L. lacustre (Brot.) Samp.) from Portugal even showing a chromosome number of 2n = 22x = 198 (docosaploid level). Leucanthemum is distributed over the whole European continent and extends into Northern Asia (the tetraploid L. ircutianum DC. is found in Siberia), while some species were also introduced into temperate regions of the Northern and Southern Hemispheres [21]. The current species delimitation of the genus is mostly based on differences in morphology, especially in general leaf shape and leaf dissection, as the flower characters are relatively invariant, and ploidy level, as well as geographical distribution [22,23] and, more recently, genetic differentiation [19].
Previous studies addressed the species delimitation and phylogeny of the diploid Leucanthemum representatives based on multicopy nuclear markers (nrDNA ETSs), AFLP fingerprinting, and single-copy nuclear markers [24,25,26,27]. Cloning of nrDNA ETS amplicons revealed that some diploid taxa exclusively possess a plesiomorphic ETS ribotype cluster closely related to ETS ribotypes of the outgroup and that others are characterised by the exclusive possession of an apomorphic ETS ribotype cluster, while a third group of taxa exhibit an additive pattern of the two types [24]. This finding was supported by Konowalik et al. [25] using AFLP fingerprinting and multilocus species-tree reconstructions based on low-copy markers of the nuclear genome, and it was demonstrated that the species with the plesiomorphic ETS ribotypes form an early-diverging paraphyletic grade, in which the monophyletic group of taxa with the apomorphic ETS ribotypes are nested. Furthermore, the authors applied coalescent-based simulations to distinguish between hybridisation and incomplete lineage sorting (ILS), revealing that for most of the diploid taxa involved (and especially for those in the second group) incongruence among gene trees could not be explained by ILS alone and that recent hybridisation or even homoploid hybrid speciation events must be assumed. Consequently, some infraspecific taxa were raised to species level to account for their assumed independent formation through homoploid hybrid speciation (i.e., L. cacuminis, L. eliasii, and L. pyrenaicum).
Species delimitation in a morphologically close-knit group of taxa in the clade with the apomorphic nrDNA ETS ribotypes was subsequently carried out by Wagner et al. [26], who used AFLP fingerprinting, sequence information from plastid and nuclear low-copy markers, and coalescent-based Bayesian delimitation methods to infer species boundaries in the L. ageratifolium group, despite the frequent presence of hybrid individuals in this group. Finally, Wagner et al. [27] presented a multilocus phylogenetic reconstruction of the subtribe Leucantheminae, in which the diversification among diploid Leucanthemum species was dated to the last 1.9 (1.1–2.9) Ma, arguing for the strong influence of Pleistocene oscillations on species formation in this genus. Most recently, Ott et al. [19] combined restriction-site-associated DNA sequencing (RADseq), ecological niche modelling (ENM), geographical patterns, and geometric morphometrics for integrative species delimitation among the diploid Leucanthemum representatives.
In contrast to the extensive investigation of species delimitation and the phylogenetic relationships of diploids, the evolutionary histories of only a few tetraploid Leucanthemum species have been the subject of previous studies. Oberprieler et al. [28] found indications of an allopolyploid origin of the tetraploid L. ircutianum subsp. ircutianum based on AFLP markers. One year later, Greiner et al. [29] showed that the tetraploid L. pseudosylvaticum is able to form fertile offspring when crossed with its diploid relative L. pluriflorum, suggesting a relatively recent diversification. Another study of the L. pluriflorum group by Greiner et al. [30] produced evidence for the allopolyploid and autopolyploid origins of L. pseudosylvaticum and L. corunnense, respectively, and raised L. pseudosylvaticum, which was formerly a subspecies of L. ircutianum, to species rank. Finally, the most recent contribution to the taxonomy of the tetraploid Leucanthemum representatives by Oberprieler et al. [31] suggested infraspecific ranks for the widely distributed L. ircutianum subsp. ircutianum and its amphi-Adriatic counterpart L. ircutianum subsp. leucolepis based on AFLP fingerprinting and sequence variations in nuclear and plastid DNA.
The present contribution aims at a comprehensive and integrative assessment of all tetraploid Leucanthemum taxa. Applying the conceptual framework for species-rank decisions proposed by Oberprieler [16], morphological, ecological, geographical, and genealogical/genetic evidence is used to devise a taxonomic treatment for eight tetraploid taxa that have been hitherto accepted as occupying specific or infraspecific ranks in Central and Southern Europe, the only exception being the NW Spanish L. corunnenese Lago, for which an autotetraploid origin based on the diploid L. pluriflorum has been shown in a previous study [30].
2. Material and Methods
2.1. Taxon Selection
Besides the widespread L. ircutianum subsp. ircutianum, the following seven geographically more restricted tetraploid taxa were considered as entities, for which an integrative taxonomic treatment was envisioned in the present contribution: these comprise the three species endemic to the Iberian Peninsula, L. cantabricum Sennen, L. crassifolium (Lange) Lange, and L. pseudosylvaticum (Vogt) Vogt & Oberpr., along with the NE Spanish and SW French L. delarbrei Timb.-Lagr. subp. delarbrei, subsp. ruscinonense (Jeanb. & Timb.-Lagr.) Vogt et al. and L. meridionale Legrand and the amphi-Adriatic L. ircutianum subsp. leucolepis (Briq. & Cavill.) Vogt & Greuter (see Figure 1 for a distribution map).
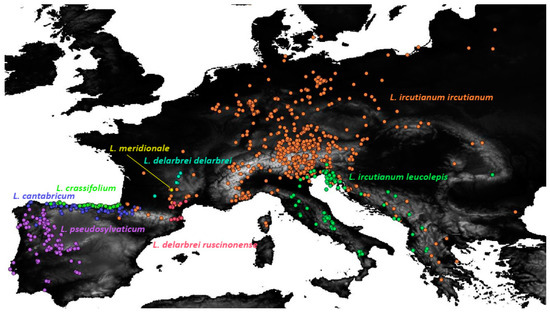
Figure 1.
Distribution of the eight tetraploid Leucanthemum morphotaxa in Southern and Central Europe based on locality information from revised herbarium specimens which was used in the ecological niche modelling and geographical overlap analyses of the present study (see Supplementary Material ES02 for the list of georeferenced accessions).
2.2. Morphological Analyses
Differences in leaf shape, an important delimitation criterion for Leucanthemum taxa, have been demonstrated in the diploids of the genus [19] and were assessed here by measuring the general leaf shape and the degree of dissection of the leaves by applying elliptic Fourier analysis (EFA; [32]) and calculating leaf dissection indices (LDIs), respectively. For this purpose, 285 images of digitised herbarium specimens were provided by the herbarium of the Berlin Botanical Museum (B; see Supplementary Material ES01). Using these images, we manually annotated 1,338 intact leaves with polygons around the leaves’ outlines and polylines along the leaves’ main veins using the Computer Vision Annotation Tool (CVAT; https://github.com/openvinotoolkit/cvat (accessed on 31 January 2023)) according to Ott et al. [19]. Subsequently, we straightened the leaves to reduce the influence of deformations either caused by developmental irregularities or introduced by the drying process and extracted the leaf contours as binary masks.
Using the binary masks, we conducted EFA and calculated LDIs using the Python packages scikit-image [33], numpy [34], scikit-learn [35], and PyEFD (https://github.com/hbldh/pyefd (accessed on 31 January 2023)). For the EFA, we used 20 harmonics, normalised the descriptors, and applied principal component analysis (PCA) for decorrelation and feature extraction to the non-constant descriptors (normalisation causes the three descriptors A1, B1, and C1 to be constant).
To find differences in general leaf shape, we used the first 15 principal components (PCs), explaining 50% of the total variance with two different testing strategies: (1) the permutation-based test for differences in Euclidean distance in PC space as applied by [19]; and (2) non-parametric multivariate analysis of variance (NPMANOVA; [36]) implemented in the R package ‘vegan’ v2.6 (function ‘adonis’; [37]). The number of permutations was set to 5000 and 100 for the former and the latter, respectively. To assess differences in leaf dissection, we subjected the LDI values to Welch’s tests. All tests were corrected using Bonferroni´s method.
2.3. Ecological Niche Modelling
Ecoclimatological and edaphic niches of the tetraploid Leucanthemum taxa were reconstructed using ecological niche modelling (ENM) and compared using permutation-based statistical tests. For this purpose, we retrieved the collection locations of 1470 individuals from several herbaria (Supplementary Material ES02) and obtained rasters of 19 bioclimatic and 10 edaphic variables at depth levels of 0–5 cm, 5–15 cm, and 15–30 cm (Supplementary Material ES03) from Worldclim Bioclim [38] and SoilGrids [39], respectively. The rasters were cropped to encompass Central Europe (longitude -10.0°–26.0°; latitude 35.9°–52°) and scaled to a resolution of 2.5 arc minutes using the R package ‘raster’ v3.5.15 [40] (https://github.com/rspatial/raster (accessed on 31 January 2023)). In addition to the climate rasters for presence, we retrieved paleoclimate rasters for the last glacial maximum (LGM; CCSM4) and the last interglacial (LIG; lig_30s) from Worldclim Bioclim and subjected them to the same preprocessing, but with additional recoding of temperature, since LGM and LIG datasets used Bioclim 1 temperature encoding. For the edaphic variables, we averaged the raster values of the three mentioned depth levels to obtain a single raster for each soil variable. Since it was computationally not tractable to work with all 29 rasters, we applied principal component analysis (PCA) with standardisation implemented in the R package ‘ENMtools’ v1.0.6 [41] for feature extraction to the recent Bioclim and SoilGrids rasters, separately. The first three principal-component (PC) rasters for each of the datasets were selected for the ENM.
To compare the ecological niches of the different taxa, we reconstructed potential distribution ranges for all tetraploid taxa except L. meridionale using MaxEnt v3.4.4 [42], with 5000 iterations and 6-fold cross-validation, and applied niche-equivalency tests implemented in ‘ENMTools’ for all combinations of tetraploid taxa with MaxEnt as species distribution, 200 replicates, a species range of 50 km, and 1000 background points with a range of 20 km. Leucanthemum meridionale was excluded from these comparisons because this species is endemic to a very limited area, causing all collection points to fall into the same raster cell and thus rendering ENM impossible. Potential niches at LGM and LIG were also reconstructed using MaxEnt with the same parameters as for the PC rasters, but this time with LGM and LIG as projection rasters and including collection data for the diploid species provided by Ott et al. [19].
2.4. Geographical Distribution
The geographic co-distribution of the tetraploid taxa, excluding L. meridionale again due to its restricted sampling area as a point endemic, was assessed by evaluating the overlap of spatial distributions using the same data as for the ENM analyses. Unfortunately, there are no comprehensive distribution rasters available for the tetraploid Leucanthemum representatives, which is why we had to resort to approximating the geographic distribution from the available sampling points. We applied the method proposed by Ott et al. [19], which approximates the true geographical distribution by reconstructing the potential area using an ENM (in this case, MaxEnt v3.4.4; [42]) and subsequently removes unconnected and unsampled regions (i.e., regions that are separated from collection points by areas of low probability). To test for sympatry, we applied the permutation approach of Ott et al. [19] with 400 simulated datasets and a threshold of 0.25; the resulting p-values were corrected for multiple testing using Bonferroni´s method.
2.5. RADseq Assembly
Double-digest RADseq (ddRADseq; [43]) was conducted based on an accession set comprising 51 individuals from all presently accepted 17 diploid Leucanthemum species (20 taxa; [19]) and 35 individuals from 8 tetraploid Leucanthemum taxa (see the table in Supplementary Material ES04), the latter being the focal group of the present study. ddRADseq reads of the diploid samples were taken from Ott et al. [19], while for the tetraploid individuals, genomic DNA was extracted from silica-dried specimens according to the CTAB DNA extraction protocol of Doyle and Dickson [44]. For assembly-quality assessment, replicates were generated by repeating the extraction for two samples (L. ircutianum DC. subsp. ircutianum: accession L055-03/-031; L. ircutianum subsp. leucolepis: accession 170-02/-021). ddRAD Illumina sequencing (2 x 150 bp; NextSeq 500, Illumina Inc., San Diego, CA, USA) using the restriction enzymes Pst1 and ApeK1, including demultiplexing and adapter clipping, was conducted by LGC Genomics (Berlin, Germany).
Demultiplexed and adapter clipped reads from the diploid and tetraploid samples were assembled using iPyrad v0.9.81 [45] against the reference provided by Ott et al. [19]; the minimum number of samples per locus (min_samples_locus) was set to 20, while the remaining parameters were kept at default values. For detection of assembly errors, locus, allele, and SNP error rates were calculated (see [19,46]).
2.6. RADseq Network Analysis
Neighbor-nets based on SNP-based Nei distances ([47]; https://github.com/simjoly/pofad (accessed on 31 January 2023)) were calculated using SplitsTree4 v4.15.1 [48] for (a) the complete dataset and (b) for the tetraploid accessions exclusively. Distances were calculated based on the variant output (VCF file) of iPyrad using a custom Python and C tool (https://github.com/TankredO/nei_vcf (accessed on 31 January 2023)). With SNP-based Nei distances, differences in variant (SNP) frequencies are directly included in distance calculation, rendering this kind of distance metric particularly useful for comparing diploid and polyploid samples.
2.7. RADseq Consensus Clustering
Weighted ensemble of random (k)k-means (WKM) clustering [49] was applied to detect clusters of genetically similar individuals within the group of tetraploid Leucanthemum representatives. WKM clustering is a semi-supervised consensus (ensemble) clustering technique, allowing for a priori-defined fuzzy pairwise must-link and must-not-link constraints. Roughly, WKM clustering fits a number of k-means clusterings (e.g., 1000), each for a random subset of features (‘variables’) and data points (‘samples’), and with a random number of clusters (k). For each k-means run, clustering-level and cluster-level consistencies are calculated, which measure deviations from the fuzzy constraints concerning the whole clustering and single clusters, respectively. In addition, the mean silhouette index is calculated as an internal clustering-quality metric. The three values are non-linearly combined to obtain a clustering weight (i.e., a value determining the clustering quality). The clustering co-association matrix, i.e., the matrix determining which samples were clustered together (this can be thought of as a similarity matrix with only 1 and 0 entries; 1 if the samples were placed in the same cluster and 0 otherwise), is multiplied by the clustering weight. All weighted co-association matrices are summed (and optionally scaled) to obtain a consensus (co-association) matrix. Finally, a consensus clustering is calculated using this consensus matrix via hierarchical or spectral clustering.
For clustering of the tetraploid Leucanthemum representatives, the previously calculated SNP-based Nei distances (see above, under Network analysis) were subjected to a principal coordinate analysis (PCoA) with Lingoes negative eigenvalue correction, and the first principal coordinates (PCos) explaining at least 50% of the variance were selected. Based on the PCo scores, WKM clustering was applied with must-link constraints for the individuals of each of the taxa L. cantabricum, L. crassifolium, L. delarbrei subsp. delarbrei, L. delarbrei subsp. ruscinonense, L. ircutianum subsp. leucolepis, and L. meridionale and must-not-link constraints among individuals from L. delarbrei subsp. delarbrei and L. meridionale. The fraction of random features and samples was set to 0.60 and 0.80, respectively. The number of k-means runs was set to 5000, and k was allowed to take values from 2 to 14, including the lower and upper bounds. Finally, the consensus matrix was subjected to an average-linkage hierarchical clustering for each of the k-values, and the clustering scoring that was best according to the Bayesian information criterion (BIC) and the Calinski–Harabasz criterion (CH) was selected. All analyses were performed using pyckmeans v0.9.4 (https://github.com/TankredO/pyckmeans (accessed on 31 January 2023)).
2.8. Genealogical Species Delimitation
To find the potential diploid parental species of the tetraploid taxa under study, we applied a custom version of SNiPloid [50], as proposed by Wagner et al. [51]. Very similar to the original SNiPloid algorithm, our script compares two parent individuals (i.e., diploids) with one child individual (i.e., a tetraploid) on a per-SNP basis, distinguishing and counting the frequencies of five different SNP categories: categories 1 and 2 count so-called inter-specific SNPs, where exactly one parental SNP is identical to the child SNP and the other not. Categories 3 and 4 are so-called derived SNPs, meaning that an SNP is unique to the child individual, while the parents are homozygous for the same variant; patterns 3 and 4 cannot be distinguished based on unphased SNP data. Category 5 SNPs are homeo-SNPs, where the child is heterozygous (polymorphic), combining both parents’ homologous and monomorphic alleles. For all categories, only SNPs where both parents were homozygous (monomorphic) were considered.
We applied the SNiPloid approach to look for signs of allopolyploid speciation and expected recently formed allotetraploids to express a high number of category 5 SNPs, while longer established allotetraploid species should express a higher frequency of derived SNPs (i.e., category 3 and 4 SNPs). Finally, category 1 and 2 SNPs may indicate gene flow to the putative parents or alternatively be an additional signature of recent polyploidisation. For each tetraploid taxon and each possible pair of diploid parent species, therefore, we applied our custom Python implementation of the SNiPloid algorithm to the concatenated SNP output returned by iPyrad (.snps ouput). For each triplet, we tested all individuals and calculated the arithmetic means of SNP patterns 1 through 5.
3. Results
3.1. Morphology
The degree of leaf dissection varied significantly at an alpha level of p-value < 0.01 for all but three taxon pairs (Table 1, upper triangle). Differences in general leaf shape, measured as Euclidean distances among principal-component (PC)-transformed Fourier descriptors, were significant for 14 of the 28 taxon pairs (Table 1, lower triangle, left number). When applying NPMANOVA to the PC scores, 16 of the 28 taxon pairs showed significant differences in leaf-outline shape (Table 1, lower tringle, right number).

Table 1.
Bonferroni-corrected p-values for pairwise tests of morphological similarity. The upper triangle represents p-values of the LDI-based Welch’s tests, determining divergence in the dissection of the leaves. The lower triangle comprises p-values of the permutation tests of Euclidean distances in PC space (left) and NPMANOVA results (right), quantifying differences in general leaf shape. Corrected p-values are truncated to 1.0.
3.2. Ecological Niche Modelling and Geographical Range Overlap
All pairwise niche-equivalency tests except those for the two taxon pairs L. cantabricum–L. crassifolium (both N Spain) and L. cantabricum (N Spain)–L. delarbrei subsp. ruscinonense (NE Spain, SW France) were significant at an alpha level of 0.01 (see Table 2, where p-values of 0 indicate no niche overlap at all). Maps depicting predicted potential distribution ranges of the diploids and tetraploids based on recent, LGM, and LIG bioclimatic variables are provided in the supporting information (Supplementary Material ES05, ES06).

Table 2.
Bonferroni-corrected p-values for pairwise niche equivalency tests based on the two test statistics, D (upper triangle) and I (lower triangle).
In the tests on geographical range overlap (Table 3), all taxon pairs except for L. cantabricum–L. crassifolium (both N Spain) and L. delarbrei subsp. ruscinonense (NE Spain, SW France)–L. ircutianum subsp. ircutianum (large parts of Europe) were significant, suggesting a deviation from sympatric distributions for most of the tetraploid taxa.

Table 3.
Bonferroni-corrected p-values for pairwise tests of geographical overlap, i.e., sympatry. Corrected p-values are truncated to 1.0.
3.3. RADseq Assembly and Analysis
Of 216,551,342 raw reads, 81,517,011 demultiplexed, adapter-clipped, and restriction-enzyme-filtered reads were mapped against the reference of Ott et al. [19]. The resulting assembly comprised 7,342 loci with 156,057 SNPs, of which 83,375 were parsimony-informative. The percentages of missing data for sequence and SNP matrices were 36.9% and 34.5%, respectively. The trustworthiness of the RADseq fingerprinting procedure was confirmed by the high degree of similarity between re-extracted and reanalysed accessions (L055-031, 170-021) and their counterparts (L055-03, 170-02) in the subsequent analyses.
The network reconstruction based on SNP-based Nei distances for the complete dataset (i.e., all diploid and tetraploid individuals) placed all tetraploid taxa into the so-called L. vulgare group (Figure 2A). While all diploid samples were found to be clustered according to species membership, the majority of the tetraploid accessions followed this pattern except representatives of L. ircutianum subsp. ircutianum and L. delarbrei subsp. ruscinonense. Regarding the former, the accessions were observed to form two independent clusters comprising two and five accessions (the latter including L055-03 and its replicate L055-031), and a single individual (437-01) was ungrouped with any cluster. For L. delarbrei subsp. ruscinonense, two clusters with two (accessions 139-01 and 355-03) and three (accessions 101-01, 110-01, and 349-01) individuals were observed in quite distant positions in the network. When subjecting the tetraploid accessions alone to Neighbor-net clustering (Figure 2B), sample 437-01 was found to be grouped with the larger of the two L. ircutianum subsp. ircutianum clusters. As a consequence, this cluster comprises accessions of the taxon from Austria (L062-04), Corsica (437-01), Germany (L052-02, L055-03), Italy (87-01), and Montenegro (177-01), while the second one, closer to L. cantabricum, contains the two accessions from SW France (106-01 and 343-01). Additionally, all accessions of L. delarbrei subsp. ruscinonense now cluster together, and the network reconstruction indicates that there are closer genetic similarities between (a) L. delarbrei subsp. delarbrei and L. meridionale (both S France) and (b) L. crassifolium (N Spain), L. pseudosylvaticum (NW Spain), and L. delarbrei subsp. ruscinonense (SW France, NE Spain), while accessions of L. ircutianum subsp. leucolepis (Italy, Balkan Peninsula) form a cluster relatively isolated from these.
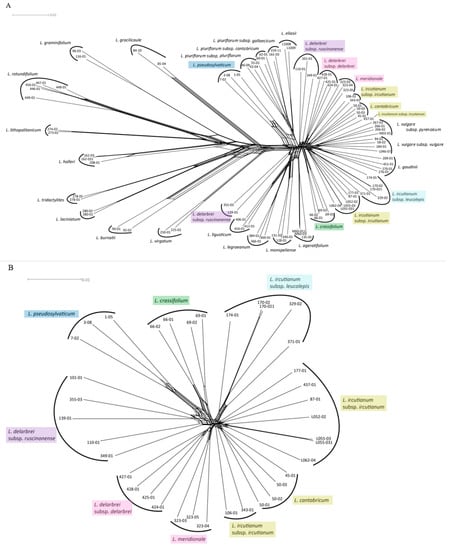
Figure 2.
NeighborNet network reconstructions based on SNP-level Nei distances of the full dataset (A), comprising individuals from all 17 diploid species and eight tetraploid taxa (in coloured boxes), and of the tetraploids only (B). For the full dataset, all tetraploid taxa are found within the so-called L. vulgare-group (right). The L. vulgare-group is separated from the remaining diploid species by relatively long branches (center). All diploid samples are clustered according to their species membership (A). The same is the case for the tetraploid taxa, except for L. ircutianum subsp. ircutianum, which is separated into two groups ((B), lower right).
The optimal number of clusters according to the Bayesian information criterion (BIC; ‘lower is better’) was found to be six (Figure 3B), while the Calinksi–Harabasz (CH; ‘higher is better’) score was optimal for k = 2, but also had a local optimum at six clusters. The consensus clustering with the co-association matrix at k = 6 (Figure 3A) merged L. meridionale and L. delarbrei subsp. delarbrei (pink cluster) and L. ircutianum subsp. ircutianum and L. cantabricum (yellow cluster), while the remaining assignments of accessions to clusters followed their taxonomic classifications.
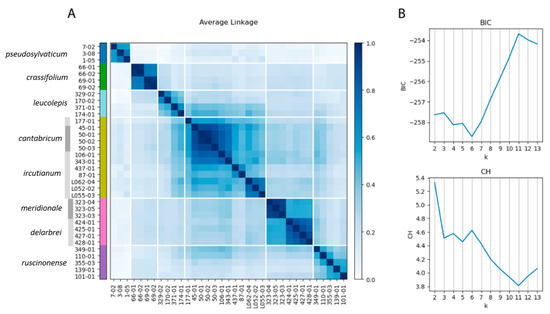
Figure 3.
(A) Co-association matrix for the tetraploid Leucanthemum taxa based on weighted ensemble of random (k)k-Means (WKM) clustering and SNP-based Nei distances. Consensus clusters for k = 6 are shown, with colour bars depicting consensus cluster membership. (B) BIC and CH metrics for number of clusters ranging from k = 2 to k = 13.
3.4. Genealogical Species Delimitation
Plots of the category proportions for all taxon triplets surveyed in the SNiPloid approach to infer parental diploids are provided in the supporting information (Supplementary Material ES07). We found that SNP categories 1 and 2 increased with the genetic distances of the corresponding parental species from the child taxa. The sum of categories 3 and 4 generally decreased with genetic distance. For the analysed taxa, we could not find any irregularity in this pattern. The frequency of category 5 was relatively constant, but also slightly decreased with the genetic distances of the parental species. There seemed to be slight shifts in category frequencies, with at least one parent outside of the L. vulgare group (i.e., L. vulgare, L. gaudinii, L. pluriflorum, L. ageratifolium, L. monspeliense, L. legraenum, and L. ligusticum) included as a parental species.
4. Discussion
Having provided an integrative classification scheme for the diploid representatives of the genus [19], the present contribution aims at a comparable taxonomic treatment of tetraploid Leucanthemum taxa based on the same sources of evidence for species delimitation: genetic diversification, morphological discontinuities, ecological differentiation, and information on the geographical distribution of taxa. In addition to the methodological approach taken in the diploid case, we additionally tried to incorporate genealogical aspects into this integrative taxonomy of Leucanthemum tetraploids: by trying to infer the parentage of tetraploid taxa from RADseq-based single-nucleotide polymorphism (SNP) data, we hoped for additional arguments for a classification scheme that incorporates the evolutionary history of the study group. This follows the rationale that, by elucidating the combination of diploid genomes in the polyploids, the disentangling of auto- and alloploid formations of these taxa and the knowledge of their independent vs. non-independent evolutionary trajectories could be used in taxonomic decisions.
4.1. Genealogical and Genetic Patterns
Unfortunately, the present analyses provide us only with a quite limited notion about the evolutionary origins of the tetraploid Leucanthemum taxa under study. This disappointing result is somewhat unexpected in consideration of the huge amount of genomic information received from the GBS fingerprinting, with over 150,000 SNPs from over 7000 loci. However, our present analyses simultaneously point towards the probable explanation for the unresolved evolutionary patterns in this plant group: all eight surveyed tetraploid taxa appear to be closely related to the close-knit species group of diploids around L. vulgare (Figure 2A), for which already at the diploid level species delimitation and reconstruction of phylogenetic relationships were found to be problematic [19]. Owing to the fact that the radiation of the whole genus Leucanthemum was dated to the last 1.9 (1.1–2.9) Ma [27] and the diversification of the L. vulgare group is presumably not older than 400 ka, polyploid species formation from these diploid ancestors has to be assumed to have occurred not earlier than the Middle (Chibanian, 0.77–0.13 Ma) or Late Pleistocene (0.13–0.01 Ma) with its glaciation cycles of the Mindel, Elster, Riss, and Würm eras.
It is comprehensible, therefore, that our present efforts to trace the parenthood of diploid species for the formation of tetraploid taxa by application of the SNiPloid-based strategy proposed by Wagner et al. [51] for Salix polyploids revealed no patterns in Leucanthemum. We think that the observed failure of the mentioned strategy, in contrast to its successful application in Salix tetraploids, is not due to a lack of sufficient loci and sampled SNPs (23,393 loci and 320,010 SNPs in Salix vs. 7342 loci and 156,057 SNPs in Leucanthemum) but due to the fact that the putative parental diploid species in Salix are much older than in Leucanthemum. He et al. [52] provide a dated phylogeny for the Salix subg. Chamaetia/Vetrix clade, in which the two diploids S. purpurea L. and S. repens L.—the two putative parents of the tetraploid S. caesia Vill. analysed by [51] using SNiPloid—are dated as having diverged from each other in the Early Miocene, around 20 Ma ago. It appears obvious that in a constellation like this, with the long evolutionary independence of diploid precursor lineages and a relatively recent formation (Pleistocene) of a tetraploid taxon, the SNiPloid-based strategy proposed by Wagner et al. [51] will reveal a clear signal. However, this cannot be expected in the Leucanthemum case, with its comparatively young diploid lineages.
There may be methodological concerns that the SNiPloid approach is strongly influenced by the filtering of RADseq reads gained from polyploids. This is due to the fact that, when parameters (cluster thresholds) are optimised in iPyrad to avoid under- and oversplitting of loci and to minimise paralogy, paralogy caused by whole-genome duplication (i.e., homoeology) will be also reduced and this could diminish potential additive allelic signals of allopolyploidy. However, we circumvented this problem by applying a reference-based assembly of reads from tetraploids (mapped against the diploid reference from Ott et al. [19]) instead of performing a de novo assembly of reads from both diploid and tetraploid accessions. As a consequence, we are confident that our negative SNiPloid results are mainly due to the young age of polyploidisation events in Leucanthemum.
Nevertheless, despite the failure to pinpoint potential diploid parental taxa of the tetraploids using the SNiPloid-based strategy, some relationships among taxa at the two ploidy levels were revealed by the SNP-based network reconstruction (Figure 2). The most obvious connection of a tetraploid taxon with diploid species is the case of the NW Iberian L. pseudosylvaticum, which was found to cluster with the group comprising the N Spanish L. eliasii and the three subspecies of the NW Spanish L. pluriflorum. This position supports former reconstructions based on AFLP fingerprinting and sequence variation in cpDNA intergenic spacers and nrDNA ETSs [24,30] that all pointed towards the contribution of L. pluriflorum subsp. pluriflorum to an alleged allopolyploid origin of L. pseudosylvaticum, while they remained equivocal with respect to the donor of the other diploid genome to the latter. The allopolyploid nature of L. pseudosylvaticum receives considerable support from our present SNP-based analysis due to the obvious position of the three accessions of the species (1-05, 3-08, and 7-02) at the vertex of a parallelogram at their base in the NeighborNet network of Figure 2A. Following an interpretation of this parallelogram as an indication for L. pseudosylvaticum sharing one edge (genome) with L. pluriflorum/L. eliasii, one may feel justified in hypothesising the sharing of the other edge (genome) with one of the diploid species making up the left side of the network. Owing to the fact that all other members of this subgroup are presently not found in the Iberian Peninsula, L. gracilicaule—a diploid endemic to the region around Valencia in SE Spain—could then be the most probable candidate for the other parental taxon of L. pseudosylvaticum, if an extinct diploid may not have acted as such. The latter scenario receives plausibility from studies on other polyploid complexes that have encountered so-called ‘ghost (sub)genomes’ of extinct diploids in polyploid taxa (e.g., in Viola [53], in Fragaria [54], and in Brachypodium [55]).
In contrast to the evolutionary history of L. pseudosylvaticum, hypotheses about the formation of the other seven tetraploid taxa included in the present study remain even more obscure due to the nearly star-like structure in the right part of the NeighborNet network of Figure 2A. The position of the closely related diploid species L. gaudinii and L. vulgare (with its two subspecies L. vulgare subsp. vulgare and subsp. pyrenaicum) amongst accessions of the tetraploid taxa L. ircutianum (with its two subspecies L. ircutianum subsp. ircutianum and subsp. leucolepis) and L. cantabricum may point towards the participation of either of these two diploids (or a joint ancestor of both) in the formation of the latter. In the case of L. ircutianum subsp. ircutianum, it has been shown by Oberprieler et al. [28] that L. vulgare subsp. vulgare may have acted as the paternal partner in this allopolyploidisation and L. virgatum as the maternal partner [56]. At least the latter parentage receives little support from our present SNP-based reconstructions that locate L. virgatum quite distantly from all members of the L. vulgare cluster. Strangely enough, however, the cpDNA-based analysis of Greiner et al. [56] demonstrated that chloroplast haplotypes of L. virgatum (or haplotypes closely related to these) could not only be found in accessions of L. ircutianum subsp. ircutianum but also in some or even all representatives of L. ircutianum subsp. leucolepis, L. cantabricum, and L. crassifolium included in the mentioned study. Finally, chloroplast haplotypes characterised for L. delarbrei subsp. delarbrei and subsp. ruscinonense (sub L. monspeliense) were found to be closely related to L. halleri and L. vulgare, respectively [56]. In summary, these findings may either indicate real maternal parentages in the allopolyploidisation events (with subsequent assimilation of the nuclear genome towards the paternal parent) or events of chloroplast capture caused by hybridisation at the tetraploid level or between diploids and polyploids (tetraploids or taxa with even higher ploidy levels), indeed questioning the possibility of a comprehensive reconstruction of phylogenetic relationships among all taxa of the ploidy complex of Leucanthemum.
In genetic terms, SNP-based species-delimitation analyses carried out by weighted ensemble of random (k)k-means (WKM) clustering [49] and determination of the optimal number of clusters according to the Bayesian information criterion (BIC) and Calinski–Harbaz (CH) scores revealed significant discontinuities among six of the eight morphotaxon hypotheses (Figure 3). A lack of sufficient genetic differentiation was inferred for L. cantabricum and L. ircutianum subsp. ircutianum and for L. meridionale and L. delarbrei subsp. delarbrei, respectively. In both cases, this was found to correspond to the close positions of accessions from these taxon pairs in the NeighborNet network for the tetraploids (Figure 2B). In methodological respects, WKM clustering is a pattern-based, phenetic species-delimitation procedure not equivalent to process-based species-delimitation methods resting on the multispecies coalescent (MSC), which are used in many present-day delimitation studies based on molecular data in diploid taxon groups (e.g., [19,26,27] in Leucanthemum and [57] in Rhodanthemum). The lack of an MSC-based model adapted for polyploids hampers coalescent-based species delimitation here. However, since we have demonstrated in Leucanthemum diploids that pattern-based methods, such as consensus k-means (CKM; [58]) clustering, revealed genetic differentiation patterns equivalent to those derived according to a coalescent-based species-delimitation approach [19], we are confident that the differentiation patterns among tetraploids inferred with WKM clustering are trustworthy approximations to their genealogical structures.
It is obvious from both the NeighborNet network (Figure 2) and the WKM clustering (Figure 3) that L. delarbrei subsp. delarbrei and L. meridionale are closely related to each other. Despite the lack of significant genetic differentiation between them, as indicated by the BIC and CH scores, the two taxa appear to be genetically homogenous. This is remarkable due to the dispersed origins of accessions of the former taxon from four different locations on two volcanic mountain massifs in the southern Massif Central, France (Puy de Sancy, Monts du Cantal), while the latter is endemic to Puy de Wolf, a serpentine mountain c. 65 km SW of the Monts du Cantal. The situation is different for the other taxon pair, for which genetic differentiation was found to be not significant according to BIC and CH scores. In this case, L. cantabricum, with four accessions from two populations in NW Spain (Galicia), is also distinct but nested in a highly diverse cluster representing L. ircutianum subsp. ircutianum, with eight accessions from eight populations sampled throughout the European range of the taxon.
Two circumstances, however, are considered noteworthy here in evaluating the genetic/genealogical relationship of the two taxa: (a) The two populations sampled for L. cantabricum in the present analysis are the westernmost ones in the distributional range of the species that is mainly distributed in the W Pyrenean mountains and the Cordillera Cantábrica in N Spain [23], the locus classicus of the taxon being around 250 km to the east of the sampled populations. (b) In his revision of the genus for the Iberian Peninsula, Vogt [22] considered the stronger dissected leaves of L. cantabricum (sub L. ircutianum subsp. cantabricum) as being the main difference between this taxon and L. ircutianum subsp. ircutianum. The author, however, reports also that the morphological variation observed in the former taxon is considerable and that morphologically intermediate individuals with respect to the latter are observed frequently. Sampling of accessions from the centre of the distributional range of L. cantabricum (especially from the locus classicus) may reveal more pronounced genetic differences to L. ircutianum subsp. ircutianum, and the two populations sampled for the present study may turn out to be intermediate forms rather than pure representatives of L. cantabricum. Nevertheless, the undeniable closeness of the two taxa and their connectedness via intermediates will remain an unequivocal genealogical pattern.
4.2. Morphological Patterns
Leaf morphology—especially leaf outline and leaf dissection—is of paramount importance for taxon delimitation and taxon determination in Leucanthemum. As demonstrated by many taxonomic treatments of the genus in floras of European countries (e.g., Flora Iberica [23], Flora Gallica [59], and Flora d´Italia [60]), other morphological features, such as the colour of margins of involucral bracts, plant size, number of capitula per stalk, and achene size, come only second to these characters. Therefore, we felt justified in utilising leaf morphology as the sole proxy for morphological variation in the study group, especially because leaf-morphological features could be easily and objectively inferred using the applied machine-learning techniques and images of digitised herbarium specimens. These techniques allowed us to analyse 285 representative specimens and more than 1300 individual leaves across the study group in terms of leaf dissection and leaf shape in a time-effective and reproducible manner and to test for significant differences between pairs of the eight morphotaxa. We are fully aware of the fact that significant differences in these tests do not indicate the suitability of these characteristics as sole taxon-diagnostic features, as all taxa show considerable variation and overlap broadly in leaf characteristics. However, the main motivation for the inclusion of morphological characteristics in species-delimitation analyses is not for practical reasons of applicability (determinability), but—as in the case of genetic/genealogical features—for the assessment of morphological discontinuities as potential proxies for the evolutionary independence of lineages and their reciprocal reproductive isolation.
When compared across all possible pair-wise test combinations, leaf dissection (as measured by LDI) discriminates among the eight morphotaxa more profoundly than leaf outline (measured in elliptic Fourier analyses and tested for significance either with a permutation or an NPMANOVA approach): while only three (11%) of the 28 possible pair-wise comparisons of LDIs revealed no significant differences between taxa, 14 (50%) and 12 (43%) of the tests addressing leaf-shape differences showed non-significant results. We think that these contrasting results do not only have a biological reason, but also a methodological one: in its present and here-applied formulation (comparison of the length of leaf outline with leaf area), LDI does not only measure the intensity of leaf incision, but also the deviation of the leaf shape from a perfect circle. That means that it also captures leaf shape. In contrast, elliptic Fourier analysis (EFA) not only exclusively captures the outline of leaves, but also (with higher harmonics) the subdivision of the leaf lamina. However, since parameters gained with the latter harmonics of the EFA are down-weighted against parameters from earlier ones in a principal component analysis (PCA), it may be justified to consider both measures applied here (i.e., LDI and EFA) as overlapping with respect to what they capture in terms of leaf morphology, but with a tendency towards assessment of leaf dissection with the former and leaf outline with the latter.
Discrepancies between LDIs and EFA descriptors of leaf morphology were found to be especially pronounced when L. meridionale was included in the pair-wise comparisons: while six of the seven comparisons concerning LDIs revealed significance, the majority of EFA-based comparisons resulted in non-significant results. The former is quite reasonable when the habit of the species is studied both in its natural habitat and in a herbarium: compared with all other tetraploid taxa of Leucanthemum, L. meridionale, with its reduced scape size and its filigree leaves, resembles more the diploid representatives of the genus than the tetraploid ones. This led Tison and de Foucault [59] among others, who did not know about the tetraploid nature of this taxon, to speculate on the close (and allegedly) hybridogenic relationship of the species to L. vulgare and L. graminifolium. The somewhat reduced habit of L. meridionale may be a consequence of the exceptional habitat preference of the species for serpentine soils; the lack of a significant difference in LDIs with respect to L. delarbrei subsp. delarbrei, which is found on volcanic soils further north and is genetically closely related with it, however, may also point towards a close phylogenetic relationship between these two tetraploid Leucanthemum taxa.
A further noteworthy similarity in terms of leaf-morphological descriptors was observed between L. ircutianum subsp. ircutianum and subsp. leucolepis, for which all three tests remained non-significant (Table 1). As described by Oberprieler et al. [31], the former wide-spread taxon and the latter amphi-Adriatic one (including the S Italian L. ircutianum subsp. asperulum as a synonym) are allopatrically distributed in the Apennine Peninsula but sympatrically distributed in the Balkan Peninsula and form morphologically intermediate forms through hybridisation in overlapping regions of their distribution ranges. The main difference between the two taxa is in the colouration of the margins of involucral bracts, which are black to dark-brown in subsp. ircutianum and hyaline in subsp. leucolepis. In genetic respects, the two taxa represent two significantly distinct (Figure 3), albeit closely related (Figure 2), clusters.
4.3. Ecological and Geographical Patterns
As in the case of pair-wise statistical testing of leaf-morphological aspects, testing for significantly non-overlapping distribution ranges (Table 3) and ecological niches (Table 2) among the tetraploid Leucanthemum morphotaxa did not reveal complete and therefore diagnostic separation in these respects (strict allopatry or non-overlapping ecological niches); it rather led to the detection of patterns of bimodality in the spatial and ecoclimatological distributions of the species pairs compared. Therefore, the overwhelming number of significant test results (even after Bonferroni correction for multiple testing) for both geographical (2 of 21 tests) or ecological (also 2 of 21 tests) differences between species pairs may just represent tendencies in spatial or ecoclimatological differentiation and not complete discontinuities. Nevertheless, when looking at the distribution ranges of the eight tetraploid morphotaxa under study (Figure 1) and adding information from taxon descriptions concerning their ecological behaviour (e.g., [22] for the taxa of the Iberian Peninsula), it becomes obvious that the mentioned test results understate rather than hyperbolise differentiation patterns.
In geographical respects, the distribution ranges of the eight morphotaxa clearly follow an allopatric pattern, with exceptions being the observed non-significant differentiation between L. ircutianum subsp. ircutianum and L. delarbrei subsp. ruscinonense on the one hand and L. cantabricum and L. crassifolium on the other. As in the case of the point-endemic L. meridionale, for which statistical testing for geographical and ecological differences was not possible due to the restricted distribution range of the species, we think, however, that these test results are strongly scale-dependent and more on the conservative than on the oversensitive side. In the case of the taxon pair of L. cantabricum and L. crassifolium, the two species show a clear allopatric distribution on a small scale, with the former being restricted to the mountains of the Cordillera Cantábrica and adjacent regions [22,23] and the latter to littoral habitats of the N Spanish coast. In the other cases, the wide-spread (and constantly further spreading) L. ircutianum subsp. ircutianum encloses the geographically restricted L. delarbrei subsp. ruscinonense and L. meridionale in a more parapatric than allopatric pattern, which may be responsible for the non-significant test result. For all three taxon pairs, however, ecological differences were either revealed by our tests on niche overlap (L. ircutianum subsp. ircutianum vs. L. delarbrei subsp. ruscinonense) or by small-scale habitat differences—especially in edaphic factors—that have not been captured by the geography-based ecoclimatological niche modelling underlying the test-setup, L. meridionale being adapted to serpentine soils and L. crassifolium to saliferous coastal habitats.
4.4. Integration of Sources of Evidence
Species delimitation in plants—especially in hybridising plant groups (syngameons) and polyploid complexes—is a problematic matter [18]. The applicability of a strict biological species concept (BSC) has been denied by the majority of botanists owing to the sheer frequency of hybridisation in the plant kingdom and the numerous groups with agamospermic (apomictic) reproduction. Additionally, the usage of actual (hybridisation) or potential interbreeding (crossability) as a proxy for the evolutionary independence of lineages is deceptive. The lack of correlation between hybrid-formation capability and phylogenetic proximity in plants is exemplified by many examples of old and well-characterised lineages (species, sometimes even from different genera) that easily hybridise when brought into contact on the one hand and extremely young lineages (like autopolyploids) that are reproductively isolated instantly on the other, demonstrating that interfertility is a plesiomorphic character state [61]. Other species concepts, as enumerated by Zachos [62], share the problem that the criteria entertained in the different concepts differ in temporal sequence and relative importance across the tree of life due to the fact that speciation is a continuous process “over a timeframe that is too long to study from start to finish” (the ‘speciation continuum’; [63]). With these unsatisfactory consequences for biological classification, taxonomic decisions following a ‘unified species concept’ sensu De Queiroz [5] have considerable attraction because this concept shifts the focus away from properties being used for defining species towards their usage as indicators for independently evolving metapopulation lineages (species). Additionally, it allows for and demands the integration of multiple sources of evidence for taxonomic ranking.
Here, we follow a procedural protocol for species-rank decisions designed by Oberprieler [16] that is based on the integration of morphology, ecology, and geography, as proposed by von Wettstein [64], and expanded for the inclusion of an additional genetic/genealogical axis (the ‘Wettstein tesseract’). Conceptually, it follows the evolutionary species concept (EvoSC) of Wiley [65], who defined species as “a single lineage of ancestral-descendent populations of organisms which maintains its identity from other such lineages and which has its own evolutionary tendencies and historical fate”, but addresses the major drawback of that definition, namely, that genealogy-based, multispecies-coalescent species-delimitation methods tend to mistake population structures for species boundaries [66]. By adding geographical, ecological, and morphological sources of evidence, the ‘Wettstein tesseract’ provides a tool for conceptualising decisions on species or subspecific ranks and has been successfully applied to the diploids of Leucanthemum—the ”warps and wefts” of this polyploid complex [19].
Despite the somewhat pessimistic outlook on the phylogenetics of the Leucanthemum polyploidy complex, the present study is extremely helpful in terms of providing a pattern-based species delimitation at the tetraploid level and its integrative taxonomical conceptualisation. Aiming at a reproducible, objective classification of eight morphotaxon hypotheses in terms of delimitation and ranking, we have analysed genetic, ecological, and morphological variation together with information from taxon distributions, summarised in Figure 4. In the diagram, statistically significant differences found between pairs of taxon hypotheses are depicted in red, while the lack of significant discontinuities is shown in green. Owing to the restricted distribution range of L. meridionale, which is a point endemic of a serpentine mountain in S France (Aveyron, Rodez, Decazeville, Firmi, and Puy de Wolf), this taxon has been omitted from pair-wise comparisons that are dependent on modelling procedures based on spatial distribution data comprising more than a single raster cell (scaled to a resolution of 2.5 arc minutes). Due to its described endemicity, L. meridionale (accessions 323-03, -04, and -05) is allopatrically distributed with all other tetraploid Leucanthemum taxa except L. ircutianum subsp. ircutianum, which is found in parapatry on non-serpentine soils surrounding Puy de Wolf (and sampled with accession 343-01). Its ecological niche is uniquely determined by edaphic factors connected to serpentine soils (only paralleled by the diploid L. pluriflourum subsp. gallaecicum in NW Spain and the decaploid L. pachyphyllum in N Italy) rather than climatic ones.
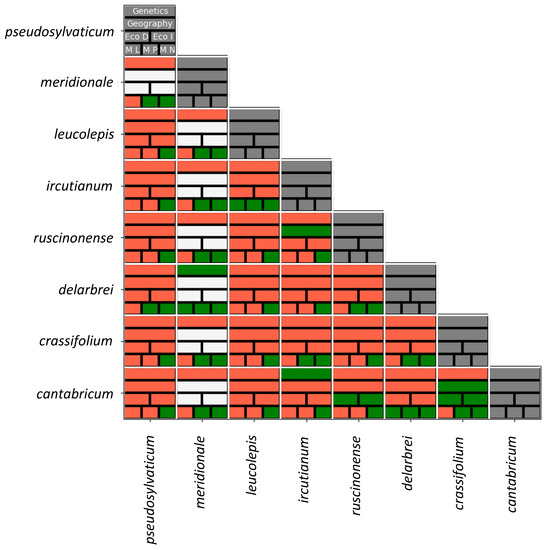
Figure 4.
Taxon pair-wise differences in genealogy, geography, ecology (EcoD: Schoener´s D; EcoI: Warren´s I), and morphology [ML: Welch’s test on leaf-dissection indices (LDI); MP: permutation test on leaf shape (Elliptic Fourier Analysis, EFA), MN: NPMANOVA on leaf shape (EFA)]. Significant differences are marked in red, non-significant ones in green; white cells indicate lack of testability due to the restricted geographical range of L. meridionale.
Application of the ‘Wettstein tesseract’ tool to the present study group of Leucanthemum tetraploids in seeking the most reasonable ranking for closely related species hypotheses (morphotaxa) leads to the following reasoning:
(a) Lecanthemum crassifolium (N Spain), L. delarbrei subsp. ruscinonense (E Pyrenees and northeastern foothills), and L. pseudosylvaticum (W Iberian Peninsula) are best considered as being genetically closely related but independent lineages (Figure 2 and Figure 3) that merit species ranking due to their significant ecological (Table 2) and leaf-morphological differences (Table 1) that have evolved in strict allopatry. While for L. pseudosylvaticum at least one parental diploid species is known (L. pluriflorum subsp. pluriflorum; [24,30]), the matter is unsettled as yet for the other two tetraploid lineages. Following haplotype-network reconstructions for the whole genus [56], L. pluriflorum subsp. pluriflorum, as the maternal diploid ancestor of these, could be excluded definitively, and L. virgatum appears to be the most probable candidate for this role in L. crassifolium, while L. delarbrei subsp. ruscinonense (sub L. monspeliense in [56]) shares its chloroplast haplotype with a large number of diploid taxa: L. ageratifolium (NE Spain, SW France; sub L. vulgare subsp. pujiulae in [56]), L. burnatii (S France), L. eliasii (N Spain; sub L. vulgare subsp. eliasii in [56]), L. gaudinii (Alps, Carpathian Mountains), L. graminifolium (S France), L. pluriflorum subsp. cantabricum (N Spain; sub L. gaudinii subsp. cantabricum in [56]), and both subspecies of L. vulgare (subsp. vulgare, widespread; subsp. barrelieri, Pyrenees).
(b) Leucanthemum delarbrei subsp. delarbrei (northern Massif Central, France) and L. meridionale (western Massif Central, France) are allopatrically distributed sister groups (Figure 2) that lack sufficient genetic differentiation to merit species ranking (Figure 3) and show non-significantly different leaf morphology (Table 1) but exhibit edaphic differences (the former growing on siliceous rocks of old volcano cones, the latter on serpentine soils). Therefore, subspecific ranking appears appropriate for these two taxa. The chloroplast haplotype found in one of two accessions of L. delarbrei subsp. delarbrei surveyed by Greiner et al. [56] was found to be closely related to the diploid L. halleri and may point towards a phylogenetic relationship with this Alpine species, while information on a chloroplast haplotype in L. meridionale is still lacking.
(c) Finally, L. cantabricum and the two subpecies of L. ircutianum also show close genetic relationships in the NeighborNet network of Figure 2. Here, however, genetic differentiation patterns (Figure 3) do not correspond to the hitherto proposed classification because L. ircutianum subsp. ircutianum shows a closer genetic similarity with L. cantabricum than with L. ircutianum subsp. leucolepis. When following the conceptual framework of species-rank decision making with the ‘Wettstein tesseract’, the significant differentiations between L. ircutianum subsp. ircutianum and subsp. leucolepis in genetic, geographical, and ecoclimatological aspects would argue for an acknowledgement of the two taxa at species level (the lack of leaf-morphological differences is counterbalanced by differences in the colours of the margins of the involucral bracts). The main argument for treating the two entities as independent species, following the logic of von Wettstein [64], is that the ecoclimatiological differences between the two would allow them to keep their lineage identity when allopatry would change into sympatry in the future. Oberprieler et al. [31] have shown, however, that there are mixed stands of the two taxa in Central Italy, where hybridisation and backcrossing leads to introgressive hybrid swarms, with a complete blurring of taxon limits. Since the situation seems comparable in the Western Balkan Peninsula, where the two taxa grow sympatrically and intermediate forms are found [31], we propose to keep the two entities at subspecific rank for conservative reasons aiming at minimizing taxonomic and nomenclatural disruptions if not unequivocally demanded by the underlying data and analyses.
Subspecific ranking under L. ircutianum, on the other hand, is less equivocal for L. cantabricum (Cordillera Cantábrica in N Spain) following the here-presented results: the two taxa are allopatrically distributed and show ecoclimatological and morphological differences (more strongly dissected leaves in L. cantabrica) but lack genetic differentiation (Figure 4). The observation of morphologically intermediates by Vogt [22] further argues for hybridisation between the two taxa and the appropriateness of subspecific ranking.
5. Taxonomic Treatment
With the described conceptual framework at hand, species delimitation in the group of tetraploid Leucanthemum taxa under study could be put into effect as follows:
(1) Leucanthemum crassifolium (Lange) Lange in Willk. & Lange, Prodr. Fl. Hispan. 2: 96. 1865 ≡Leucanthemum pallens var. crassifolium Lange in Vidensk. Meddel. Dansk Naturhist. Foren. Kjøbenhavn, ser. 2, 3: 77. 1861 ≡Leucanthemum ircutianum subsp. crassifolium (Lange) Vogt in Ruizia 10: 127. 1991—Lectotype (Vogt, Ruizia 10: 128. 1991): In rupibus maritimis ad Portugalete, Cantabria, Oct. 1851, Herb. Joh. Lange (C! (C10007128)).
Notes.—Endemic to the Cantabrian coast between Asturias and the Basque region (NE Spain and SW France). Habitats are coastal rocks and salt-influenced coastal slopes from sea level to 20 m. Leucanthemum crassifolium is characterised by succulent leaves (Figure 5) and involucral bracts with dark-brown hyaline margins of involucral bracts. It was first described as a variety of L. pallens by Lange [67] and subsequently raised to subspecific [22] and specific rank [23].
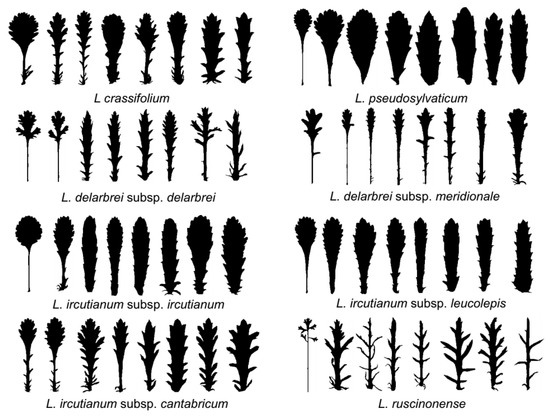
Figure 5.
Silhouettes of basal and cauline leaves from different accessions of the eight tetraploid Leucanthemum taxa under study.
(2a) Leucanthemum delarbrei Timb.-Lagr. (subsp. delarbrei) in Mém. Acad. Sci. Clermont-Ferrand 20: 508. 1878.—Lectotype: Not yet designated. =Chrysanthemum leucanthemum var. pinnatifidum Lecoq. & Lamotte, Cat. Pl. Plateau Central: 227. 1847 ≡Leucanthemum vulgare var. pinnatifidum (Lecoq. & Lamotte) Briq. & Cavill. in Burnat, Fl. Alpes Marit. 6: 91. 1916 ≡Leucanthemum ircutianum var. pinnatifidum (Lecoq. & Lamotte) D. Löve & J.-P. Bernard in Svensk. Bot. Tidskr. 53: 444. 1959.—Ind. loc.: “AR.- Mont Dore; pâturages et pentes herbeuses de Chaudefour, bords du chemin de Sancy à Vassivière. Bozat! AR.”—Lectotype (designated here by Vogt & Oberprieler): Mont Dore, Sancy, 15 (..) 1844, leg. H. Lecoq & M. Lamotte (P! (P00729975)).
Notes.—Endemic to the siliceous rocks of the old volcano cones of the central Massif Central in France (Monts Dore, Monts du Cantal). Its habitats are siliceous rocks and meadows between 1550 m and 1750 m. Leucanthemum delarbrei subsp. delarbrei is characterised by strongly dissected leaves, from pinnatifid to bipinnatisect (Figure 5), and dark- to light-brown hyaline margins of involucral bracts.
(2b) Leucanthemum delarbrei subsp. meridionale (Legrand) Oberpr., T. Ott & Vogt, comb. nov. ≡Leucanthemum meridionale Legrand in Bull. Soc. Bot. France 28: 56. 1881 (basionym) ≡Leucanthemum vulgare var. (‘χ’) meridionale (Legrand) Rouy, Fl. France 8: 274. 1903 ≡Chrysanthemum leucanthemum f. (‘g’) meridionale (Legrand) Fiori in Fiori & Béguinot, Fl. Italia 3: 239. 1903 ≡Leucanthemum vulgare subsp. meridionale (Legrand) Nyman, Consp. Fl. Eur., Suppl. 2: 169. 1889.—Ind loc.: “Habite dans les interstices des rochers serpentineuses du puy de Wolf, près de Firmy (Aveyron); fleurit de fin mai à juillet. Je reçus cette plante en 1879 ... leg. F(rère) Saltel (Baenitz, Herbarium eur. n° 4184).”—Lectotype (designated here by Vogt & Oberprieler): Puy de Wolf, pr. Firmy; mai, juin 1879 (Aveyron).—Gallia merid., Saltel.—Comm. Le Grand (P! (P00729957)).
Notes.—Endemic to the Puy de Wolf (France, Aveyron, Rodez, Decazeville, Firmi), a serpentine mountain in the western Massif Central. It is found in dry and open, south-facing grassland on serpentine soil between 400 m and 600 m. Leucanthemum meridionale is characterised by its lanky habitus resembling the diploid L. vulgare or a member of the genus Leucanthemopsis with quite narrow leaves and cuneate leaf bases (Figure 5).
(3a) Leucanthemum ircutianum DC. (subsp. ircutianum), Prodr. 6: 47. 1838.—Lectotype (Vogt, Ruizia 10: 119. 1991): In pratis, 1828, Turczaninoff a Irkoutsk, Turcz.: 1830 (G-DC! (G00451151)).
Notes.—Besides the diploid L. vulgare Lam., this taxon is the most widely distributed species of the genus [22]. It is present in nearly all countries of the Euro + Med region [20] but has also been introduced into all continents except Antarctica. Habitats are anthropogenetically influenced and include meadows and roadsides. Leucanthemum ircutianum subsp. ircutianum is morphologically very similar to L. ircutianum subsp. leucolepis (see Figure 5) but can be differentiated by the dark-brown hyaline margins of involucral bracts.
(3b) Leucanthemum ircutianum subsp. cantabricum (Sennen) Vogt in Ruizia 10: 121. 1991 ≡Leucanthemum cantabricum Sennen, Diagn. Nouv.: 50. 1936 ≡Leucanthemum vulgare subsp. cantabricum Sennen, Diagn. Nouv. 50. 1936 (nom. altern.).—Lectotype (Vogt, Ruizia 10: 121. 1991): Santander: La Calda de Besaya, rochers silliceux humides, 5.6.1927, E. Leroy (BC-Sennen!).
Notes.—Distributed between the western foothills of the Pyrenees (Spain and France), along the northern slopes of the Cantabrian Mountains to Asturias and Galicia in the west. Habitats include road and meadow margins, meadows, and pastures; the elevational distribution ranges from sea level to 800 m. Leucanthemum cantabricum is characterised by dissected (pinnatisect to pinnatipartite), non-succulent leaves (Figure 5) and dark-brown hyaline margins of involucral bracts. It was described as a species by Sennen [68] but reduced to subspecific rank under L. ircutianum by Vogt [22] before being reacknowledged at species rank by Vogt [23].
(3c) Leucanthemum ircutianum subsp. leucolepis (Briq. & Cavill.) Vogt & Greuter in Willdenowia 33: 41. 2003 ≡Leucanthemum leucolepis (Briq. & Cavill.) Gajić in Josifović, Fl. SR Srbije 9: 185. 1977 ≡Leucanthemum vulgare subsp. leucolepis Briq. & Cavill. in Burnat, Fl. Alpes Marit. 6: 93. 1916 ≡Chrysanthemum leucanthemum subsp. leucolepis (Briq. & Cavill.) Schinz & Thell., Fl. Schweiz, ed. 4, 1: 685. 1923 ≡Leucanthemum leucolepis (Briq. & Cavill.) Horvatić in Acta Bot. Croat. 22: 214. 1963, nom. inval. (Turland et al. 2018: Art. 41.5) ≡Leucanthemum pallens subsp. leucolepis (Briq. & Cavill.) Faverger in Anales Inst. Bot. Cavanilles 32: 1236. 1975.—Lectotype (Oberprieler et al., Bot. J. Linn. Soc. 199: 844. 2022): Flora Italica Exsiccata, curantibus Adr. Fiori, A. Béguinot, R. Pampanini, number 175—Etruria, Prov. di Firenze, Vallombrosa, in pratis, alt. 900–1000 m., solo pingui, siliceo, 19.6.1904, A. Fiori (G-BU! (G00848032)).
Notes.—A taxon with an amphi-Adriatic distribution, with populations throughout Italy and along the Adriatic coast of the Balkan Peninsula. The taxon was described as a subspecies of the diploid L. vulgare but was subsequently considered a subspecies of the hexaploid L. pallens due to its white hyaline margins of involucral bracts or seen as an independent species. The observation of an allopatric distribution with L. ircutianum subsp. ircutianum as more Mediterranean facies of the species, together with the observation of hybrid individuals and hybrid swarms in areas of joint occurrence of the two tetraploid taxa in the Apennine and Balkan Peninsulas by Oberprieler et al. [31] argued for treatment as a subspecies of L. ircutianum. In contrast to L. ircutianum subsp. ircutianum, it has paler hyaline margins of the involucral bracts.
(4) Leucanthemum pseudosylvaticum (Vogt) Vogt & Oberpr. in Ann. Bot. (Oxford), n.s., 111: 1121. 2013 ≡Leucanthemum ircutianum subsp. pseudosylvaticum Vogt in Ruizia 10: 134. 1991.—Holotype: Portugal, Distrito Porto, Serra do Marão, Mesão Frio-Amarante, feuchter Hang südlich der Paßhöhe, ca. 800 m, 20.07.1986, R. Vogt 4711 & E. Bayón (M! (M-0030173)).
Notes.—Distributed throughout the western part of the Iberian Peninsula (Portugal and Spain). Its habitats comprise road margins and slopes, margins of creeks, and ditches, at an elevational range from 100 m to 1600 m. Leucanthemum pseudosylvaticum is characterised by leaves with proximally (sub)entire margins (Figure 5) and involucral bracts with pallid to light-brown hyaline margins. It was described by Vogt [22] as a subspecies of L. ircutianum but shown to merit species rank due to its evolutionary independence from the latter species by Oberprieler et al. [24] and Greiner et al. [29,30].
(5) Leucanthemum ruscinonense (Jeanb. & Timb.-Lagr.) Oberpr., T.Ott & Vogt, comb. et stat. nov. ≡Leucanthemum palmatum var. ruscinonense Jeanb. & Timb.-Lagr. in Mém. Acad. Sci. Toulouse, ser 8, 1(2): 192. 1879 (basionym) ≡Leucanthemum monspeliense var. ruscinonense (Jeanb. & Timb.-Lagr.) O. Bolòs & Vigo in Collect. Bot. (Barcelona) 17: 91. 1988 ≡Leucanthemum cebennense var. ruscinonense (Jeanb. & Timb.-Lagr.) Gaut., Catal. Rais. Fl. Pyr. Orient.: 232. 1898 ≡Leucanthemum delarbrei subsp. ruscinonense (Jeanb. & Timb.-Lagr.) Vogt, Florian Wagner & Oberpr., Fl. Iber. 16(3): 1866. 2019—Ind loc.: “... les Alberes ...”.—Holotype: ... la tour de la Massane dans la altura, Pyr. Orient., 21.5.1877, E. Timbal-Lagrave (TL-Timbal-Lagrave!).
Notes.—Endemic to the eastern Pyrenees (Spain and France) and the SW parts of the Massif Central (Haut Languedoc, Montagne Noire). Habitats comprise road margins, creeks, and stony slopes, between 250 m and 1900 m. It is characterised by strongly dissected leaves, pinnatifid to bipinnatisect (Figure 5), and dark- to light-brown hyaline margins. High variability in terms of leaf dissection is speculated to be the result of potential hybridisation. The taxon was, for a long time, considered part of L. monspeliense L. (e.g., [22]), which is also distributed in the Massif Central (Cevennes), but has a diploid chromosome number.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology12020288/s1. File ES01: List of herbarium specimens of Leucanthemum tetraploids housed at the Berlin Botanical Museum (B) and used for the morphological analyses of the present study. File ES02: List of collection localities of Leucanthemum tetraploids used in the ecological niche modelling and geographical overlap analyses of the present study. File ES03: Descriptions of the bioclimatological and edaphic variables used for ecological niche modelling. File ES04: List of samples used for the ddRAD analysis with information on voucher specimens in the Botanical Museum Berlin (B) and ploidy, collection localities, coordinates, and collectors. File ES05: Maps depicting predicted potential distribution ranges of the Leucanthemum diploids based on recent, last glacial maximum (LGM), and last interglacial (LIG) bioclimatic variables. File ES06: Maps depicting predicted potential distribution ranges of the Leucanthemum tetraploids based on recent, last glacial maximum (LGM), and last interglacial (LIG) bioclimatic variables. File ES07: Plots of the SNiPloid category proportions for all triplets (two diploid potential parent species and one tetraploid daughter species).
Author Contributions
Conceptualisation, C.O., T.O., and R.V.; methodology, T.O.; software, T.O.; validation, T.O. and C.O.; formal analysis, T.O.; investigation, T.O. and C.O.; resources, C.O. and R.V.; data curation, C.O. and R.V.; writing—original draft preparation, C.O., T.O., and R.V.; writing—review and editing, C.O. and R.V.; visualisation, C.O. and T.O.; supervision, C.O.; project administration, C.O.; funding acquisition, C.O. and R.V. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by grants from the German Research Foundation (DFG) in the frame of the SPP 1991 “Taxon-omics—New Approaches for Discovering and Naming Biodiversity” to C.O. (OB 155/13-1) and R.V. (VO 1595/3-1).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw ddRAD reads were deposited at NCBI (Bioproject PRJNA884628).
Acknowledgments
We thank Anja Heuschneider for preparing the samples for ddRADseq sequencing. The suggestions from Agnes Scheunert and Ulrich Lautenschlager, considering the analyses and the manuscript, were, as always, welcome and are very much appreciated. Points raised by the three reviewers of the present contribution are thankfully acknowledged and improved the manuscript considerably.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Stuessy, T.F. Ultrastructural data for the practicing plant systematist. Am. Zool. 1979, 19, 621. [Google Scholar] [CrossRef]
- Zachos, F.E.; Christidis, L.; Garnett, S.T. Mammalian species and the twofold nature of taxonomy: A comment on Taylor et al. Mammalia 2020, 84, 1. [Google Scholar] [CrossRef]
- Ence, D.D.; Carstens, B.C. SpedeSTEM: A rapid and accurate method for species delimitation. Mol. Ecol. Resour. 2011, 11, 473. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.L.; Carstens, B.C. Species delimitation using molecular data. In Species Problems and Beyond–Contemporate Issues in Philosophy and Practice; Wilkins, J.S., Zachos, F.E., Pavlinov, I.Y., Eds.; CRC Press: Boca Raton, FL, USA, 2022; pp. 145–159. [Google Scholar]
- De Queiroz, K. Species concepts and species delimitation. Syst. Biol. 2007, 56, 879. [Google Scholar] [CrossRef] [PubMed]
- Dayrat, B. Towards integrative taxonomy. Biol. J. Linn. Soc. 2005, 85, 407. [Google Scholar] [CrossRef]
- Will, K.W.; Mishler, B.D.; Wheeler, Q.D. The perils of DNA barcoding and the need for integrative taxonomy. Syst. Biol. 2005, 54, 844. [Google Scholar] [CrossRef]
- Guillot, G.; Renaud, S.; Ledevin, R.; Michaux, J.; Claude, J. A unifying model for the analysis of phenotypic, genetic, and geographical data. Syst. Biol. 2012, 61, 897. [Google Scholar] [CrossRef] [PubMed]
- Zapata, F.; Jiménez, I. Species delimitation: Inferring gaps in morphology across geography. Syst. Biol. 2012, 61, 179. [Google Scholar] [CrossRef]
- Vásquez-Cruz, M.; Vovides, A.P.; Sosa, V. Disentangling species limits in the Vauquelinia corymbosa complex (Pyreae, Rosaceae). Syst. Bot. 2017, 42, 1. [Google Scholar] [CrossRef]
- Solís-Lemus, C.; Knowles, L.L.; Ané, C. Bayesian species delimitation combining multiple genes and traits in a unified framework. Evolution 2015, 69, 492. [Google Scholar] [CrossRef]
- Hausdorf, B.; Hennig, C. Species delimitation and geography. Mol. Ecol. Resour. 2020, 20, 950. [Google Scholar] [CrossRef] [PubMed]
- Doyen, J.T.; Slobodchikoff, C.N. An operational approach to species classification. Syst. Zool. 1974, 23, 239. [Google Scholar] [CrossRef]
- Schlick-Steiner, B.C.; Steiner, F.M.; Seifert, B.; Stauffer, C.; Christian, E.; Crozier, R.H. Integrative taxonomy: A multisource approach to exploring biodiversity. Annu. Rev. Entomol. 2010, 55, 421. [Google Scholar] [CrossRef] [PubMed]
- Padial, J.M.; Miralles, A.; De la Riva, I.; Vences, M. The integrative future of taxonomy. Front. Zool. 2010, 7, 16. [Google Scholar] [CrossRef]
- Oberprieler, C. The Wettstein tesseract: A tool for conceptualising species-rank decisions and illustrating speciation trajectories. Taxon. [CrossRef]
- Sukumaran, J.; Holder, M.T.; Knowles, L.L. Incorporating the speciation process into species delimitation. PLoS Comput. Biol. 2021, 17, e1008924. [Google Scholar] [CrossRef] [PubMed]
- Hörandl, E. Novel approaches for species concepts and delimitation in polyploids and hybrids. Plants 2022, 11, 204. [Google Scholar] [CrossRef]
- Ott, T.; Schall, M.; Vogt, R.; Oberprieler, C. The warps and wefts of a polyploid complex: Integrative species delimitation of the diploid Leucanthemum (Compositae, Anthemideae) representatives. Plants 2022, 11, 1878. [Google Scholar] [CrossRef]
- Euro+Med (2006): Euro+Med PlantBase-the information resource for Euro-Mediterranean plant diversity. Available online: http://ww2.bgbm.org/EuroPlusMed/ (accessed on 24 March 2022).
- Meusel, H.; Jäger, E.J. Vergleichende Chorologie der Zentraleuropäischen Flora; Gustav Fischer Verlag: Jena, Germany; Stuttgart, Germany; New York, NY, USA, 1992; Volume 3. [Google Scholar]
- Vogt, R. Die Gattung Leucanthemum Mill. (Compositae—Anthemideae) auf der Iberischen Halbinsel. Ruizia 1991, 10, 1–261. [Google Scholar]
- Vogt, R. Leucanthemum Mill. In Flora Iberica; Benedí, C., Buira, A., Rico, E., Crespo, M.B., Quintanar, A., Aedo, C., Eds.; Real Jardín Botánico, CSIC: Madrid, Spain, 2019; Volume 16, pp. 1848–1880. [Google Scholar]
- Oberprieler, C.; Greiner, R.; Konowalik, K.; Vogt, R. The reticulate evolutionary history of the polyploid NW Iberian Leucanthemum pluriflorum clan (Compositae, Anthemideae) as inferred from nrDNA ETS sequence diversity and eco-climatological niche-modelling. Mol. Phylogenet. Evol. 2014, 70, 478. [Google Scholar] [CrossRef]
- Konowalik, K.; Wagner, F.; Tomasello, S.; Vogt, R.; Oberprieler, C. Detecting reticulate relationships among diploid Leucanthemum Mill. (Compositae, Anthemideae) taxa using multilocus species tree reconstruction methods and AFLP fingerprinting. Mol. Phylogenet. Evol. 2015, 92, 308. [Google Scholar] [CrossRef] [PubMed]
- Wagner, F.; Härtl, S.; Vogt, R.; Oberprieler, C. ‘Fix Me Another Marguerite!’: Species delimitation in a group of intensively hybridizing lineages of ox-eye daisies (Leucanthemum Mill., Compositae-Anthemideae). Mol. Ecol. 2017, 26, 4260. [Google Scholar] [CrossRef] [PubMed]
- Wagner, F.; Ott, T.; Zimmer, C.; Reichhart, V.; Vogt, R.; Oberprieler, C. ‘At the crossroads towards polyploidy’: Genomic divergence and extent of homoploid hybridization are drivers for the formation of the ox-eye daisy polyploid complex (Leucanthemum, Compositae-Anthemideae). New Phytol. 2019, 223, 2039. [Google Scholar] [CrossRef]
- Oberprieler, C.; Eder, C.; Meister, J.; Vogt, R. AFLP fingerprinting suggests an allopolyploid origin of two members of the Leucanthemum vulgare aggregate (Compositae, Anthemideae) in central Europe. Nord. J. Bot. 2011, 29, 370. [Google Scholar] [CrossRef]
- Greiner, R.; Oberprieler, C. The role of inter-ploidy block for reproductive isolation of the diploid Leucanthemum pluriflorum Pau (Compositae, Anthemideae) and its tetra- and hexaploid relatives. Flora 2012, 207, 629. [Google Scholar] [CrossRef]
- Greiner, R.; Vogt, R.; Oberprieler, C. Evolution of the polyploid north-west Iberian Leucanthemum pluriflorum clan (Compositae, Anthemideae) based on plastid DNA sequence variation and AFLP fingerprinting. Ann. Bot. 2013, 111, 1109. [Google Scholar] [CrossRef]
- Oberprieler, C.; Conti, F.; Dorfner, M.; Eder, S.M.; Heuschneider, A.; Ott, T.; Scheunert, A.; Vogt, R. The taxonomy of Leucanthemum ircutianum (Asteraceae, Anthemideae) in the Apennine Peninsula based on AFLP fingerprinting, plastid DNA sequence variation and eco-climatological niche reconstruction. Bot. J. Linn. Soc. 2022, 194, 830. [Google Scholar] [CrossRef]
- Kuhl, F.P.; Giardina, C.R. Elliptic Fourier features of a closed contour. Comput. Graph. Image Process. 1982, 18, 236. [Google Scholar] [CrossRef]
- Van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. scikit-image: Image processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef] [PubMed]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.; Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Solymos, P.; Stevens, M.; Szoecs, E.; et al. Vegan: Community ecology package–R package version 2.6-4, 2022. Available online: https://CRAN.R-project.org/package=vegan (accessed on 31 January 2023).
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302. [Google Scholar] [CrossRef]
- Hengl, T.; Jesus, J.M.; de Heuvelink, G.B.M.; Gonzalez, M.R.; Kilibarda, M.; Blagotić, A.; Shangguan, W.; Wright, M.N.; Geng, X.; Bauer-Marschallinger, B.; et al. SoilGrids250m: Global gridded soil information based on machine learning. PLoS ONE 2017, 12, e0169748. [Google Scholar] [CrossRef]
- Hijmans, R. Raster: Geographic Data Analysis and Modeling–R package Version 3.6-11. 2022. Available online: https://CRAN.R-project.org/package=raster (accessed on 31 January 2023).
- Warren, D.L.; Matzke, N.J.; Cardillo, M.; Baumgartner, J.B.; Beaumont, L.J.; Turelli, M.; Glor, R.E.; Huron, N.A.; Simões, M.; Iglesias, T.L.; et al. ENMTools 1.0: An R package for comparative ecological biogeography. Ecography 2021, 44, 504. [Google Scholar] [CrossRef]
- Phillips, S.; Dudík, M.; Schapire, R. Maxent Software for Modeling Species Niches and Distributions, Version 3.4.4. 2021. Available online: http://biodiversityinformatics.amnh.org/open_source/maxent/ (accessed on 31 January 2023).
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Dickson, E.E. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon 1987, 36, 715. [Google Scholar] [CrossRef]
- Eaton, D.A.R.; Overcast, I. ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 2020, 36, 2592. [Google Scholar] [CrossRef]
- Mastretta-Yanes, A.; Arrigo, N.; Alvarez, N.; Jorgensen, T.H.; Piñero, D.; Emerson, B.C. Restriction site-associated DNA sequencing, genotyping error estimation and de novo assembly optimization for population genetic inference. Mol. Ecol. Resour. 2015, 15, 28. [Google Scholar] [CrossRef]
- Joly, S.; Bruneau, A. Incorporating allelic variation for reconstructing the evolutionary history of organisms from multiple genes: An example from Rosa in North America. Syst. Biol. 2006, 55, 623. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; He, S.; Lin, Z.; Yang, F.; Zhou, Q.; Zhou, X. An adaptive robust semi-supervised clustering framework using weighted consensus of random k-means ensemble. IEEE Trans. Knowl. Data Eng. 2019, 33, 1877–1890. [Google Scholar] [CrossRef]
- Peralta, M.; Combes, M.C.; Cenci, A.; Lashermes, P.; Dereeper, A. SNiPloid: A utility to exploit high-throughput SNP data derived from RNA-Seq in allopolyploid species. Int. J. Plant Genom. 2013, 2013, 890123. [Google Scholar] [CrossRef] [PubMed]
- Wagner, N.D.; He, L.; Hörandl, E. Phylogenomic relationships and evolution of polyploid Salix species revealed by RAD sequencing data. Front. Plant Sci. 2020, 11, 1077. [Google Scholar] [CrossRef]
- He, L.; Wagner, N.D.; Hörandl, E. Restriction-site associated DNA sequencing data reveal a radiation of willow species (Salix L., Salicaceae) in the Hengduan Mountains and adjacent areas. J. Syst. Evol. 2021, 59, 44. [Google Scholar] [CrossRef]
- Marcussen, T.; Heier, L.; Brysting, A.K.; Oxelman, B.; Jakobsen, K.S. From gene trees to a dated allopolyploid network: Insights from the angiosperm genus Viola (Violaceae). Syst. Biol. 2015, 64, 84. [Google Scholar] [CrossRef]
- Kamneva, O.K.; Syring, J.; Liston, A.; Rosenberg, N.A. Evaluating allopolyploid origins in strawberries (Fragaria) using haplotypes generated from target capture sequencing. BMC Evol. Biol. 2017, 17, 1. [Google Scholar] [CrossRef]
- Sancho, R.; Inda, L.A.; Dáz-Pérez, A.; Des Marais, D.L.; Gordon, S.; Vogel, J.P.; Lusinska, J.; Hasterok, R.; Contreras-Moreira, B.; Catalán, P. Tracking the ancestry of known and ‘ghost’ homeologous subgenomes in model grass Brachypodium polyploids. Plant J. 2022, 109, 1535. [Google Scholar] [CrossRef]
- Greiner, R.; Vogt, R.; Oberprieler, C. Phylogenetic studies in the polyploid complex of the genus Leucanthemum (Compositae, Anthemideae) based on cpDNA sequence variation. Plant Syst. Evol. 2012, 298, 1407. [Google Scholar] [CrossRef]
- Wagner, F.; Ott, T.; Schall, M.; Lautenschlager, U.; Vogt, R.; Oberprieler, C. Taming the Red Bastards: Hybridisation and species delimitation in the Rhodanthemum arundanum-group (Compositae, Anthemideae). Mol. Phylogenet. Evol. 2020, 144, 106702. [Google Scholar] [CrossRef]
- Monti, S.; Tamayo, P.; Mesirov, J.; Golub, T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn. 2003, 52, 91. [Google Scholar] [CrossRef]
- Tison, J.M.; de Foucault, B. Flora Gallica: Flore de France; Biotope: Mèze, France, 2014. [Google Scholar]
- Lo Presti, R.M.; Oberprieler, C.; Vogt, R. Tribus VI–Anthemideae (gen. 56, 58-73, 75-79). In Flora d´Italia, 2nd, 3rd ed.; Pignatti, S., Ed.; Edagricole: Milano, Italy, 2018; pp. 815–881. [Google Scholar]
- Seberg, O. Genome analysis, phylogeny, and classification. Plant Syst. Evol. 1989, 166, 159. [Google Scholar] [CrossRef]
- Zachos, F.E. Species Concepts in Biology; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Stankowski, S.; Ravinet, M. Defining the speciation continuum. Evolution 2021, 75, 1256. [Google Scholar] [CrossRef] [PubMed]
- Von Wettstein, R. Grundzüge der Geographisch-Morphologischen Methode der Pflanzensystematik; Gustav Fischer Verlag: Jena, Germany, 1898. [Google Scholar]
- Wiley, E.O. The evolutionary species concept reconsidered. Syst. Biol. 1978, 27, 17. [Google Scholar] [CrossRef]
- Sukumaran, J.; Knowles, L.L. Multispecies coalsecent delimits structure, not species. Proc. Natl. Acad. Sci. USA 2017, 114, 1607. [Google Scholar] [CrossRef] [PubMed]
- Lange, J.M. Pugillus plantarum imrimis hispanicarum quas in itinere 1851-52 legit Joh. Lange, Vidensk. Meddel. Dan. Naturhist. Kjöbenhaven 1861, 3, 33–116. [Google Scholar]
- Sennen, F. Diagnoses des Nouveautés Parues dans les Exsiccata Plantes d´Espagne et du Maroc de 1928 à Imp; Anglada: Vic, Spain, 1936. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).