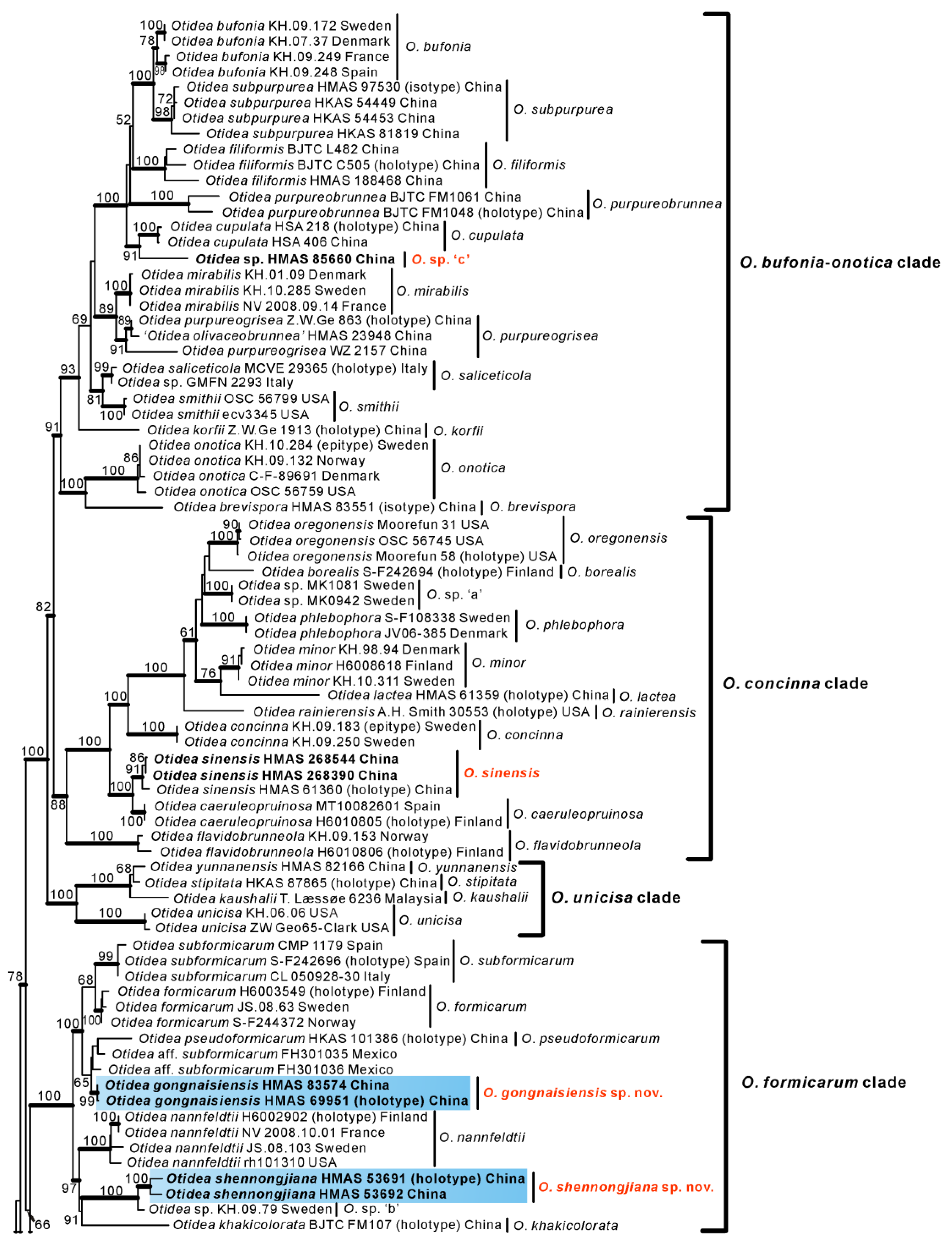
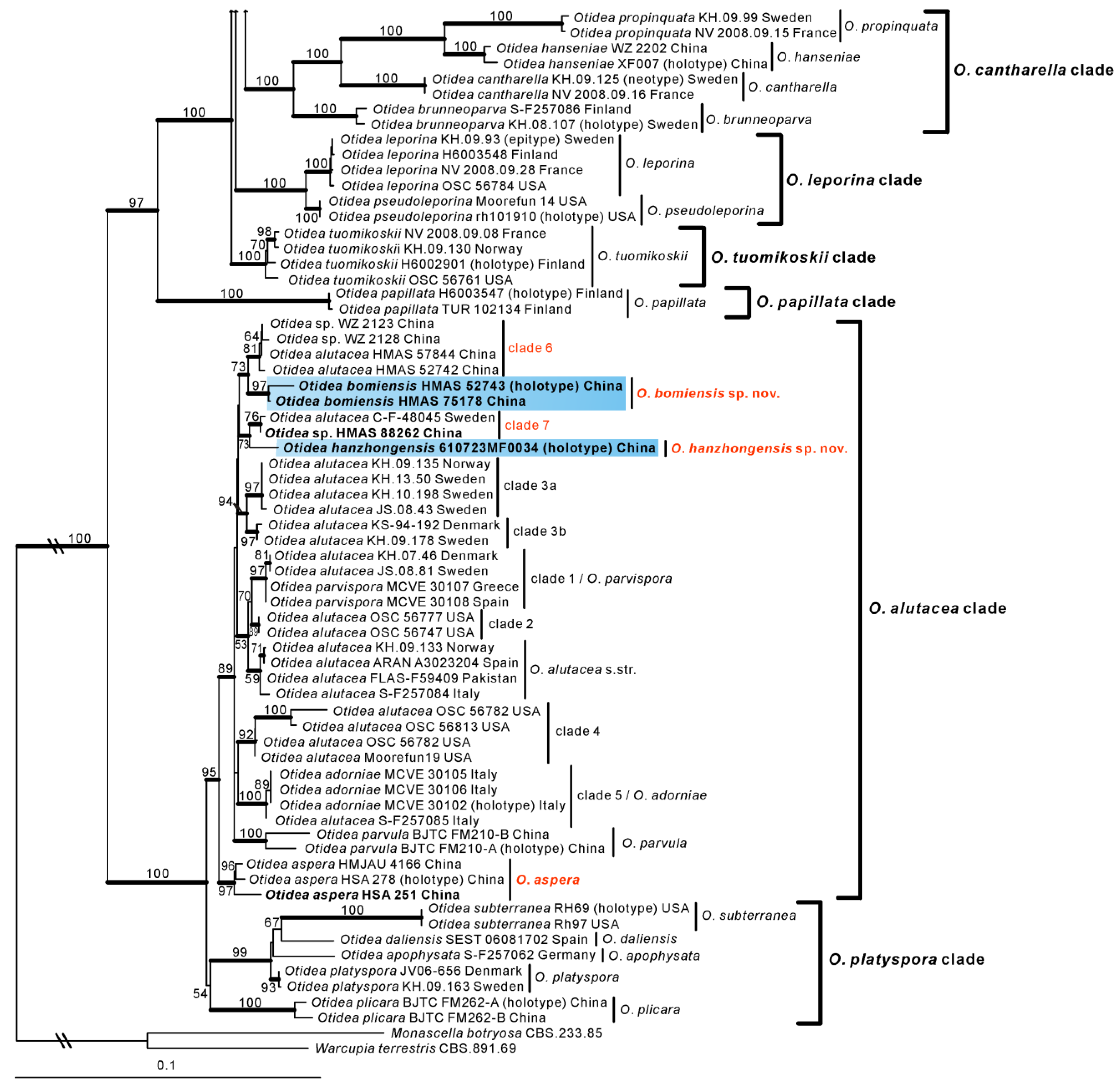
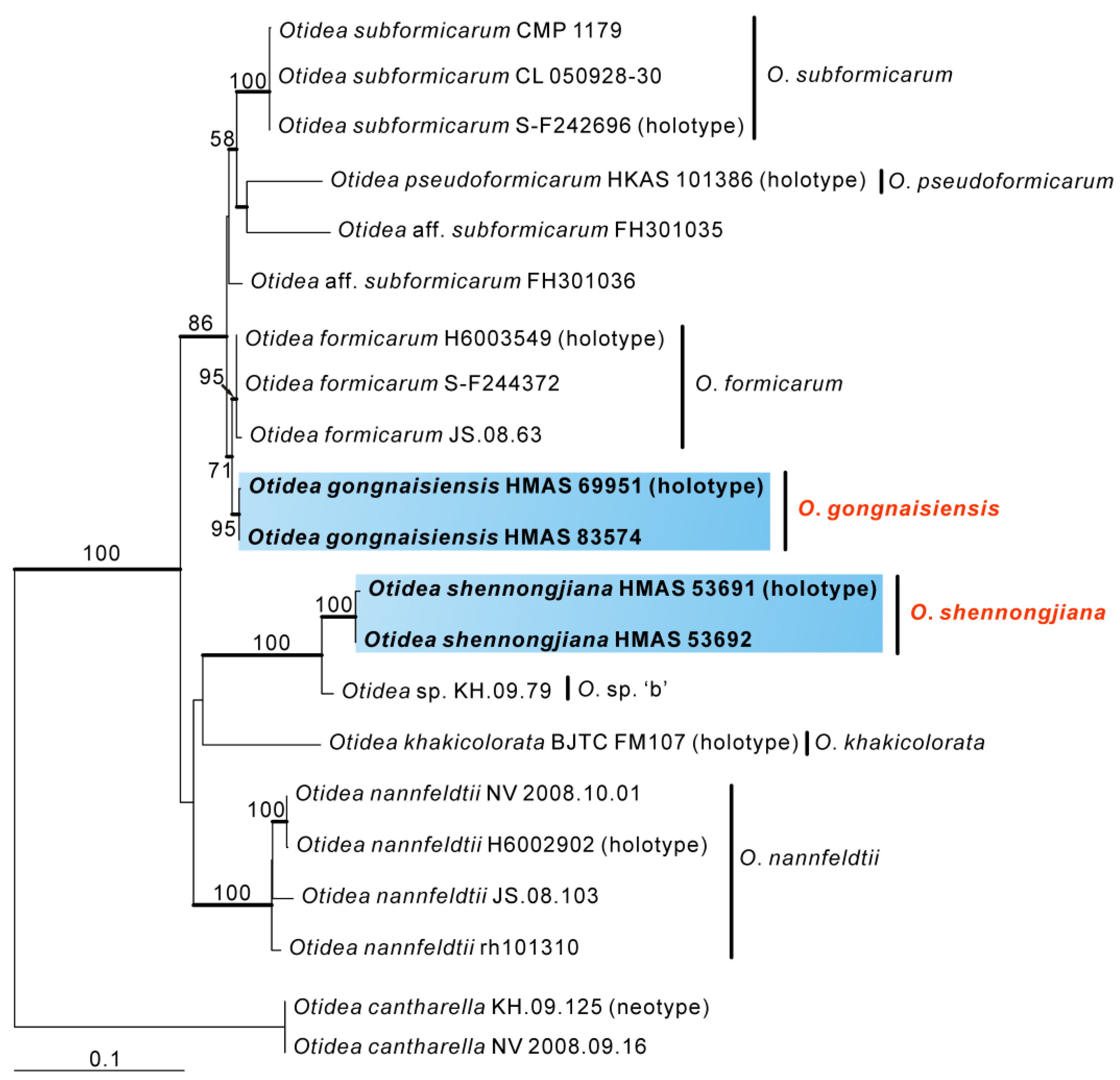
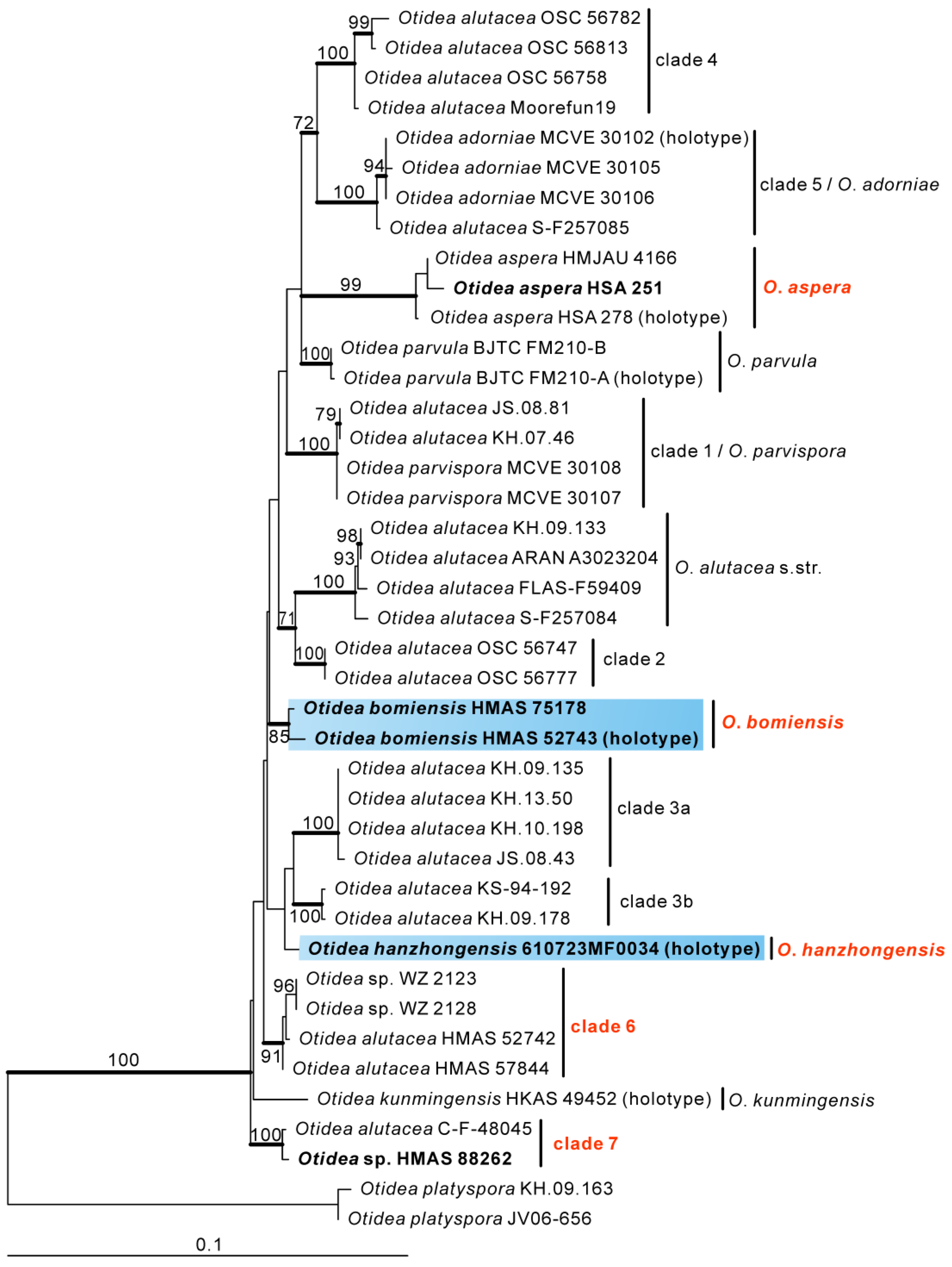
3.2. Taxonomy
Based on our phylogenies and morphological data, four new species and two known species of Otidea from China were described and illustrated here.
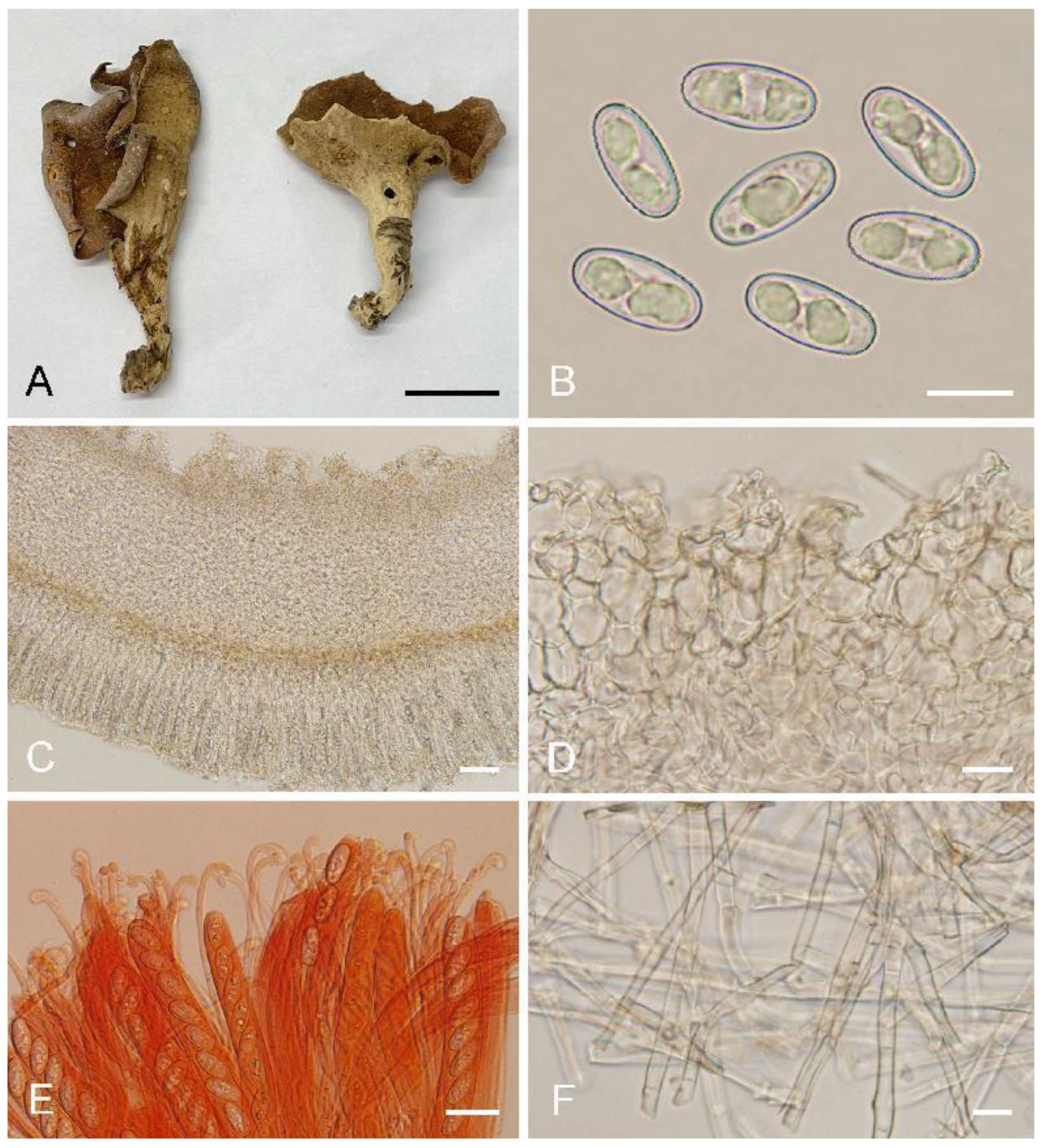
Otideabomiensis L. Fan & Y.Y. Xu, sp. nov. (
Figure 4)
MycoBank: MB844136
Etymology: bomiensis, referring to the locality where the type specimen was collected.
Holotype: China. Tibet Autonomous Region, Nyingchi City, Bomi County, Zhamu Town, Daxing Village, alt. 2900–3000 m, on soil in mixed forest, 28 August 1983, X.L. Mao M1430 (HMAS 52743).
Saprobic on soil. Apothecia solitary or gregarious, 10–40 mm high, 5–25 mm wide, broadly ear-shaped to shallowly or deeply cup-shaped, split, stipitate. Hymenium light brown to brown when fresh, dark brown when dry, margin with purpure tones, subsmooth. Receptacle surface pale yellow when fresh, slightly hygrophanous, pale yellow brown to yellowish brown when dry, furfuraceous to finely warty. Stipe 7–15 × 5–7 µm. Basal tomentum and mycelium white. Apothecial section 700–1000 µm thick. Ectal excipulum of textura angularis, 80–120 µm thick, cells thin-walled to slightly thick-walled, pale brown, 12–30 × 10–21 µm. Medullary excipulum of textura intricata, 300–550 µm thick, hyphae 3–9 µm wide, thin-walled, septate, hyaline to light brown. Subhymenium c. 40–60 µm thick, visible as a brown zone, of densely arranged cylindrical to swollen cells. Paraphyses septate, bent to curved, a few straight, of uniform width or slightly enlarged at the apices to 3–4.5 µm wide, without or with a low notch. Asci 150–200 × 10–14 µm, 8-spored, unitunicate, operculate, cylindrical, hyaline, non-amyloid, long pedicellate, arising from croziers, ascospores released from an eccentric split at the apical apex. Ascospores overlapping uniseriate, ellipsoid to oblong ellipsoid, hyaline, with one to two large guttules, smooth, (14.5–)15–16.8(–17.2) × (6.5–)7.1–7.9(–8.3) µm (Lm × Wm = 15.9 × 7.5 µm, Q = 1.9–2.2, Qm = 2, n = 50). Receptacle surface with broad conical warts, 30–50 µm high, formed by short, fasciculate, hyphoid hairs, of 4–6 subglobose to elongated cells, constricted at septa, 5–9 µm wide. Resinous exudates absent to scarce. Basal mycelium of interwoven, 3–6 µm wide, septate, hyaline to pale brown hyphae, unchanged in KOH, smooth.
Additional specimens examined: China. Sichuan Province, Xiangcheng County, Daxueshan, alt. 4300 m, on soil under Rhododendron sp., 25 July 1998, Z. Wang, WZ185 (HMAS 75178).
Notes:
Otidea bomiensis is characterized by the stipitate, light brown to brown hymenium, pale yellow receptacle surface, the lack of resinous exudates on the ectal excipulum and basal mycelium, ellipsoid to oblong ellipsoid ascospores and bent to curved paraphyses. The hymenium color of
O. adorniae Agnello, M. Carbone & P. Alvarado and
O. alutacea (Pers.) Massee is similar to that of
O. bomiensis, but
O. adorniae differs from
O. bomiensis in having smaller ascospores (10.5–)11–12(–12.5) × 6–6.5(–7) µm,
O. alutacea differs in darker receptacle surface color, which is yellowish brown, sometimes with purplish brown tones [
8,
10].
Otidea aspera also has pale yellow receptacle surface similar to
O. bomiensis, but
O. aspera differs in having shorter ascospores (12–)12.8–15(–15.5) × (5.8)6.5–7.5(–8) µm and high warts 50–80 µm on the receptacle surface [
15].
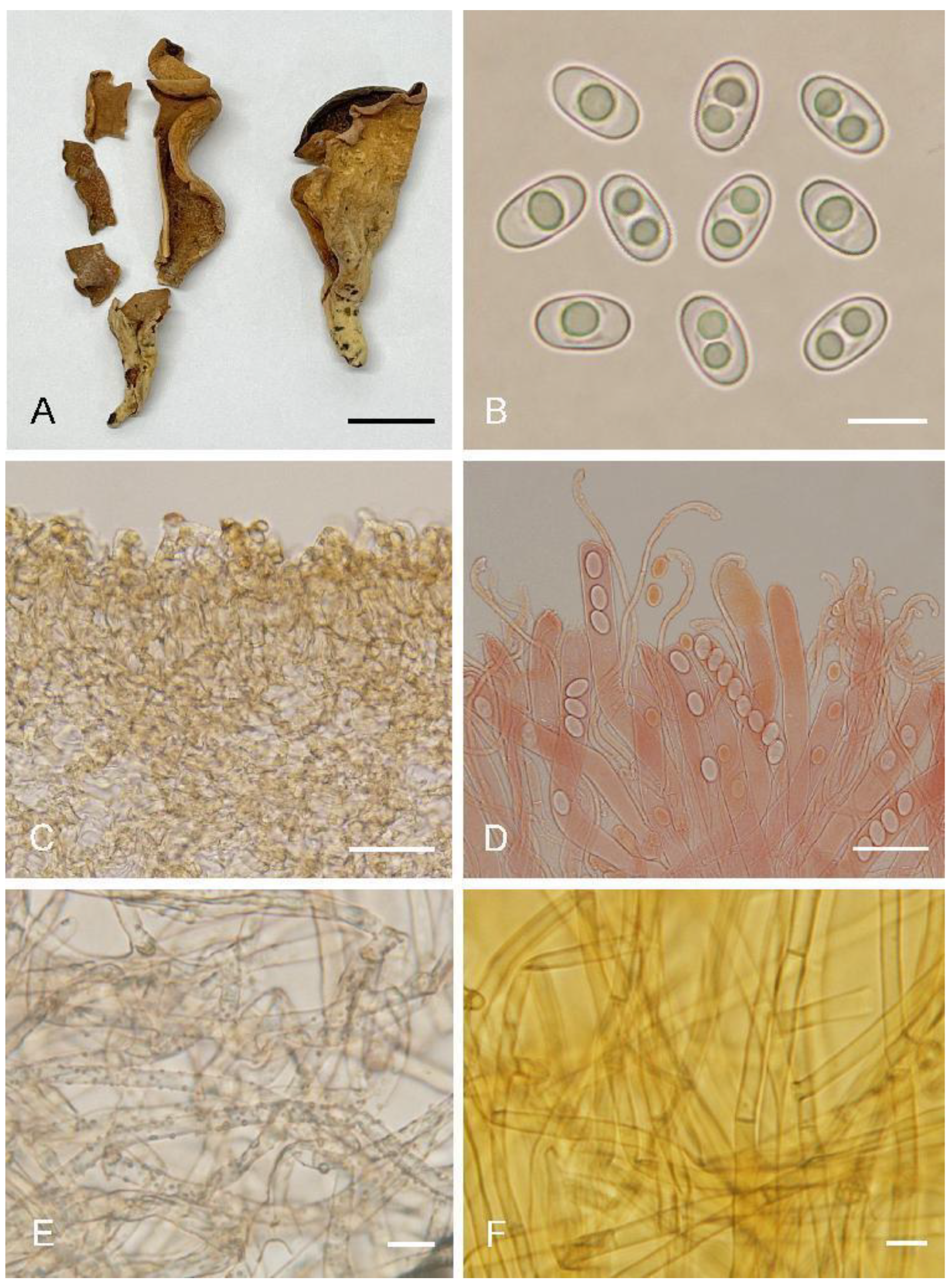
Otideagongnaisiensis L. Fan & Y.Y. Xu, sp. nov. (
Figure 5)
MycoBank: MB844137
Etymology: gongnaisiensis, referring to the locality where the type specimen was collected.
Holotype: China. Xinjiang Autonomous Region, Gongnaisi National Forest Park, August 1994, J.Y. Wang, 132 (HMAS 69951).
Saprobic on cone or soil. Apothecia solitary or gregarious, 15–40 mm high, 10–25 mm wide, long ear-shaped or broadly ear-shaped, split, stipitate. Hymenium yellowish beige when fresh, dirty yellow with pale brown tones when dry, subsmooth. Receptacle surface concolorous with hymenium when fresh, slightly hygrophanous, dirty yellow when dry, furfuraceous. Stipe 5–15 × 3–7 mm. Basal tomentum and mycelium whitish. Apothecial section 700–1000 µm thick. Ectal excipulum of textura angularis, 75–150 µm thick, cells thin-walled, brownish, 12–30 × 10–26 µm. Medullary excipulum of textura intricata, 300–500 µm thick, hyphae 3–9 µm wide, thin-walled, septate, hyaline to light brown. Subhymenium c. 75–120 µm thick, visible as a yellowish brown zone of densely arranged cylindrical to swollen cells. Paraphyses septate, curved to hooked of uniform width or slightly enlarged at the apices to 2.5–4 (5) µm wide, without or with 1–2 low notches, sometimes with 1–2 slightly swollen areas near the apex. Asci 150–200 × 8–12 µm, 8-spored, unitunicate, operculate, cylindrical, hyaline, non-amyloid, long pedicellate, arising from croziers, ascospores release from an eccentric split at the apical apex. Ascospores overlapping uniseriate, ellipsoid, hyaline, with one to two large guttules, smooth, (10.2–)10.6–12.2(–12.5) × (5.7–)6–6.8(–7.1) µm (Lm × Wm = 11.3 × 6.4 µm, Q = 1.6–1.9, Qm = 1.8, n = 50). Receptacle surface almost seldom warts, sometimes with scattered hyphoid hairs 20–30 µm high, of 2–3 subglobose to elongated cells, slightly constricted at septa, 6–11 µm wide. Resinous exudates present on the receptacle surface, brown to dark brown, dissolving into amber drops in MLZ, partially dissolving and turning yellowish brown in KOH. Basal mycelium of 3–5 µm wide, septate, hyaline to pale brown hyphae, unchanged in KOH, with many, small, irregularly, brown, resinous exudates on the surface, dissolving in MLZ, partially and more slowly in KOH.
Additional specimens examined: China. Xinjiang Autonomous Region, Jimusa’er, alt. 1700 m, on cone of Picea sp., 2 August 2003, W.Y. Zhuang & Y. Nong, 4670 (HMAS 83574).
Notes:
Otidea gongnaisiensis is characterized by the stipitate, yellowish beige, broadly ear-shaped apothecia, seldom warts on receptacle surface, and excipular resinous exudates partially dissolving and turning yellowish brown in KOH.
Otidea formicarum Harmaja and
O. pseudoformicarum A.H. Ekanayaka, Q. Zhao & K.D. Hyde are similar in apothecial shape and color to
O. gongnaisiensis, but
O. formicarum can be distinguished by its relatively small, reddish brown apothecia, comparatively shorter ascospores (L
m = 10–10.7 µm), and unique habitat, often occurring on anthills [
8].
Otidea pseudoformicarum differs in shorter asci (115–150 × 7–10 µm) and smaller ascospores (8–10 × 5–7 µm) [
13]. The ascospores size of
O. subformicarum Olariaga, Van Vooren, Carbone & K. Hansen is similar to
O.
gongnaisiensis, but it can be distinguished by orange brown to reddish brown apothecial color, and high warts (45–65 µm) on the receptacle surface [
8].
Otidea shennongjiana can also be distinguished from
O. gongnaisiensis by orange tones of apothecia, smooth basal mycelium, and the resinous exudate in the ectal excipulum partly dissolving into drops in KOH.
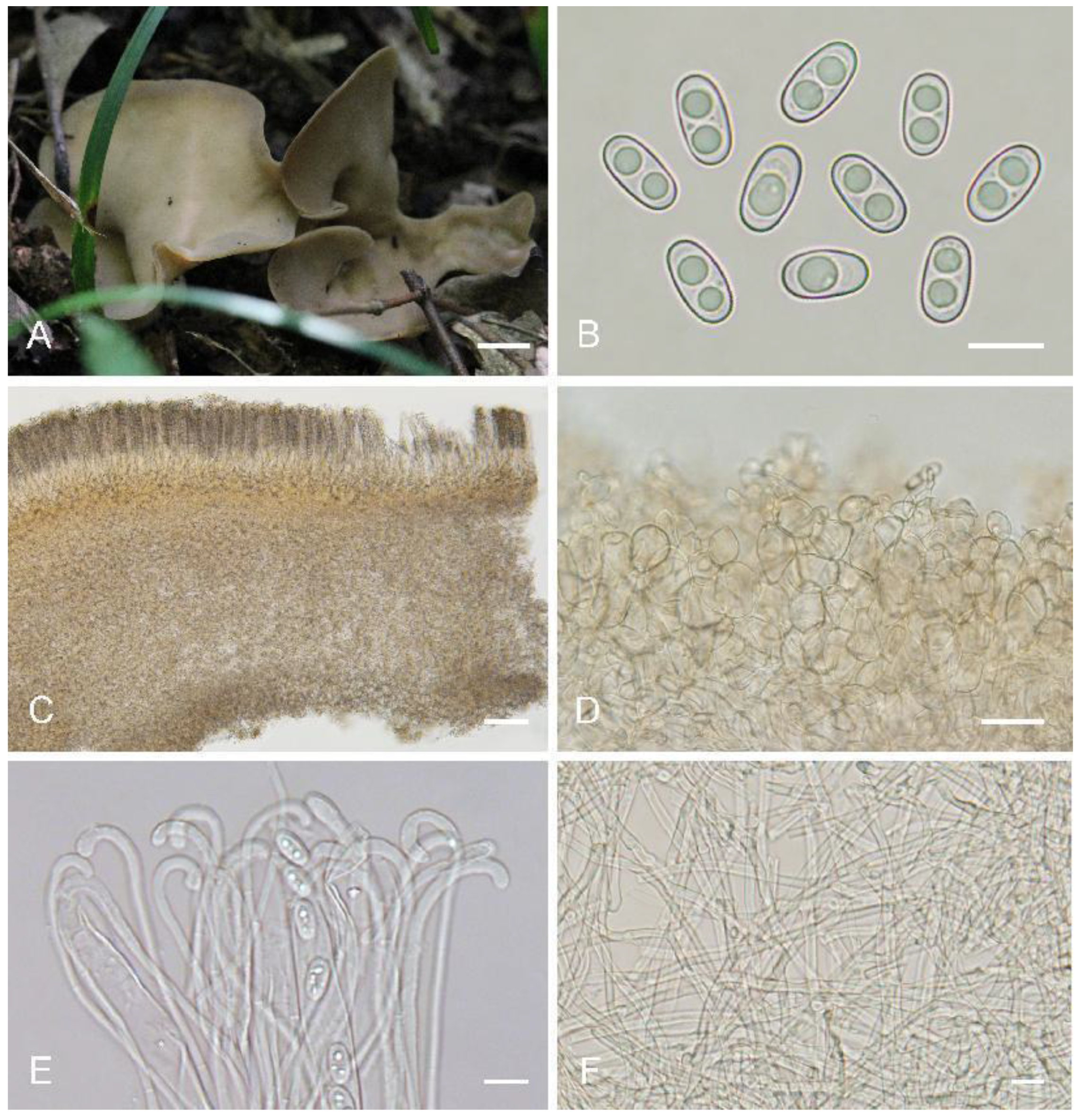
Otideahanzhongensis L. Fan, M.Q. Zhu & Y.Y. Xu, sp. nov. (
Figure 6)
MycoBank: MB844138
Etymology: hanzhongensis, referring to the locality where the type specimen was collected.
Holotype: China. Shaanxi Province, Hanzhong City, Zuoxi River Management Station, alt. 1200 m, 2 October 2016, M.Q. Zhu, (610723MF0034).
Saprobic on soil. Apothecia solitary or gregarious, 10–40 mm high, 20–50 mm wide, broadly ear-shaped to shallowly cup-shaped or shallowly disc, broader above, split, sessile or shortly stipitate. Hymenium pale greyish yellow to pale ochre when fresh, dark yellowish brown to brown when dry, subsmooth. Receptacle surface pale yellow when fresh, slightly hygrophanous, yellowish brown when dry, furfuraceous to finely warty. If present, the stipe is very short. Basal tomentum and mycelium whitish to grayish white. Apothecial section 600–850 µm thick. Ectal excipulum of textura angularis, 80–130 µm thick, cells thin-walled, pale brown, 11–30 × 9–22 µm. Medullary excipulum of textura intricata, 250–450 µm thick, hyphae 3–8.5 µm wide, thin-walled, septate, hyaline to light brown. Subhymenium ca. 50–100 µm thick, visible as a yellowish brown zone of densely arranged cylindrical to swollen cells. Paraphyses septate, curved to hooked, usually enlarged at the apices, 3.5–5 μm wide at apex, 2–2.5 μm below, without notch. Asci 120–160 × 8.5–11.5 µm, 8-spored, unitunicate, operculate, cylindrical, hyaline, non-amyloid, long pedicellate, arising from croziers, ascospores released from an eccentric split at the apical apex. Ascospores overlapping uniseriate, ellipsoid, hyaline, with one to two large guttules, smooth, (10.5–)11–13(–13.5) × (5.5–)6–6.8(–7) µm (Lm × Wm = 12 × 6.5 µm, Q = 1.7–2, Qm = 1.85, n = 50). Receptacle surface with broadly conical warts, 40–60 µm high, formed by hyphoid hairs, of 3–5 subglobose to elongated cells, constricted at septa, 5–9 µm wide. Resinous exudates absent to scarce. Basal mycelium of 2.5–5 µm wide, septate, hyaline to pale brown hyphae, unchanged in KOH, smooth.
Notes:
Otidea hanzhongensis is characterized by the pale greyish yellow to pale ochre hymenium, pale yellow receptacle color, small ascospores, short asci, and the lack of resinous exudates on the ectal excipulum and basal mycelium.
Otidea parvispora (Parslow & Spooner) M. Carbone, Agnello, Kautmanová, Z.W. Ge & P. Alvarado and
O. aspera also share similar light-colored apothecia, but
O. parvispora can be distinguished by light ochraceous-buff hymenium, pale fawn receptacle surface, and longer asci 165–185 × 9–10 µm [
11].
Otidea aspera differs from having longer ascospores (12–)12.8–15(–15.5) × (5.8–)6.5–7.5(–8) µm and longer asci 150–200 × 9–13 µm [
15]. In the
O. alutacea clade, the other new collected species,
O. bomiensis, is also easily distinguished from
O. hanzhongensis by the light brown to brown hymenium, bigger ascospores (14.5–)15–16.8(–17.2) × (6.5–)7.1–7.9(–8.3) µm and longer asci 150–200 × 10–14 µm.
Otideashennongjiana L. Fan & Y.Y. Xu, sp. nov. (
Figure 7)
MycoBank: MB844139
Etymology: shennongjiana, referring to the locality where the type specimen was collected.
Holotype: China. Hubei Province, Shennongjia National Forest Park, changyanwu, 29 July 1984, J.X. Tian, 75 (HMAS 53691).
Saprobic on rotten wood and roots. Apothecia gregarious to caespitose, 8–38 mm high, 5–15 mm wide, broadly ear-shaped, sometimes cup-shaped, split, stipitate. Hymenium light yellow with orange tones to pale orange when fresh, orange brown to reddish brown when dry, subsmooth. Receptacle surface pale yellow when fresh, slightly hygrophanous, pale yellowish brown, margin pale reddish brown when dry, furfuraceous to finely warty. Stipe 5–15 × 3–5 mm. Basal tomentum and mycelium white. Apothecial section 550–900 µm thick. Ectal excipulum of textura angularis, 80–120 µm thick, cells thin-walled, hyaline to brown, 11–26 × 9–16 µm. Medullary excipulum of textura intricata, 200–350 µm thick, hyphae 3–8 µm wide, thin to slightly thick walled, hyaline to light brown. Subhymenium c. 75–150 µm thick, visible as a brown zone, of densely arranged cylindrical to swollen cells. Paraphyses septate, straight to curved, a few hooked, of uniform width at the apices to 2.2–3.5 µm wide, without or a few with 1–2 low notches. Asci 135–175 × 8–12 µm, 8-spored, unitunicate, operculate, cylindrical, hyaline, non-amyloid, long pedicellate, arising from croziers, ascospores released from an eccentric split at the apical apex. Ascospores overlapping uniseriate, ellipsoid, hyaline, with one to two large guttules, smooth, (10.5–)11–13(–13.5) × (5.8–)6.2–7.2(–7.5) µm (Lm × Wm = 12 × 6.8 µm, Q = 1.6–1.9, Qm = 1.76, n = 50). Receptacle surface with warts, 30–50 µm high, formed by short, fasciculate, hyphoid hairs, of 3–6 subglobose to elongated cells, constricted at septa, 5–10 µm wide. Resinous exudates abundant on the receptacle surface, yellow brown to dark brown, dissolving into amber drops in MLZ, and partly dissolving into amber drops in KOH. Basal mycelium of 2.5–5.5 µm wide, septate, hyaline to pale brown hyphae, unchanged in KOH, smooth.
Additional specimens examined: China. Hubei Province, Shennongjia National Forest Park, changyanwu, 4 August 1984, J.X. Tian, 95 (HMAS 53692).
Notes:
Otidea shennongjiana is characterized by the broadly ear-shaped apothecia, light yellow with orange tones hymenium, pale yellow receptacle surface, big ascospores, smooth basal mycelium, and by its habitat, often occurring on rotten wood and roots. Same as with
O. shennongjiana, the apothecia of
O. nannfeldtii Harmaja, and
O. subformicarum all have orange tones, but
O. nannfeldtii differs from
O. shennongjiana by the smaller ascospores (9–)9.5–10.5(–11.5) × 5.5–6.5(–7) µm, excipular resinous exudates turning reddish brown in KOH and with yellow resinous exudates on basal mycelium [
8].
Otidea subformicarum differs in narrow ascospores (10.5–12 × 6–6.5 µm), long asci (184–237 × 11–11.5 µm), and pale yellow drops on basal mycelium surface [
8]. In the
O. formicarun clade,
Otidea khakicolorata L. Fan & Y.Y. Xu also have smooth basal mycelium, but it can be distinguished by khaki to pale ochre apothecia, smaller ascospores (8.5–)9–10(–10.5) × 4.5(5)–6(–6.5) µm, and excipular resinous exudates turning reddish brown in KOH [
15]. Phylogenetic analyses revealed that
O. shennongjiana was grouped with an undescribed species
Otidea sp. ‘b’, with high support values (
Figure 1 and
Figure 2). Mean ascospores length and width of
Otidea sp. ‘b’ is 11.1 × 6.7 µm, which is smaller than
O. shennongjiana. DNA analysis showed that they shared only 95.45% ITS sequence similarity, suggesting that
O. shennongjiana and
Otidea sp. ‘b’ may be different species.
Otidea aspera L. Fan & Y.Y. Xu, Journal of Fungi 8 (3, no. 272): 9 (2022) (
Figure 8E,F)
Specimens examined: China, Shanxi Province, Pangquangou National Nature Reserve, Badaogou, alt. 2000 m, 28 August 2018, J.Z. Cao, LH251 (HSA 251).
Notes: This species has been recently described in Shanxi Province and Inner Mongolia, China [
15]. The specimen (HSA 251) collected from Shanxi Province was confirmed to be
O. aspera by morphological and molecular evidence in this study. As shown in
Figure 8E, the color of the hymenium of HSA 251 is grayish brown to brown, which is darker than the original descriptions (greyish yellow to light brown) of
O. aspera. This may be due to the different maturity stages at which different fruit-bodies were collected. Therefore, the color of the hymenium of
O. aspera was re-described as follows: hymenium surface grayish yellow, light brown to brown when fresh.
Otidea sinensis J.Z. Cao & L. Fan, Mycologia 82(6): 736 (1990) (
Figure 8C)
Specimens examined: China, Sichuan Province, Xiaojin County, National Scenic Area of Four Girls Mountain, alt. 2014 m, 15 August 2013, W.L. Lu, 1734 (HMAS 268390); ibid., 15 August 2013, W.L. Lu, 1728 (HMAS 268544).
Saprobic on soil. Apothecia gregarious to caespitose, 10–30 mm high, 5–25 mm wide, long ear-shaped, or broadly ear-shaped, split, stipitate. Hymenium cream to greyish yellow when dry, subsmooth. Receptacle surface pale yellowish brown when dry, finely warty. Stipe 5–10 × 3–6 mm. Basal tomentum and mycelium white. Apothecial section 600–1000 µm thick. Ectal excipulum of textura angularis, 100–150 µm thick, cells thin-walled, hyaline to brown, 12–35 × 10–24 µm. Medullary excipulum of textura intricata, 400–500 µm thick, hyphae 3–9 µm wide, thin-walled, hyaline. Subhymenium c. 50–80 µm thick, visible as a pale brown zone, of densely arranged cylindrical to swollen cells. Paraphyses septate, curved to hooked, slightly enlarged at the apices to 3–5.5 µm wide, without or a few with 1–2 low notches. Asci 150–180 × 8–11 µm, 8-spored, unitunicate, operculate, cylindrical, hyaline, non-amyloid, long pedicellate, arising from croziers, ascospores released from an eccentric split at the apical apex. Ascospores overlapping uniseriate, ellipsoid to slightly fusoid, hyaline, with one to two large guttules, smooth, 10.5–13 × 5.5–6.7 µm (Lm × Wm = 11.8 × 6.3 µm, Q = 1.7–2, Qm = 1.88, n = 50). Receptacle surface with warts, 50–85 µm high, formed by short, fasciculate, hyphoid hairs, of 4–6 subglobose to elongated cells, constricted at septa, 4–9 µm wide. Resinous exudates abundant on the receptacle surface, yellow to yellow brown, partially dissolving and turning slightly reddish in MLZ, unchanged in KOH. Basal mycelium of 3–7 µm wide, septate, hyaline to pale brown hyphae, turning yellow in KOH, with small, irregularly, brown, resinous exudates on the surface, partially dissolving in KOH, partially dissolving in MLZ.
Notes: The two specimens (HMAS 268390 and HMAS 268544) grouped together with the type specimen of
O. sinensis with strong a support value (BS = 91%, PP = 1.00). We examined the morphology of these two specimens, which are consistent with the original description by Cao et al. [
31]. The two specimens were, therefore, confirmed to be
O. sinensis. The type specimen of
O. sinensis was collected from Heilongjiang Province in northeast China. Our results demonstrate that
O. sinensis is also distributed in southwest China.