Tissue-Specific Transcriptomes in the Secondary Cell Wall Provide an Understanding of Stem Growth Enhancement in Solidago canadensis during Invasion
Abstract
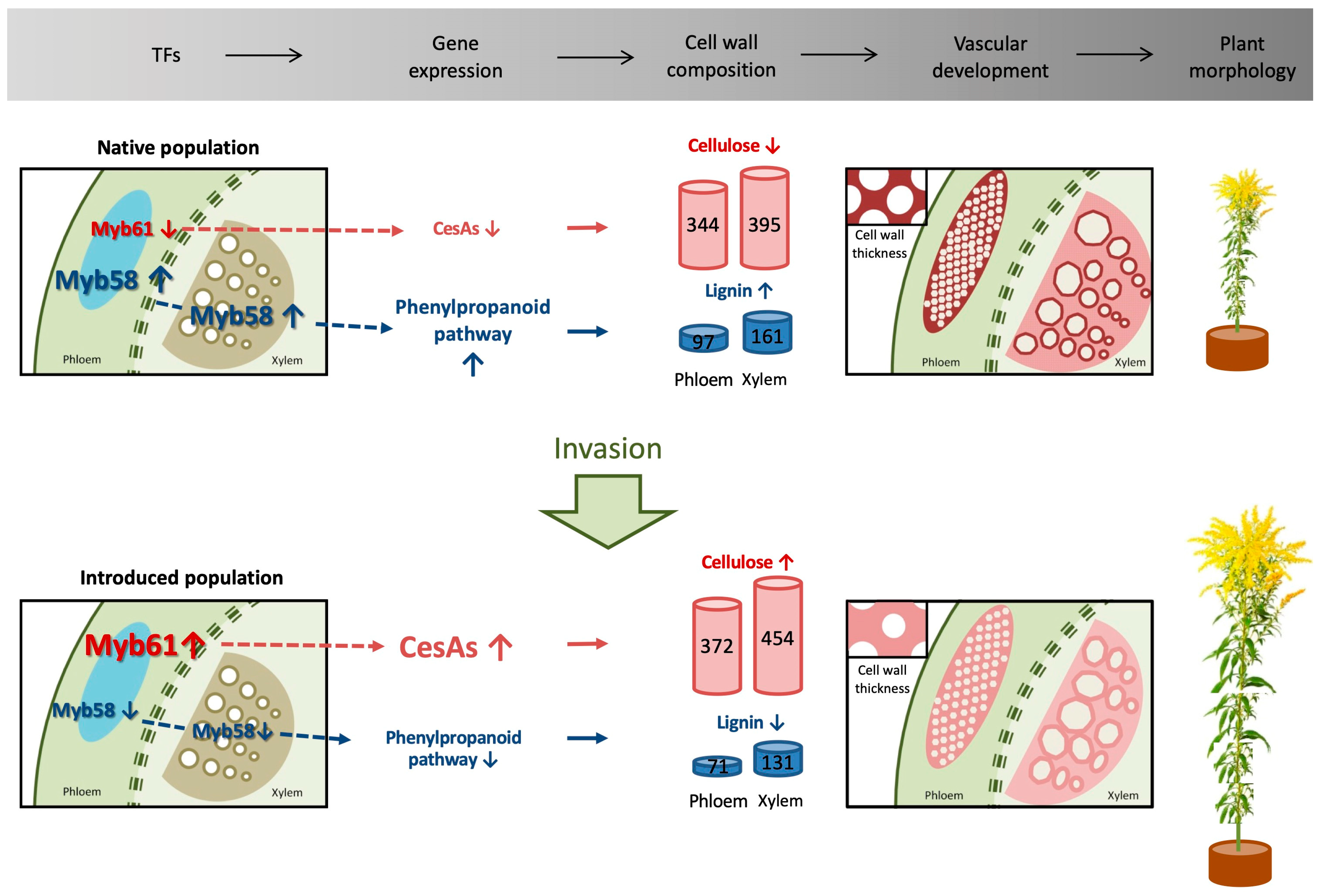
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of S. canadensis Populations
2.2. Common Garden Experiments
2.3. Stem Biomass Allocation and Anatomical Analysis
2.4. RNA Extraction, cDNA Library Preparation, and RNA-Seq
2.5. De Novo Assembly and Functional Annotation
2.6. Differentially Expressed Gene Analysis
2.7. Quantitative RT-PCR
2.8. Statistical Analysis
3. Results
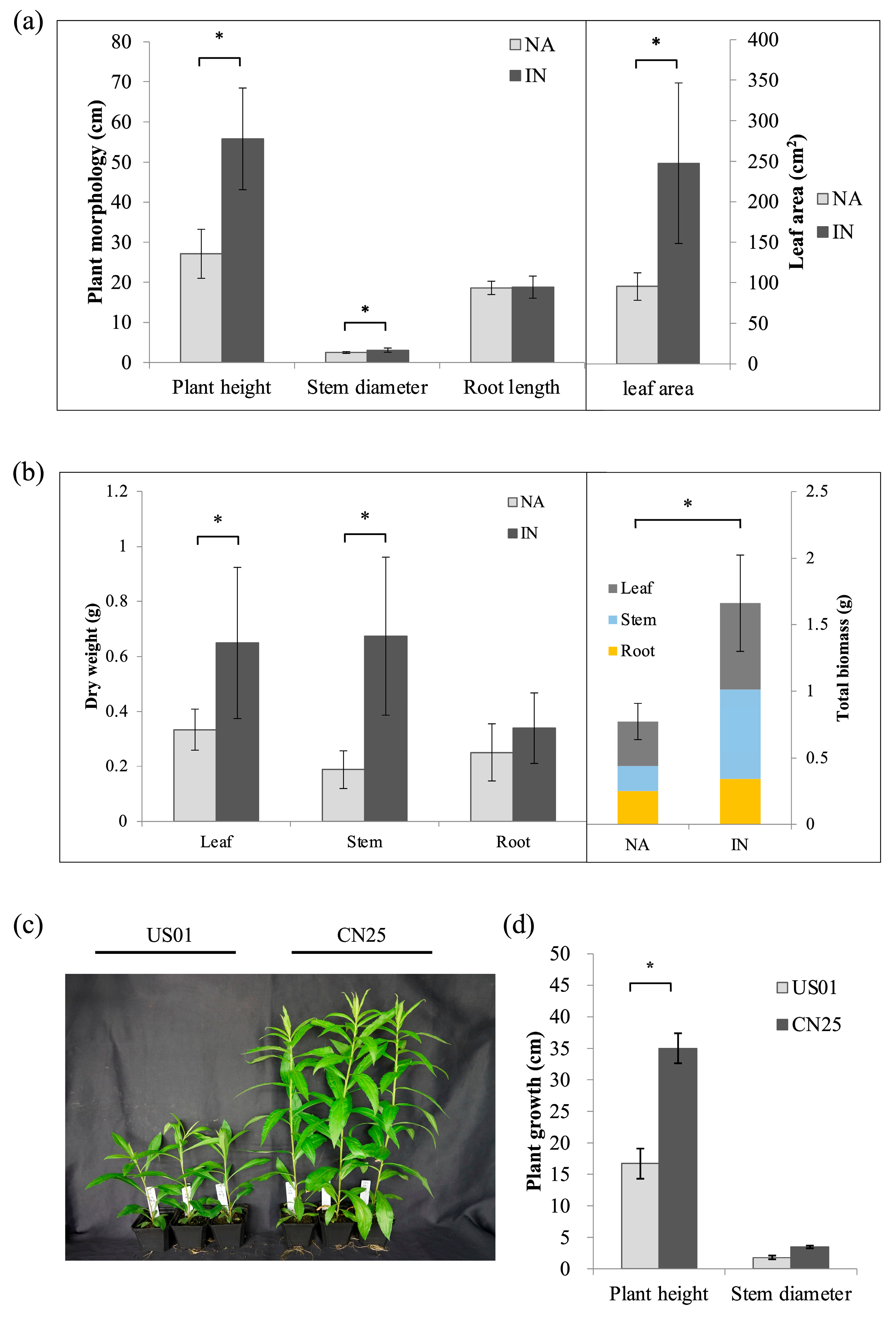
3.1. Invasive Population of S. canadensis Displayed a Stronger Aboveground Growth Ability
3.2. RNA-Sequencing and De Novo Assembly
3.3. Functional Annotation
3.4. DEG Identification between CN25 and US01 Populations Shows Tissue-Specific Expression
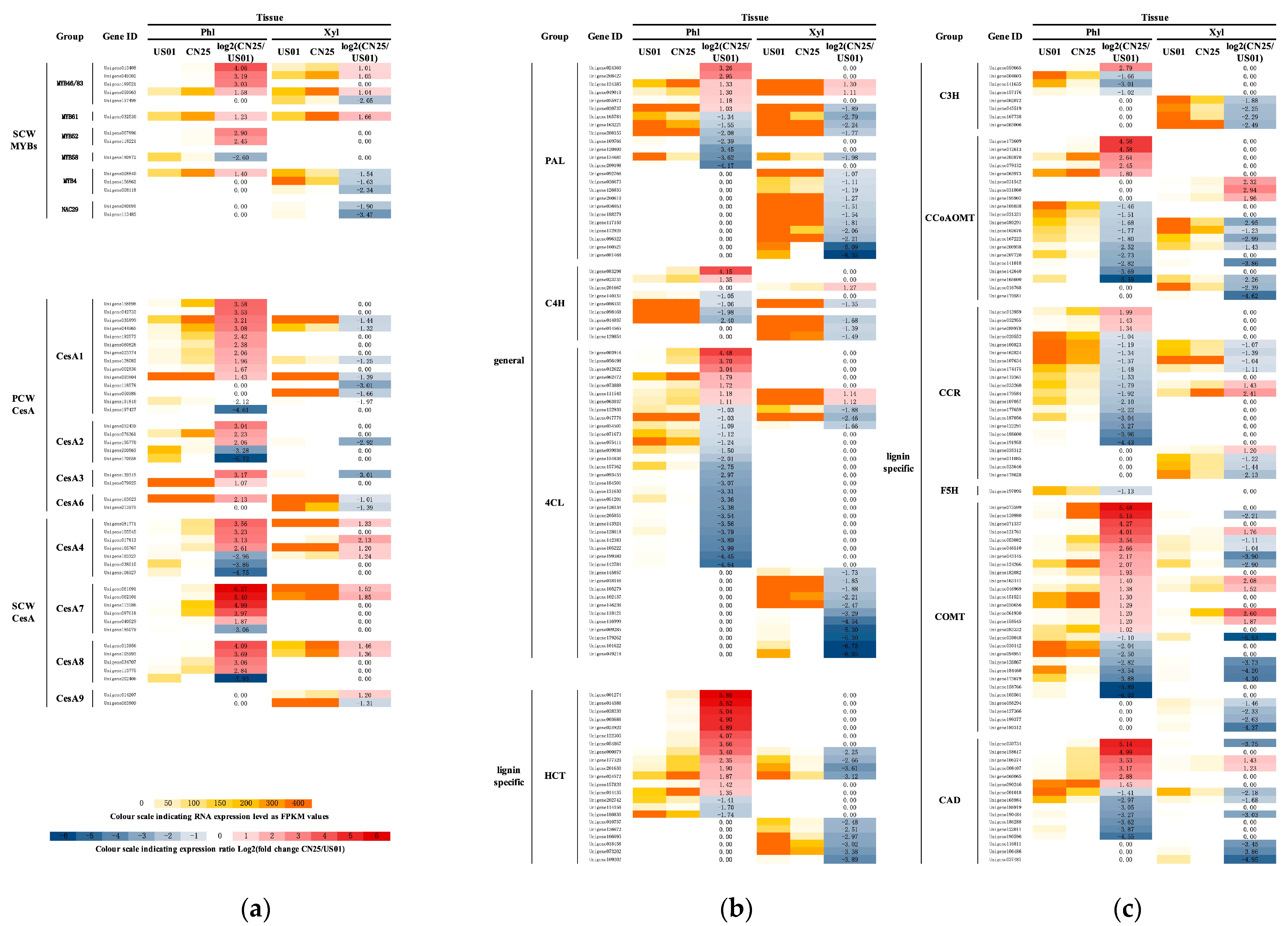
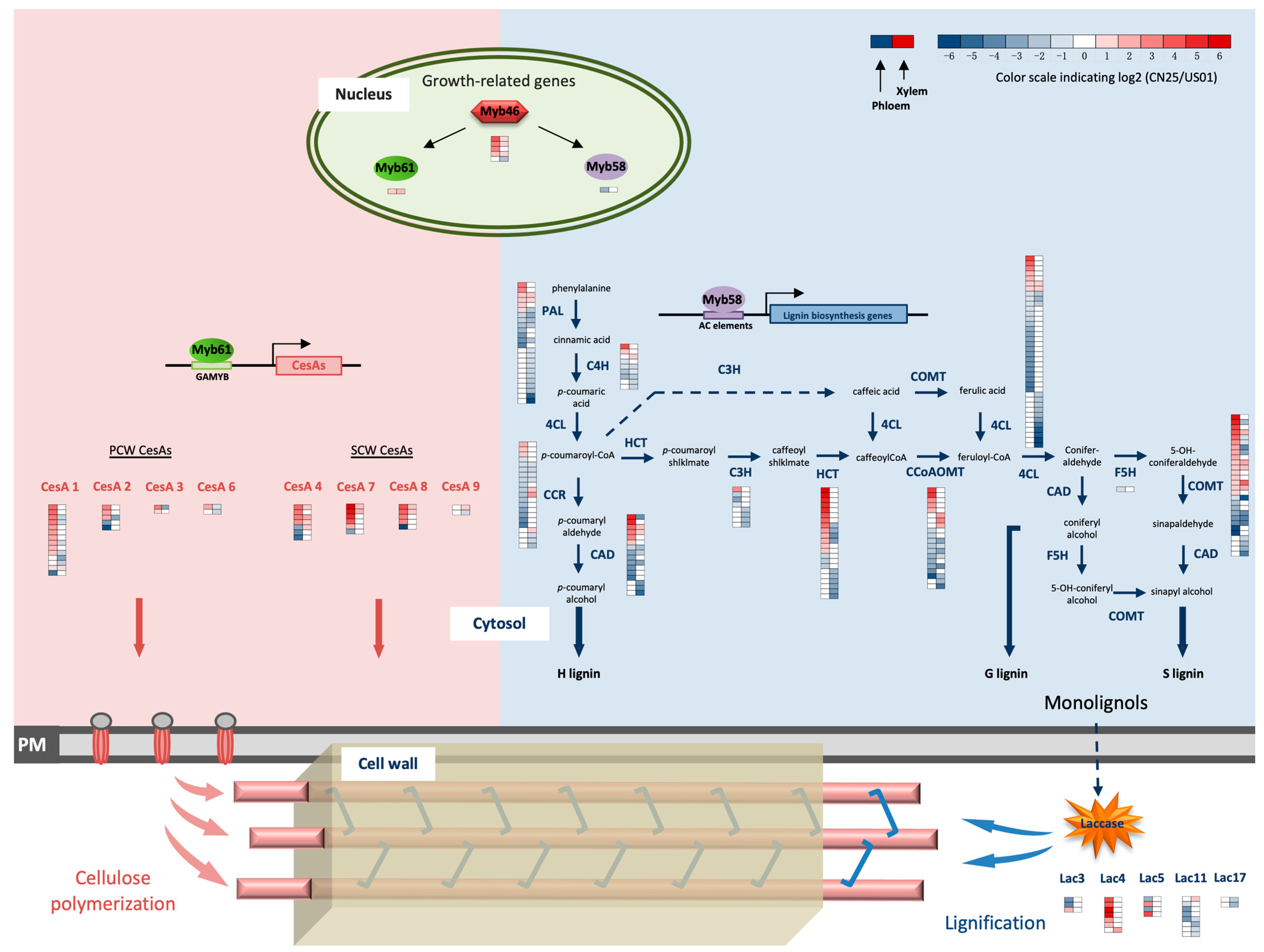
3.5. MYB Transcription Factors Are Likely to Be Regulators of SCW Development in S. canadensis
3.6. Cellulose Biosynthesis Genes Are Highly Expressed in Invasive Populations of S. canadensis
3.7. Lignin-Related Genes Are Weakly Expressed in Invasive Populations of S. canadensis
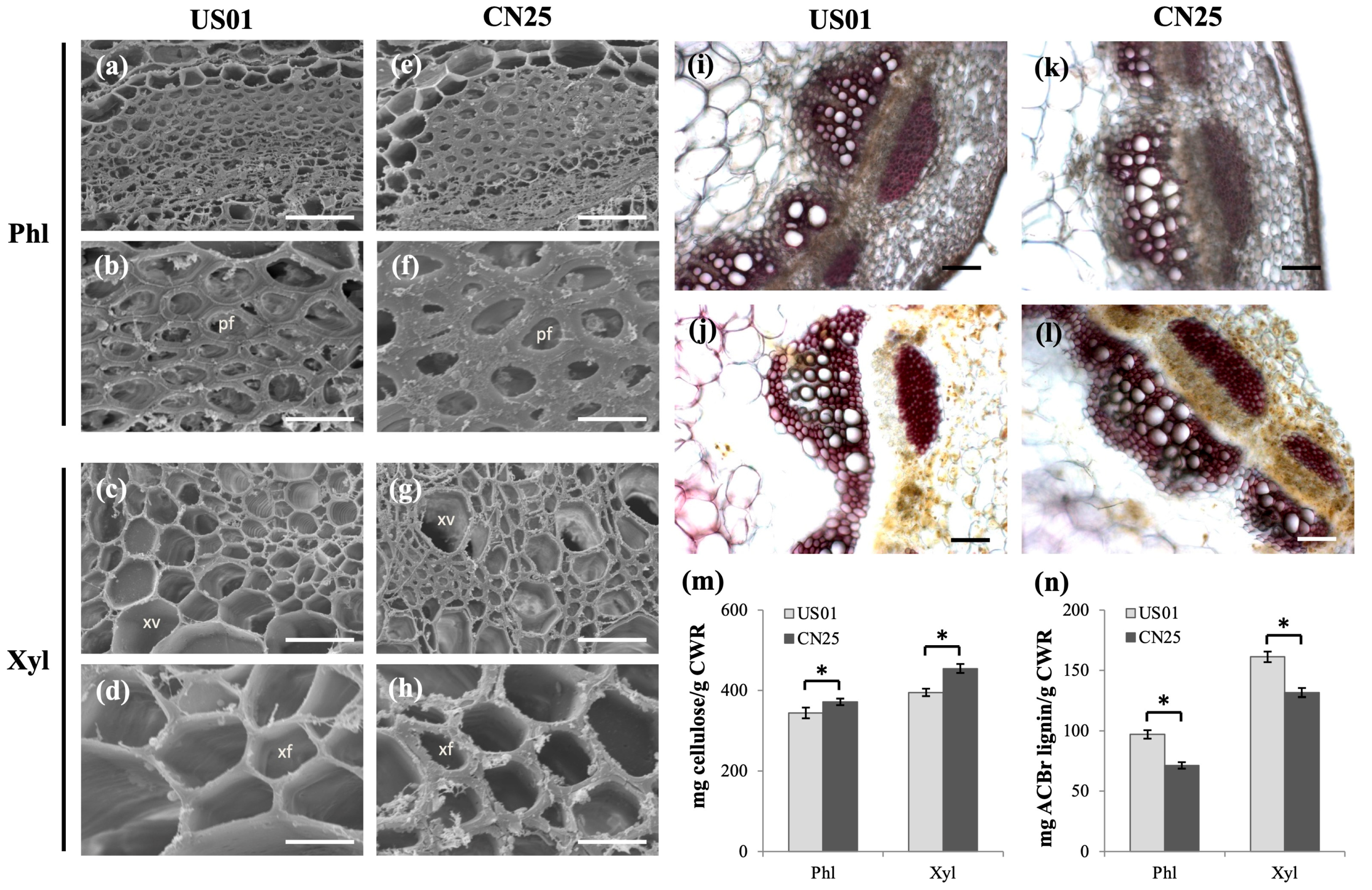
3.8. Confirmation of Anatomical and Biochemical Traits of US01 and CN25
3.9. qRT-PCR Validation of RNA-Seq Data
4. Discussion
4.1. Tissue-Specific Transcriptome Remodeling Underlies the Drastic Differences between Stem Growth in US01 and CN25 Populations
4.2. Transcriptomic Networks Underlying the Up-Regulation of Cellulose Biosynthesis in the IN Population
4.3. Transcriptomic Networks Underlying the Down-Regulation of Lignin Biosynthesis in the IN Population
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prieur-Richard, A.H.; Lavorel, S. Invasions: The perspective of diverse plant communities. Austral. Ecol. 2010, 25, 1–7. [Google Scholar] [CrossRef]
- Wu, H.; Carrillo, J.; Ding, J. Invasion by alligator weed, Alternanthera philoxeroides, is associated with decreased species diversity across the latitudinal gradient in China. J. Plant Ecol. 2015, 9, 311–319. [Google Scholar] [CrossRef]
- Richardson, D.M. Forest trees as invasive aliens. Conserv. Biol. 1998, 12, 18–26. [Google Scholar] [CrossRef]
- Romero, A.; Chamorro, L.; Sans, F.X. Weed diversity in crop edges and inner fields of organic and conventional dryland winter cereal crops in NE Spain. Agric. Ecosyst. Environ. 2008, 124, 97–104. [Google Scholar] [CrossRef]
- Leger, E.A.; Rice, K.J. Invasive California poppies (Eschscholzia californica Cham.) grow larger than native individuals under reduced competition. Ecol. Lett. 2010, 6, 257–264. [Google Scholar] [CrossRef]
- Kumschick, S.; Hufbauer, R.A.; Alba, C.; Blumenthal, D.M. Evolution of fast-growing and more resistant phenotypes in introduced common mullein (Verbascum thapsus). J. Ecol. 2013, 101, 378–387. [Google Scholar] [CrossRef]
- Scheibe, K.M.; Dehnhard, M.; Meyer, H.H.D.; Scheibe, A. Noninvasive monitoring of reproductive function by determination of faecal progestagens and sexual behaviour in a herd of Przewalski mares in a semireserve. Acta Theriol. 1999, 44, 451–463. [Google Scholar] [CrossRef]
- Liu, G.; Huang, Q.-Q.; Lin, Z.-G.; Huang, F.-F.; Liao, H.-X.; Peng, S.-L.; Van Kleunen, M. High Tolerance to Salinity and Herbivory Stresses May Explain the Expansion of Ipomoea cairica to Salt Marshes. PLoS ONE 2012, 7, e48829. [Google Scholar] [CrossRef]
- Erskine-Ogden, J.; Grotkopp, E.; Rejmánek, M. Mediterranean, invasive, woody species grow larger than their less-invasive counterparts under potential global environmental change. Am. J. Bot. 2016, 103, 613–624. [Google Scholar] [CrossRef]
- Grotkopp, E.; Rejmánek, M. High seedling relative growth rate and specific leaf area are traits of invasive species: Phylogenetically independent contrasts of woody angiosperms. Am. J. Bot. 2007, 94, 526–532. [Google Scholar] [CrossRef]
- Sun, Y.; Roderick, G.K. Rapid evolution of invasive traits facilitates the invasion of common ragweed, Ambrosia artemisiifolia. J. Ecol. 2019, 107, 2673–2687. [Google Scholar] [CrossRef]
- Lucas, W.J.; Groover, A.; Lichtenberger, R.; Furuta, K.; Yadav, S.-R.; Helariutta, Y.; He, X.-Q.; Fukuda, H.; Kang, J.; Brady, S.M.; et al. The plant vascular system: Evolution, development and functions. J. Integr. Plant Biol. 2013, 55, 294–388. [Google Scholar] [CrossRef]
- Mathias, S.; Rebecca, S.; Brian, E. Xylem tissue specification, patterning, and differentiation mechanisms. J. Exp. Bot. 2013, 64, 11–31. [Google Scholar]
- Barlow, P. From Cambium to Early Cell Differentiation Within the Secondary Vascular System. In Vascular Transport in Plants; Academic Press: Cambridge, MA, USA, 2005; pp. 279–306. [Google Scholar]
- Wang, H.L.; Offler, C.E.; Patrick, J.W. The cellular pathway of photosynthate transfer in the developing wheat grain. II. A structural analysis and histochemical studies of the pathway from the crease phloem to the endosperm cavity. Plant Cell Environ. 1995, 18, 373–388. [Google Scholar] [CrossRef]
- Ruan, Y.L.; Patrick, J.W. The cellular pathway of postphloem sugar transport in developing tomato fruit. Planta 1995, 196, 434–444. [Google Scholar] [CrossRef]
- Fu, M.; Xu, M.; Zhou, T.; Wang, D.; Tian, S.; Han, L.; Dong, H.; Zhang, C. Transgenic expression of a functional fragment of harpin protein Hpa1 in wheat induces the phloem-based defence against English grain aphid. J. Exp. Bot. 2014, 65, 1439–1453. [Google Scholar] [CrossRef]
- Zhai, y.; Li, P.; Mei, Y.; Chen, M.; Chen, X.; Xu, H.; Zhou, X.; Dong, H.; Zhang, C.; Jiang, W. Three MYB genes co-regulate the phloem-based defence against English grain aphid in wheat. J. Exp. Bot. 2017, 68, 4153–4169. [Google Scholar] [CrossRef]
- Uchida, N.; Lee, J.S.; Horst, R.J.; Lai, H.H.; Kajita, R.; Kakimoto, T.; Tasaka, M.; Torii, K.U. Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. USA 2012, 109, 6337. [Google Scholar] [CrossRef] [PubMed]
- Brutch, N.; Chabbert, B.; Deyholos, M.; Hayashi, T.; Lev-Yadun, S.; Mellerowicz, E.J.; Morvan, C.; Neutelings, G.; Pilate, G.; Gorshkova, T. Plant Fiber Formation: State of the Art, Recent and Expected Progress, and Open Questions. Crit. Rev. Plant Sci. 2012, 31, 201–228. [Google Scholar]
- Bashline, L.; Lei, L.; Li, S.; Gu, Y. Cell Wall, Cytoskeleton, and Cell Expansion in Higher Plants. Mol. Plant 2014, 7, 586–600. [Google Scholar] [CrossRef]
- Kumar, M.; Atanassov, I.; Turner, S. Functional Analysis of Cellulose Synthase (CESA) Protein Class Specificity. Plant Physiol. 2017, 173, 970–983. [Google Scholar] [CrossRef]
- Grimapettenati, J.; Soler, M.; Camargo, E.L.O.; Wang, H. Transcriptional Regulation of the Lignin Biosynthetic Pathway Revisited: New Players and Insights. Adv. Bot. Res. 2012, 61, 173–218. [Google Scholar]
- Braun, D.M.; Wang, L.; Ruan, Y.-L. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J. Exp. Bot. 2014, 65, 1713–1735. [Google Scholar] [CrossRef] [PubMed]
- Maris, A.; Suslov, D.; Fry, S.C.; Verbelen, J.P.; Vissenberg, K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 2009, 60, 3959–3972. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.-C.; Ko, J.-H.; Kim, J.-Y.; Kim, J.; Bae, H.-J.; Han, K.-H. MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 2013, 73, 26–36. [Google Scholar] [CrossRef]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Genome-Wide Survey and Expression Analysis of the Plant-Specific NAC Transcription Factor Family in Soybean During Development and Dehydration Stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z.H. MYB58 and MYB63 Are Transcriptional Activators of the Lignin Biosynthetic Pathway during Secondary Cell Wall Formation in Arabidopsis. Plant Cell Online 2009, 21, 248–266. [Google Scholar] [CrossRef]
- Geng, P.; Zhang, S.; Liu, J.; Zhao, C.; Zhao, Q. MYB20, MYB42, MYB43 and MYB85 Regulate Phenylalanine and Lignin Biosynthesis during Secondary Cell Wall Formation. Plant Physiol. 2020, 182, 1272–1283. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.H. Global Analysis of Direct Targets of Secondary Wall NAC Master Switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar] [CrossRef]
- Szymura, M.; Szymura, T.H. Growth, phenology, and biomass allocation of alien Solidago species in central Europe. Plant Species Biol. 2015, 30, 245–256. [Google Scholar] [CrossRef]
- Cheng, J.; Yang, X.; Xue, L.; Yao, B.; Lu, H.; Tian, Z.; Li, J.; Zhou, X.; Zhang, Y.; Haq, M.Z.U.; et al. Polyploidization contributes to evolution of competitive ability: A long term common garden study on the invasive Solidago canadensis in China. Oikos 2020, 129, 700–713. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, L.; Chen, S.; Qiang, S. Transcription-mediated tissue-specific lignification of vascular bundle causes trade-offs between growth and defence capacity during invasion of Solidago canadensis. Plant Sci. 2020, 301, 110638. [Google Scholar] [CrossRef]
- Effland, M.J. Modified procedure to determine acid-insoluble lignin in wood and pulp. Tappi 1977, 60, 6482484. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Walck, J.L.; Baskin, J.M.; Baskin, C.C. Effects of competition from introduced plants on establishment, survival, growth and reproduction of the rare plant Solidago shortii (Asteraceae). Biol. Conserv. 1999, 88, 213–219. [Google Scholar] [CrossRef]
- Xu, C.; Li, Z.; Wang, J. Linking heat and adaptive responses across temporal proteo-transcriptome and physiological traits of Solidago canadensis. Environ. Exp. Bot. 2020, 175, 104035. [Google Scholar] [CrossRef]
- Romano, J.M.; Dubos, C.; Prouse, M.B.; Wilkins, O.; Campbell, M.M. AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytol. 2012, 195, 774–786. [Google Scholar] [CrossRef]
- Huang, D.; Wang, S.; Zhang, B.; Shang-Guan, K.; Zhou, Y. A Gibberellin-Mediated DELLA-NAC Signaling Cascade Regulates Cellulose Synthesis in Rice. Plant Cell 2015, 27, 1681–1696. [Google Scholar] [CrossRef]
- Desprez, T.; Vernhettes, S.; Fagard, M.; Refrégier, G.; Desnos, T.; Aletti, E.; Py, N.; Pelletier, S.; Höfte, H. Resistance against Herbicide Isoxaben and Cellulose Deficiency Caused by Distinct Mutations in Same Cellulose Synthase Isoform CESA6. Plant Physiol. 2002, 128, 482–490. [Google Scholar] [CrossRef]
- Taylor, N.G.; Gardiner, J.C.; Whiteman, R.; Turner, S.R. Cellulose synthesis in the Arabidopsis secondary cell wall. Cellulose 2004, 11, 329–338. [Google Scholar] [CrossRef]
- Wang, D.; Qin, Y.; Fang, J.; Yuan, S.; Peng, L.; Zhao, J.; Li, X. A Missense Mutation in the Zinc Finger Domain of OsCESA7 Deleteriously Affects Cellulose Biosynthesis and Plant Growth in Rice. PLoS ONE 2016, 11, e0153993. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Xu, B.; Pan, W.; Gong, C.; Wang, Q.; Tian, J.; Li, B.; Zhang, D. Allelic Variation in a Cellulose Synthase Gene (PtoCesA4) Associated with Growth and Wood Properties in Populus tomentosa. G3 Genesgenetics 2013, 3, 2069–2084. [Google Scholar] [CrossRef]
- Petrik, D.L.; Cass, C.L.; Dharshana, P.; Foster, C.E.; Vogel, J.P.; Karlen, S.D.; John, R.; Sedbrook, J.C. BdCESA7, BdCESA8, and BdPMT Utility Promoter Constructs for Targeted Expression to Secondary Cell-Wall-Forming Cells of Grasses. Front. Plant Sci. 2016, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Takshak, S.; Agrawal, S.B. Secondary metabolites and phenylpropanoid pathway enzymes as influenced under supplemental ultraviolet-B radiation in Withania somnifera Dunal, an indigenous medicinal plant. J. Photochem. Photobiol. Biol. 2014, 140, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Hudgins, J.W.; Franceschi, V.R. Methyl jasmonate-induced ethylene production is responsible for conifer phloem defense responses and reprogramming of stem cambial zone for traumatic resin duct formation. Plant Physiol. 2004, 135, 2134–2149. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Linker, R.; Gepstein, S.; Tanimoto, E.; Yamamoto, R.; Neumann, P.M. Progressive Inhibition by Water Deficit of Cell Wall Extensibility and Growth along the Elongation Zone of Maize Roots Is Related to Increased Lignin Metabolism and Progressive Stelar Accumulation of Wall Phenolics. Plant Physiol. 2006, 140, 603–612. [Google Scholar] [CrossRef]
- Bellaloui, N. Soybean Seed Phenol, Lignin, and Isoflavones and Sugars Composition Altered by Foliar Boron Application in Soybean under Water Stress. Food Nutr. Sci. 2012, 3, 579–590. [Google Scholar] [CrossRef]

| Population NO. | Longitude | Latitude | Location |
|---|---|---|---|
| Invasive populations (IN) | |||
| CN05 | 118.83 | 32.05 | Jiangsu, CN |
| CN11 | 121.24 | 31.93 | Jiangsu, CN |
| CN30 | 121.54 | 29.87 | Zhejiang, CN |
| CN17 | 119.45 | 34.85 | Jiangsu, CN |
| CN10 | 121.07 | 32.07 | Jiangsu, CN |
| CN14 | 120.83 | 31.32 | Jiangsu, CN |
| CN65 | 118.88 | 32.11 | Jiangsu, CN |
| CN25 | 119.44 | 32.46 | Jiangsu, CN |
| CN38 | 120.17 | 30.90 | Anhui, CN |
| CN47 | 116.55 | 31.66 | Anhui, CN |
| Native populations (NA) | |||
| US01 | 79.23 | 37.15 | 765#, Altavista, VA, USA |
| US52 | 73.24 | 43.64 | 529# Fair Haven, VT, USA |
| US28 | 73.79 | 43.03 | 250# Maltca, NY, USA |
| US31 | 79.32 | 42.47 | Arrowhead Dr. Dunkirk, NY, USA |
| US34 | 87.94 | 42.92 | 20th St. Oak Creek, WI, USA |
| US44 | 87.33 | 41.42 | Crown Point Church, MO, USA |
| US59 | 94.27 | 45.54 | 2398 76th Ave St. Cloud, MN, USA |
| US60 | 95.88 | 45.61 | US59&HWY28 Morris, MN, USA |
| US06 | 81.65 | 38.35 | 2 Cantley Dr. Charleston, WV, USA |
| US02 | 79.70 | 34.25 | 2591N Williston Rd. Florence, SC, USA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Tian, Z.; Shi, J.; Yu, R.; Zhang, S.; Qiang, S. Tissue-Specific Transcriptomes in the Secondary Cell Wall Provide an Understanding of Stem Growth Enhancement in Solidago canadensis during Invasion. Biology 2023, 12, 1347. https://doi.org/10.3390/biology12101347
Zhang Y, Tian Z, Shi J, Yu R, Zhang S, Qiang S. Tissue-Specific Transcriptomes in the Secondary Cell Wall Provide an Understanding of Stem Growth Enhancement in Solidago canadensis during Invasion. Biology. 2023; 12(10):1347. https://doi.org/10.3390/biology12101347
Chicago/Turabian StyleZhang, Yu, Zhongsai Tian, Jiaqi Shi, Ruoyu Yu, Shuxin Zhang, and Sheng Qiang. 2023. "Tissue-Specific Transcriptomes in the Secondary Cell Wall Provide an Understanding of Stem Growth Enhancement in Solidago canadensis during Invasion" Biology 12, no. 10: 1347. https://doi.org/10.3390/biology12101347
APA StyleZhang, Y., Tian, Z., Shi, J., Yu, R., Zhang, S., & Qiang, S. (2023). Tissue-Specific Transcriptomes in the Secondary Cell Wall Provide an Understanding of Stem Growth Enhancement in Solidago canadensis during Invasion. Biology, 12(10), 1347. https://doi.org/10.3390/biology12101347






