Assessing the Effectiveness of Correlative Ecological Niche Model Temporal Projection through Floristic Data
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Environmental Variables
2.2. Study Species and Occurrence Data
2.3. Algorithms and Packages
2.4. Modelling Procedures
2.5. Model Evaluation Metrics
2.6. Final Evaluations of Procedures Response
2.7. Data Analysis
3. Results
3.1. Performance of the Predictions
3.1.1. Set I: Three Environmental Variables
3.1.2. Set II: 35 Environmental Variables and Dimensionality Reduction via PCA
3.2. Evaluation Metrics of the Predictions
3.3. Evaluation of the Transferability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peterson, A.T.; Soberón, J. Species distribution modeling and ecological niche modeling: Getting the concepts right. Nat. Conserv. 2012, 10, 102–107. [Google Scholar] [CrossRef]
- Peterson, A.T.; Papeş, M.; Soberón, J. Mechanistic and Correlative Models of Ecological Niches. Eur. J. Ecol. 2015, 1, 28–38. [Google Scholar] [CrossRef]
- Soberón, J.M. Niche and area of distribution modeling: A population ecology perspective. Ecography 2010, 33, 159–167. [Google Scholar] [CrossRef]
- Edwards, J.L. Research and societal benefits of the Global Biodiversity Information Facility. BioScience 2004, 54, 485–486. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Schmitt, S.; Pouteau, R.; Justeau, D.; De Boissieu, F.; Birnbaum, P. ssdm: An R package to predict distribution of species richness and composition based on stacked species distribution models. Methods Ecol. Evol. 2017, 8, 1795–1803. [Google Scholar] [CrossRef]
- Feria, A.T.P.; Peterson, A.T. Prediction of bird community composition based on point-occurrence data and inferential algorithms: A valuable tool in biodiversity assessments. Divers. Distrib. 2002, 8, 49–56. [Google Scholar] [CrossRef]
- Raxworthy, C.J.; Martinez-Meyer, E.; Horning, N.; Nussbaum, R.A.; Schneider, G.E.; Ortega-Huerta, M.A.; Peterson, A.T. Predicting distributions of known and unknown reptile species in Madagascar. Nature 2003, 426, 837–841. [Google Scholar] [CrossRef]
- Bosso, L.; Smeraldo, S.; Russo, D.; Chiusano, M.L.; Bertorelle, G.; Johannesson, K.; Butlin, R.K.; Danovaro, R.; Raffini, F. The rise and fall of an alien: Why the successful colonizer Littorina saxatilis failed to invade the Mediterranean Sea. Biol. Invasions 2022, 6, 1–19. [Google Scholar] [CrossRef]
- Ferreira, R.B.; Parreira, M.R.; de Arruda, F.V.; Falcão, M.J.; de Freitas Mansano, V.; Nabout, J.C. Combining ecological niche models with experimental seed germination to estimate the effect of climate change on the distribution of endangered plant species in the Brazilian Cerrado. Environ. Monit. Assess. 2022, 194, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, S.J.; White, M.D.; Newell, G.R. How useful are species distribution models for managing biodiversity under future climates? Ecol. Soc. 2010, 15, 1–13. [Google Scholar] [CrossRef]
- Peterson, A.T.; Cobos, M.E.; Jiménez-García, D. Major challenges for correlational ecological niche model projections to future climate conditions. Ann. N. Y. Acad. Sci. 2018, 1429, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Melo-Merino, S.M.; Reyes-Bonilla, H.; Lira-Noriega, A. Ecological niche models and species distribution models in marine environments: A literature review and spatial analysis of evidence. Ecol. Model. 2020, 415, 108837. [Google Scholar] [CrossRef]
- Escobar, L.E.; Craft, M.E. Advances and limitations of disease biogeography using ecological niche modeling. Front. Microbiol. 2016, 7, 1174. [Google Scholar] [CrossRef]
- Smeraldo, S.; Bosso, L.; Salinas-Ramos, V.B.; Ancillotto, L.; Sánchez-Cordero, V.; Gazaryan, S.; Russo, D. Generalists yet different: Distributional responses to climate change may vary in opportunistic bat species sharing similar ecological traits. Mammal Rev. 2021, 51, 571–584. [Google Scholar] [CrossRef]
- Lombardi, J.V.; Perotto-Baldivieso, H.L.; Hewitt, D.G.; Scognamillo, D.G.; Campbell, T.A.; Tewes, M.E. Assessment of appropriate species-specific time intervals to integrate GPS telemetry data in ecological niche models. Ecol. Inform. 2022, 70, 101701. [Google Scholar] [CrossRef]
- Wiens, J.A.; Stralberg, D.; Jongsomjit, D.; Howell, C.A.; Snyder, M.A. Niches, models, and climate change: Assessing the assumptions and uncertainties. Proc. Natl. Acad. Sci. USA 2009, 106, 19729–19736. [Google Scholar] [CrossRef] [PubMed]
- Karger, D.N.; Conrad, O.; Böhner, J.; Kawohl, T.; Kreft, H.; Soria-Auza, R.W.; Zimmermann, N.E.; Linder, H.P.; Kessler, M. Climatologies at high resolution for the earth’s land surface areas. Sci. Data 2017, 4, 1–20. [Google Scholar] [CrossRef]
- Dyderski, M.K.; Paź, S.; Frelich, L.E.; Jagodziński, A.M. How much does climate change threaten European forest tree species distributions? Glob. Chang. Biol. 2018, 24, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.T.; Soberón, J.; Pearson, R.G.; Anderson, R.P.; Martínez-Meyer, E.; Nakamura, M.; Araújo, M.B. Ecological Niches and Geographic Distributions (MPB-49); Princeton University Press: Princeton, NJ, USA, 2011; pp. 1–328. [Google Scholar] [CrossRef]
- Franklin, J. Mapping Species Distributions: Spatial Inference and Prediction; Cambridge University Press: Cambridge, UK, 2010; pp. 1–339. [Google Scholar] [CrossRef]
- Sillero, N. What does ecological modelling model? A proposed classification of ecological niche models based on their underlying methods. Ecol. Model. 2011, 222, 1343–1346. [Google Scholar] [CrossRef]
- Hirzel, A.H.; Hausser, J.; Chessel, D.; Perrin, N. Ecological-niche factor analysis: How to compute habitat-suitability maps without absence data? Ecology 2002, 83, 2027–2036. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Barbet-Massin, M.; Jiguet, F.; Albert, C.H.; Thuiller, W. Selecting pseudo-absences for species distribution models: How, where and how many? Methods Ecol. Evol. 2012, 3, 327–338. [Google Scholar] [CrossRef]
- Sillero, N.; Barbosa, A.M. Common mistakes in ecological niche models. Int. J. Geogr. Inf. Sci. 2020, 35, 213–226. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M.; Elith, J.; Graham, C.H.; Lehmann, A.; Leathwick, J.; Ferrier, S. Sample selection bias and presence-only distribution models: Implications for background and pseudo-absence data. Ecol. Appl. 2009, 19, 181–197. [Google Scholar] [CrossRef]
- Iturbide, M.; Bedia, J.; Gutiérrez, J.M. Background sampling and transferability of species distribution model ensembles under climate change. Glob. Planet Chang. 2018, 166, 19–29. [Google Scholar] [CrossRef]
- Hallgren, W.; Santana, F.; Low-Choy, S.; Zhao, Y.; Mackey, B. Species distribution models can be highly sensitive to algorithm configuration. Ecol. Model. 2019, 408, 108719. [Google Scholar] [CrossRef]
- Graham, C.H.; Ferrier, S.; Huettman, F.; Moritz, C.; Peterson, A.T. New developments in museum based informatics and applications in biodiversity analysis. Trends Ecol. Evol. 2004, 19, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Guisan, A.; Thuiller, W.; Zimmermann, N.E. Habitat Suitability and Distribution Models: With Applications in R; Cambridge University Press: Cambridge, UK, 2017; pp. 1–462. [Google Scholar] [CrossRef]
- Elith, J. Quantitative methods for modeling species habitat: Comparative performance and an application to Australian plants. In Quantitative Methods for Conservation Biology; Ferson, S., Burgman, M., Eds.; Springer: New York, NY, USA, 2002; pp. 39–58. [Google Scholar] [CrossRef]
- Booth, T.H.; Nix, H.A.; Busby, J.R.; Hutchinson, M.F. BIOCLIM: The first species distribution modelling package, its early applications and relevance to most current MAXENT studies. Divers. Distrib. 2014, 20, 1–9. [Google Scholar] [CrossRef]
- Stockwell, D. The GARP modelling system: Problems and solutions to automated spatial prediction. Int. J. Geogr. Inf. Sci. 1999, 13, 143–158. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M.; Schapire, R.E. A maximum entropy approach to species distribution modeling. In Proceedings of the Twenty-First International Conference on Machine Learning, Banff, AB, Canada, 4–8 July 2004; 2004; pp. 83–91. [Google Scholar] [CrossRef]
- Guisan, A.; Thuiller, W. Predicting species distribution: Offering more than simple habitat models. Ecol. Lett. 2005, 8, 993–1009. [Google Scholar] [CrossRef] [PubMed]
- Norberg, A.; Abrego, N.; Blanchet, F.G.; Adler, F.R.; Anderson, B.J.; Anttila, J.; Araújo, M.B.; Dallas, T.; Dunson, D.; Elith, J.; et al. A comprehensive evaluation of predictive performance of 33 species distribution models at species and community levels. Ecol. Monogr. 2019, 89, e01370. [Google Scholar] [CrossRef]
- De Araújo, C.B.; Marcondes-Machado, L.O.; Costa, G.C. The importance of biotic interactions in species distribution models: A test of the Eltonian noise hypothesis using parrots. J. Biogeogr. 2014, 41, 513–523. [Google Scholar] [CrossRef]
- Gillard, M.; Thiébaut, G.; Deleu, C.; Leroy, B. Present and future distribution of three aquatic plants taxa across the world: Decrease in native and increase in invasive ranges. Biol. Invasions 2017, 19, 2159–2170. [Google Scholar] [CrossRef]
- Mylne, A.Q.; Pigott, D.M.; Longbottom, J.; Shearer, F.; Dude, K.A.; Messina, J.P.; Weiss, D.J.; Moyes, C.L.; Golding, N.; Hay, S.I. Mapping the zoonotic niche of Lassa fever in Africa. Trans. R. Soc. Trop. Med. Hyg. 2015, 109, 483–492. [Google Scholar] [CrossRef]
- Patsiou, T.S.; Conti, E.; Zimmermann, N.E.; Theodoridis, S.; Randin, C.F. Topo-climatic microrefugia explain the persistence of a rare endemic plant in the Alps during the last 21 millennia. Glob. Chang. Biol. 2014, 20, 2286–2300. [Google Scholar] [CrossRef]
- Randin, C.F.; Dirnböck, T.; Dullinger, S.; Zimmermann, N.E.; Zappa, M.; Guisan, A. Are niche-based species distribution models transferable in space? J. Biogeogr. 2006, 33, 1689–1703. [Google Scholar] [CrossRef]
- Wenger, S.J.; Olden, J.D. Assessing transferability of ecological models: An underappreciated aspect of statistical validation. Methods Ecol. Evol. 2012, 3, 260–267. [Google Scholar] [CrossRef]
- Roberts, D.R.; Hamann, A. Predicting potential climate change impacts with bioclimate envelope models: A palaeoecological perspective. Glob. Ecol. Biogeogr. 2012, 21, 121–133. [Google Scholar] [CrossRef]
- Veloz, S.D.; Williams, J.W.; Blois, J.L.; He, F.; Otto-Bliesner, B.; Liu, Z. No-analog climates and shifting realized niches during the late quaternary: Implications for 21st-century predictions by species distribution models. Glob. Chang. Biol. 2012, 18, 1698–1713. [Google Scholar] [CrossRef]
- Duque-Lazo, J.; Van Gils, H.; Groen, T.A.; Navarro-Cerrillo, R.M. Transferability of species distribution models: The case of Phytophthora cinnamomi in Southwest Spain and Southwest Australia. Ecol. Model. 2016, 320, 62–70. [Google Scholar] [CrossRef]
- Qiao, H.; Feng, X.; Escobar, L.E.; Peterson, A.T.; Soberón, J.; Zhu, G.; Papeş, M. An evaluation of transferability of ecological niche models. Ecography 2019, 42, 521–534. [Google Scholar] [CrossRef]
- Busby, J.R. BIOCLIM—A bioclimate analysis and prediction system. In Nature Conservation: Cost Effective Biological Surveys and Data Analysis; Csiro Publishing: Clayton, Australia, 1991; pp. 64–68. [Google Scholar]
- Busby, J.R. BIOCLIM—A bioclimate analysis and prediction system. Plant Prot. Q. 1991, 6, 8–9. [Google Scholar]
- Stockwell, D.R. Improving ecological niche models by data mining large environmental datasets for surrogate models. Ecol. Model. 2006, 192, 188–196. [Google Scholar] [CrossRef]
- Baselga, A.; Araújo, M.B. Individualistic vs community modelling of species distributions under climate change. Ecography 2009, 32, 55–65. [Google Scholar] [CrossRef]
- Warren, D.L.; Matzke, N.J.; Iglesias, T.L. Evaluating presence-only species distribution models with discrimination accuracy is uninformative for many applications. J. Biogeogr. 2020, 47, 167–180. [Google Scholar] [CrossRef]
- Title, P.O.; Bemmels, J.B. ENVIREM: An expanded set of bioclimatic and topographic variables increases flexibility and improves performance of ecological niche modeling. Ecography 2018, 41, 291–307. [Google Scholar] [CrossRef]
- Barbet-Massin, M.; Jetz, W. A 40-year, continent-wide, multispecies assessment of relevant climate predictors for species distribution modelling. Divers. Distrib. 2014, 20, 1285–1295. [Google Scholar] [CrossRef]
- Karger, D.N.; Zimmermann, N.E. CHELSAcruts—High resolution temperature and precipitation timeseries for the 20th century and beyond. EnviDat 2018. Available online: https://doi.org/10.16904/envidat.159 (accessed on 5 April 2019).
- Hijmans, R.J.; Phillips, S.; Leathwick, J.; Elith, J.; Hijmans, M.R.J. R Package, Version 1.1-4; Dismo. 2017. Available online: https://cran.r-project.org/web/packages/dismo/index.html (accessed on 7 April 2019).
- Petitpierre, B.; Broennimann, O.; Kueffer, C.; Daehler, C.; Guisan, A. Selecting predictors to maximize the transferability of species distribution models: Lessons from cross-continental plant invasions. Glob. Ecol. Biogeogr. 2017, 26, 275–287. [Google Scholar] [CrossRef]
- Gardner, A.S.; Maclean, I.M.; Gaston, K.J. Climatic predictors of species distributions neglect biophysiologically meaningful variables. Divers. Distrib. 2019, 25, 1318–1333. [Google Scholar] [CrossRef]
- Leutner, B.; Horning, N.; Schwalb-Willmann, J. R Package, Version 0.2.6.9999; RStoolbox: Tools for Remote Sensing Data Analysis. 2020. Available online: https://cran.r-project.org/web/packages/RStoolbox/index.html (accessed on 15 April 2019).
- Bartolucci, F.; Peruzzi, L.; Galasso, G.; Albano, A.; Alessandrini, A.; Ardenghi, N.M.G.; Astuti, G.; Bacchetta, G.; Ballelli, S.; Banfi, E.; et al. An updated checklist of the vascular flora native to Italy. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2018, 152, 179–303. [Google Scholar] [CrossRef]
- Peruzzi, L.; Conti, F.; Bartolucci, F. An inventory of vascular plants endemic to Italy. Phytotaxa 2014, 168, 1–75. [Google Scholar] [CrossRef]
- Peruzzi, L.; Domina, G.; Bartolucci, F.; Galasso, G.; Peccenini, S.; Raimondo, F.M.; Albano, A.; Alessandrini, A.; Banfi, E.; Barberis, G.; et al. An inventory of the names of vascular plants endemic to Italy, their loci classici and types. Phytotaxa 2015, 196, 1–217. [Google Scholar] [CrossRef]
- Pignatti, S. Flora d’Italia; New Business Media: Milano, Italy, 2017; Volume 1, pp. 1–1064. [Google Scholar]
- Pignatti, S. Flora d’Italia; New Business Media: Milano, Italy, 2017; Volume 2, pp. 1–1178. [Google Scholar]
- Pignatti, S. Flora d’Italia; New Business Media: Milano, Italy, 2018; Volume 3, pp. 1–1286. [Google Scholar]
- Lihová, J.; Tribsch, A.; Stuessy, T.F. Cardamine apennina: A new endemic diploid species of the C. pratensis group (Brassicaceae) from Italy. Plant Syst. Evol. 2004, 245, 69–92. [Google Scholar] [CrossRef]
- FlorItaly—The Portal to the Flora of Italy. 2021. Available online: http:/dryades.units.it/floritaly (accessed on 6 April 2021).
- Peruzzi, L.; Astuti, G.; Carta, A.; Roma-Marzio, F.; Dolci, D.; Caldararo, F.; Bartolucci, F.; Bernardo, L. Nomenclature, morphometry, karyology and SEM cypselae analysis of Carduus brutius (Asteraceae) and its relatives. Phytotaxa 2015, 202, 237–249. [Google Scholar] [CrossRef]
- Peruzzi, L.; Roma-Marzio, F.; Dolci, D.; Flamini, G.; Braca, A.; De Leo, M. Phytochemical data parallel morpho-colorimetric variation in Polygala flavescens DC. Plant Biosyst. 2019, 153, 817–834. [Google Scholar] [CrossRef]
- Conti, F.; Bartolucci, F.; Bacchetta, G.; Pennesi, R.; Lakušić, D.; Niketić, M. A taxonomic revision of the Siler montanum group (Apiaceae) in Italy and the Balkan Peninsula. Willdenowia 2021, 51, 321–347. [Google Scholar] [CrossRef]
- Bedini, G.; Peruzzi, L. Wikiplantbase #Italia v1.0. 2019. Available online: https://bot.biologia.unipi.it/wpb/italia/index (accessed on 19 March 2019).
- GBIF. The Global Biodiversity Information Facility. 2019. Available online: https://www.gbif.org (accessed on 19 March 2019).
- JACQ Consortium Virtual Herbaria Website. 2019. Available online: https://www.jacq.org/ (accessed on 19 March 2019).
- Osorio-Olvera, L.; Lira-Noriega, A.; Soberón, J.; Peterson, A.T.; Falconi, M.; Contreras-Díaz, R.G.; Martínez-Meyer, E.; Brave, V.; Brave, N. NTBOX: An R package with graphical user interface for modelling and evaluating multidimensional ecological niches. Methods Ecol. Evol. 2020, 11, 1199–1206. [Google Scholar] [CrossRef]
- Cobos, M.E.; Osorio-Olvera, L.; Soberon, J.; Peterson, A.T.; Brave, V.; Brave, N. ellipsenm: Ecological Niche’s Characterizations Using Ellipsoids. R. Package, Ver. 0.3.4. 2020. Available online: https://github.com/marlonecobos/ellipsenm (accessed on 1 June 2019).
- Carpenter, G.; Gillison, A.N.; Winter, J. DOMAIN: A flexible modelling procedure for mapping potential distributions of plants and animals. Biodivers. Conserv. 1993, 2, 667–680. [Google Scholar] [CrossRef]
- McCullagh, P.; Nelder, J.A. Generalized Linear Models; Chapman and Hall: London, UK, 1989; pp. 1–526. [Google Scholar] [CrossRef]
- Guisan, A.; Edwards Jr, T.C.; Hastie, T. Generalized linear and generalized additive models in studies of species distributions: Setting the scene. Ecol. Model. 2002, 157, 89–100. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R. Generalized Additive Models; Chapman and Hall: London, UK, 1990; pp. 1–325. [Google Scholar] [CrossRef]
- Friedman, J.H. Multivariate adaptive regression splines. Ann. Stat. 1991, 19, 1–67. [Google Scholar] [CrossRef]
- Friedman, J.H.; Roosen, C.B. An introduction to multivariate adaptive regression splines. Stat. Methods Med. Res. 1995, 4, 197–217. [Google Scholar] [CrossRef] [PubMed]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference and Prediction; Springer: New York, NY, USA, 2001; pp. 1–758. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Buja, A. Flexible discriminant analysis by optimal scoring. J. Am. Stat. Assoc. 1994, 89, 1255–1270. [Google Scholar] [CrossRef]
- Hastie, T.; Buja, A.; Tibshirani, R. Penalized discriminant analysis. Ann. Stat. 1995, 23, 73–102. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, NY, USA, 2009; pp. 1–745. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Stone, C.J.; Olshen, R.A. Classification and Regression Trees; Routledge: Boca Raton, FL, USA, 1984; pp. 1–368. [Google Scholar] [CrossRef]
- Ripley, B.D. Pattern Recognition and Neural Networks; Cambridge University Press: Cambridge, UK, 1996; pp. 1–403. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, B.D. Exploratory multivariate analysis. In Modern Applied Statistics with S; Springer: New York, NY, USA, 2002; pp. 301–330. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by random Forest. R News 2002, 2, 18–22. [Google Scholar]
- Schölkopf, B.; Platt, J.C.; Shawe-Taylor, J.; Smola, A.J.; Williamson, R.C. Estimating the support of a high-dimensional distribution. Neural Comput. 2001, 13, 1443–1471. [Google Scholar] [CrossRef]
- Drake, J.M.; Randin, C.; Guisan, A. Modelling ecological niches with support vector machines. J. Appl. Ecol. 2006, 43, 424–432. [Google Scholar] [CrossRef]
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. R Package, Version.1.7-0.1; e1071. 2015. Available online: https://cran.r-project.org/web/packages/e1071/index.html (accessed on 7 April 2019).
- Phillips, S.J.; Anderson, R.P.; Dudík, M.; Schapire, R.E.; Blair, M.E. Opening the black box: An open-source release of Maxent. Ecography 2017, 40, 887–893. [Google Scholar] [CrossRef]
- Blonder, B.; Lamanna, C.; Violle, C.; Enquist, B.J. The n-dimensional hypervolume. Glob. Ecol. Biogeogr. 2014, 23, 595–609. [Google Scholar] [CrossRef]
- Blonder, B.; Morrow, C.B.; Maitner, B.; Harris, D.J.; Lamanna, C.; Violle, C.; Enquist, B.J.; Kerkhoff, A.J. New approaches for delineating n-dimensional hypervolumes. Methods Ecol. Evol. 2018, 9, 305–319. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing; 2017; Available online: www.r-project.org (accessed on 1 November 2018).
- Naimi, B.; Araújo, M.B. sdm: A reproducible and extensible R platform for species distribution modelling. Ecography 2016, 39, 368–375. [Google Scholar] [CrossRef]
- Thuiller, W. BIOMOD–optimizing predictions of species distributions and projecting potential future shifts under global change. Glob. Chang. Biol. 2003, 9, 1353–1362. [Google Scholar] [CrossRef]
- Thuiller, W.; Lafourcade, B.; Engler, R.; Araújo, M.B. BIOMOD—A platform for ensemble forecasting of species distributions. Ecography 2009, 32, 369–373. [Google Scholar] [CrossRef]
- Thuiller, W.; Georges, D.; Engler, R.; Breiner, F.; Georges, M.D.; Thuiller, C.W. R Package, Version 3.4.11; biomod2. 2016. Available online: https://cran.r-project.org/web/packages/biomod2/index.html (accessed on 7 April 2019).
- Leroy, B.; Meynard, C.N.; Bellard, C.; Courchamp, F. virtualspecies, an R package to generate virtual species distributions. Ecography 2016, 39, 599–607. [Google Scholar] [CrossRef]
- Schoener, T.W. The Anolis lizards of Bimini: Resource partitioning in a complex fauna. Ecology 1968, 49, 704–726. [Google Scholar] [CrossRef]
- Warren, D.L.; Glor, R.E.; Turelli, M. Environmental niche equivalency versus conservatism: Quantitative approaches to niche. Evolution 2008, 62, 2868–2883. [Google Scholar] [CrossRef]
- Elith, J.; Kearney, M.; Phillips, S. The art of modelling range-shifting species. Methods Ecol. Evol. 2010, 1, 330–342. [Google Scholar] [CrossRef]
- Owens, H.L.; Campbell, L.P.; Dornak, L.L.; Saupe, E.E.; Barve, N.; Soberón, J.; Ingenloff, K.; Lira-Noriega, A.M.; Hensz, C.; Myers, C.E.; et al. Constraints on interpretation of ecological niche models by limited environmental ranges on calibration areas. Ecol. Model. 2013, 263, 10–18. [Google Scholar] [CrossRef]
- Cobos, M.E.; Peterson, A.T.; Barve, N.; Osorio-Olvera, L. kuenm: An R package for detailed development of ecological niche models using Maxent. PeerJ. 2019, 7, e6281. [Google Scholar] [CrossRef] [PubMed]
- Warren, D.L.; Seifert, S.N. Ecological niche modeling in Maxent: The importance of model complexity and the performance of model selection criteria. Ecol. Appl. 2011, 21, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.G.; Raxworthy, C.J.; Nakamura, M.; Peterson, A.T. Predicting species distributions from small numbers of occurrence records: A test case using cryptic geckos in Madagascar. J. Biogeogr. 2007, 34, 102–117. [Google Scholar] [CrossRef]
- Peterson, A.T.; Papeş, M.; Soberón, J. Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecol. Model. 2008, 213, 63–72. [Google Scholar] [CrossRef]
- Lobo, J.M.; Jiménez-Valverde, A.; Real, R. AUC: A misleading measure of the performance of predictive distribution models. Global Ecol. Biogeogr. 2008, 17, 145–151. [Google Scholar] [CrossRef]
- Jiménez-Valverde, A. Insights into the area under the receiver operating characteristic curve (AUC) as a discrimination measure in species distribution modelling. Glob. Ecol. Biogeogr. 2012, 21, 498–507. [Google Scholar] [CrossRef]
- Hirzel, A.H.; Le Lay, G.; Helfer, V.; Randin, C.; Guisan, A. Evaluating the ability of habitat suitability models to predict species presences. Ecol. Model. 2006, 199, 142–152. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Wilcoxon, F. Individual comparisons by ranking methods. Biometr. Bull. 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Williams, J.N.; Seo, C.; Thorne, J.; Nelson, J.K.; Erwin, S.; O’Brien, J.M.; Schwartz, M.W. Using species distribution models to predict new occurrences for rare plants. Divers. Distrib. 2009, 15, 565–576. [Google Scholar] [CrossRef]
- Mi, C.; Huettmann, F.; Guo, Y.; Han, X.; Wen, L. Why choose Random Forest to predict rare species distribution with few samples in large undersampled areas? Three Asian crane species models provide supporting evidence. PeerJ 2017, 5, e2849. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Soberón, J.; Peterson, A.T. No silver bullets in correlative ecological niche modelling: Insights from testing among many potential algorithms for niche estimation. Methods Ecol. Evol. 2015, 6, 1126–1136. [Google Scholar] [CrossRef]
- Baele, C.M.; Lennon, J.J. Incorporating uncertainty in predictive species distribution modelling. Philos. Trans. R. Soc. B 2012, 367, 247–258. [Google Scholar] [CrossRef]
- Chen, X.; Dimitrov, N.B.; Meyers, L.A. Uncertainty analysis of species distribution models. PLoS ONE 2019, 14, e0214190. [Google Scholar] [CrossRef] [PubMed]
- Gelman, A.; Hill, J. Data Analysis Using Regression and Multilevel/Hierarchical Models; Cambridge University Press: Cambridge, UK, 2006; pp. 1–648. [Google Scholar] [CrossRef]
- Fitzpatrick, M.C.; Hargrove, W.W. The projection of species distribution models and the problem of non-analog climate. Biodivers. Conserv. 2009, 18, 2255–2261. [Google Scholar] [CrossRef]
- Fourcade, Y.; Besnard, A.G.; Secondi, J. Paintings predict the distribution of species, or the challenge of selecting environmental predictors and evaluation statistics. Glob. Ecol. Biogeogr. 2018, 27, 245–256. [Google Scholar] [CrossRef]
- Lee-Yaw, J.A.; McCune, J.L.; Pironon, S.; Sheth, S.N. Species distribution models rarely predict the biology of real populations. Ecography 2022, 2022, e05877. [Google Scholar] [CrossRef]
- Qazi, A.W.; Saqib, Z.; Zaman-ul-Haq, M. Trends in species distribution modelling in context of rare and endemic plants: A systematic review. Ecol. Process. 2022, 11, 40. [Google Scholar] [CrossRef]
- Moudrý, V. Modelling species distributions with simulated virtual species. J. Biogeogr. 2015, 42, 1365–1366. [Google Scholar] [CrossRef]
- Guillera-Arroita, G.; Lahoz-Monfort, J.J.; Elith, J.; Gordon, A.; Kujala, H.; Lentini, P.E.; McCarthy, M.A.; Tingley, R.; Wintle, B.A. Is my species distribution model fit for purpose? Matching data and models to applications. Glob. Ecol. Biogeogr. 2015, 24, 276–292. [Google Scholar] [CrossRef]
- Radosavljevic, A.; Anderson, R.P. Making better Maxent models of species distributions: Complexity, overfitting and evaluation. J. Biogeogr. 2014, 41, 629–643. [Google Scholar] [CrossRef]
- Valavi, R.; Elith, J.; Lahoz-Monfort, J.J.; Guillera-Arroita, G. blockCV: An r package for generating spatially or environmentally separated folds for k-fold cross-validation of species distribution models. Methods Ecol. Evol. 2018, 10, 225–232. [Google Scholar] [CrossRef]
- Muscarella, R.; Galante, P.J.; Soley-Guardia, M.; Boria, R.A.; Kass, J.M.; Uriarte, M.; Anderson, R.P. ENM eval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol. Evol. 2014, 5, 1198–1205. [Google Scholar] [CrossRef]
- Pacifici, M.; Foden, W.B.; Visconti, P.; Watson, J.E.; Butchart, S.H.; Kovacs, K.M.; Scheffers, B.R.; Hole, D.G.; Martin, T.G.; Akçakaya, H.R.; et al. Assessing species vulnerability to climate change. Nat. Clim. Chang. 2015, 5, 215–224. [Google Scholar] [CrossRef]
- Brodie, S.; Smith, J.A.; Muhling, B.A.; Barnett, L.A.K.; Carroll, G.; Fiedler, P.; Bograd, S.J.; Hazen, E.L.; Jacox, M.G.; Andrews, K.S.; et al. Recommendations for quantifying and reducing uncertainty in climate projections of species distributions. Glob. Chang. Biol. 2022. [Google Scholar] [CrossRef]
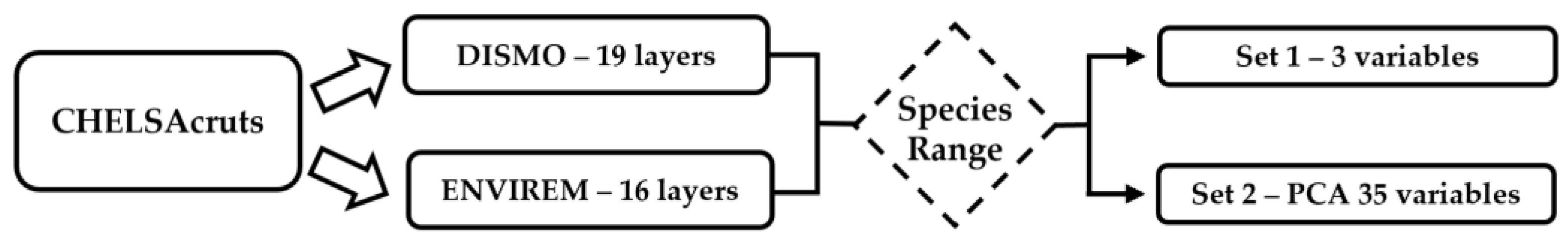
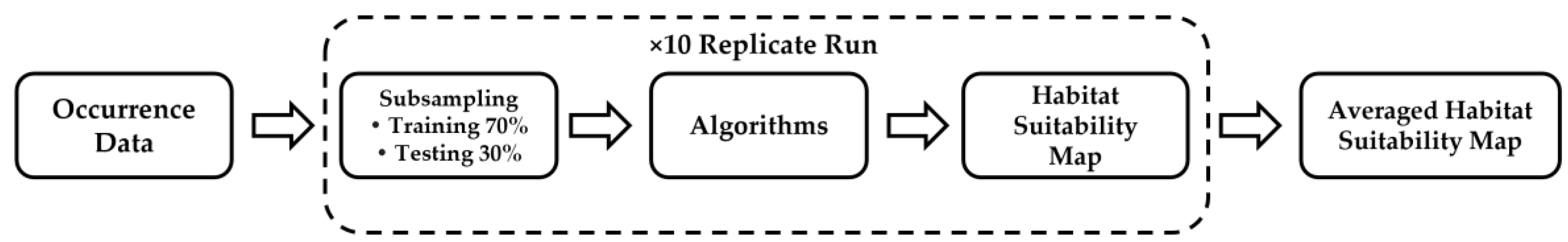
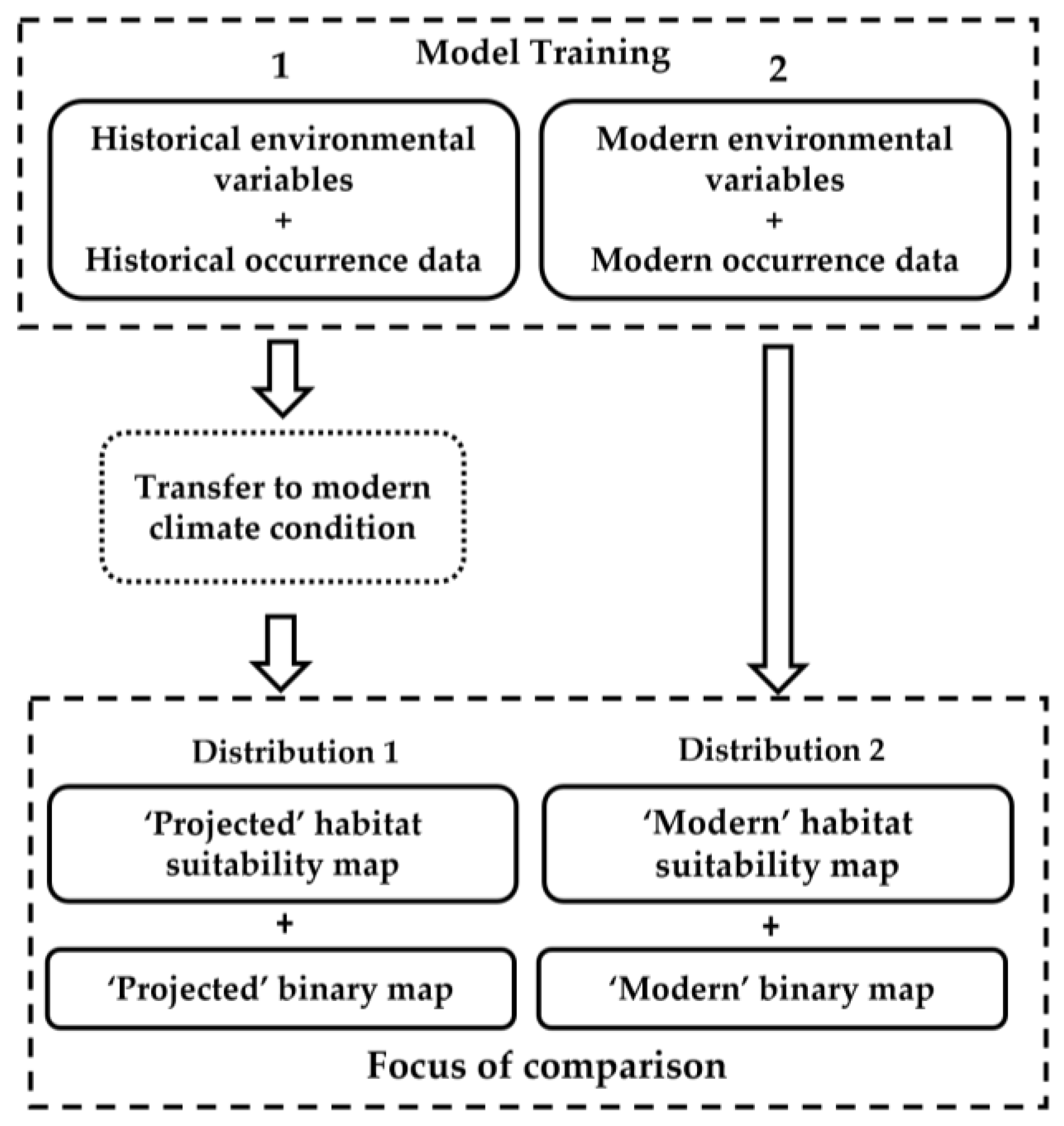
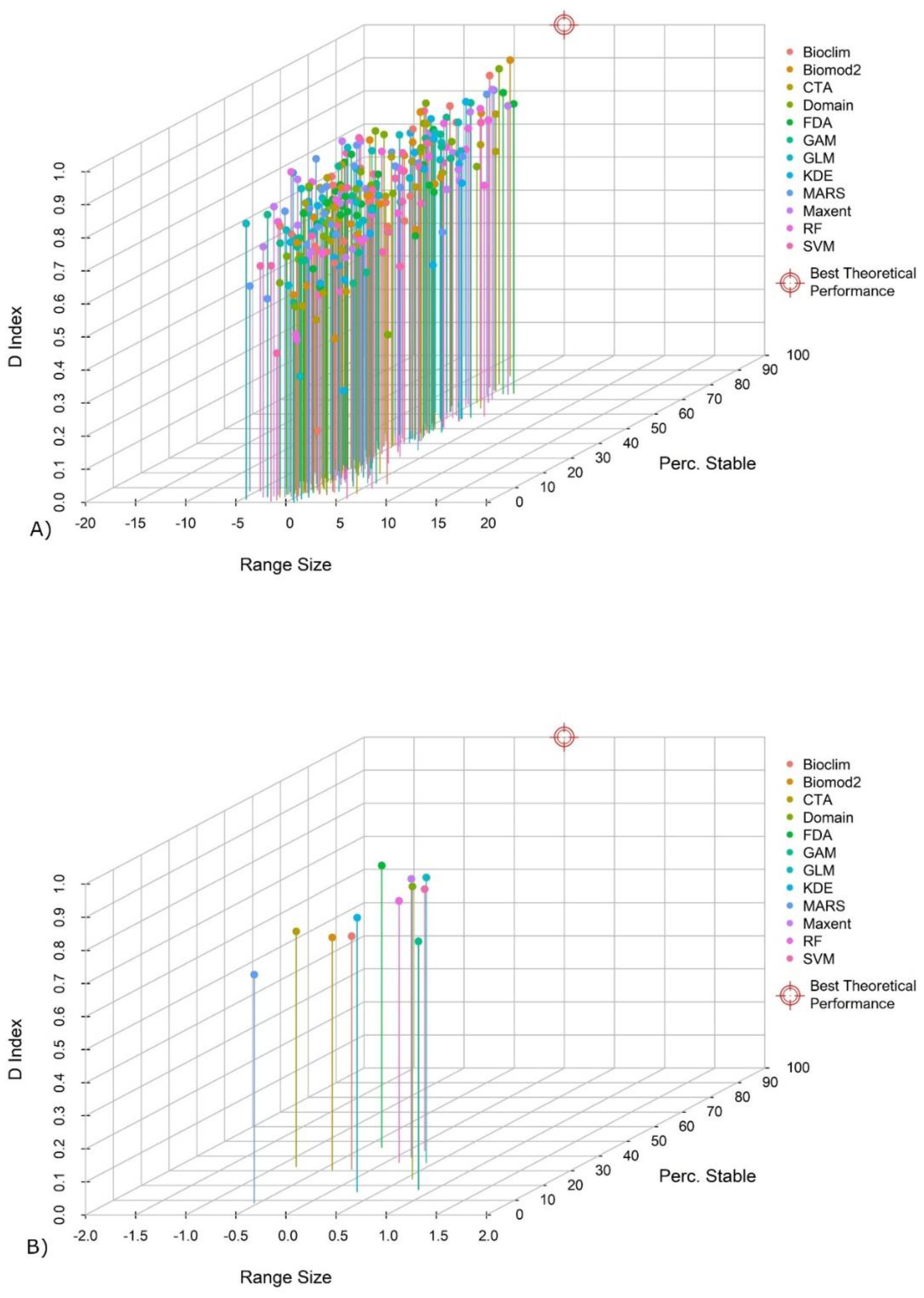

| Reference | Algorithms | Species/Data | Transferability Test |
|---|---|---|---|
| Randin et al. [42] | GLM, GAM | 54 species with more than 30 occurrences from vegetation plots | Evaluation metrics; Kulczynski’s coefficient |
| Wenger and Olden [43] | GLMM, ANN, R | Salvelinus fontinalis (Mitchill, 1814); Salmo trutta Linnaeus, 1758 | Evaluation metrics combined with resampling methods |
| Roberts and Hamann [44] | RF | Modern ecosystem types | Validation based on palaeoecological records |
| Veloz et al. [45] | BRT, MARS, MARS-COM, GAM, GLM | Fossil-pollen data | Tests of niche equivalency (D) and niche similarity (I) |
| Duque-Lazo et al. [46] | ANN, BRT, CART, FDA, GAM, GLM, MaxEnt, MARS, RF, SRE | Presence-absence data for Phytophthora cinnamomi Rands (presence n = 599; absence n = 1193) | Evaluation metrics; transferability index |
| Qiao et al. [47] | BIOCLIM, ENFA, CONVEXHULL, MVE, GLM, GAM, BRT, GARP, Maxent, KDE, MA | 16 virtual species distributed across mainland Eurasia | Sensitivity, specificity and TSS plus volume ratio of estimated niches |
| Procedure | Type and Algorithm | Data Input | Package (Version) | Background/Pseudoabsence Cells |
|---|---|---|---|---|
| BIOCLIM | Single (BIOCLIM) | Presence only | Dismo (1.1-4) | 1000 background cells |
| Domain | Single (Domain) | Presence only | Dismo (1.1-4) | 1000 background cells |
| GLM | Single (GLM) | Presence-pseudoabsence | SSDM (0.2.8) | 1000 pseudoabsence cells |
| GAM | Single (GAM) | Presence-pseudoabsence | SSDM (0.2.8) | 1000 pseudoabsence cells |
| MARS | Single (MARS) | Presence-pseudoabsence | SSDM (0.2.8) | 1000 pseudoabsence cells |
| FDA | Single (FDA) | Presence-pseudoabsence | sdm (1.0-89) | Pseudoabsences = presence cells |
| CTA | Single (CTA) | Presence-pseudoabsence | sdm (1.0-89) | Pseudoabsences = presence cells |
| RF | Single (RF) | Presence-pseudoabsence | SSDM (0.2.8) | Pseudoabsences = presence cells |
| SVM | Single (SVM) | Presence-pseudoabsence | SSDM (0.2.8) | Pseudoabsences = presence cells |
| Maxent | Single (Maxent) | Presence background | Dismo (1.1-4) | 10,000 background cells |
| Biomod2 | Ensemble (GLM, GBM, GAM, MARS, Maxent, RF, CTA, ANN, and FDA) | Presence-pseudoabsence | biomod2 (3.4.11) | Pseudoabsences = presence cells × 10 |
| KDE | Single (KDE) | Presence only | Hypervolume (2.0.12) | 1000 background cells |
| Bioclim | Biomod2 | CTA | Domain | FDA | GAM | GLM | KDE | MARS | Maxent | RF | SVM | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioclim | ||||||||||||
| Biomod2 | - | |||||||||||
| CTA | - | - | ||||||||||
| Domain | - | - | - | |||||||||
| FDA | - | - | - | - | ||||||||
| GAM | - | - | - | - | × | |||||||
| GLM | - | - | - | - | - | × | ||||||
| KDE | - | - | - | - | × | - | × | |||||
| MARS | × | × | × | × | × | × | × | - | ||||
| Maxent | - | - | - | - | - | × | - | × | × | |||
| RF | - | - | - | - | - | - | - | - | × | - | ||
| SVM | - | - | - | - | - | - | - | × | × | - | - |
| Bioclim | Biomod2 | CTA | Domain | FDA | GAM | GLM | KDE | MARS | Maxent | RF | SVM | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioclim | ||||||||||||
| Biomod2 | - | |||||||||||
| CTA | - | - | ||||||||||
| Domain | - | - | - | |||||||||
| FDA | - | - | - | - | ||||||||
| GAM | - | - | - | - | - | |||||||
| GLM | - | - | - | - | - | - | ||||||
| KDE | - | - | - | - | × | - | - | |||||
| MARS | × | × | × | × | × | × | × | - | ||||
| Maxent | - | - | - | - | - | - | - | - | × | |||
| RF | - | - | - | - | - | - | - | - | × | - | ||
| SVM | - | - | - | - | - | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dolci, D.; Peruzzi, L. Assessing the Effectiveness of Correlative Ecological Niche Model Temporal Projection through Floristic Data. Biology 2022, 11, 1219. https://doi.org/10.3390/biology11081219
Dolci D, Peruzzi L. Assessing the Effectiveness of Correlative Ecological Niche Model Temporal Projection through Floristic Data. Biology. 2022; 11(8):1219. https://doi.org/10.3390/biology11081219
Chicago/Turabian StyleDolci, David, and Lorenzo Peruzzi. 2022. "Assessing the Effectiveness of Correlative Ecological Niche Model Temporal Projection through Floristic Data" Biology 11, no. 8: 1219. https://doi.org/10.3390/biology11081219
APA StyleDolci, D., & Peruzzi, L. (2022). Assessing the Effectiveness of Correlative Ecological Niche Model Temporal Projection through Floristic Data. Biology, 11(8), 1219. https://doi.org/10.3390/biology11081219







