Impact of c-di-GMP on the Extracellular Proteome of Rhizobium etli
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Protein Sample Preparation
2.3. Proteomics
2.4. Western Blots
2.5. Membrane Vesicles Isolation and Quantification
2.6. Transmission Electron Microscopy Immunolabeling
2.7. Bacterial Growth Curves and Cell Viability Determination
2.8. Bioinformatics Analyses
3. Results
3.1. Cyclic Diguanylate-Dependent Extracellular Proteome of Rhizobium etli
3.2. High cdG Levels Do Not Enhance Cell Lysis nor MV Formation
3.3. cdG Promotes Exportation of the Cytoplasmic Proteins EF-Tu and Gap
3.4. Subcellular Localization of R. etli Gap Protein
3.5. Extracellular Gap Proteoforms Promoted by c-di-GMP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The First 25 Years of a Universal Bacterial Second Messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Galperin, M. Discovery of the Second Messenger Cyclic di-GMP. In c-di-GMP Signaling: Methods and Protocols; Sauer, K., Ed.; Springer: New York, NY, USA, 2017; Volume 1657, pp. 1–8. [Google Scholar] [CrossRef]
- Hengge, R.; Häussler, S.; Pruteanu, M.; Stülke, J.; Tschowri, N.; Turgay, K. Recent Advances and Current Trends in Nucleotide Second Messenger Signaling in Bacteria. J. Mol. Biol. 2019, 431, 908–927. [Google Scholar] [CrossRef]
- Petchiappan, A.; Naik, S.Y.; Chatterji, D. Tracking the homeostasis of second messenger cyclic-di-GMP in bacteria. Biophys. Rev. 2020, 12, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Hengge, R. High-specificity local and global c-di-GMP signaling. Trends Microbiol. 2021, 29, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mendoza, D.; Sanjuán, J. Exploiting the commons: Cyclic diguanylate regulation of bacterial exopolysaccharide production. Curr. Opin. Microbiol. 2016, 30, 36–43. [Google Scholar] [CrossRef]
- Fong, J.N.C.; Yildiz, F.H. Biofilm Matrix Proteins. Microbiol. Spectr. 2015, 3, 201–222. [Google Scholar] [CrossRef]
- Cooley, R.B.; Smith, T.J.; Leung, W.; Tierney, V.; Borlee, B.R.; O’Toole, G.A.; Sondermann, H. Cyclic Di-GMP-Regulated Periplasmic Proteolysis of a Pseudomonas aeruginosa Type Vb Secretion System Substrate. J. Bacteriol. 2016, 198, 66–76. [Google Scholar] [CrossRef]
- Lindenberg, S.; Klauck, G.; Pesavento, C.; Klauck, E.; Hengge, R. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J. 2013, 32, 2001–2014. [Google Scholar] [CrossRef]
- Pérez-Mendoza, D.; Coulthurst, S.J.; Humphris, S.; Campbell, E.; Welch, M.; Toth, I.K.; Salmond, G.P.C. A multi-repeat adhesin of the phytopathogen, Pectobacterium atrosepticum, is secreted by a Type I pathway and is subject to complex regulation involving a non-canonical diguanylate cyclase. Mol. Microbiol. 2011, 82, 719–733. [Google Scholar] [CrossRef]
- Roelofs, K.G.; Jones, C.J.; Helman, S.R.; Shang, X.; Orr, M.W.; Goodson, J.R.; Galperin, M.; Yildiz, F.H.; Lee, V.T. Systematic Identification of Cyclic-di-GMP Binding Proteins in Vibrio cholerae Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems. PLoS Pathog. 2015, 11, e1005232. [Google Scholar] [CrossRef] [PubMed]
- Trampari, E.; Stevenson, C.E.; Little, R.H.; Wilhelm, T.; Lawson, D.M.; Malone, J.G. Bacterial Rotary Export ATPases Are Allosterically Regulated by the Nucleotide Second Messenger Cyclic-di-GMP. J. Biol. Chem. 2015, 290, 24470–24483. [Google Scholar] [CrossRef] [PubMed]
- Baena, F.J.L.; Vinardell, J.-M.; Medina, C. Regulation of Protein Secretion Systems Mediated by Cyclic Diguanylate in Plant-Interacting Bacteria. Front. Microbiol. 2019, 10, 1289. [Google Scholar] [CrossRef] [PubMed]
- Curtis, P.D.; Atwood, J.; Orlando, R.; Shimkets, L.J. Proteins Associated with the Myxococcus xanthus Extracellular Matrix. J. Bacteriol. 2007, 189, 7634–7642. [Google Scholar] [CrossRef]
- Jiao, Y.; D’Haeseleer, P.; Dill, B.D.; Shah, M.; VerBerkmoes, N.C.; Hettich, R.L.; Banfield, J.F.; Thelen, M.P. Identification of Biofilm Matrix-Associated Proteins from an Acid Mine Drainage Microbial Community. Appl. Environ. Microbiol. 2011, 77, 5230–5237. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Roschitzki, B.; Riedel, K.; Eberl, L. Identification of Proteins Associated with the Pseudomonas aeruginosa Biofilm Extracellular Matrix. J. Proteome Res. 2012, 11, 4906–4915. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, L.; Toyofuku, M.; Hynen, A.L.; Kurosawa, M.; Pessi, G.; Petty, N.K.; Osvath, S.R.; Cárcamo-Oyarce, G.; Gloag, E.S.; Shimoni, R.; et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 2016, 7, 11220. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, L.; Fu, G.; Zhou, W.; Sun, Y.; Zheng, P.; Sun, J.; Zhang, D. A novel strategy for protein production using non-classical secretion pathway in Bacillus subtilis. Microb. Cell Factories 2016, 15, 69. [Google Scholar] [CrossRef]
- Ebner, P.; Rinker, J.; Nguyen, M.T.; Popella, P.; Nega, M.; Luqman, A.; Schittek, B.; Di Marco, M.; Stevanovic, S.; Götz, F. Excreted Cytoplasmic Proteins Contribute to Pathogenicity in Staphylococcus aureus. Infect. Immun. 2016, 84, 1672–1681. [Google Scholar] [CrossRef]
- Götz, F.; Yu, W.; Dube, L.; Prax, M.; Ebner, P. Excretion of cytosolic proteins (ECP) in bacteria. Int. J. Med. Microbiol. 2015, 305, 230–237. [Google Scholar] [CrossRef]
- Wang, G.; Xia, Y.; Song, X.; Ai, L. Common Non-classically Secreted Bacterial Proteins with Experimental Evidence. Curr. Microbiol. 2015, 72, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, L.; Ferreira, E.; Giménez, R.; Fernández, F.J.; Taulés, M.; Aguilar, J.; Vega, M.C.; Badia, J.; Baldomà, L. Secretion of the housekeeping protein glyceraldehyde-3-phosphate dehydrogenase by the LEE-encoded type III secretion system in enteropathogenic Escherichia coli. Int. J. Biochem. Cell Biol. 2012, 44, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Krol, E.; Schäper, S.; Becker, A. Cyclic di-GMP signaling controlling the free-living lifestyle of alpha-proteobacterial rhizobia. Biol. Chem. 2020, 401, 1335–1348. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mendoza, D.; Romero-Jiménez, L.; Rodríguez-Carvajal, M.; Lorite, M.J.; Muñoz, S.; Olmedilla, A.; Sanjuán, J. The Role of Two Linear β-Glucans Activated by c-di-GMP in Rhizobium etli CFN42. Biology 2022, 11, 1364. [Google Scholar] [CrossRef]
- Beringer, J.E. R Factor Transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef]
- Bravo, A.; Mora, J. Ammonium assimilation in Rhizobium phaseoli by the glutamine synthetase-glutamate synthase pathway. J. Bacteriol. 1988, 170, 980–984. [Google Scholar] [CrossRef]
- Hurkman, W.J.; Tanaka, C.K. Solubilization of Plant Membrane Proteins for Analysis by Two-Dimensional Gel Electrophoresis. Plant Physiol. 1986, 81, 802–806. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Encarnación, S.; Hernández, M.; Martínez-Batallar, G.; Contreras, S.; Vargas, M.D.C.; Mora, J. Comparative proteomics using 2-D gel electrophoresis and mass spectrometry as tools to dissect stimulons and regulons in bacteria with sequenced or partially sequenced genomes. Biol. Proced. Online 2005, 7, 117–135. [Google Scholar] [CrossRef]
- Candiano, G.; Bruschi, M.; Musante, L.; Santucci, L.; Ghiggeri, G.M.; Carnemolla, B.; Orecchia, P.; Zardi, L.; Righetti, P.G. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 2004, 25, 1327–1333. [Google Scholar] [CrossRef]
- Ciordia, S.; Alvarez-Sola, G.; Rullán, M.; Urman, J.M.; Ávila, M.A.; Corrales, F.J. Digging deeper into bile proteome. J. Proteom. 2021, 230, 103984. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Fernández, A.; Paradela, A.; Navajas, R.; Albar, J.P. Generalized Method for Probability-based Peptide and Protein Identification from Tandem Mass Spectrometry Data and Sequence Database Searching. Mol. Cell Proteom. 2008, 7, 1748–1754. [Google Scholar] [CrossRef] [PubMed]
- López-Serra, P.; Marcilla, M.; Villanueva, A.; Ramos-Fernández, A.; Palau, A.; Leal, L.; Wahi, J.E.; Setien-Baranda, F.; Szczesna, K.; Moutinho, C.; et al. A DERL3-associated defect in the degradation of SLC2A1 mediates the Warburg effect. Nat. Commun. 2014, 5, 3608. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Pérez-Cruz, C.; Cañas, M.-A.; Giménez, R.; Badia, J.; Mercade, E.; Baldomà, L.; Aguilera, L. Membrane Vesicles Released by a hypervesiculating Escherichia coli Nissle 1917 tolR Mutant Are Highly Heterogeneous and Show Reduced Capacity for Epithelial Cell Interaction and Entry. PLoS ONE 2016, 11, e0169186. [Google Scholar] [CrossRef]
- Frias, A.; Manresa, A.; de Oliveira, E.; López-Iglesias, C.; Mercade, E. Membrane Vesicles: A Common Feature in the Extracellular Matter of Cold-Adapted Antarctic Bacteria. Microb. Ecol. 2010, 59, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Masco, L.; Crockaert, C.; Van Hoorde, K.; Swings, J.; Huys, G. In Vitro Assessment of the Gastrointestinal Transit Tolerance of Taxonomic Reference Strains from Human Origin and Probiotic Product Isolates of Bifidobacterium. J. Dairy Sci. 2007, 90, 3572–3578. [Google Scholar] [CrossRef]
- Vozza, N.F.; Abdian, P.; Russo, D.M.; Mongiardini, E.J.; Lodeiro, A.R.; Molin, S.; Zorreguieta, A. A Rhizobium leguminosarum CHDL- (Cadherin-Like-) Lectin Participates in Assembly and Remodeling of the Biofilm Matrix. Front. Microbiol. 2016, 7, 1608. [Google Scholar] [CrossRef]
- Krehenbrink, M.; Downie, J.A. Identification of protein secretion systems and novel secreted proteins in Rhizobium leguminosarum bv. viciae. BMC Genom. 2008, 9, 55. [Google Scholar] [CrossRef]
- Pérez-Mendoza, D.; Aragón, I.M.; Prada-Ramírez, H.A.; Romero-Jiménez, L.; Ramos, C.; Gallegos, M.-T.; Sanjuán, J. Responses to Elevated c-di-GMP Levels in Mutualistic and Pathogenic Plant-Interacting Bacteria. PLoS ONE 2014, 9, e91645. [Google Scholar] [CrossRef]
- Schäper, S.; Steinchen, W.; Krol, E.; Altegoer, F.; Skotnicka, D.; Søgaard-Andersen, L.; Bange, G.; Becker, A. AraC-like transcriptional activator CuxR binds c-di-GMP by a PilZ-like mechanism to regulate extracellular polysaccharide production. Proc. Natl. Acad. Sci. USA 2017, 114, E4822–E4831. [Google Scholar] [CrossRef] [PubMed]
- Schäper, S.; Krol, E.; Skotnicka, D.; Kaever, V.; Hilker, R.; Søgaard-Andersen, L.; Becker, A. Cyclic Di-GMP Regulates Multiple Cellular Functions in the Symbiotic Alphaproteobacterium Sinorhizobium meliloti. J. Bacteriol. 2016, 198, 521–535. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Jeffery, C.J. An analysis of surface proteomics results reveals novel candidates for intracellular/surface moonlighting proteins in bacteria. Mol. Biosyst. 2016, 12, 1420–1431. [Google Scholar] [CrossRef] [PubMed]
- Abarca-Grau, A.M.; Burbank, L.P.; de Paz, H.D.; Crespo-Rivas, J.C.; Marco-Noales, E.; López, M.M.; Vinardell, J.M.; von Bodman, S.B.; Penyalver, R. Role for Rhizobium rhizogenes K84 Cell Envelope Polysaccharides in Surface Interactions. Appl. Environ. Microbiol. 2012, 78, 1644–1651. [Google Scholar] [CrossRef] [PubMed]
- Mazur, A.; Król, J.E.; Wielbo, J.; Urbanik-Sypniewska, T.; Skorupska, A. Rhizobium leguminosarum bv. trifolii PssP Protein Is Required for Exopolysaccharide Biosynthesis and Polymerization. Mol. Plant-Microbe Interact. 2002, 15, 388–397. [Google Scholar] [CrossRef]
- Tun-Garrido, C.; Bustos, P.; González, V.; Brom, S. Conjugative Transfer of p42a from Rhizobium etli CFN42, Which Is Required for Mobilization of the Symbiotic Plasmid, Is Regulated by Quorum Sensing. J. Bacteriol. 2003, 185, 1681–1692. [Google Scholar] [CrossRef]
- Zorreguieta, A.; Finnie, C.; Downie, J.A. Extracellular Glycanases of Rhizobium leguminosarum Are Activated on the Cell Surface by an Exopolysaccharide-Related Component. J. Bacteriol. 2000, 182, 1304–1312. [Google Scholar] [CrossRef]
- Pidutti, P.; Federici, F.; Brandi, J.; Manna, L.; Rizzi, E.; Marini, U.; Cecconi, D. Purification and characterization of ribosomal proteins L27 and L30 having antimicrobial activity produced by the Lactobacillus salivarius SGL. J. Appl. Microbiol. 2017, 124, 398–407. [Google Scholar] [CrossRef]
- Yegorov, Y.; Sendersky, E.; Zilberman, S.; Nagar, E.; Ben-Asher, H.W.; Shimoni, E.; Simkovsky, R.; Golden, S.S.; LiWang, A.; Schwarz, R. A Cyanobacterial Component Required for Pilus Biogenesis Affects the Exoproteome. mBio 2021, 12, e03674-20. [Google Scholar] [CrossRef]
- Harvey, K.L.; Jarocki, V.M.; Charles, I.G.; Djordjevic, S.P. The Diverse Functional Roles of Elongation Factor Tu (EF-Tu) in Microbial Pathogenesis. Front. Microbiol. 2019, 10, 2351. [Google Scholar] [CrossRef]
- Widjaja, M.; Harvey, K.L.; Hagemann, L.; Berry, I.J.; Jarocki, V.M.; Raymond, B.B.A.; Tacchi, J.L.; Gründel, A.; Steele, J.R.; Padula, M.P.; et al. Elongation factor Tu is a multifunctional and processed moonlighting protein. Sci. Rep. 2017, 7, 11227. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 2008, 20, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Oeda, K.; Watanabe, E.; Mikami, T.; Fukita, Y.; Nishimura, K.; Komai, K.; Matsuda, K. Protein function. Chaperonin turned insect toxin. Nature 2001, 411, 44. [Google Scholar] [CrossRef] [PubMed]
- Carter, R.A.; Worsley, P.S.; Sawers, G.; Challis, G.L.; Dilworth, M.J.; Carson, K.C.; Lawrence, J.A.; Wexler, M.; Johnston, A.W.B.; Yeoman, K.H. The vbs genes that direct synthesis of the siderophore vicibactin in Rhizobium leguminosarum: Their expression in other genera requires ECF σ factor RpoI. Mol. Microbiol. 2002, 44, 1153–1166. [Google Scholar] [CrossRef]
- Vercruysse, M.; Fauvart, M.; Jans, A.; Beullens, S.; Braeken, K.; Cloots, L.; Engelen, K.; Marchal, K.; Michiels, J. Stress response regulators identified through genome-wide transcriptome analysis of the (p)ppGpp-dependent response in Rhizobium etli. Genome Biol. 2011, 12, R17–R19. [Google Scholar] [CrossRef] [PubMed]
- Yurgel, S.N.; Qu, Y.; Rice, J.T.; Ajeethan, N.; Zink, E.M.; Brown, J.M.; Purvine, S.; Lipton, M.S.; Kahn, M.L. Specialization in a Nitrogen-Fixing Symbiosis: Proteome Differences Between Sinorhizobium medicae Bacteria and Bacteroids. Mol. Plant-Microbe Interact. 2021, 34, 1409–1422. [Google Scholar] [CrossRef] [PubMed]
- Meneses, N.; Taboada, H.; Dunn, M.F.; Vargas, M.D.C.; Buchs, N.; Heller, M.; Encarnación, S. The naringenin-induced exoproteome of Rhizobium etli CE3. Arch. Microbiol. 2017, 199, 737–755. [Google Scholar] [CrossRef]
- Meneses, N.; Mendoza-Hernández, G.; Encarnación, S. The extracellular proteome of Rhizobium etli CE3 in exponential and stationary growth phase. Proteome Sci. 2010, 8, 51. [Google Scholar] [CrossRef]
- Egea, L.; Aguilera, L.; Giménez, R.; Sorolla, M.; Aguilar, J.; Badía, J.; Baldoma, L. Role of secreted glyceraldehyde-3-phosphate dehydrogenase in the infection mechanism of enterohemorrhagic and enteropathogenic Escherichia coli: Interaction of the extracellular enzyme with human plasminogen and fibrinogen. Int. J. Biochem. Cell Biol. 2007, 39, 1190–1203. [Google Scholar] [CrossRef]
- Giménez Claudio, R.; Aguilera Gil, M.L.; Ferreira, E.; Aguilar Piera, J.; Baldomà Llavinés, L.; Badía Palacín, J. Glyceraldehyde-3-phosphate dehydrogenase as a moonlighting protein in bacteria. In Recent Advances in Pharmaceutical Sciences IV; Diego Muñoz Torrero, M.V.-C.J.E.i.L., Ed.; Research Signpost: Kerala, India, 2014; pp. 165–180. [Google Scholar]
- Sirover, M.A. Structural analysis of glyceraldehyde-3-phosphate dehydrogenase functional diversity. Int. J. Biochem. Cell Biol. 2014, 57, 20–26. [Google Scholar] [CrossRef]
- Henderson, B. Moonlighting Proteins: Novel Virulence Factors in Bacterial Infections; Wiley Blackwell, John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017. [Google Scholar]
- Marcos, C.M.; De Oliveira, H.C.; Silva, J.D.F.D.; Assato, P.A.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. The multifaceted roles of metabolic enzymes in the Paracoccidioides species complex. Front. Microbiol. 2014, 5, 719. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. Why study moonlighting proteins? Front. Genet. 2015, 6, 211. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.; Martin, A. Bacterial Moonlighting Proteins and Bacterial Virulence. Curr. Top. Microbiol. Immunol. 2013, 358, 155–213. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. Intracellular/surface moonlighting proteins that aid in the attachment of gut microbiota to the host. AIMS Microbiol. 2019, 5, 77–86. [Google Scholar] [CrossRef]
- Forrest, S.; Welch, M. Arming the troops: Post-translational modification of extracellular bacterial proteins. Sci. Prog. 2020, 103, 36850420964317. [Google Scholar] [CrossRef] [PubMed]
- Macek, B.; Forchhammer, K.; Hardouin, J.; Weber-Ban, E.; Grangeasse, C.; Mijakovic, I. Protein post-translational modifications in bacteria. Nat. Rev. Microbiol. 2019, 17, 651–664. [Google Scholar] [CrossRef]
- Leutert, M.; Entwisle, S.W.; Villén, J. Decoding Post-Translational Modification Crosstalk with Proteomics. Mol. Cell Proteom. 2021, 20, 100129. [Google Scholar] [CrossRef]
- Zhao, Y.; Jensen, O.N. Modification-specific proteomics: Strategies for characterization of post-translational modifications using enrichment techniques. Proteomics 2009, 9, 4632–4641. [Google Scholar] [CrossRef]
- Jeffery, C.J. Protein species and moonlighting proteins: Very small changes in a protein’s covalent structure can change its biochemical function. J. Proteom. 2016, 134, 19–24. [Google Scholar] [CrossRef]
- Yagüe, P.; Gonzalez-Quiñonez, N.; Fernández-García, G.; Alonso-Fernández, S.; Manteca, A. Goals and Challenges in Bacterial Phosphoproteomics. Int. J. Mol. Sci. 2019, 20, 5678. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, H.; Zhang, X.; Jiang, H.; Liu, C.; Wu, F.; Qian, L.; Hao, B.; Czajkowsky, D.M.; Guo, S.; et al. Interplay between the bacterial protein deacetylase CobB and the second messenger c-di- GMP. EMBO J. 2019, 38, e100948. [Google Scholar] [CrossRef] [PubMed]
- Grenga, L.; Little, R.H.; Chandra, G.; Woodcock, S.D.; Saalbach, G.; Morris, R.; Malone, J.G. Control of mRNA translation by dynamic ribosome modification. PLoS Genet. 2020, 16, e1008837. [Google Scholar] [CrossRef] [PubMed]
- Little, R.H.; Grenga, L.; Saalbach, G.; Howat, A.M.; Pfeilmeier, S.; Trampari, E.; Malone, J.G. Adaptive Remodeling of the Bacterial Proteome by Specific Ribosomal Modification Regulates Pseudomonas Infection and Niche Colonisation. PLoS Genet. 2016, 12, e1005837. [Google Scholar] [CrossRef] [PubMed]
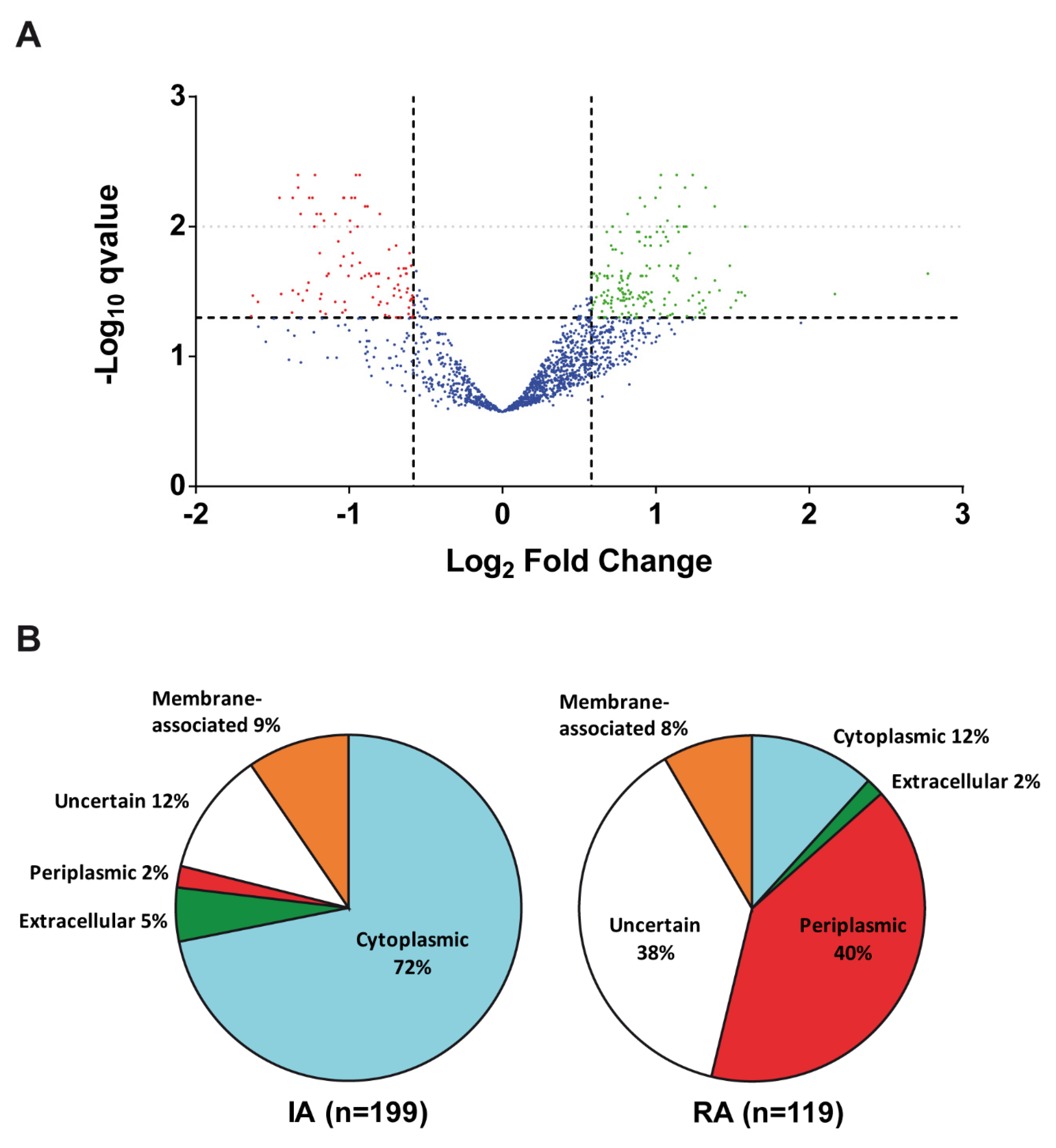
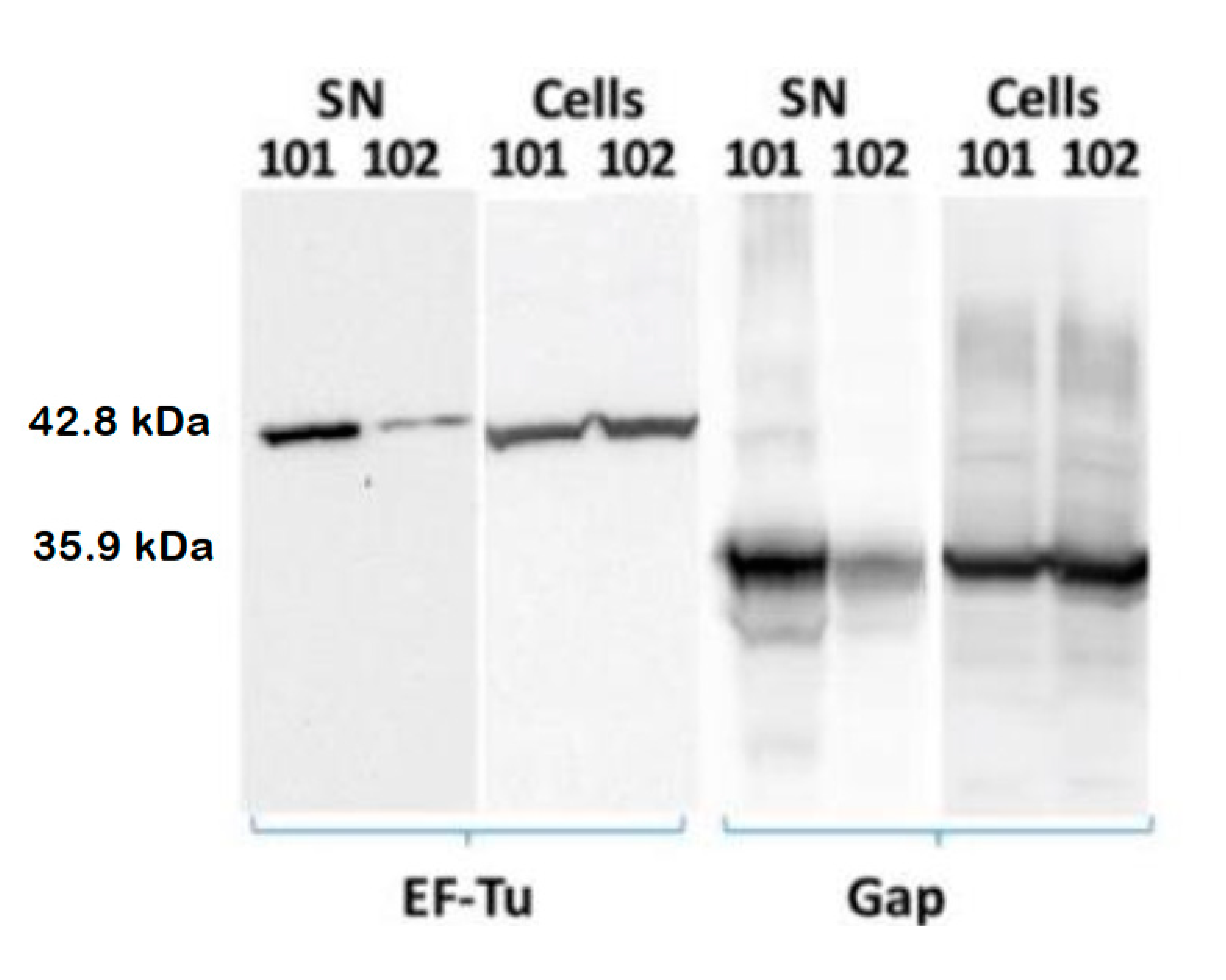
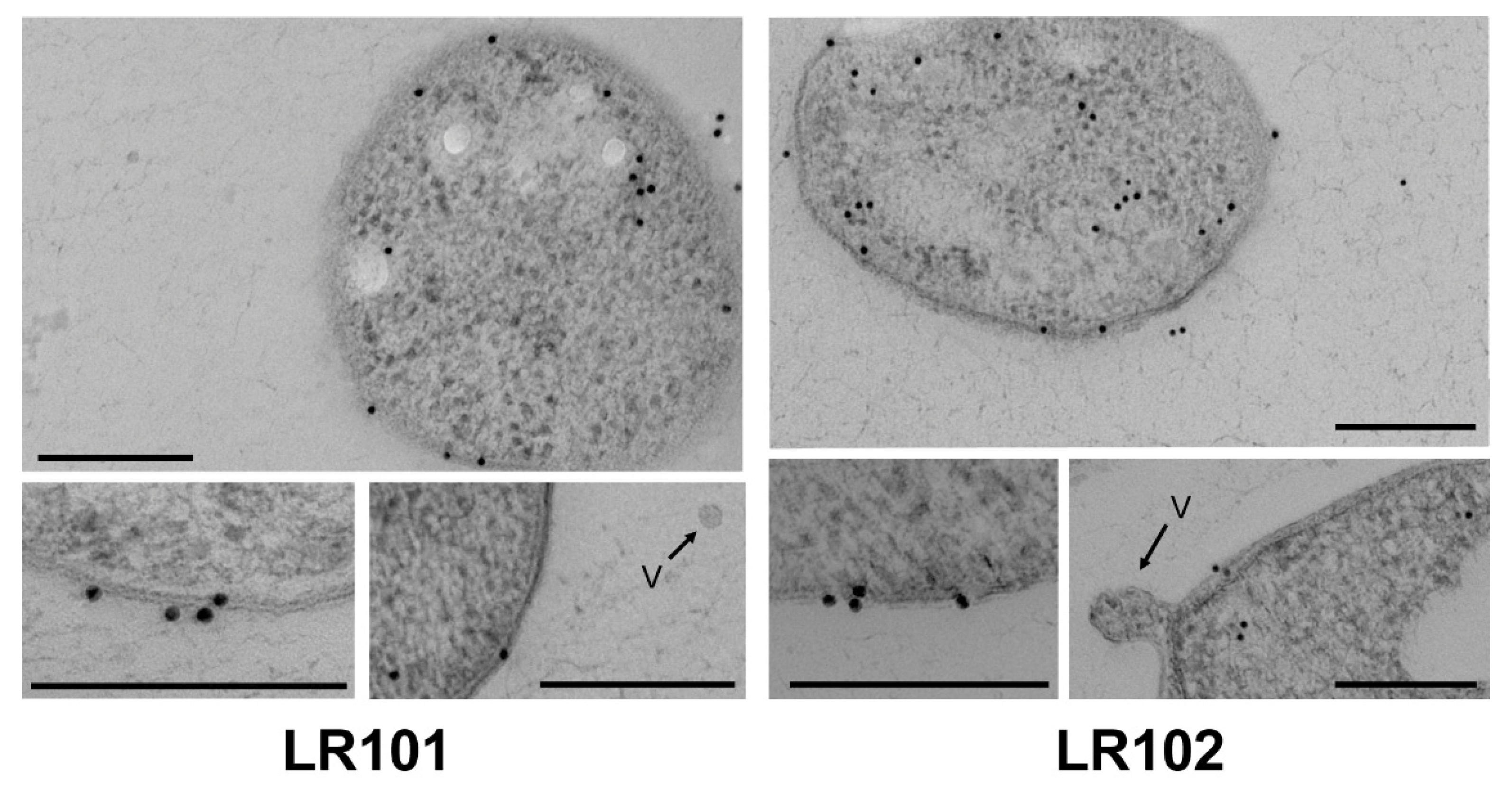
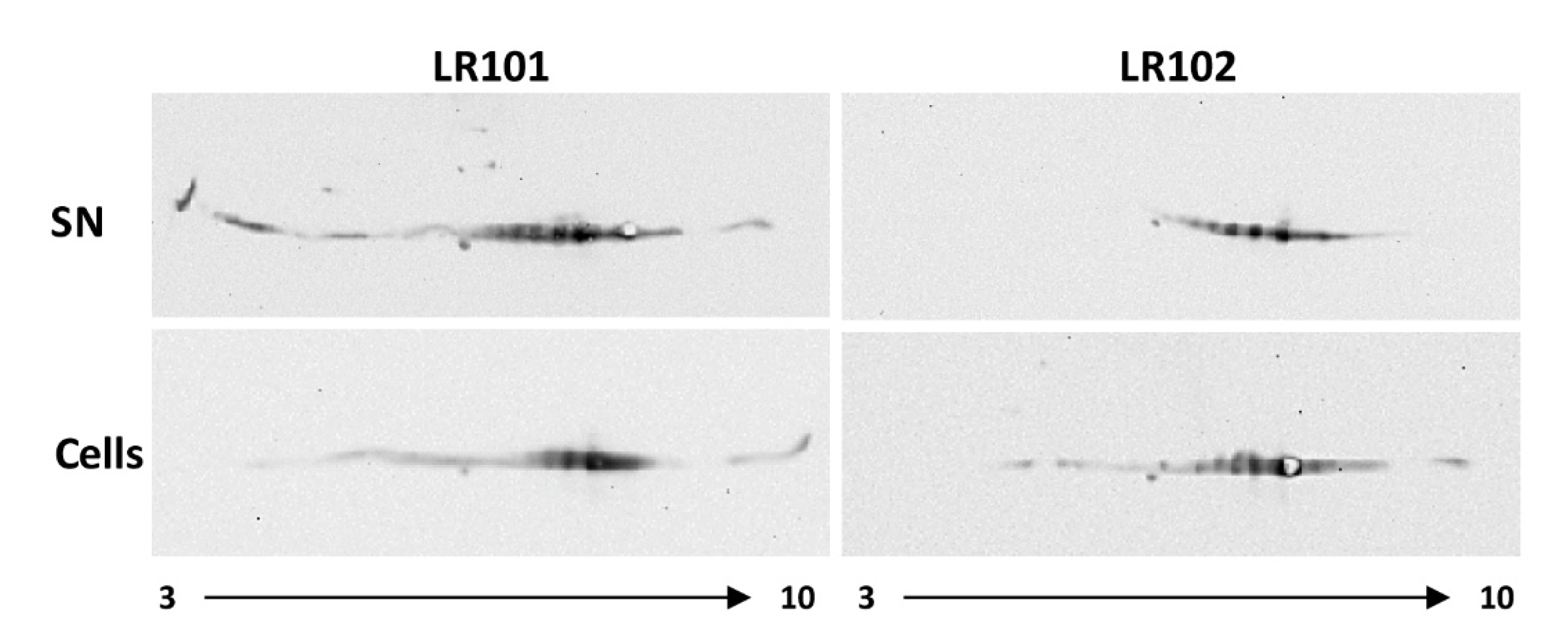
| Category | Function | Differential Abundance Proteins | |
|---|---|---|---|
| Increased Abundance (IA) | Reduced Abundance (RA) | ||
| Extracellular Function and Structural | Aggregation/Adhesion | RapAch, RHE_CH02633, RHE_CH02634 | RapB1, RapB2 |
| Flagella/Motility | FlaCch2, FlaCch3, FlaCch4, FlaCch5, FlaCe, FlgKch, FlgEch, FlgD, FlgG, FlgL | ||
| Peptidoglycan | RHE_CH02869, RHE_CH04101, RHE_CH01507 | ||
| Polysaccharide | RkpK, PssP | PlyA1, PssO | |
| Conjugation | TrbB, TrbE | ||
| Protein Synthesis and Modifications | Chaperones | GroL1, GroL2, GroL4, Tig, YidC, DnaK, | PpiD2 |
| Proteases | ClpA, ClpP2, ClpP3, ClpX, FtsH, HflC, HflK, HslU, HslV, Lon, PtrB | RHE_CH01654 | |
| Translation | GatA, GatB, Tuf, FusA, GltX, RpsA, RpsB, RpsC, RpsD, RpsE, RpsH, RpsK, RpsQ, RpsU1, RplA, RplB, RplC, RplD, RplE, RplI, RplN, RplP, RplS, RplU, RplV, RplX, RplY, RpmA, YchF | Fmt | |
| Carbon Metabolism | Glycolysis/TCA | Eno, Frk, Pfk, FbaB, Gap, Glk, PdhA1, PdhA2, PdhB, LpdAch1 SucA, SucB, SucC, SucD, Mdh, GltA, FumC | |
| Others | PckA, GlgC, Eda, KdgK, XylA, RHE_CH03723 | GalE2, CynT, RHE_PD00192 | |
| Nucleid Acid Metabolism | Transcription | NusA, RpoA, RpoB, RpoC, RpoD, Mfd, Pnp | GreA, IalA, |
| Purin and Pyrimidine Metabolism | PurA, PurB, GuaB, Upp, PyrH, CodAb | CpdB, CyaJ | |
| Replication/Recombination | DnaN, MgpS, GyrA, GyrB, ParC, RecA | ||
| Others | Hrm | ||
| Nitrogen Metabolism | Amino Acid Metabolism | AldA, DadA, ArgF, SerA, SerC, ThrA, TyrC, HisB, IlvC, IlvDch2, IlvE2, MetK, MetZ, CysK1, MdeAf, AhcY, AatAch, AatCch, | RHE_PF00434 |
| BNF-Symbiosis | NifA, NodTch, NtrY, NoeJ, ChvI | NodD-like RHE_PD00316 | |
| Nitrogen Assimilation | GlnA1, GlnA2, AmtB | ||
| Lipid Metabolism | Lipid biosynthesis and degradation | HbdA, PhbAch, PhbB, FabA, FabF1, FabI1, Acd1, RHE_CH01048, RHE_CH03352, RHE_CH01307 | |
| Energy Production/Electron transfer | ATP synthesis and electron transfer | AtpA, AtpC, AtpD, AtpF2, AtpG, AtpH, FbcC, FbcF, RrpP, EtfAf, CtaC, NdhCh | CycF, RHE_CH02618 |
| Cofactors–Metals | Biosynthesis of cofactors and metal metabolism | Bfr, CobN, CobS, ThiO, RHE_CH00859 | |
| Biosynthesis of siderophores | VbsA, VbsL | ||
| Stress | Stress responses | OsmC | NerA |
| Transport | Transporters | AglK, SecD1, PstB, ZnuC, MntH, TolQ, SufC, RHE_CH00008, RHE_CH02346, RHE_PB00094 | PstS, TolB, UgpBc, UgpBch1, XylF, DppAch1, DppAch2, DppAch3, RbsBch3, AapJ, BraC1, BraC2, OppA, AfuA1, AfuA3, HmuT, OccT, ModA, PotF, MexE1, ZnuA, RHE_CH00971, RHE_PC00008, RHE_PC00118, RHE_PC00160, RHE_CH02418, RHE_PF00186, RHE_CH01465, RHE_CH04006, RHE_PF00410, RHE_CH02293, RHE_CH03027, RHE_CH03963, RHE_CH03445, RHE_PB00126, RHE_CH00175, RHE_PF00269, RHE_CH02683, RHE_CH02890, RHE_PE00259, RHE_PF00068, RHE_CH00492, RHE_CH01210, RHE_CH02898, RHE_PF00091, RHE_PB00025, RHE_PF00321, RHE_PB00139, RHE_PF00395, RHE_CH00485, RHE_PC00167, RHE_CH03866, RHE_CH02084 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorite, M.J.; Casas-Román, A.; Girard, L.; Encarnación, S.; Díaz-Garrido, N.; Badía, J.; Baldomá, L.; Pérez-Mendoza, D.; Sanjuán, J. Impact of c-di-GMP on the Extracellular Proteome of Rhizobium etli. Biology 2023, 12, 44. https://doi.org/10.3390/biology12010044
Lorite MJ, Casas-Román A, Girard L, Encarnación S, Díaz-Garrido N, Badía J, Baldomá L, Pérez-Mendoza D, Sanjuán J. Impact of c-di-GMP on the Extracellular Proteome of Rhizobium etli. Biology. 2023; 12(1):44. https://doi.org/10.3390/biology12010044
Chicago/Turabian StyleLorite, María J., Ariana Casas-Román, Lourdes Girard, Sergio Encarnación, Natalia Díaz-Garrido, Josefa Badía, Laura Baldomá, Daniel Pérez-Mendoza, and Juan Sanjuán. 2023. "Impact of c-di-GMP on the Extracellular Proteome of Rhizobium etli" Biology 12, no. 1: 44. https://doi.org/10.3390/biology12010044
APA StyleLorite, M. J., Casas-Román, A., Girard, L., Encarnación, S., Díaz-Garrido, N., Badía, J., Baldomá, L., Pérez-Mendoza, D., & Sanjuán, J. (2023). Impact of c-di-GMP on the Extracellular Proteome of Rhizobium etli. Biology, 12(1), 44. https://doi.org/10.3390/biology12010044









