Non-Ionic Osmotic Stress Induces the Biosynthesis of Nodulation Factors and Affects Other Symbiotic Traits in Sinorhizobium fredii HH103
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. β-Galactosidase Assay
2.3. Nod Factor Purification, Biological Activity, and Structural Determination by UHPLC-HRMS/MS
2.4. RNA Extraction and Sequencing
2.5. RNA-seq Data Analysis
2.6. Quantification of Indole Acetic Acid (IAA) Production
2.7. Motility Assay
2.8. PAGE Analysis of LPS and KPS
2.9. AHL Well Diffusion Assay
2.10. HPLC and Mass Spectrometry Analysis of AHL
3. Results
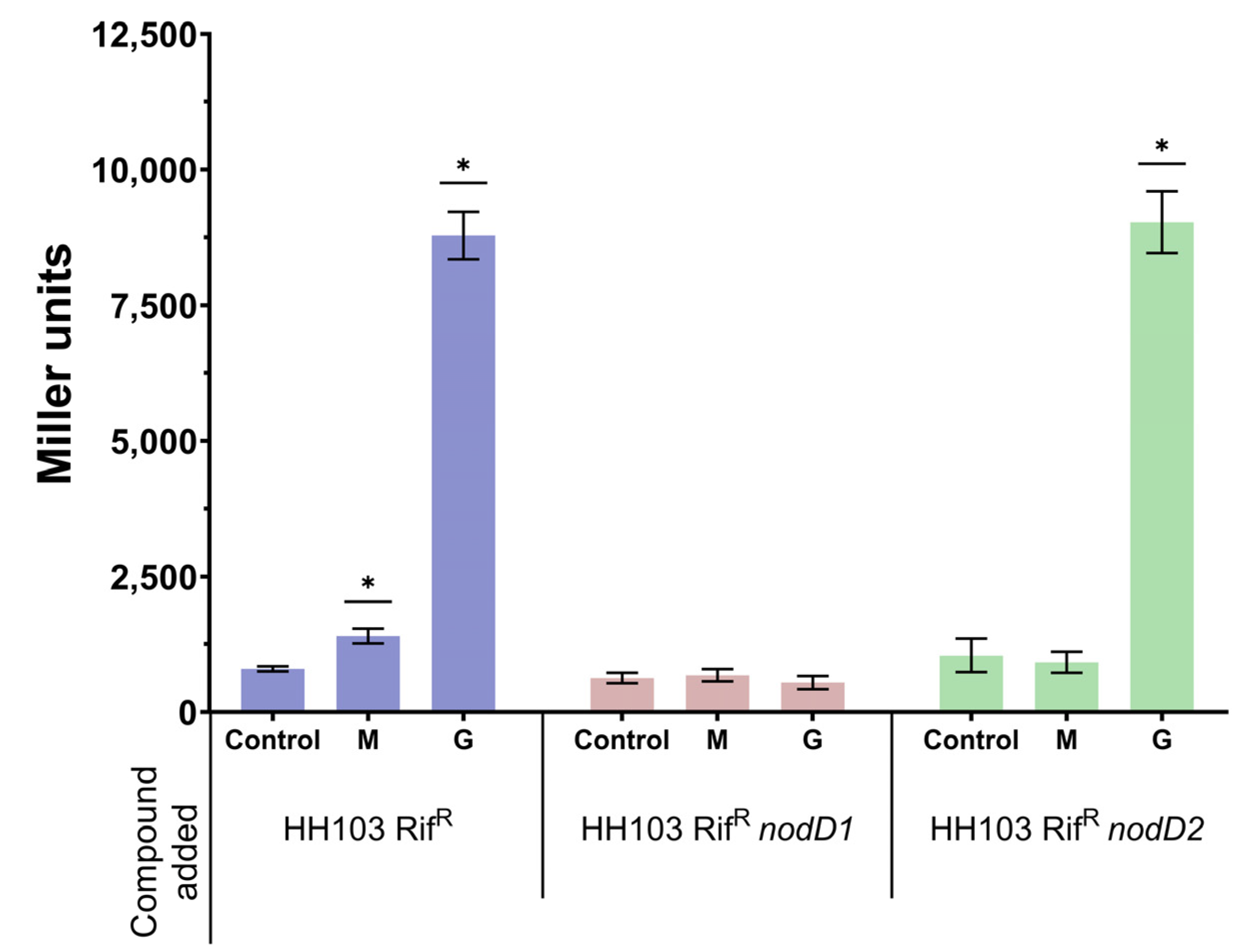
3.1. Expression of S. fredii HH103 Nod Genes Is Induced by the Presence of 400 mM Mannitol
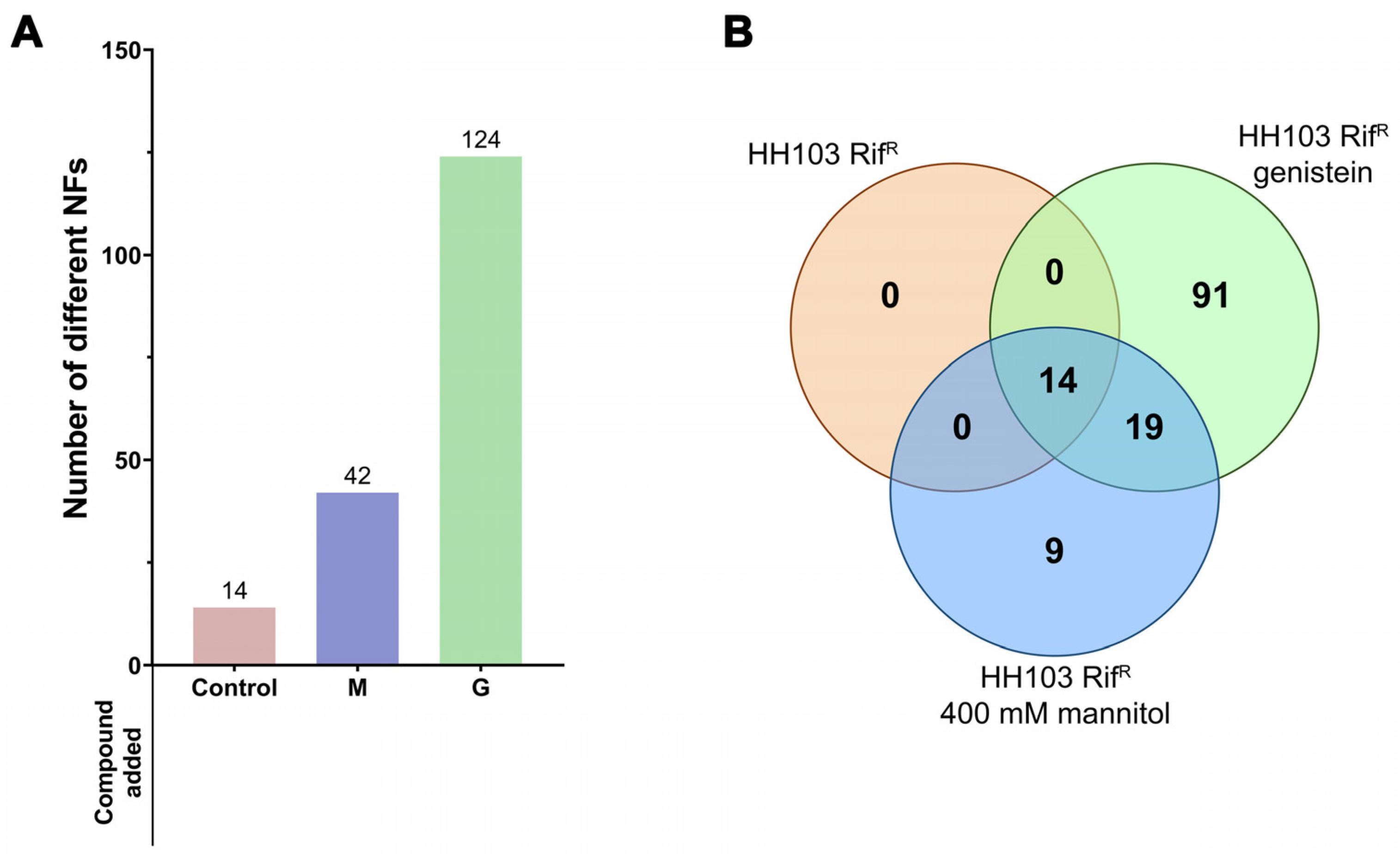
3.2. S. fredii HH103 Produces Biologically Active Nod Factors in the Presence of 400 mM Mannitol
3.3. A High Concentration of Mannitol Affects the Expression of Hundreds of S. fredii HH103 Genes
3.4. The Presence of a High Concentration of Mannitol Induces the Expression of 350 S. fredii HH103 Genes
3.5. The Presence of a High Concentration of Mannitol Represses the Expression of 393 S. fredii HH103 Genes
3.6. The Presence of a High Concentration of Mannitol Affects different Physiological Traits in S. fredii HH103 Genes
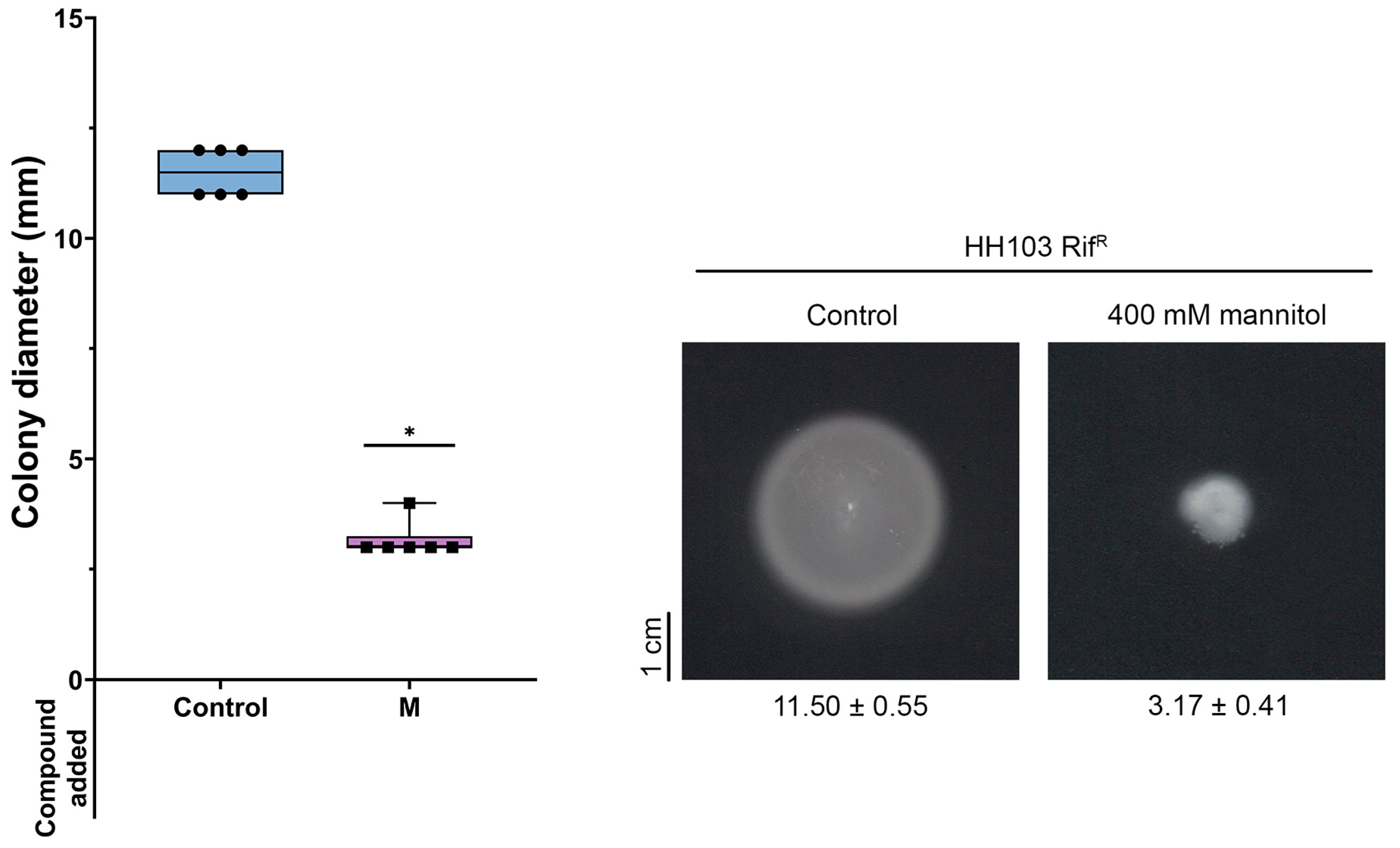
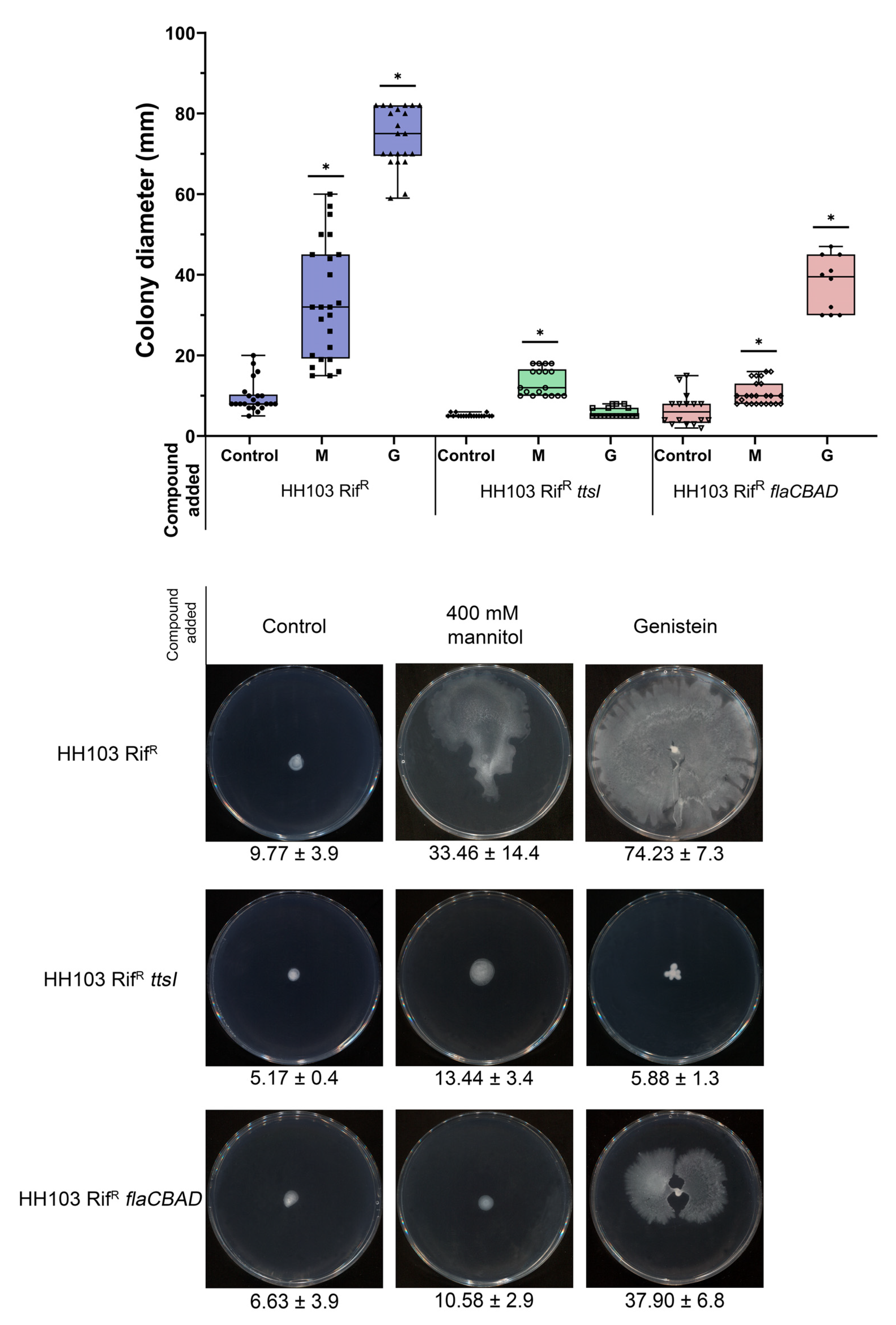
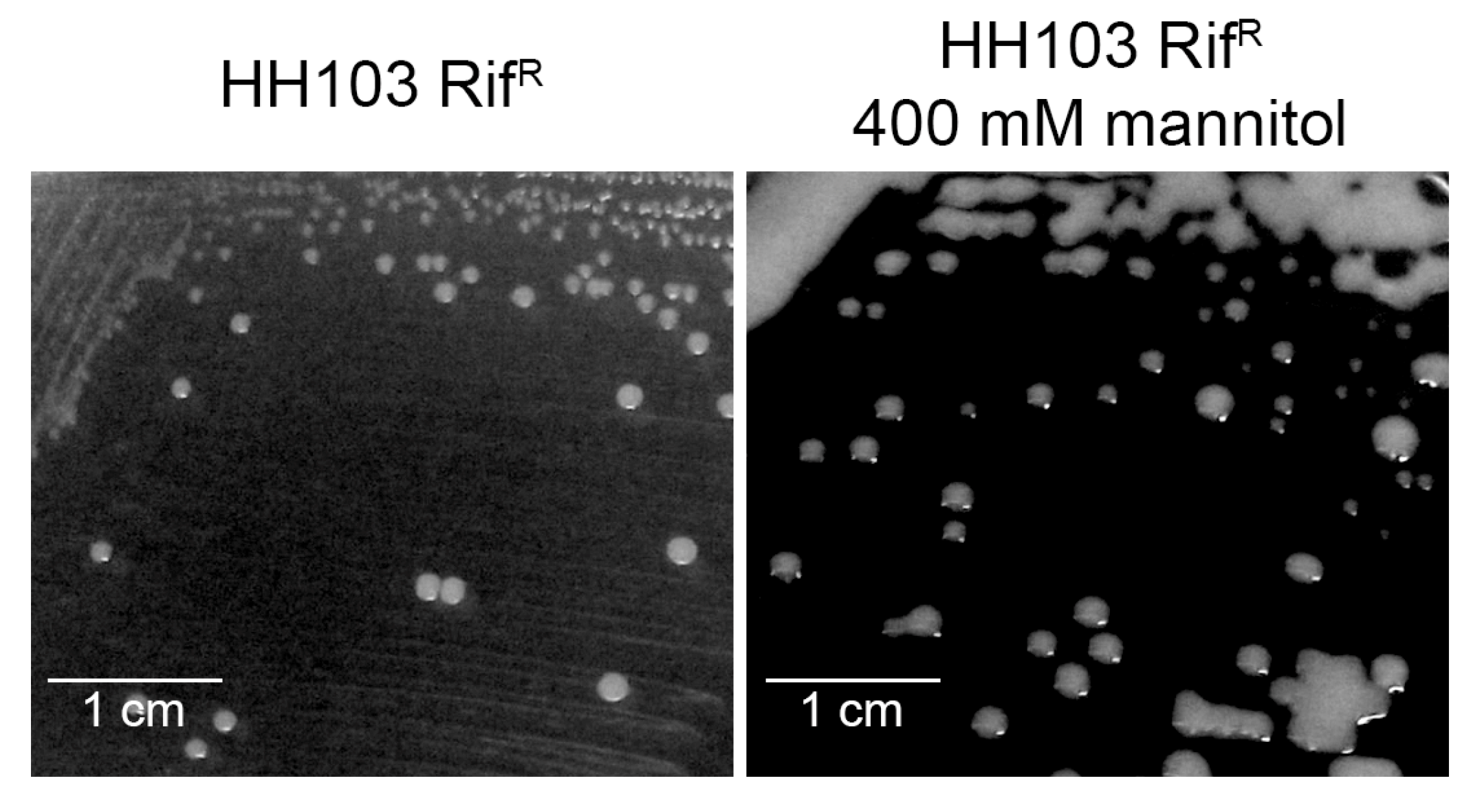
3.6.1. Motility
3.6.2. Surface Polysaccharides Biosynthesis
3.6.3. Production of IAA
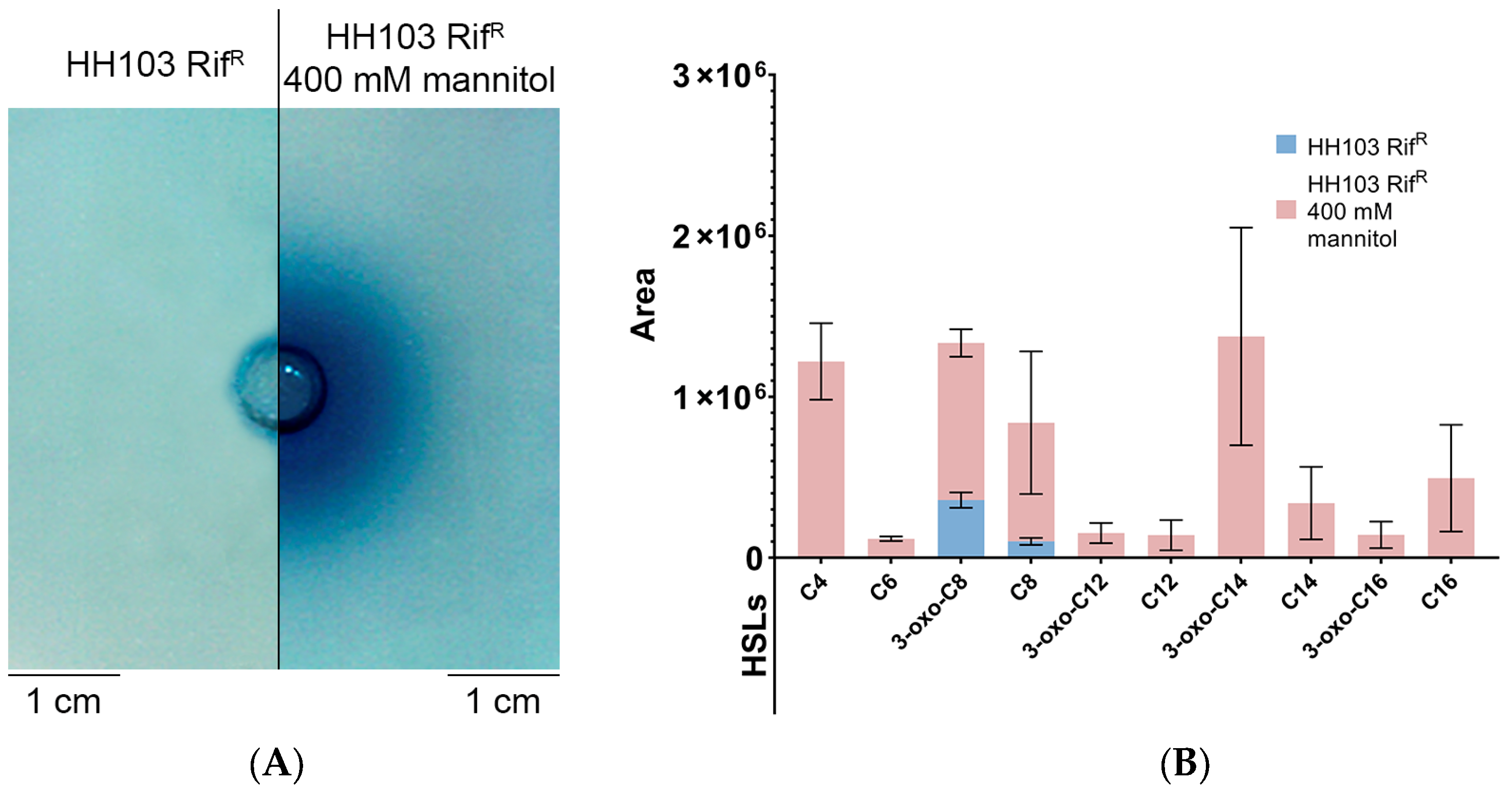
3.6.4. Production of AHLs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- López-Baena, F.J.; Ruiz-Sainz, J.E.; Rodríguez-Carvajal, M.A.; Vinardell, J.M. Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 2016, 17, 755. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell. 2020, 32, 15–41. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lan, L.; Jin, Y.; Yu, N.; Wang, D.; Wang, E. Mechanisms underlying legume-rhizobium symbioses. J. Integr. Plant Biol. 2022, 64, 244–267. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, F.; Wall, L.; Fabra, A. Starting points in plant-bacteria nitrogen-fixing symbioses: Intercellular invasion of the roots. J. Exp. Bot. 2017, 68, 1905–1918. [Google Scholar] [CrossRef]
- Janczarek, M.; Rachwał, K.; Marzec, A.; Grzadziel, J.; Palusińska-Szysz, M. Signal molecules and cell-surface components involved in early stages of the legume-rhizobium interactions. Appl. Soil Ecol. 2015, 85, 94–113. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Fuentes-Romero, F.; Ruiz-Sainz, J.E.; Janzarek, M.; Vinardell, J.M. Rhizobial exopolysaccharides: Genetic regulation of their synthesis and relevance in symbiosis with legumes. Int. J. Mol.Sci. 2021, 22, 6233. [Google Scholar] [CrossRef]
- Fraysse, N.; Couderc, F.; Poinsot, V. Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur. J. Biochem. 2003, 270, 1365–1380. [Google Scholar] [CrossRef]
- Jiménez-Guerrero, I.; Medina, C.; Vinardell, J.M.; Ollero, F.J.; López-Baena, F.J. The rhizobial type 3 secretion system: The Dr. Jekyll and Mr. Hyde in the rhizobium-legume symbiosis. Int. J. Mol. Sci. 2022, 23, 11089. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; del Cerro, P.; Jiménez-Guerrero, I.; López-Baena, G.J.; Cubo, M.T.; Hungria, M.; Megías, M.; Ollero, F.J. RNA-seq analysis of the Rhizobium tropici CIAT 899 transcriptome shows similarities in the activation patterns of symbiotic genes in the presence of apigenin and salt. BMC Genom. 2016, 17, 198. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Guasch-Vidal, B.; González-Barroso, S.; López-Baena, F.J.; Cubo, T.; Ollero, F.J.; Gil-Serrano, A.M.; Rodríguez-Carvajal, M.A.; Bellogín, R.A.; Espuny, M.R. Nodulation-gene-inducing flavonoids increase overall production of autoinducers and expression of N-acyl homoserine lactone synthesis genes in rhizobia. Res. Microbiol. 2011, 162, 715–723. [Google Scholar] [CrossRef]
- Del Cerro, P.; Rolla-Santos, A.A.P.; Valderrama-Fernández, R.; Gil-Serrano, A.M.; Bellogín, R.A.; Gomes, D.F.; Pérez-Montaño, F.; Megías, M.; Hungria, M.; Ollero, F.J. NrcR, a new transcriptional regulator of Rhizobium tropici CIAT 899 involved in the legume root-nodule symbiosis. PLoS ONE 2016, 11, e0154029. [Google Scholar] [CrossRef]
- del Cerro, P.; Megías, M.; López-Baena, F.J.; Gil-Serrano, M.; Pérez-Montaño, F.; Ollero, F.J. Osmotic stress activates nif and fix genes and induces the Rhizobium tropici CIAT 899 Nod factor production via NodD2 by up-regulation of the nodA2 operon and the nodA3 gene. PLoS ONE 2019, 14, e0213298. [Google Scholar] [CrossRef]
- del Cerro, P.; Rolla-Santos, A.A.P.; Gomes, D.F.; Marks, B.; Pérez-Montaño, F.; Rodríguez-Carvajal, M.A.; Gil-Serrano, A.; Megías, M.; Ollero, F.J.; Hungría, M. Regulatory nodD1 and nodD2 genes of Rhizobium tropici strain CIAT 899 and their role in early steps of molecular signaling and host nodulation. BMC Genom. 2015, 16, 251. [Google Scholar] [CrossRef]
- del Cerro, P.; Rolla-Santos, A.A.; Gomes, D.F.; Marks, B.B.; Espuny, M.R.; Rodríguez-Carvajal, M.A.; Soria-Díaz, M.E.; Nakatani, A.S.; Hungría, M.; Ollero, F.J.; et al. Opening the “black box” of nodD3, nodD4 and nodD5 genes of Rhizobium tropici strain CIAT 899. BMC Genom. 2015, 16, 864. [Google Scholar] [CrossRef]
- Del Cerro, P.; Pérez-Montaño, F.; Gil-Serrano, A.; López-Baena, F.J.; Megías, M.; Hungria, M.; Ollero, F.J. The Rhizobium tropici CIAT 899 NodD2 protein regulates the production of Nod factors under salt stress in a flavonoid-independent manner. Sci. Rep. 2017, 7, 46712. [Google Scholar] [CrossRef]
- del Cerro, P.; Ayala-García, P.; Buzón, P.; Castells-Graells, R.; López-Baena, F.J.; Ollero, F.J.; Pérez-Montaño, F. OnfD, an AraC-type transcriptional regulator encoded by Rhizobium tropici CIAT 899 and involved in Nod factor synthesis and symbiosis. Appl. Environ. Microbiol. 2020, 86, e01297-20. [Google Scholar] [CrossRef]
- Dowdle, S.F.; Bohlool, B.B. Predominance of fast-growing Rhizobium japonicum in a soybean field in the People’s Republic of China. Appl. Environ. Microbiol. 1985, 50, 1171–1176. [Google Scholar] [CrossRef]
- Margaret, I.; Becker, A.; Blom, J.; Bonilla, I.; Groesmann, A.; Göttfert, M.; Lloret, J.; Mittard-Runte, V.; Rückert, C.; Ruiz-Sainz, J.E.; et al. Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean. J. Biotechnol. 2011, 155, 11–19. [Google Scholar] [CrossRef]
- Gil-Serrano, A.M.; Franco-Rodríguez, G.; Tejero-Mateo, P.; Thomas-Oates, J.; Spaink, H.P.; Ruiz-Sainz, J.E.; Megías, M.; Lamrabet, Y. Structural determination of the lipo-chitinoligosaccharide nodulation signals produced by Rhizobium fredii HH103. Carbohydr. Res. 1997, 303, 435–443. [Google Scholar] [CrossRef]
- Vinardell, J.M.; López-Baena, F.J.; Hidalgo, A.; Ollero, F.J.; Bellogín, R.; Espuny, M.R.; Temprano, F.; Romero, F.; Krishnan, H.B.; Pueppke, S.G.; et al. The effect of FITA mutations on the symbiotic properties of Sinorhizobium fredii varies in a chromosomal-background-dependent manner. Arch. Microbiol. 2004, 181, 144–154. [Google Scholar] [CrossRef]
- Fuentes-Romero, F.; Navarro-Gómez, P.; Ayala-García, P.; Moyano-Bravo, I.; López-Baena, F.J.; Pérez-Montaño, F.; Ollero, F.J.; Acosta-Jurado, S.; Vinardell, J.M. The nodD1 gene of Sinorhizobium fredii HH103 restores nodulation capacity on bean in a Rhizobium tropici CIAT 899 nodD1/nodD2 mutant, but the secondary symbiotic regulators nolR, nodD2 or syrM prevent HH103 to nodulate with this legume. Microorganisms 2022, 10, 139. [Google Scholar] [CrossRef] [PubMed]
- Machado, D.; Pueppke, S.G.; Vinardell, J.M.; Ruiz-Sainz, J.E.; Krishnan, H.B. Expression of nodD1 and nodD2 in Sinorhizobium fredii, a nitrogen-fixing symbiont of soybean and other legumes. Mol. Plant-Microbe Interact. 1998, 11, 375–382. [Google Scholar] [CrossRef]
- Vinardell, J.M.; Ollero, F.J.; Hidalgo, A.; López-Baena, F.J.; Medina, C.; Ivanov-Vangelov, K.; Parada, M.; Madinabeitia, N.; Espuny, M.R.; Bellogín, R.A.; et al. NolR regulates diverse symbiotic signals of Sinorhizobium fredii HH103. Mol. Plant-Microbe Interact. 2004, 17, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Jurado, S.; Alías-Villegas, C.; Navarro-Gómez, P.; Zehner, S.; Murdoch, P.D.; Rodríguez-Carvajal, M.A.; Soto, M.J.; Ollero, F.J.; Ruiz-Sainz, J.E.; Göttfert, M.; et al. The Sinorhizobium fredii HH103 MucR1 global regulator is connected with the nod regulon and is required for efficient symbiosis with Lotus burttii and Glycine max cv. Williams. Mol. Plant-Microbe Interact. 2016, 29, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Jurado, S.; Rodríguez-Navarro, D.N.; Kawaharada, Y.; Rodríguez-Carvajal, M.A.; Gil-Serrano, A.; Soria-Díaz, M.E.; Pérez-Montaño, F.; Fernández-Perea, J.; Niu, Y.; Alías-Villegas, C.; et al. Sinorhizobium fredii HH103 nolR and nodD2 mutants gain capacity for infection thread invasion of Lotus japonicus Gifu and Lotus burttii. Environ. Microbiol. 2019, 21, 1718–1739. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Rodríguez-Navarro, D.N.; Kawaharada, Y.; Perea, J.F.; Gil-Serrano, A.; Jin, H.; An, Q.; Rodríguez-Carvajal, M.A.; Andersen, S.U.; Sandal, N.; et al. Sinorhizobium fredii HH103 invades Lotus burttii by crack entry in a Nod factor-and surface polysaccharide-dependent manner. Mol. Plant-Microbe Interact. 2016, 29, 925–937. [Google Scholar] [CrossRef]
- Di Lorenzo, F.; Speciale, I.; Silipo, A.; Alías-Villegas, C.; Acosta-Jurado, S.; Rodríguez-Carvajal, M.A.; Dardanelli, M.S.; Palmigiano, A.; Garozzo, D.; Ruiz-Sainz, J.E.; et al. Structure of the unusual Sinorhizobium fredii HH103 lipopolysaccharide and its role in symbiosis. J. Biol. Chem. 2020, 295, 10969–10987. [Google Scholar] [CrossRef]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Monreal, J.A.; Preston, G.M.; Fones, H.; Vioque, B.; Ollero, F.J.; López-Baena, F.J. The Sinorhizobium (Ensifer) fredii HH103 type 3 secretion system suppresses early defense responses to effectively nodulate soybean. Mol. Plant-Microbe Interact. 2015, 28, 790–7909. [Google Scholar] [CrossRef]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Medina, C.; Ollero, F.J.; López-Baena, F.J. NopC is a Rhizobium-specific type 3 secretion system effector secreted by Sinorhizobium (Ensifer) fredii HH103. PLoS ONE 2015, 10, e0142866. [Google Scholar] [CrossRef]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Medina, C.; Ollero, F.J.; López-Baena, F.J. The Sinorhizobium (Ensifer) fredii HH103 nodulation outer protein NopI Is a determinant for efficient nodulation of soybean and cowpea plants. Appl. Environ. Microbiol. 2017, 83, e02770-16. [Google Scholar] [CrossRef]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Zdyb, A.; Beutler, M.; Werner, G.; Göttfert, M.; Ollero, F.J.; Vinardell, J.M.; López-Baena, F.J. GunA of Sinorhizobium (Ensifer) fredii HH103 is a T3SS-secreted cellulase that differentially affects symbiosis with cowpea and soybean. Plant Soil 2019, 435, 15–26. [Google Scholar] [CrossRef]
- Jiménez-Guerrero, I.; Acosta-Jurado, S.; Medina, C.; Ollero, F.J.; Alías-Villegas, C.; Vinardell, J.M.; Pérez-Montaño, F.; López-Baena, F.J. The Sinorhizobium fredii HH103 type III secretion system effector NopC blocks nodulation with Lotus japonicus Gifu. J. Exp. Bot. 2020, 71, 6043–6056. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Alías-Villegas, C.; Almozara, A.; Espuny, M.R.; Vinardell, J.M.; Pérez-Montaño, F. Deciphering the symbiotic significance of quorum sensing systems of Sinorhizobium fredii HH103. Microorganisms 2020, 8, 68. [Google Scholar] [CrossRef]
- Lamrabet, Y.; Bellogín, R.A.; Cubo, T.; Espuny, R.; Gil, A.; Krishnan, H.B.; Megias, M.; Ollero, F.J.; Pueppke, S.G.; Ruiz-Sainz, J.E.; et al. Mutation in GDP-fucose synthesis genes of Sinorhizobium fredii alters Nod factors and significantly decreases competitiveness to nodulate soybeans. Mol. Plant-Microbe Interact. 1999, 12, 207–217. [Google Scholar] [CrossRef]
- López-Baena, F.J.; Vinardell, J.M.; Pérez-Montaño, F.; Crespo-Rivas, J.C.; Bellogín, R.A.; Espuny, M.R.; Ollero, F.J. Regulation and symbiotic significance of nodulation outer proteins secretion in Sinorhizobium fredii HH103. Microbiol. Read. 2008, 154, 1825–1836. [Google Scholar] [CrossRef]
- Beringer, J.E. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef]
- Nutman, P.S. The Modified Fåhraeus Slide Technique. In A Manual for the Practical Study of Root Nodule Bacteria. Handbook, Number 15, Appendix III; Vincent, J.M., Ed.; Blackwell Scientific: Oxford, UK; Edinburgh, UK, 1970; pp. 144–145. [Google Scholar]
- Spaink, H.P.; Aarts, A.; Stacey, G.; Bloemberg, G.V.; Lugtenberg, B.J.J.; Kennedy, E.P. Detection and separation of Rhizobium and Bradyrhizobium Nod metabolites using thin-layer chromatography. Mol. Plant-Microbe Interact. 1992, 5, 72–80. [Google Scholar] [CrossRef]
- Robertsen, B.K.; Aiman, P.; Darwill, A.G.; Mcneil, M.; Albersheim, P. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 1981, 67, 389–400. [Google Scholar] [CrossRef]
- Sourjik, V.; Schmitt, R. Different roles of CheY1 and CheY2 in the chemotaxis of Rhizobium meliloti. Mol. Microbiol. 1996, 22, 427–436. [Google Scholar] [CrossRef]
- Farrand, S.K.; Qin, Y.; Oger, P. Quorum-sensing system of Agrobacterium plasmids: Analysis and utility. Methods Enzymol. 2002, 358, 452–484. [Google Scholar] [CrossRef]
- Miller, J.H. Experiment in Molecular Genetics; Cold Spring Harbor: New York, NY, USA, 1972. [Google Scholar]
- de Maagd, R.A.; Wijffelman, C.A.; Pees, E.; Lugtenberg, B.J.J. Detection and subcellular localization of two Sym plasmid dependent proteins of Rhizobium leguminosarum biovar viciae. J. Bacteriol. 1998, 170, 4424–4427. [Google Scholar] [CrossRef] [PubMed]
- López-Lara, I.M.; van Der Drift, K.M.; van Brussel, A.A.; Haverkamp, J.; Lugtenberg, B.J.; Thomas-Oates, J.E.; Spaink, H.P. Induction of nodule primordia on Phaseolus and Acacia by lipo-chitin oligosaccharide nodulation signals from broad-host-range Rhizobium strain GRH2. Plant Mol. Biol. 1995, 29, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Truchet, G.; Camut, S.; de Billy, F.; Odorico, R.; Vasse, J. The Rhizobium-legume symbiosis. Two methods to discriminate between nodules and other root derived structures. Protoplasma 1989, 149, 82–89. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Jiménez-Guerrero, I.; Acosta-Jurado, S.; Navarro-Gómez, P.; Ollero, F.J.; Ruiz-Sainz, J.E.; López-Baena, F.J.; Vinardell, J.M. A transcriptomic analysis of the effect of genistein on Sinorhizobium fredii HH103 reveals novel rhizobial genes putatively involved in symbiosis. Sci. Rep. 2016, 6, 31592. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map forma The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 987–2993. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Phyton framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 3166–3169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Alías-Villegas, C.; Fuentes-Romero, F.; Cuéllar, V.; Navarro-Gómez, P.; Soto, M.J.; Vinardell, J.-M.; Acosta-Jurado, S. Surface Motility regulation of Sinorhizobium fredii HH103 by plant flavonoids and the NodD1, TtsI, NolR, and MucR1 symbiotic bacterial regulators. Int. J. Mol. Sci. 2022, 23, 7698. [Google Scholar] [CrossRef]
- Hidalgo, Á.; Margaret, I.; Crespo-Rivas, J.C.; Parada, M.; Murdoch, P.D.S.; López, A.; Buendía-Clavería, A.M.; Moreno, J.; Albareda, M.; Gil-Serrano, A.M.; et al. The rkpU gene of Sinorhizobium fredii HH103 is required for bacterial K-antigen polysaccharide production and for efficient nodulation with soybean but not with cowpea. Microbiol. Read. 2010, 156, 3398–3411. [Google Scholar] [CrossRef]
- Parada, M.; Vinardell, J.M.; Ollero, F.J.; Hidalgo, A.; Gutiérrez, R.; Buendía-Clavería, A.M.; Lei, W.; Margaret, I.; López-Baena, F.J.; Gil-Serrano, A.M.; et al. Sinorhizobium fredii HH103 mutants affected in capsular polysaccharide (KPS) are impaired for nodulation with soybean and Cajanus cajan. Mol. Plant-Microbe Interact. 2006, 19, 43–52. [Google Scholar] [CrossRef]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar] [CrossRef]
- Cha, C.; Gao, P.; Chen, Y.C.; Shaw, P.D.; Farrand, S.K. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant associated bacteria. Mol. Plant-Microbe Interact. 1998, 11, 1119–1129. [Google Scholar] [CrossRef]
- Vinardell, J.M.; Acosta-Jurado, S.; Göttfert, M.; Zehner, S.; Becker, A.; Baena-Ropero, I.; Blom, J.; Crespo-Rivas, J.C.; Goesmann, A.; Jaenicke, S.; et al. The Sinorhizobium fredii HH103 genome: A comparative analysis with S. fredii strains differing in their symbiotic behaviour with soybean. Mol. Plant-Microbe Interact. 2015, 28, 811–824. [Google Scholar] [CrossRef]
- Madinabeitia, N.; Bellogín, R.A.; Buendía-Clavería, A.M.; Camacho, M.; Cubo, T.; Espuny, M.R.; Gil-Serrano, A.M.; Lyra, M.C.; Moussaid, A.; Ollero, F.J.; et al. Sinorhizobium fredii HH103 has a truncated nolO gene due to a -1 frameshift mutation that is conserved among other geographically distant S. fredii strains. Mol. Plant-Microbe Interact. 2002, 15, 150–159. [Google Scholar] [CrossRef]
- Rodríguez-Navarro, D.N.; Rodríguez-Carvajal, M.A.; Acosta-Jurado, S.; Soto, M.J.; Margaret, I.; Crespo-Rivas, J.C.; Sanjuan, J.; Temprano, F.; Gil-Serrano, A.; Ruiz-Sainz, J.E.; et al. Structure and biological roles of Sinorhizobium fredii HH103 exopolysaccharide. PLoS ONE 2014, 9, e115391. [Google Scholar] [CrossRef]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Hawkins, J.P.; Oresnik, I.J. The rhizobium-legume symbiosis: Co-opting successful stress management. Front. Plant Sci. 2022, 12, 796045. [Google Scholar] [CrossRef]
- Kumar, C.M.; Mande, S.C.; Mahajan, G. Multiple chaperonins in bacteria-novel functions and non-canonical behaviors. Cell Stress Chaperones 2015, 20, 555–574. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.; Achom, M.; Wilkinson, H.; Lagunas, B.; Gifford, M.L. Symbiotic outcome modified by the diversification from 7 to over 700 nodule-specific cysteine-rich peptides. Genes 2020, 11, 348. [Google Scholar] [CrossRef] [PubMed]
- Bittner, A.N.; Foltz, A.; Oke, V. Only one of five groEL genes is required for viability and successful symbiosis in Sinorhizobium meliloti. J. Bacteriol. 2007, 189, 1884–1889. [Google Scholar] [CrossRef]
- Wei, F.; Liu, Y.; Zhou, D.; Zhao, W.; Chen, Z.; Chen, D.; Li, Y.; Zhang, X.X. Transcriptomic identification of a unique set of nodule-specific cysteine-rich peptides expressed in the nitrogen-fixing root nodule of Astragalus sinicus. Mol. Plant-Microbe Interact. 2022, 35, 893–905. [Google Scholar] [CrossRef] [PubMed]
- Margaret-Oliver, I.; Lei, W.; Parada, M.; Rodríguez-Carvajal, M.A.; Crespo-Rivas, J.C.; Hidalgo, A.; Gil-Serrano, A.; Moreno, J.; Rodríguez-Navarro, D.N.; Buendía-Clavería, A.; et al. Sinorhizobium fredii HH103 does not strictly require KPS and/or EPS to nodulate Glycyrrhiza uralensis, an indeterminate nodule-forming legume. Arch. Microbiol. 2012, 192, 87–102. [Google Scholar] [CrossRef]
- Glazebrook, J.; Reed, J.W.; Reuber, T.L.; Walker, G.C. Genetic analyses of Rhizobium meliloti exopolysaccharides. Int. J. Biol. Macromol. 1990, 12, 67–70. [Google Scholar] [CrossRef]
- Barnett, M.J.; Long, S.R. The Sinorhizobium meliloti SyrM regulon: Effects on global gene expression are mediated by syrA and nodD3. J. Bacteriol. 2015, 197, 1792–17806. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Navarro-Gómez, P.; Murdoch, P.; Del, S.; Crespo-Rivas, J.C.; Jie, S.; Cuesta-Berrio, L.; Ruiz-Sainz, J.E.; Rodríguez-Carvajal, M.A.; Vinardell, J.M. Exopolysaccharide production by Sinorhizobium fredii HH103 is repressed by genistein in a NodD1-dependent manner. PLoS ONE 2016, 11, e0160499. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Alias-Villegas, C.; Navarro-Gómez, P.; Almozara, A.; Rodríguez-Carvajal, M.A.; Medina, C.; Vinardell, J.M. Sinorhizobium fredii HH103 syrM inactivation affects the expression of a large number of genes, impairs nodulation with soybean and extends the host-range to Lotus japonicus. Environ. Microbiol. 2020, 22, 1104–1124. [Google Scholar] [CrossRef]
- Breedveld, M.W.; Miller, K.J. Cyclic beta-glucans of members of the family Rhizobiaceae. Microbiol. Rev. 1994, 58, 145–161. [Google Scholar] [CrossRef]
- Crespo-Rivas, J.C.; Margaret, I.; Hidalgo, A.; Buendía-Clavería, A.M.; Ollero, F.J.; López-Baena, F.J.; del Socorro Murdoch, P.; Rodríguez-Carvajal, M.A.; Soria-Díaz, M.E.; Reguera, M.; et al. Sinorhizobium fredii HH103 cgs mutants are unable to nodulate determinate- and indeterminate nodule-forming legumes and overproduce an altered EPS. Mol. Plant-Microbe Interact. 2009, 22, 575–588. [Google Scholar] [CrossRef]
- Wheatley, R.M.; Ford, B.L.; Li, L.; Aroney, S.T.N.; Knights, H.E.; Ledermann, R.; East, A.K.; Ramachandran, V.K.; Poole, P.S. Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis. Proc. Natl. Acad. Sci. USA 2020, 117, 23823–23834. [Google Scholar] [CrossRef]
- Aroney, S.T.N.; Poole, P.S.; Sánchez-Cañizares, C. Rhizobial chemotaxis and motility systems at work in the soil. Front. Plant Sci. 2021, 12, 725338. [Google Scholar] [CrossRef]
- Alemneh, A.A.; Zhou, Y.; Ryder, M.H.; Denton, M.D. Mechanisms in plant growth-promoting rhizobacteria that enhance legume-rhizobial symbioses. J. Appl. Microbiol. 2020, 129, 1133–1156. [Google Scholar] [CrossRef]
- Calatrava-Morales, N.; McIntosh, M.; Soto, M.J. Regulation mediated by N-acyl homoserine lactone quorum sensing signals in the Rhizobium-legumes symbiosis. Genes 2018, 9, 263. [Google Scholar] [CrossRef]

| Gene Name | Forward | Reverse |
|---|---|---|
| d257 | acagacagctaaattctctgc | gatgttgtcatcctctggata |
| ndvA | cagctacaaccgtatcgaag | gaggatcgtcatcatcgaaat |
| nodA | cgtcatgtatccggtgctgca | cgttggcggcaggttgaga |
| nopX | gcaaatctcctccgtaca | ctcttcaatcgccttcct |
| oppD | gtctcgttccagctctatc | cgccgagaatttcagga |
| rkpI | ggcctctacatgtatttcga | ccggaccgtagaacagcgaga |
| SFHH103_1866 | gtcgaagcgttttacattac | gaggaaatccagtccatagc |
| SFHH103_3035 | gtggcgactttatggaac | gagttcgatcacctcctc |
| SFHH103_3139 | aatgatctgacctggcatt | gctcgatatactcgttgatg |
| SFHH103_6032 | cggatctgaaacgcatgt | cgcggaaaataaatgcag |
| sinI | cctgggagatatccggttcga | cactacgacaaggcca |
| 16S | gataccctggtagtccac | taaaccacatgctccacc |
| Replicon | Number of Genes | Induced | % | Repressed | % |
|---|---|---|---|---|---|
| Chromosome | 4014 | 119 | 2.96 | 292 | 7.27 |
| pSfHH103e | 1991 | 148 | 7.43 | 72 | 3.62 |
| pSfHH103d = pSym | 667 | 77 | 11.54 | 6 | 0.90 |
| pSfHH103c | 169 | 4 | 2.37 | 9 | 5.33 |
| pSfHH103b | 62 | 2 | 3.23 | 5 | 8.06 |
| pSfHH103a2 | 38 | 0 | 0 | 8 | 21.05 |
| pSfHH103a1 | 19 | 0 | 0 | 1 | 5.26 |
| Total | 6960 | 350 | 5.03 | 393 | 5.65 |
| Fold-Change | |||
|---|---|---|---|
| Gene | Description | RNA-seq | qPCR |
| SFHH103_06032 | ABC-type sugar transport systems, ATPase components | 146.7 | 33.0 |
| SFHH103_00224, rkpI | capsular polysaccharide biosynthesis\export transmembrane protein | 35.2 | 2.5 |
| SFHH103_06488, oppD | ATPase components of various ABC-type transport systems | 18.8 | 9.7 |
| psfHH103d_257 | Histidinol-phosphate aminotransferase | 18.1 | 7.0 |
| SFHH103_01572, sinI | Autoinducer synthase protein | 10.8 | 9.1 |
| psfHH103d_335, nopX | T3SS component NopX | 6.3 | 3.0 |
| psfHH103d_126, nodA | N-acyltransferase nodulation protein NodA | −1.5 | 3.9 |
| SFHH103_03433, ndvA | Glucan exporter ATP-binding protein | −5.6 | −6.3 |
| SFHH103_03035 | Conserved hypothetical protein. P pilus assembly/Cpx signaling pathway, periplasmic inhibitor/zinc-resistance associated protein | −29.0 | −25.0 |
| SFHH103_01866 | RTX toxins and related Ca2+-binding proteins | −25.6 | −16.7 |
| SFHH103_03139 | Oxidoreductase | −58.6 | −3.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuentes-Romero, F.; Moyano-Bravo, I.; Ayala-García, P.; Rodríguez-Carvajal, M.Á.; Pérez-Montaño, F.; Acosta-Jurado, S.; Ollero, F.J.; Vinardell, J.-M. Non-Ionic Osmotic Stress Induces the Biosynthesis of Nodulation Factors and Affects Other Symbiotic Traits in Sinorhizobium fredii HH103. Biology 2023, 12, 148. https://doi.org/10.3390/biology12020148
Fuentes-Romero F, Moyano-Bravo I, Ayala-García P, Rodríguez-Carvajal MÁ, Pérez-Montaño F, Acosta-Jurado S, Ollero FJ, Vinardell J-M. Non-Ionic Osmotic Stress Induces the Biosynthesis of Nodulation Factors and Affects Other Symbiotic Traits in Sinorhizobium fredii HH103. Biology. 2023; 12(2):148. https://doi.org/10.3390/biology12020148
Chicago/Turabian StyleFuentes-Romero, Francisco, Isamar Moyano-Bravo, Paula Ayala-García, Miguel Ángel Rodríguez-Carvajal, Francisco Pérez-Montaño, Sebastián Acosta-Jurado, Francisco Javier Ollero, and José-María Vinardell. 2023. "Non-Ionic Osmotic Stress Induces the Biosynthesis of Nodulation Factors and Affects Other Symbiotic Traits in Sinorhizobium fredii HH103" Biology 12, no. 2: 148. https://doi.org/10.3390/biology12020148
APA StyleFuentes-Romero, F., Moyano-Bravo, I., Ayala-García, P., Rodríguez-Carvajal, M. Á., Pérez-Montaño, F., Acosta-Jurado, S., Ollero, F. J., & Vinardell, J.-M. (2023). Non-Ionic Osmotic Stress Induces the Biosynthesis of Nodulation Factors and Affects Other Symbiotic Traits in Sinorhizobium fredii HH103. Biology, 12(2), 148. https://doi.org/10.3390/biology12020148








