Minimal Residual Disease Detected by the 7NB-mRNAs ddPCR Assay Is Associated with Disease Progression in High-Risk Neuroblastoma Patients: A Prospective Multicenter Observational Study in Japan
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
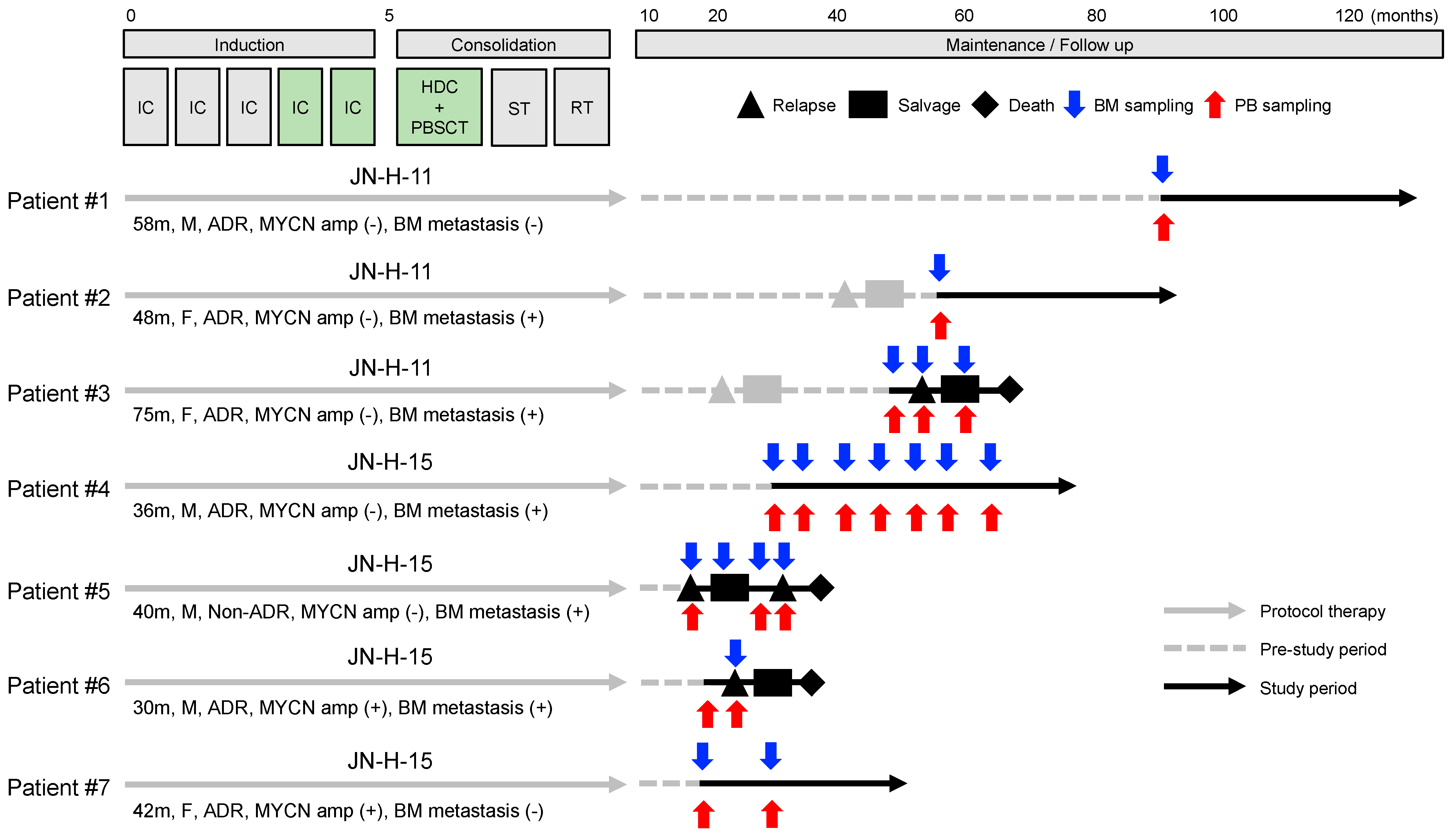
2.1. Study Design and Patient Samples
2.2. Patient Sample Processing and MRD Analysis
2.3. Disease Evaluation
2.4. Statistical Analysis
3. Results
3.1. Characteristics of Patients and Samples
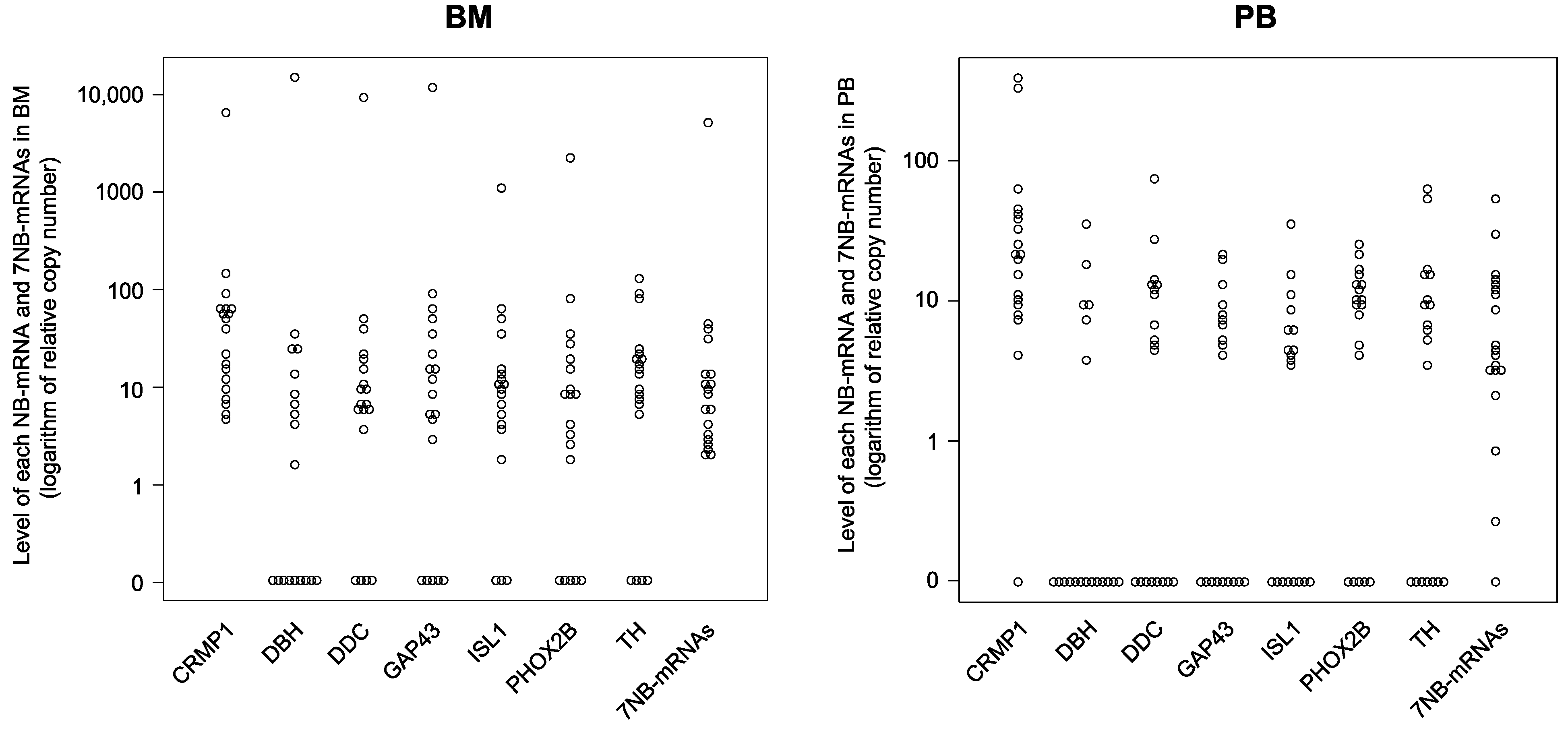
3.2. Expression Levels of Each NB-mRNA and 7NB-mRNAs in the BM and PB Samples
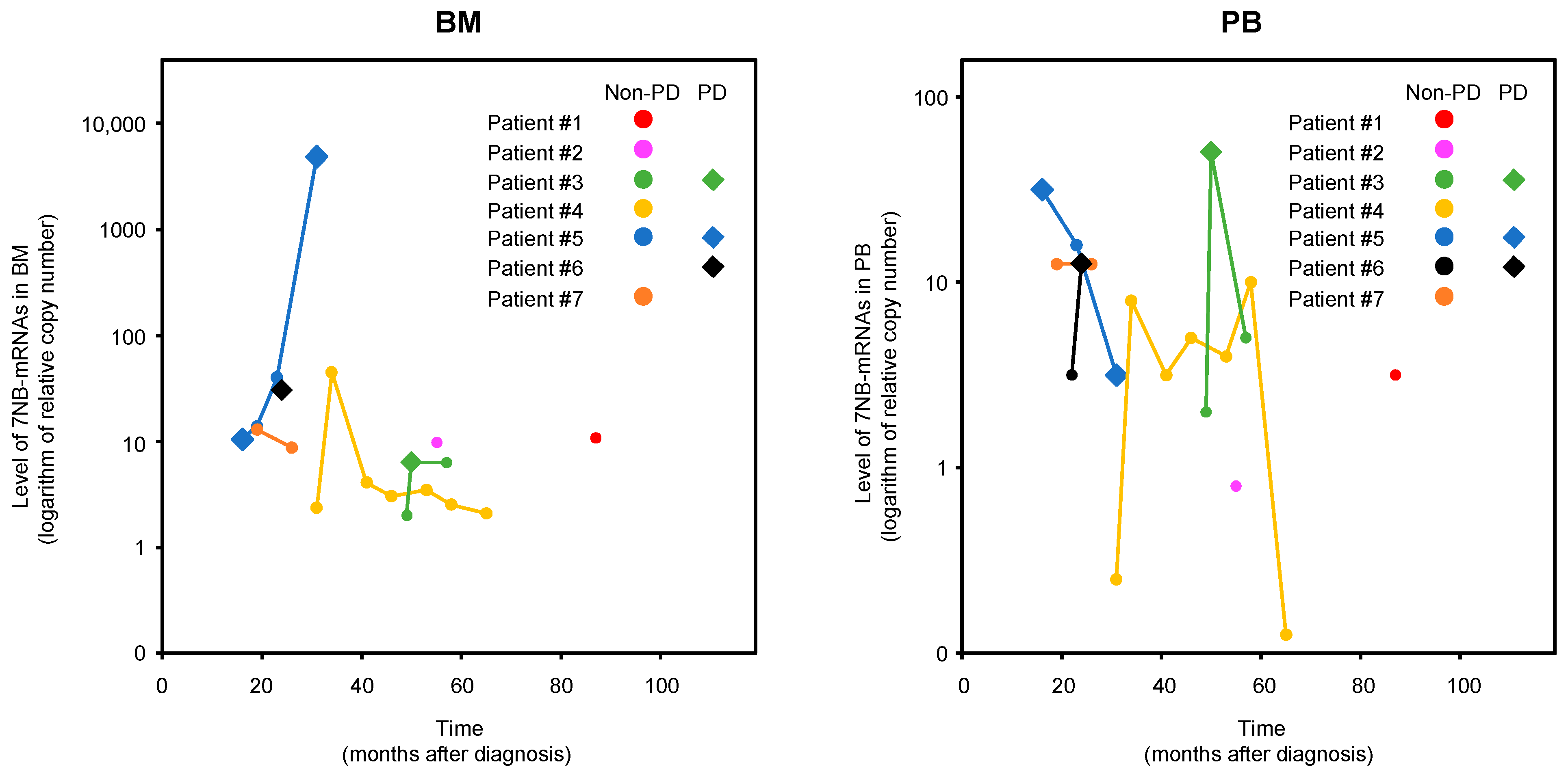
3.3. BM-MRD and PB-MRD Detected by 7NB-mRNAs ddPCR Assay
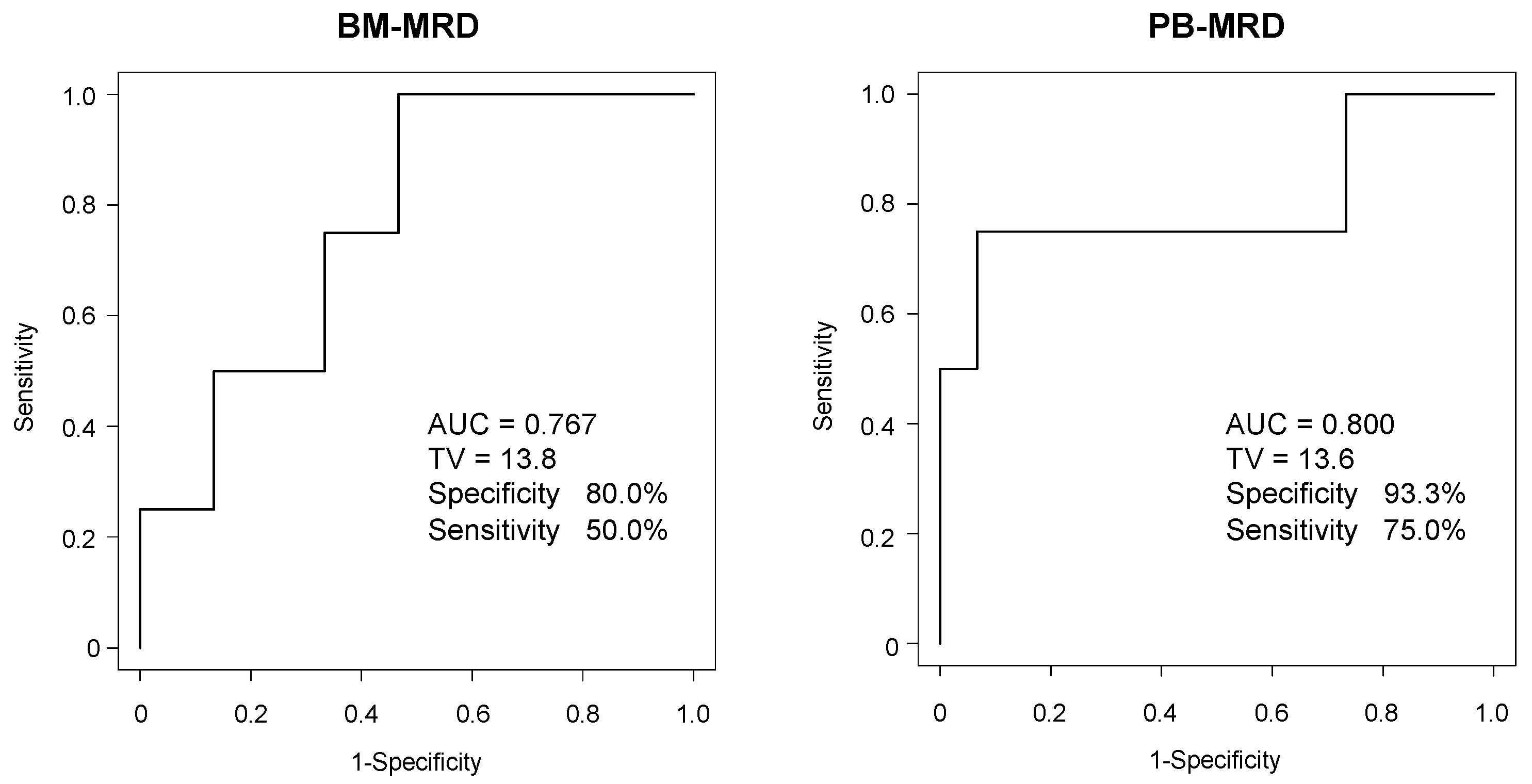
3.4. ROC Analysis of BM-MRD and PB-MRD for Disease Progression
3.5. PPV and NPV of BM-MRD and PB-MRD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Delloye-Bourgeois, C.; Castellani, V. Hijacking of Embryonic Programs by Neural Crest-Derived Neuroblastoma: From Physiological Migration to Metastatic Dissemination. Front. Mol. Neurosci. 2019, 12, 52. [Google Scholar] [CrossRef]
- Ponzoni, M.; Bachetti, T.; Corrias, M.V.; Brignole, C.; Pastorino, F.; Calarco, E.; Bensa, V.; Giusto, E.; Ceccherini, I.; Perri, P. Recent advances in the developmental origin of neuroblastoma: An overview. J. Exp. Clin. Cancer Res. 2022, 41, 92. [Google Scholar] [CrossRef]
- Brodeur, G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2120. [Google Scholar] [CrossRef] [PubMed]
- Monclair, T.; Brodeur, G.M.; Ambros, P.F.; Brisse, H.J.; Cecchetto, G.; Holmes, K.; Kaneko, M.; London, W.B.; Matthay, K.K.; Nuchtern, J.G.; et al. The International Neuroblastoma Risk Group (INRG) Staging System: An INRG Task Force Report. J. Clin. Oncol. 2009, 27, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Cohn, S.L.; Pearson, A.D.J.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The International Neuroblastoma Risk Group (INRG) Classification System: An INRG Task Force Report. J. Clin. Oncol. 2009, 27, 289–297. [Google Scholar] [CrossRef]
- Liang, W.H.; Federico, S.M.; London, W.B.; Naranjo, A.; Irwin, M.S.; Volchenboum, S.L.; Cohn, S.L. Tailoring Therapy for Children With Neuroblastoma on the Basis of Risk Group Classification: Past, Present, and Future. JCO Clin. Cancer Inform. 2020, 4, 895–905. [Google Scholar] [CrossRef]
- Park, J.R.; Bagatell, R.; London, W.B.; Maris, J.M.; Cohn, S.L.; Mattay, K.K.; Hogarty, M.; Committee COGN. Children’s Oncology Group’s 2013 blueprint for research: Neuroblastoma. Pediatr. Blood Cancer 2013, 60, 985–993. [Google Scholar] [CrossRef]
- Tolbert, V.P.; Matthay, K.K. Neuroblastoma: Clinical and biological approach to risk stratification and treatment. Cell Tissue Res. 2018, 372, 195–209. [Google Scholar] [CrossRef]
- Beiske, K.; Ambros, P.F.; Burchill, S.A.; Cheung, I.Y.; Swerts, K. Detecting minimal residual disease in neuroblastoma patients-the present state of the art. Cancer Lett. 2005, 228, 229–240. [Google Scholar] [CrossRef]
- Brownhill, S.C.; Burchill, S.A. PCR-based amplification of circulating RNAs as prognostic and predictive biomarkers—Focus on neuroblastoma. Pract. Lab. Med. 2017, 7, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Stutterheim, J.; Zappeij-Kannegieter, L.; Versteeg, R.; Caron, H.N.; van der Schoot, C.E.; Tytgat, G.A. The prognostic value of fast molecular response of marrow disease in patients aged over 1 year with stage 4 neuroblastoma. Eur. J. Cancer 2011, 47, 1193–1202. [Google Scholar] [CrossRef]
- Viprey, V.F.; Gregory, W.M.; Corrias, M.V.; Tchirkov, A.; Swerts, K.; Vicha, A.; Dallorso, S.; Brock, P.; Luksch, R.; Valteau-Couanet, D.; et al. Neuroblastoma mRNAs predict outcome in children with stage 4 neuroblastoma: A European HR-NBL1/SIOPEN study. J. Clin. Oncol. 2014, 32, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.K.; Ostrovnaya, I.; Kuk, D.; Cheung, I.Y. Bone marrow minimal residual disease was an early response marker and a consistent independent predictor of survival after anti-GD2 immunotherapy. J. Clin. Oncol. 2015, 33, 755–763. [Google Scholar] [CrossRef]
- Yanez, Y.; Hervas, D.; Grau, E.; Oltra, S.; Perez, G.; Palanca, S.; Bermudez, M.; Marquez, C.; Canete, A.; Castel, V. TH and DCX mRNAs in peripheral blood and bone marrow predict outcome in metastatic neuroblastoma patients. J. Cancer Res. Clin. Oncol. 2016, 142, 573–580. [Google Scholar] [CrossRef]
- Marachelian, A.; Villablanca, J.G.; Liu, C.W.; Liu, B.; Goodarzian, F.; Lai, H.A.; Shimada, H.; Tran, H.C.; Parra, J.A.; Gallego, R.; et al. Expression of Five Neuroblastoma Genes in Bone Marrow or Blood of Patients with Relapsed/Refractory Neuroblastoma Provides a New Biomarker for Disease and Prognosis. Clin. Cancer Res. 2017, 23, 5374–5383. [Google Scholar] [CrossRef]
- Thwin, K.K.M.; Ishida, T.; Uemura, S.; Yamamoto, N.; Lin, K.S.; Tamura, A.; Kozaki, A.; Saito, A.; Kishimoto, K.; Mori, T.; et al. Level of Seven Neuroblastoma-Associated mRNAs Detected by Droplet Digital PCR Is Associated with Tumor Relapse/Regrowth of High-Risk Neuroblastoma Patients. J. Mol. Diagn. 2020, 22, 236–246. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The digital MIQE guidelines: Minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Burchill, S.A.; Beiske, K.; Shimada, H.; Ambros, P.F.; Seeger, R.; Tytgat, G.A.; Brock, P.R.; Haber, M.; Park, J.R.; Berthold, F. Recommendations for the standardization of bone marrow disease assessment and reporting in children with neuroblastoma on behalf of the International Neuroblastoma Response Criteria Bone Marrow Working Group. Cancer 2017, 123, 1095–1105. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Pritchard, J.; Berthold, F.; Carlsen, N.L.; Castel, V.; Castelberry, R.P.; De Bernardi, B.; Evans, A.E.; Favrot, M.; Hedborg, F.; et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J. Clin. Oncol. 1993, 11, 1466–1477. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Hishiki, T.; Matsumoto, K.; Ohira, M.; Kamijo, T.; Shichino, H.; Kuroda, T.; Yoneda, A.; Soejima, T.; Nakazawa, A.; Takimoto, T.; et al. Results of a phase II trial for high-risk neuroblastoma treatment protocol JN-H-07: A report from the Japan Childhood Cancer Group Neuroblastoma Committee (JNBSG). Int. J. Clin. Oncol. 2018, 23, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Moss, T.J.; Sanders, D.G. Detection of neuroblastoma cells in blood. J. Clin. Oncol. 1990, 8, 736–740. [Google Scholar] [CrossRef]
- Seeger, R.C.; Reynolds, C.P.; Gallego, R.; Stram, D.O.; Gerbing, R.B.; Matthay, K.K. Quantitative tumor cell content of bone marrow and blood as a predictor of outcome in stage IV neuroblastoma: A Children’s Cancer Group Study. J. Clin. Oncol. 2000, 18, 4067–4076. [Google Scholar] [CrossRef]
| Enrolled Patients | |
|---|---|
| Treatment protocol | |
| JN-H-11 | 3 |
| JN-H-15 | 4 |
| Registration (months after initial diagnosis) | |
| Median | 31 |
| Range | 16–87 |
| Observation period (months) | |
| Median | 33 |
| Range | 14–47 |
| Disease evaluation at registration | |
| PD | 1 |
| Non-PD | 6 |
| Outcome | |
| Alive | 4 |
| Dead | 3 |
| Age at diagnosis (months) | |
| Median | 42 |
| Range | 30–75 |
| Sex | |
| Male | 4 |
| Female | 3 |
| Primary tumor site | |
| Adrenal gland | 6 |
| Non-adrenal gland | 1 |
| MYCN status | |
| Amplified | 2 |
| Non-amplified | 5 |
| BM metastasis at diagnosis | |
| Positive | 5 |
| Negative | 2 |
| INSS stage at diagnosis | |
| Stage 3 | 1 |
| Stage 4 | 6 |
| Collected samples | |
| BM samples | Total: 19 Per patient: 1–7 (Median 2) |
| PB samples | Total: 19 Per patient: 1–7 (Median 2) |
| BM Samples | PB Samples | |||||||
|---|---|---|---|---|---|---|---|---|
| Range | Median | IQR | n | Range | Median | IQR | n | |
| CRMP1 mRNA | 4.71–6867.47 | 37.65 | 10.55–62.86 | 19 | 0.00–396.83 | 20.77 | 9.93–40.53 | 19 |
| DBH mRNA | 0.00–14,337.35 | 1.64 | 0.00–11.08 | 19 | 0.00–35.44 | 0.00 | 0.00–5.55 | 19 |
| DDC mRNA | 0.00–9530.12 | 6.73 | 4.69–17.08 | 19 | 0.00–75.95 | 4.73 | 0.00–12.53 | 19 |
| GAP43 mRNA | 0.00–12,289.16 | 8.82 | 1.43–27.32 | 19 | 0.00–21.24 | 4.14 | 0.00–7.49 | 19 |
| ISL1 mRNA | 0.00–1168.67 | 9.94 | 3.94–14.31 | 19 | 0.00–36.73 | 3.88 | 0.00–6.15 | 19 |
| PHOX2B mRNA | 0.00–2216.87 | 8.07 | 0.94–17.53 | 19 | 0.00–25.32 | 9.52 | 2.07–13.36 | 19 |
| TH mRNA | 0.00–124.26 | 13.04 | 6.08–21.18 | 19 | 0.00–63.10 | 6.18 | 0.00–12.63 | 19 |
| 7NB-mRNAs | 2.00–4948.76 | 8.67 | 3.23–13.37 | 19 | 0.00–51.95 | 4.60 | 3.13–12.71 | 19 |
| Observation | BM-MRD Cut off = 15 | PB-MRD Cut off = 13 | ||
|---|---|---|---|---|
| PPV | NPV | PPV | NPV | |
| 0 M | 50% (2/4) | 87% (13/15) | 75% (3/4) | 93% (14/15) |
| 6 M | 75% (3/4) | 73% (11/15) | 100% (4/4) | 73% (11/15) |
| >12 M | 75% (3/4) | 67% (10/15) | 100% (4/4) | 73% (11/15) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishimura, N.; Ishida, T.; Yokota, I.; Matsumoto, K.; Shichino, H.; Fujisaki, H.; Sarashina, T.; Kamijo, T.; Takimoto, T.; Iehara, T.; et al. Minimal Residual Disease Detected by the 7NB-mRNAs ddPCR Assay Is Associated with Disease Progression in High-Risk Neuroblastoma Patients: A Prospective Multicenter Observational Study in Japan. Biology 2023, 12, 1350. https://doi.org/10.3390/biology12101350
Nishimura N, Ishida T, Yokota I, Matsumoto K, Shichino H, Fujisaki H, Sarashina T, Kamijo T, Takimoto T, Iehara T, et al. Minimal Residual Disease Detected by the 7NB-mRNAs ddPCR Assay Is Associated with Disease Progression in High-Risk Neuroblastoma Patients: A Prospective Multicenter Observational Study in Japan. Biology. 2023; 12(10):1350. https://doi.org/10.3390/biology12101350
Chicago/Turabian StyleNishimura, Noriyuki, Toshiaki Ishida, Isao Yokota, Kimikazu Matsumoto, Hiroyuki Shichino, Hiroyuki Fujisaki, Takeo Sarashina, Takehiko Kamijo, Tetsuya Takimoto, Tomoko Iehara, and et al. 2023. "Minimal Residual Disease Detected by the 7NB-mRNAs ddPCR Assay Is Associated with Disease Progression in High-Risk Neuroblastoma Patients: A Prospective Multicenter Observational Study in Japan" Biology 12, no. 10: 1350. https://doi.org/10.3390/biology12101350
APA StyleNishimura, N., Ishida, T., Yokota, I., Matsumoto, K., Shichino, H., Fujisaki, H., Sarashina, T., Kamijo, T., Takimoto, T., Iehara, T., Tajiri, T., & on behalf of the JCCG Neuroblastoma Committee. (2023). Minimal Residual Disease Detected by the 7NB-mRNAs ddPCR Assay Is Associated with Disease Progression in High-Risk Neuroblastoma Patients: A Prospective Multicenter Observational Study in Japan. Biology, 12(10), 1350. https://doi.org/10.3390/biology12101350






